Abstract
The decaying fruit in which Drosophila melanogaster feed and breed can contain ethanol in concentrations as high as 6–7%. In this cosmopolitan species, populations from temperate regions are consistently more resistant to ethanol poisoning than populations from the tropics, but little is known about the physiological basis of this difference. I show that when exposed to low levels of ethanol vapor, flies from a tropical African population accumulated 2–3 times more internal ethanol than flies from a European population, giving evidence that faster ethanol catabolism by European flies contributes to the resistance difference. Using lines differing only in the origin of their third chromosome, however, I show that faster ethanol elimination cannot fully explain the resistance difference, because relative to African third chromosomes, European third chromosomes confer substantially higher ethanol resistance, while having little effect on internal ethanol concentrations. European third chromosomes also confer higher resistance to acetic acid, a metabolic product of ethanol, than African third chromosomes, suggesting that the higher ethanol resistance conferred by the former might be due to increased resistance to deleterious effects of ethanol-derived acetic acid. In support of this hypothesis, when ethanol catabolism was blocked with an Alcohol dehydrogenase mutant, there was no difference in ethanol resistance between flies with European and African third chromosomes.
KEY WORDS: Genetic correlation, Geographic variation, Metabolic pathways, Toxin resistance
INTRODUCTION
Animals that regularly feed on decaying fruit or fermented nectar can be exposed to physiologically significant concentrations of ethanol. For example, a Malaysian tree shrew regularly consumes fermented nectar with in excess of 1% ethanol (Wiens et al., 2008), and breeding sites of the fruit fly Drosophila melanogaster can contain as much as 6–7% ethanol (McKenzie and McKechnie, 1979; Gibson et al., 1981; Oakeshott et al., 1982a). Although there is evidence that these and other species have evolved resistance to the toxic and sedating effects of ethanol (Bouletreau and David, 1981; Merçot et al., 1994; Wiens et al., 2008), little is known about the physiological basis of this resistance.
There are two general routes for evolving resistance to a toxin (Rose and Hodgson, 2001). One is to evolve reduced sensitivity to the toxin's effects, for example by altering toxin binding sites or by compensating for pathways disrupted by the toxin. The second route is to minimize the amount of toxin that reaches the target organ or tissue. Mechanisms to accomplish this are varied, and include detoxification, excretion, sequestration and reduced absorption (Rose and Hodgson, 2001). One complication of the detoxification strategy for ethanol is that the first two products of ethanol catabolism, acetaldehyde and acetate, are themselves deleterious (Israel et al., 1994; Deitrich, 2004). Also, because ethanol rapidly penetrates cell membranes and cannot be concentrated (Harris et al., 2008), excretion and sequestration are not viable mechanisms for ethanol resistance. For the same reason, options for reducing absorption, except by behavioral avoidance, are limited.
Drosophila melanogaster provides a good system in which to study the contribution of detoxification and other mechanisms to naturally evolved ethanol resistance. In addition to its well-known advantages as a model organism, D. melanogaster shows marked geographic variation in ethanol resistance, with populations from temperate latitudes being consistently more resistant than those from the tropics or subtropics (David and Bocquet, 1975; Cohan and Graf, 1985; David et al., 1986; Parkash et al., 1999; Montooth et al., 2006). This variation is correlated with allele frequency variation in the well-studied enzyme gene Alcohol dehydrogenase (Adh), whose product converts ethanol to acetaldehyde (Fig. 1): the more active Fast allele is in low frequency in most tropical populations, but reaches high frequencies in Europe, northern USA and southern Australia (Oakeshott et al., 1982b; David et al., 1986). In addition, frequencies of activity variants of two other enzymes important in ethanol detoxification, glycerol 3-phosphate dehydrogenase (GPDH) and aldehyde dehydrogenase (ALDH), also vary with latitude (David, 1982; Oakeshott et al., 1982b; Fry et al., 2008). Little is known, however, about the extent to which these and perhaps other differences between temperate and tropical populations affect internal ethanol concentrations, particularly in adult flies (see Discussion).
Fig. 1.
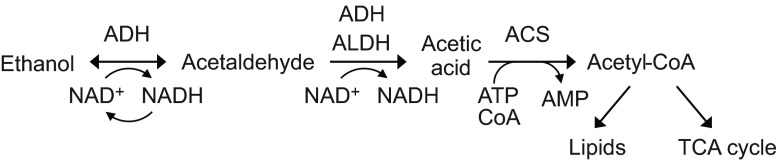
Ethanol metabolism in Drosophila melanogaster, showing the main enzymes catalyzing the first three steps. ADH, alcohol dehydrogenase; ALDH, aldehyde dehydrogenase; ACS, acetyl-CoA synthetase. Although the product of the second step is usually referred to as acetate, the immediate product is the protonated acid. Modified from Geer et al. (Geer et al., 1993).
Another relevant observation is that, both among D. melanogaster strains and among Drosophila species, resistance to ethanol is strongly correlated with resistance to acetic acid (Chakir et al., 1993; Eisses and Den Boer, 1995; Montooth et al., 2006). This could be explained in at least three ways. First, much of the mortality caused by ethanol in toxicity assays might be the result of the acetic acid generated as a product of ethanol catabolism (Fig. 1). Under this hypothesis, either variation in the ability to metabolically process acetic acid or variation in the sensitivity to a given internal concentration would appear as variation in ethanol resistance. Second, acetic acid and ethanol might have similar mechanisms of toxicity, so that genetic variation in sensitivity to one would automatically produce variation in sensitivity to the other. A third possibility is linkage disequilibrium between genes affecting the two traits, generated by correlated selection for resistance to the two toxins caused by their frequent co-occurrence in decaying fruit (McKenzie and McKechnie, 1979).
These possibilities can be distinguished by taking advantage of the fact that much of the temperate–tropical difference in ethanol and acetic acid resistance in D. melanogaster maps to the third chromosome (Chakir et al., 1996), while Adh (as well as Aldh and Gpdh) is on the second chromosome. Thus, it is possible to substitute temperate and tropical third chromosomes into a common, Adh-null background. If the difference in ethanol resistance between temperate and tropical third chromosomes is in fact due to a difference in resistance to the acetic acid generated from ethanol breakdown, blocking ethanol breakdown should eliminate the difference. Under the other hypotheses, however, blocking ethanol breakdown would be expected to have little or no effect on the resistance difference.
Here, I investigated the physiological bases of the difference in ethanol resistance between a temperate (European) and tropical (African) D. melanogaster population. First, I show that flies from the more ethanol-sensitive African population, when exposed to a non-lethal dose of ethanol vapor, accumulate 2–3 times more internal ethanol than flies from the European population, likely due in part to the observed higher ADH and ALDH activity of the European flies. Second, I confirmed the previously reported third chromosome effects on ethanol and acetic acid resistance, and show that the effect on ethanol resistance is not due to an effect on internal ethanol concentration. Finally, substituting African and European third chromosomes into an Adh-null background eliminated the difference in ethanol resistance between them, giving evidence that the third chromosome effect on ethanol resistance is due to variation in resistance to the acetic acid generated by ethanol catabolism.
RESULTS
Ethanol resistance, ADH and ALDH activity, and internal ethanol concentrations of European and African flies
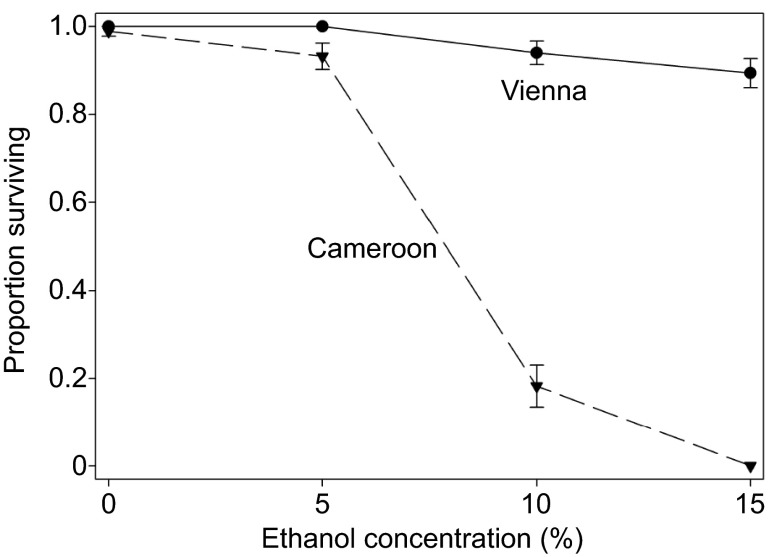
Confirming the expectation from previous studies, isofemale lines collected from a European population (Vienna region) had substantially higher survival in the presence of ethanol than lines from a tropical African population (a village in Cameroon; Fig. 2). On 5% ethanol, survival of the European lines was 100%, whereas all but two of the 16 African lines experienced some mortality. On both 10% and 15% ethanol, there was no overlap in line means between the two groups, and no African flies survived at the higher concentration (Fig. 2).
Fig. 2.
Ethanol resistance of males from isofemale lines collected near Vienna, Austria, and in Cameroon. Flies were placed in vials containing a cotton ball moistened with 1.5 ml of a solution containing 3% sucrose and the indicated concentration of ethanol. Survival was monitored after 2 days. Error bars show s.e.m. among line means (N=10 lines per location).
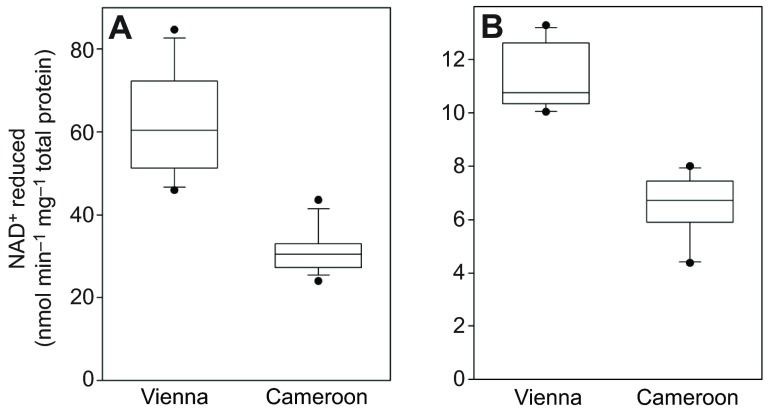
The European lines had higher activities of the enzymes alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) than the African lines, with no overlap between the two groups for either trait (Fig. 3). The twofold average difference in ADH activity is consistent with previous results showing that the AdhFast allele, homozygotes for which typically have 2- to 3-fold higher ADH activity than homozygotes for the Slow allele (Heinstra, 1993; Stam and Laurie, 1996), is in high frequency in Europe but uncommon in tropical Africa (Oakeshott et al., 1982b; David et al., 1986). In contrast, the higher activity AldhPhe479 allele described by Fry et al. (Fry et al., 2008) was present in only five of the 16 Vienna lines, and not fixed in any of the five. Because there are no other common replacement polymorphisms in Aldh (Fry et al., 2008), the activity difference shown in Fig. 3B must therefore be largely the result of greater ALDH protein abundance in the European lines.
Fig. 3.
Activities of ADH and ALDH in whole-fly homogenates of isofemale line males. The activity of ADH (A) and ALDH (B) was assessed by monitoring NAD+ reduction in a spectrophotometer. The distribution of isofemale line means (N=16 per location) is illustrated by box plots (broad lines, quartiles; narrow lines, 10th and 90th percentiles; circles, extremes).
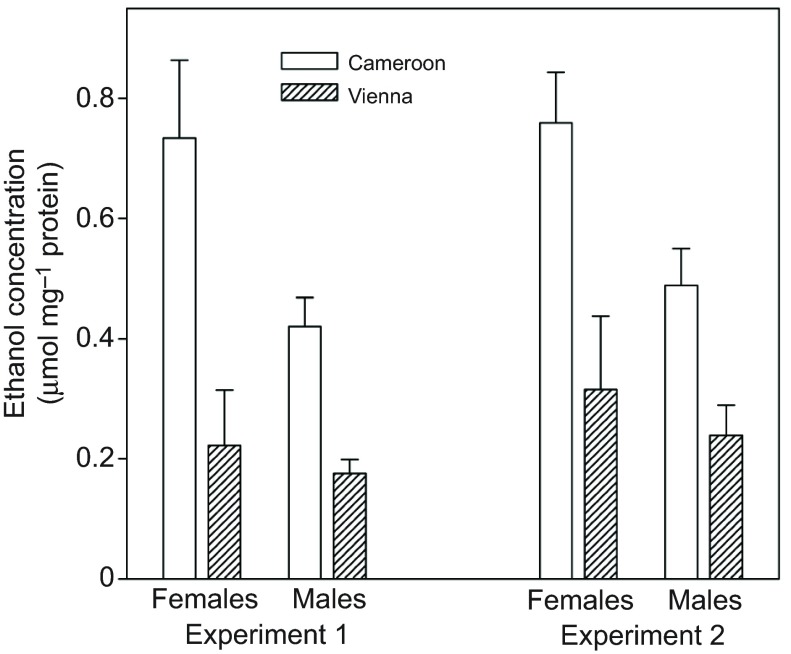
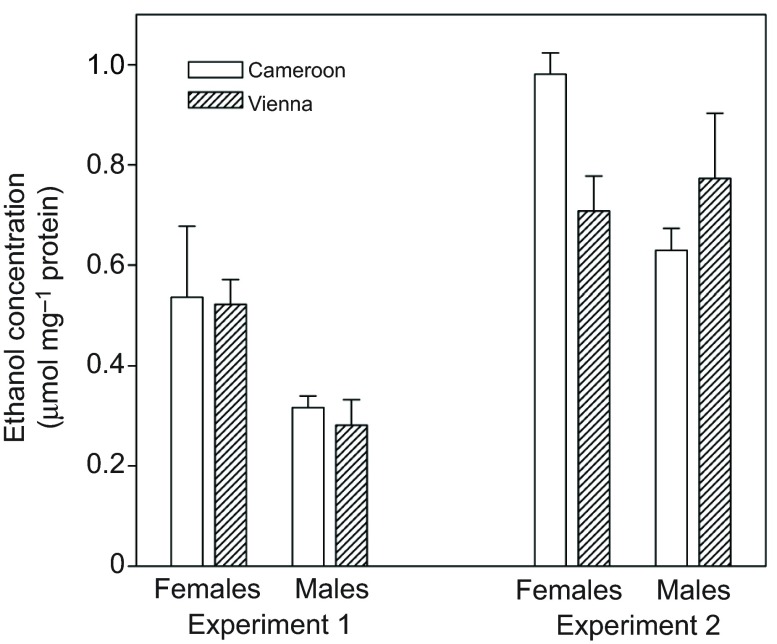
To determine whether the difference in ethanol resistance shown in Fig. 2 might be explained, at least in part, by faster metabolism of ethanol in the European flies, I compared internal ethanol concentrations of African and European flies exposed to a non-lethal dose of ethanol vapor. After either 24 h (Fig. 4A) or 4 h (Fig. 4B), African flies had ~2–3 times as much internal ethanol as European flies (main effect of region: P<0.002 in each experiment).
Fig. 4.
Mean and s.e.m. ethanol concentration of homogenates of flies exposed to a non-lethal dose of ethanol vapor. Flies came from populations formed by hybridizing 16–20 isofemale lines from either Austria or Cameroon; different populations were used for the two experiments. Experiment 1 (24 h exposure): region, P<0.0001; sex, P>0.2; interaction, P=0.01 (region effect remained significant when sexes were analyzed separately: females, P<0.001; males, P=0.003). Experiment 2 (4 h exposure): region, P<0.002; sex, P=0.06; interaction, P>0.2.
Ethanol resistance and internal ethanol concentrations of third chromosome lines
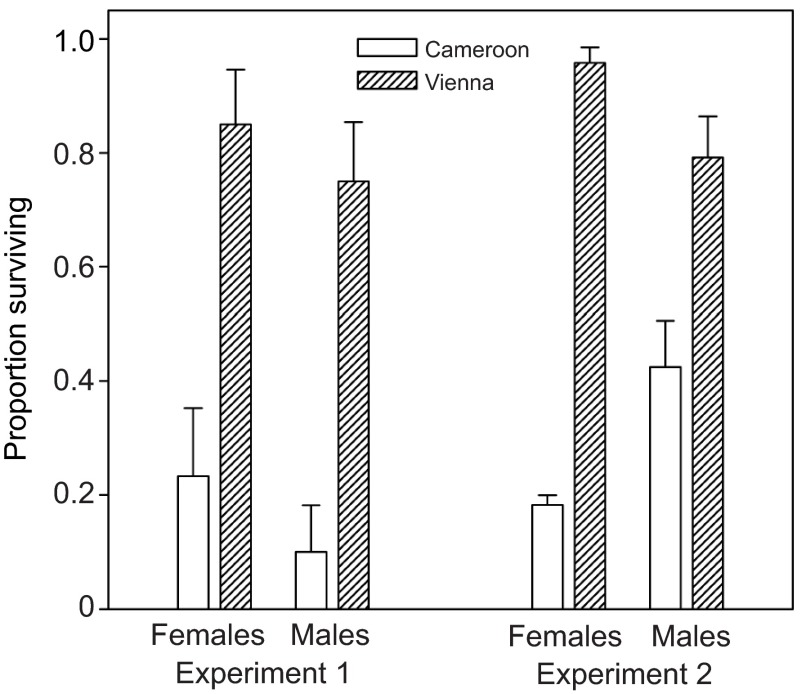
The above results give evidence that faster breakdown of ethanol in European than in African flies makes a contribution to the higher ethanol resistance of the former. A previous study (Chakir et al., 1996), however, showed that the third chromosome accounts for at least 50% of the European–African difference in ethanol resistance, even though the genes for ADH, ALDH and GPDH (another key enzyme for ethanol metabolism; see Discussion) are on the second chromosome. This suggests that European third chromosomes may confer one or more mechanisms of ethanol resistance in addition to faster ethanol breakdown. To investigate this possibility, I measured ethanol resistance and internal ethanol concentrations of lines with European and African-derived third chromosomes in an isogenic African background.
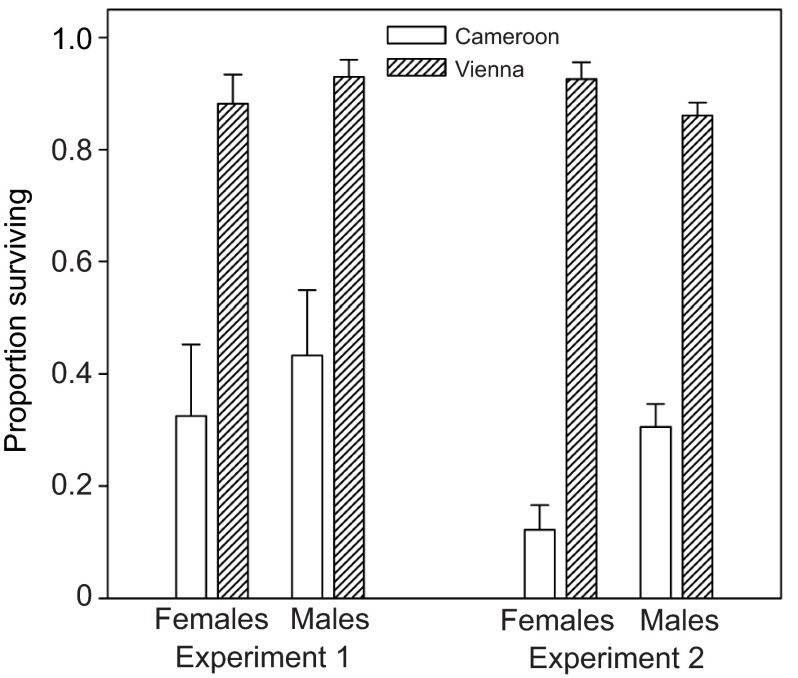
As expected, European third chromosome lines were substantially more ethanol resistant than African ones (Fig. 5). However, the two sets of lines showed no consistent difference in internal ethanol concentrations after exposure to a non-lethal ethanol dose (Fig. 6). In experiment 1, both the region effect and the sex×region interaction were non-significant (P>0.7). In experiment 2, there was no main effect of region (P>0.4), but there was a significant sex×region interaction (P=0.02). Testing the sexes separately revealed no significant difference in ethanol concentration between European and African males (P>0.2), but African females had significantly higher ethanol concentration than European females (P=0.008). In the latter case, however, the difference was small compared with that between the parental isofemale lines (Fig. 4), and there was no tendency toward a similar female-specific difference in experiment 1 (Fig. 6). These observations, together with the lack of difference in ethanol concentrations between European and African third chromosome line males, indicate that differences in the rate of ethanol detoxification or absorption make at most a minor contribution to the third chromosome effect on ethanol resistance.
Fig. 5.
Ethanol resistance of flies with third chromosomes from either Austria or Cameroon placed into an isogenic Cameroon genetic background. Flies were placed in vials containing 1 ml of sucrose solution; 550 μl of 20% ethanol was subsequently added to a dry cotton plug in the middle of the vial, and survival was monitored after 2 days. Experiment 1: means and s.e.m. of three iso-third chromosome lines per location. Region, P<0.01; sex, P>0.2; interaction, P>0.7. Experiment 2: means and s.e.m. of populations formed by hybridizing four iso-third chromosome lines per location. Region, P<0.0001; sex, P>0.3; interaction, P<0.001 (region effect remained significant at P<0.001 when sexes were analyzed separately).
Fig. 6.
Ethanol concentrations of flies with third chromosomes from Austria or Cameroon in the Cameroon genetic background. Stocks were the same as those used for the corresponding survival assays (Fig. 5). Experiment 1 (24 h exposure): region, P>0.7; sex, P=0.01; interaction, P>0.8. Experiment 2 (4 h exposure): region, P>0.4; sex, P=0.09; interaction, P=0.02 (see Results for further statistical analysis).
Relationship between ethanol and acetic acid resistance
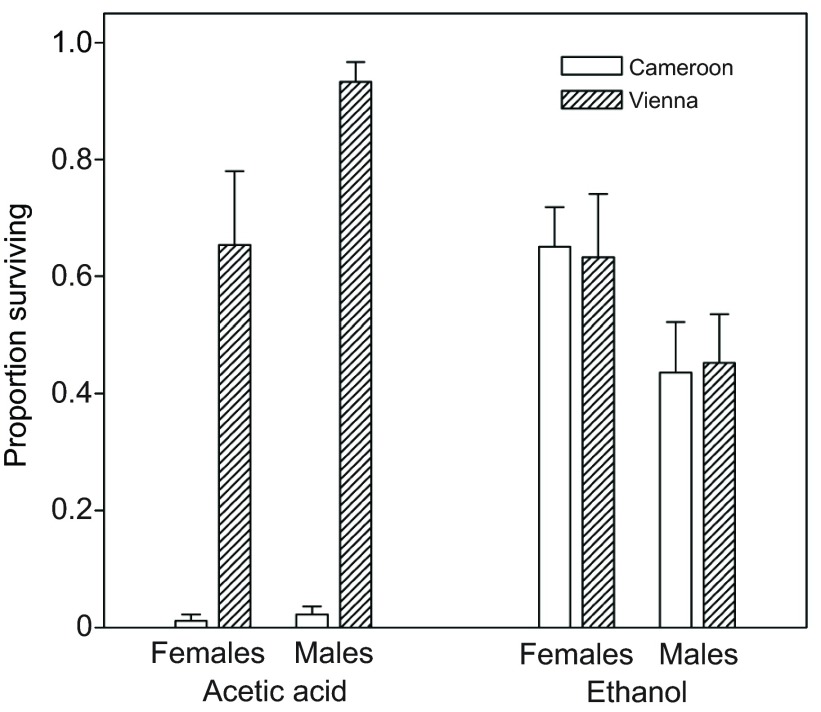
In the African genetic background, European third chromosome lines were more resistant to acetic acid than were African lines (Fig. 7; P<0.0001 in each experiment), as expected (Chakir et al., 1996). To investigate whether the ethanol resistance difference between European and African third chromosomes might in fact be due to a difference in resistance to the acetic acid generated by ethanol catabolism, I substituted one European and one African third chromosome into an Adh-null genetic background, thereby blocking most ethanol oxidation (Geer et al., 1993). The resulting lines still differed strongly in acetic acid resistance (Fig. 8A; P<0.001), but were indistinguishable from each other in ethanol resistance (Fig. 8B; P>0.5). Note that the means in Fig. 8B cannot be compared with those in Fig. 5, because the amount of ethanol used for the assays was reduced by more than fourfold to accommodate the increased ethanol sensitivity of the Adh-null lines. Similarly, the means in Fig. 8A cannot be compared with those in Fig. 7, because the amount of acetic acid was adjusted to account for the somewhat higher acetic acid resistance of flies with the Adh-null background than those with the Cameroon background (see Materials and methods).
Fig. 7.
Acetic acid resistance of flies with third chromosomes from either Austria or Cameroon in the Cameroon genetic background. Methods and stocks were the same as used for the ethanol resistance assays (see Fig. 5), with 125 μl of 40% glacial acetic acid added to the vials in place of ethanol. Experiment 1: region, P<0.0001; sex, P=0.17; interaction, P>0.8. Experiment 2: region, P<0.0001; sex, P>0.9; interaction, P=0.006 (region effect remained significant when sexes were analyzed separately: females, P<0.001; males, P=0.01).
Fig. 8.
Acetic acid and ethanol resistance of flies with a third chromosome from Austria or Cameroon in a genetic background lacking ADH activity. Acetic acid (275 μl of 40%): region, P<0.0001; sex, P=0.04; interaction, P=0.09. Ethanol (125 μl of 20%): region, P>0.9; sex, P=0.005; interaction, P>0.8.
DISCUSSION
This study has two key findings. First, when exposed to a non-debilitating concentration of ethanol vapor, flies from a relatively ethanol-sensitive African population accumulated 2–3 times as much internal ethanol as those from a more ethanol-resistant European population. Second, European third chromosomes confer greater resistance to ethanol than African third chromosomes, primarily by increasing resistance to one or more metabolites of ethanol, of which acetic acid is a strong candidate.
One caveat is that the work reported here used flies from only one European and one African location. Nonetheless, extensive surveys have shown that variation in ethanol resistance among strains collected from a given latitude is small relative to the temperate–tropical difference (David and Bocquet, 1975; David et al., 1986; Parkash et al., 1999). It seems reasonable to assume that the same is true of the underlying resistance mechanisms, particularly given that genetic differentiation is very low among both West African and European D. melanogaster populations (Pool and Aquadro, 2006).
Below, the two main findings will be discussed in turn.
Evidence for enhanced ethanol metabolism in European flies
With only a few exceptions (e.g. Montooth et al., 2006; Fry et al., 2008), most previous studies relevant to understanding the physiological basis of natural variation in ethanol resistance in D. melanogaster have focused exclusively on the Adh polymorphism (reviewed in Heinstra, 1993; Eanes, 1999). Homozygotes for the Fast allele, which is common in temperate regions but relatively rare in the tropics, reproducibly have 2- to 3-fold higher ADH activity (Vmax) than those for the Slow allele. This difference stems in part from the mobility-altering amino acid substitution itself, and in part from greater ADH protein levels in Fast homozygotes, apparently caused by non-coding differences in linkage disequilibrium with the replacement polymorphism (Stam and Laurie, 1996). In spite of many studies by several different groups, however, no strong consensus emerged about the in vivo significance of the activity difference. Estimates of the flux of carbon from ethanol to lipids and/or CO2 generally showed only a small effect of the polymorphism, or in some cases no effect. In a review of this literature, Heinstra (Heinstra, 1993) concluded that: ‘genetic variation in ADH may influence flux in third-instar larvae, but apparently it does not in adults’.
The emphasis on flux in these studies seems to have stemmed from the implicit view that ethanol's main significance for Drosophila is as an energy source. If instead ethanol is viewed primarily as a toxin, internal concentration becomes a more relevant measure of the in vivo significance of enzyme activity differences. Two studies reported internal ethanol concentrations of larvae differing in natural Adh genotype and indeed found significant differences, with FF strains having 2- to 3-fold higher concentrations than SS strains after being immersed in a 5% ethanol solution for several hours (Heinstra et al., 1987; Freriksen et al., 1994). These studies, however, used only a small number of strains (all from Europe or northern USA), and did not control for genetic background differences, so it is not clear to what extent the results can be attributed to Adh genotype. Nonetheless, the studies show that substantial differences in internal ethanol concentration can occur between natural strains. Surprisingly, the only similar study of adults (Middleton and Kacser, 1983) found no correlation between Adh genotype and the rate of disappearance of ethanol in flies exposed to a pulse of ethanol vapor, leading Heinstra (Heinstra, 1993) to make the statement quoted above. Middleton and Kacser's (Middleton and Kacser, 1983) conclusions can be called into question on at least two grounds, however. First, the single AdhF and AdhS chromosomes they used showed substantially smaller differences in ADH activity than normal for these genotypes. Second, their statistical method for estimating the rate of ethanol elimination, regressing the log of ethanol concentration against time since exposure, may have made the estimates noisier than necessary.
In the present study, genetically diverse samples from a European population and an African population were compared in terms of internal ethanol concentration when exposed to a non-lethal dose of ethanol vapor, as well as ADH and ALDH activity. European flies had substantially higher activity of the two enzymes and accumulated less than half as much internal ethanol than the African flies. Ethanol concentrations were roughly consistent between two experiments differing in exposure time (4 h versus 24 h; Fig. 4), suggesting that concentrations had reached steady state, as occurred with larvae in the experiment of Heinstra et al. (Heinstra et al., 1987). The higher ADH and ALDH activities of the European flies suggests that some, if not all, of the ethanol concentration difference was the result of faster metabolism of ethanol by the European flies. Perhaps not coincidentally, the 2- to 3-fold differences in ethanol concentration reported here are similar to those reported between AdhF and AdhS homozygous larvae by Heinstra et al. (Heinstra et al., 1987), who were able to rule out differences in absorption. [Although I did not genotype the flies for Adh, extensive surveys have shown that the frequency of the Fast allele is >0.9 in most European populations, and <<0.1 in most tropical African populations (Oakeshott et al., 1982b; David et al., 1986).]
Another novel finding is that the European isofemale lines had higher ALDH activity than the African lines, with no overlap between the two groups. Although we previously reported an amino acid polymorphism affecting ALDH enzyme activity in which the more active variant increases in frequency with latitude (Fry et al., 2008), the majority of the European lines used for the enzyme assays were fixed for the same variant as found in all of the African lines. The consistently elevated ALDH activity of the European lines relative to the African lines therefore gives evidence that the former express higher levels of ALDH protein. Higher ALDH activity is expected to contribute to ethanol resistance by reducing levels of acetaldehyde, which is more toxic than ethanol (Leal and Barbancho, 1992; Deitrich, 2004). Although higher ALDH activity could also contribute to reduced internal ethanol concentrations because of the reversibility of the conversion of ethanol to acetaldehyde (Fig. 1), acetaldehyde is in very low concentration relative to ethanol in flies metabolizing ethanol (Leal and Barbancho, 1992), suggesting that this contribution is slight.
It is also possible that other enzymes contribute to the apparent faster metabolism of ethanol by European than African flies. Catalase and one or more unidentified cytochrome p450 oxidases account for about 10% of ethanol oxidation in Drosophila (Geer et al., 1993). An enzyme that makes an important indirect contribution is GPDH, which regenerates NAD+ needed for the reaction catalyzed by ADH (Cavener and Clegg, 1978; Nelson and Cox, 2008). Mutants with impaired GPDH activity substantially reduce ethanol resistance (Eanes et al., 2009). Interestingly, the derived GpdhS allele, like the AdhF allele, increases in frequency with latitude (David, 1982; Oakeshott et al., 1982b), and some evidence suggests it increases ethanol resistance (Cavener and Clegg, 1978).
Whatever its precise cause, the substantially lower internal ethanol concentration of European than African flies is likely to contribute to the higher ethanol resistance of the former. Estimating the magnitude of this contribution would require additional experiments.
Evidence that European third chromosomes confer resistance to metabolites of ethanol
Our results confirm those of Chakir et al. (Chakir et al., 1996), who found that much of the difference in ethanol resistance between a French and West African population mapped to the third chromosome, with a parallel effect on acetic acid resistance. These authors also found that selecting each population for resistance to one of the chemicals increased resistance to the other, giving evidence that the traits share a common genetic basis. The physiological basis of this correlation has remained obscure, however.
In this study, blocking ethanol metabolism with a null mutant of Adh eliminated the difference in ethanol resistance between a European and an African third chromosome line, giving evidence that European third chromosomes increase ethanol resistance primarily by increasing resistance to metabolites of ethanol, rather than ethanol per se. Although it is possible that flies with European third chromosomes are less sensitive to acetaldehyde than those with African third chromosomes, this by itself would not explain the higher acetic acid resistance of the former, because the conversion of acetaldehyde to acetate is essentially irreversible in vivo (Weiner, 1979). Therefore, it is likely that European third chromosomes increase resistance to acetic acid or something downstream of it (Fig. 1).
Studies in mammals have shown that acetate is responsible for some of the incoordination and sedation caused by ethanol administration, and is in fact as potent in these respects as ethanol itself (Israel et al., 1994; McLaughlin et al., 2008). A recent study showed that acetate may even be the primary cause of hangover headache (Maxwell et al., 2010). At least some of these effects appear to result from the accumulation of adenosine, which can be produced from the AMP generated when acetate is ligated to coenzyme A by acetyl-CoA synthetase (Fig. 1). In mammals, adenosine is a widespread signaling molecule with at least four different receptors, one of which promotes reduced heart rate, reduced respiration and sleep (Chen et al., 2013). In contrast, there appears to be only a single adenosine receptor in Drosophila, and comparatively little is known about its function (Dolezelova et al., 2007; Wu et al., 2009).
Another potential adverse effect of acetic acid is acidification, either directly or by inhibition of the pyruvate dehydrogenase complex by acetyl-CoA, causing pyruvate to be shunted into lactic acid production (Nelson and Cox, 2008). Excess acetyl-CoA can also lead to ketoacidosis (Nelson and Cox, 2008). Although acidosis is commonly observed in intoxicated emergency room patients, the precise cause of the acidosis is controversial (Zehtabchi et al., 2005), and it is not known whether any of these effects are important in Drosophila.
Chakir et al. (Chakir et al., 1996) hypothesized that the correlation between ethanol and acetic acid resistance in Drosophila could be the result of variation in the activity of acetyl-CoA synthetase (ACS), the gene for which (AcCoAS) is on the third chromosome. The authors argued that increased activity of the enzyme could increase resistance to both ethanol and acetic acid by increasing flux through their shared pathway. As noted above, however, flux is not synonymous with fitness. Moreover, with the exception of direct acidification by acetic acid, all of the hypothesized or documented adverse effects of acetate result from excess acetyl-CoA or AMP, suggesting that higher ACS activity would not necessarily be beneficial in flies exposed to high levels or ethanol or acetic acid. Consistent with this, a null mutation in AcCoAS, although reducing ethanol-induced hyperactivity, did not increase sensitivity to ethanol sedation (Kong et al., 2010). Furthermore, in a microarray study, no increase in AcCoAS expression was observed in populations selected for ethanol resistance (Yampolsky et al., 2012), nor do temperate and tropical populations appear to differ in ACS activity (Montooth et al., 2006). However, AcCoAS is inducible by ethanol in both adults (Kong et al., 2010) and larvae (Yampolsky et al., 2012), which could be maladaptive if excess AMP or acetyl-CoA is deleterious.
I also showed that European and African third chromosome lines showed little or no difference in internal ethanol concentration when in an ADH-positive background. Differences in the rate at which ethanol is absorbed or metabolized appear to make at most a minor contribution to the third chromosome effect on ethanol resistance.
Conclusions
This study gives evidence that both enhanced ethanol metabolism and increased resistance to downstream products of ethanol underlie the higher ethanol resistance of temperate compared with tropical D. melanogaster. The manner in which metabolites of ethanol reduce fitness and how European flies have adapted to these effects remain to be determined in future work.
MATERIALS AND METHODS
Fly stocks and rearing conditions
The European sample consisted of 22 isofemale lines (descendants of single, wild-caught inseminated females) collected in a forested area near Vienna, Austria, in 2004, kindly provided by C. Schlötterer. The African sample consisted of 29 lines collected in a village in Cameroon [‘CD’ lines of Pool and Aquadro (Pool and Aquadro, 2006)], also in 2004, kindly provided by J. Pool. Experiments were conducted on the isofemale lines and on stocks derived from them, as described in the sections below.
All flies were reared in shell vials on standard cornmeal-dead yeast–molasses medium. Stocks were maintained by mass-transfer on 3 week generations at 21°C. Flies used for experiments were reared at 25°C, with 2–8 females allowed to lay eggs per vial, adjusted inversely according to the fertility of the strains being used to avoid overcrowding and maintain a roughly standard larval density. Within a particular experiment, the same density was used for all strains. Flies were handled under light CO2 anesthesia.
Ethanol resistance, enzyme activity and ethanol concentrations of isofemale lines
To confirm the previously reported European–African difference in ethanol resistance, I measured the survival of adults exposed to varying concentrations of ethanol in 10 lines from each location. For each replicate, 10 males, 2–6 days post-emergence, were transferred without anesthesia from a holding vial to a vial containing a cotton ball moistened with 1.5 ml of a 3% sucrose solution containing 5%, 10% or 15% ethanol. Three vials per concentration, and one control vial lacking ethanol, were set up per line. Vials were corked, and live and dead flies counted after 2 days. Although flies could ingest ethanol by feeding on the sugar water, ethanol is volatile, and an experiment in our laboratory showed that respiration, rather than feeding, is the main route of ethanol entry when flies are placed in bottles with ethanol-supplemented food (Zhu, 2013).
I also measured the activity of ADH and ALDH in crude extracts of 2–6 day old males from the isofemale lines, using previously reported methods (Fry et al., 2004; Fry and Saweikis, 2006). Sixteen lines per location were assayed, including those used for the survival assays, with two independent replicates of 16 males each per line. The results are expressed as nmol NAD+ reduced min−1 mg−1 total protein in the extracts. The lines were also genotyped for the Aldh replacement polymorphism as described previously (Fry et al., 2008).
To measure internal ethanol concentrations of flies, I used an assay kit based on yeast ADH (Genzyme Diagnostics, Charlottetown, PE, Canada). Because the assays were relatively labor intensive, rather than measure individual isofemale lines, I established two sets of populations by hybridizing multiple lines from each location (N=16 lines per location in set 1, N=20 in set two). Populations were maintained at a large size (>500 flies) for several generations prior to the assays. To expose flies to ethanol, single-sex groups of 2–6 day old flies were placed in vials containing a cotton plug moistened with 1 ml 5% sucrose. A second cotton plug was then pushed into the middle of the vial, trapping flies between the plugs, and the vials corked. Flies were allowed to recover from anesthesia for 1 day, at which time 200 μl of 20% ethanol was pipetted onto the middle cotton plug, and the vials recorked. In this method, ethanol absorption is expected to be almost entirely by respiration, rather than feeding; the method also has some practical advantages over the more traditional method of dissolving ethanol in sugar water. The ethanol amount used caused no mortality or visible incoordination of flies during the assay period.
After being exposed to ethanol for 24 h (experiment with population set 1; referred to hereafter as ‘experiment 1’) or 4 h (experiment 2), flies were removed and immediately ground in cold 50 mmol l−1 Tris HCl buffer, pH 7.5 (12 females or 18 males per 120 μl of buffer in experiment 1, N=3–6 samples per sex per region; 6 females or 9 males per 100 μl in experiment 2, N=4 samples per sex per region). After centrifugation, 20 μl of supernatant was used for the ethanol assay. Separately, the protein concentration of 10 μl of each fly extract was estimated as described elsewhere (Fry et al., 2004), to provide a relative measure of the amount of fly biomass in solution.
Ethanol resistance and internal ethanol concentrations of third chromosome lines
I used the crossover suppressing balancer chromosome TM3 (Lindsley and Zimm, 1992) to place individual third chromosomes from the isofemale lines into a shared African genetic background. First, following standard procedures (Greenspan, 1997), third chromosomes were extracted into a non-isogenic background using the balancer stock +NA; +NA; TM3, Sb/H, where Sb and H are the dominant markers Stubble and Hairless, respectively, and the ‘+NA’ represent wild-type first (X) and second chromosomes from an outbred stock of North American origin. Next, third chromosomes with good viability and fertility as homozygotes in the North American background were placed into the African genetic background using a second, specially constructed balancer stock +C; bw; TM3, Sb/H. The X and second chromosomes of this stock were derived from a Cameroon isofemale line, except that the recessive visible marker bw (brown eyes) had been crossed onto the tip of the second chromosome. Molecular checks using the methods of Montooth et al. (Montooth et al., 2006), Fry et al. (Fry et al., 2008) and Andolfatto et al. (Andolfatto et al., 1999), respectively, showed that the second chromosome was homozygous for the ancestral AdhSlow and AldhLeu alleles, as well as the inversion In(2L)t, all consistent with its African origin. The resulting lines (four to five per region, each derived from a different isofemale line) had X and second chromosomes, as well as cytoplasm, derived entirely from the balancer stock.
I first measured ethanol resistance and internal ethanol concentrations of three third chromosome lines per region. After some of these lines proved to be difficult to rear in large numbers, probably due to their high level of inbreeding, I established hybrid populations (hereafter, ‘C3 hybrids’) by intercrossing four lines per region, and repeated the measurements on these. The ethanol concentration assays were done simultaneously and in the same manner as those described above, with the individual lines measured in experiment 1 (N=3 samples per sex per line; one Cameroon line was excluded because of poor rearing success) and the C3 hybrids measured in experiment 2 (N=6 samples per sex per population). For the ethanol resistance assays, instead of adding ethanol directly to the sugar water, I used a procedure similar to that for the ethanol concentration assays, adding 550 μl of 20% ethanol to the middle cotton plug. Both sexes were included, with four and nine vials per sex and strain in the tests of individual lines (10 flies per vial) and C3 hybrids (20 flies per vial), respectively. In this and all subsequent resistance assays, 2–6 day old flies were used, and survival was monitored after 2 days.
Relationship between ethanol and acetic acid resistance
To determine whether Vienna third chromosomes conferred higher acetic acid resistance than Cameroon third chromosomes, individual third chromosome lines and the C3 hybrids were measured for acetic acid resistance, using similar methods to those for ethanol resistance. For the assays using the individual lines, 125 μl of 40% glacial acetic acid was pipetted onto the cotton plug, with two vials of 10 flies each per sex and line. For the C3 hybrid populations, 150 μl was used, with six vials of 20 flies each per sex and population. Survival was measured after 2 days.
To determine whether the third chromosome effect on ethanol resistance persists when ethanol breakdown is blocked, I transferred the third chromosomes of one European and one African line from the African background, where they showed the typical difference in ethanol resistance, into a background lacking ADH activity. The crosses made use of a specially constructed balancer stock Adhfn6cn; TM3, Sb/H. The resulting lines were homozygous for the Adhfn6 allele, which codes for a truncated protein and produces little or no mature mRNA (Brogna, 1999). Acetic acid and ethanol resistance of the lines were measured using similar methods to those for the Adh+ third chromosome lines, with six and 10 vials per sex and line, respectively, and 15 flies per vial. The Adhfn6 homozygous flies, not surprisingly, were more sensitive to ethanol than flies with an active ADH, so the amount of 20% ethanol used was decreased by more than fourfold, to 125 μl. The Adhfn6 flies were somewhat more resistant to acetic acid than those with the Cameroon genetic background, possibly due to the temperate origin of their first and second chromosomes (cf. Chakir et al., 1996), so the amount of 40% acetic acid used was approximately doubled, to 275 μl.
Statistical analysis
Data were analyzed using the MIXED procedure in SAS (Littell et al., 1996). Region, sex and their interaction were treated as fixed effects. Where relevant, line within region and assay date (for experiments in which assays were performed on more than one day) were included as random effects. All possible interactions between fixed and random effects were also included. Random effects that failed to explain any variation were dropped from the models; those that yielded positive variance component estimates, even if non-significant, were retained. P-values of F-tests of fixed effects are reported in the Results; full analysis results, including random-effects variance components, are given in supplementary material Table S1.
All survival proportions were arcsin-square root transformed before analysis. Untransformed proportions and their standard errors were used for graphical purposes.
Supplementary Material
ACKNOWLEDGEMENTS
I thank J. Bucukovski, J. Budnik, A. Clarke, K. Donlon, T. Ivy, J. Zhu and M. Zink for their able help with the experiments. I also thank J. Pool, C. Schlötterer and the Bloomington Drosophila Stock Center for providing fly strains, and the anonymous reviewers for helpful comments.
FOOTNOTES
Competing interests
The author declares no competing financial interests.
Funding
This work was supported by the National Institutes of Health [grant number R01AA016178]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://jeb.biologists.org/lookup/suppl/doi:10.1242/jeb.110510/-/DC1
References
- Andolfatto P., Wall J. D., Kreitman M. (1999). Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153, 1297-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouletreau M., David J. R. (1981). Sexually dimorphic response to host habitat toxicity in Drosophila parasitic wasps. Evolution 35, 395-399. [DOI] [PubMed] [Google Scholar]
- Brogna S. (1999). Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA 5, 562-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener D. R., Clegg M. T. (1978). Dynamics of correlated genetic systems. IV. Multilocus effects of ethanol stress environments. Genetics 90, 629-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakir M., Peridy O., Capy P., Pla E., David J. R. (1993). Adaptation to alcoholic fermentation in Drosophila: a parallel selection imposed by environmental ethanol and acetic acid. Proc. Natl. Acad. Sci. USA 90, 3621-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakir M., Capy P., Genermont J., Pla E., David J. R. (1996). Adaptation to fermenting resources in Drosophila melanogaster: ethanol and acetic acid tolerances share a common genetic basis. Evolution 50, 767-776. [DOI] [PubMed] [Google Scholar]
- Chen J. F., Eltzschig H. K., Fredholm B. B. (2013). Adenosine receptors as drug targets – what are the challenges? Nat. Rev. Drug Discov. 12, 265-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan F. M., Graf J.-D. (1985). Latitudinal cline in Drosophila melanogaster for knockdown resistance to ethanol fumes and for rates of response to selection for further resistance. Evolution 39, 278-293. [DOI] [PubMed] [Google Scholar]
- David J. R. (1982). Latitudinal variability of Drosophila melanogaster: allozyme frequencies divergence between European and Afrotropical populations. Biochem. Genet. 20, 747-761. [DOI] [PubMed] [Google Scholar]
- David J. R., Bocquet C. (1975). Similarities and differences in latitudinal adaptation of two Drosophila sibling species. Nature 257, 588-590. [DOI] [PubMed] [Google Scholar]
- David J., Merçot H., Capy P., McEvey S., Van Herrewege J. (1986). Alcohol tolerance and Adh gene frequencies in European and African populations of Drosophila melanogaster. Genet. Sel. Evol. 18, 405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitrich R. A. (2004). Acetaldehyde: déjà vu du jour. J. Stud. Alcohol 65, 557-572. [DOI] [PubMed] [Google Scholar]
- Dolezelova E., Nothacker H. P., Civelli O., Bryant P. J., Zurovec M. (2007). A Drosophila adenosine receptor activates cAMP and calcium signaling. Insect Biochem. Mol. Biol. 37, 318-329. [DOI] [PubMed] [Google Scholar]
- Eanes W. F. (1999). Analysis of selection on enzyme polymorphisms. Annu. Rev. Ecol. Syst. 30, 301-326. [Google Scholar]
- Eanes W. F., Merritt T. J. S., Flowers J. M., Kumagai S., Zhu C.-T. (2009). Direct evidence that genetic variation in glycerol-3-phosphate and malate dehydrogenase genes (Gpdh and Mdh1) affects adult ethanol tolerance in Drosophila melanogaster. Genetics 181, 607-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisses K. T., Den Boer A. A. (1995). Acetic acid tolerance in Drosophila is a prerequisite for ethanol tolerance. J. Evol. Biol. 8, 481-491. [Google Scholar]
- Freriksen A., de Ruiter B. L. A., Groenenberg H.-J., Scharloo W., Heinstra P. W. H. (1994). A multilevel approach to the significance of genetic variation in alcohol dehydrogenase of Drosophila. Evolution 48, 781-790. [DOI] [PubMed] [Google Scholar]
- Fry J. D., Saweikis M. (2006). Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet. Res. 87, 87-92. [DOI] [PubMed] [Google Scholar]
- Fry J. D., Bahnck C. M., Mikucki M., Phadnis N., Slattery W. C. (2004). Dietary ethanol mediates selection on aldehyde dehydrogenase activity in Drosophila melanogaster. Integr. Comp. Biol. 44, 275-283. [DOI] [PubMed] [Google Scholar]
- Fry J. D., Donlon K., Saweikis M. (2008). A worldwide polymorphism in aldehyde dehydrogenase in Drosophila melanogaster: evidence for selection mediated by dietary ethanol. Evolution 62, 66-75. [DOI] [PubMed] [Google Scholar]
- Geer B. W., Heinstra P. W. H., McKechnie S. W. (1993). The biological basis of ethanol tolerance in Drosophila. Comp. Biochem. Physiol. 105B, 203-229. [DOI] [PubMed] [Google Scholar]
- Gibson J. B., May T. W., Wilks A. V. (1981). Genetic variation at the alcohol dehydrogenase locus in Drosophila melanogaster in relation to environmental variation: ethanol levels in breeding sites and allozyme frequencies. Oecologia 51, 191-198. [DOI] [PubMed] [Google Scholar]
- Greenspan R. J. (1997). Fly Pushing: The Theory and Practice of Drosophila Genetics. Plainview, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Harris R. A., Trudell J. R., Mihic S. J. (2008). Ethanol's molecular targets. Sci. Signal. 1, re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinstra P. W. H. (1993). Evolutionary genetics of the Drosophila alcohol dehydrogenase gene-enzyme system. Genetica 92, 1-22. [DOI] [PubMed] [Google Scholar]
- Heinstra P. W. H., Scharloo W., Thörig G. E. W. (1987). Physiological significance of the alcohol dehydrogenase polymorphism in larvae of Drosophila. Genetics 117, 75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel Y., Orrego H., Carmichael F. J. (1994). Acetate-mediated effects of ethanol. Alcohol. Clin. Exp. Res. 18, 144-148. [DOI] [PubMed] [Google Scholar]
- Kong E. C., Allouche L., Chapot P. A., Vranizan K., Moore M. S., Heberlein U., Wolf F. W. (2010). Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol. Clin. Exp. Res. 34, 302-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal J. F. M., Barbancho M. (1992). Acetaldehyde detoxification mechanisms in Drosophila melanogaster adults involving aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH) enzymes. Insect Biochem. Mol. Biol. 22, 885-892. [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Zimm G. G. (1992). The Genome of Drosophila melanogaster. San Diego, CA: Academic Press. [Google Scholar]
- Littell R. C., Milliken G. A., Stroup W. W., Wolfinger R. D. (1996). SAS System for Mixed Models. Cary, NC: SAS Institute Inc. [Google Scholar]
- Maxwell C. R., Spangenberg R. J., Hoek J. B., Silberstein S. D., Oshinsky M. L. (2010). Acetate causes alcohol hangover headache in rats. PLoS ONE 5, e15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie J. A., McKechnie S. W. (1979). A comparative study of resource utilization in natural populations of Drosophila melanogaster and D. simulans. Oecologia 40, 299-309. [DOI] [PubMed] [Google Scholar]
- McLaughlin P. J., Chuck T. L., Arizzi-LaFrance M. N., Salamone J. D., Correa M. (2008). Central vs. peripheral administration of ethanol, acetaldehydeand acetate in rats: effects on lever pressing and response initiation. Pharmacol. Biochem. Behav. 89, 304-313. [DOI] [PubMed] [Google Scholar]
- Merçot H., Defaye D., Capy P., Pla E., David J. R. (1994). Alcohol tolerance, ADH activity, and ecological niche of Drosophila species. Evolution 48, 746-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R. J., Kacser H. (1983). Enzyme variation, metabolic flux and fitness: alcohol dehydrogenase in Drosophila melanogaster. Genetics 105, 633-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Siebenthall K. T., Clark A. G. (2006). Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J. Exp. Biol. 209, 3837-3850. [DOI] [PubMed] [Google Scholar]
- Nelson D. L., Cox M. M. (2008). Lehninger Principles of Biochemistry. New York, NY: W. H. Freeman. [Google Scholar]
- Oakeshott J. G., May T. W., Gibson J. B., Willcocks D. A. (1982a). Resource partitioning in five domestic Drosophila species and its relationship to ethanol metabolism. Aust. J. Zool. 30, 547-556. [Google Scholar]
- Oakeshott J. G., Gibson J. B., Anderson P. R., Knibb W. R., Anderson D. G., Chambers G. K. (1982b). Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution 36, 86-96. [DOI] [PubMed] [Google Scholar]
- Parkash R., Karan D., Munjal A. K. (1999). Geographical variation in AdhF and alcoholic resource utilization in Indian populations of Drosophila melanogaster. Biol. J. Linn. Soc. Lond. 66, 205-214. [Google Scholar]
- Pool J. E., Aquadro C. F. (2006). History and structure of sub-Saharan populations of Drosophila melanogaster. Genetics 174, 915-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R. L., Hodgson E. (2001). Adaptation to toxicants. In Introduction to Biochemical Toxicology, 3rd edn (ed. Hodgson E., Smart R. C.), pp. 163-198 New York, NY: Wiley. [Google Scholar]
- Stam L. F., Laurie C. C. (1996). Molecular dissection of a major gene effect on a quantitative trait: the level of alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 144, 1559-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. (1979). Aldehyde dehydrogenase: mechanism of action and possible physiological roles. In Biochemistry and Pharmacology of Ethanol, Vol. 1 (ed. Majchrowicz E., Noble E. P.), pp. 107-124 New York, NY: Plenum. [Google Scholar]
- Wiens F., Zitzmann A., Lachance M. A., Yegles M., Pragst F., Wurst F. M., von Holst D., Guan S. L., Spanagel R. (2008). Chronic intake of fermented floral nectar by wild treeshrews. Proc. Natl. Acad. Sci. USA 105, 10426-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. N., Ho K., Crocker A., Yue Z., Koh K., Sehgal A. (2009). The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J. Neurosci. 29, 11029-11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky L. Y., Glazko G. V., Fry J. D. (2012). Evolution of gene expression and expression plasticity in long-term experimental populations of Drosophila melanogaster maintained under constant and variable ethanol stress. Mol. Ecol. 21, 4287-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehtabchi S., Sinert R., Baron B. J., Paladino L., Yadav K. (2005). Does ethanol explain the acidosis commonly seen in ethanol-intoxicated patients? Clin. Toxicol. 43, 161-166. [PubMed] [Google Scholar]
- Zhu J. (2013). Costs and benefits of sexual selection in Drosophila. PhD dissertation, University of Rochester, Rochester, NY, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.