Abstract
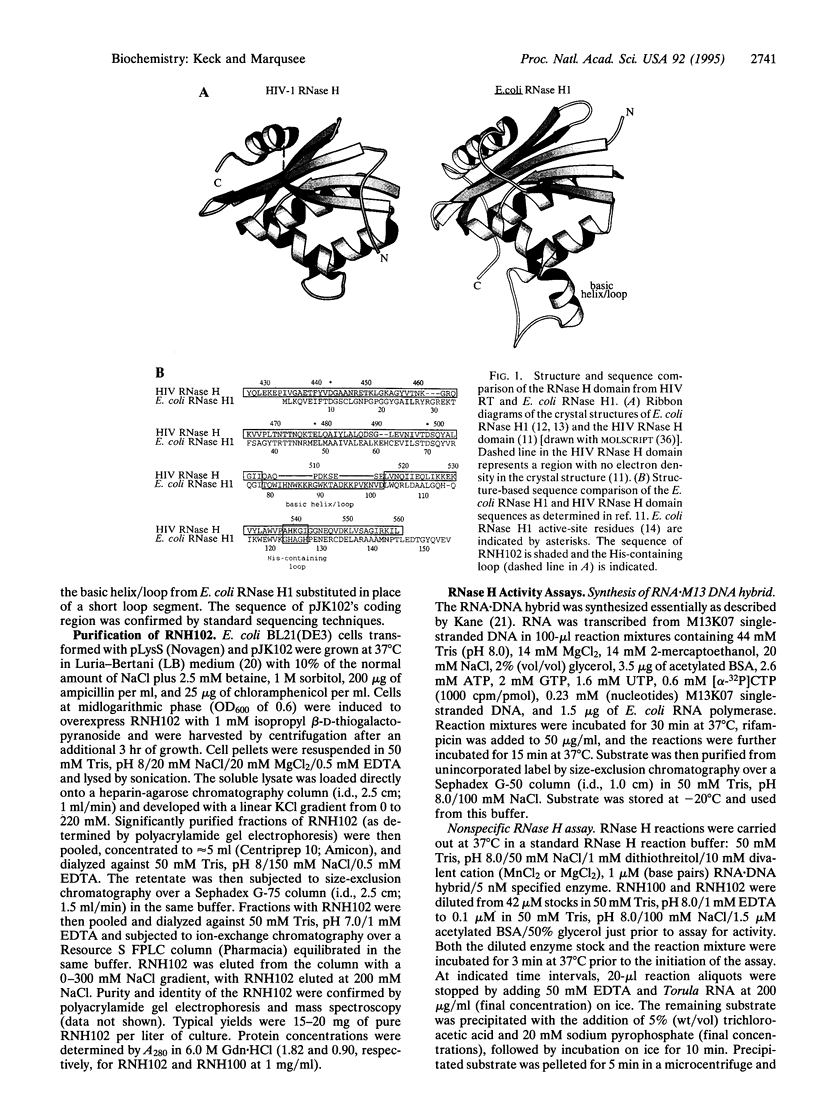
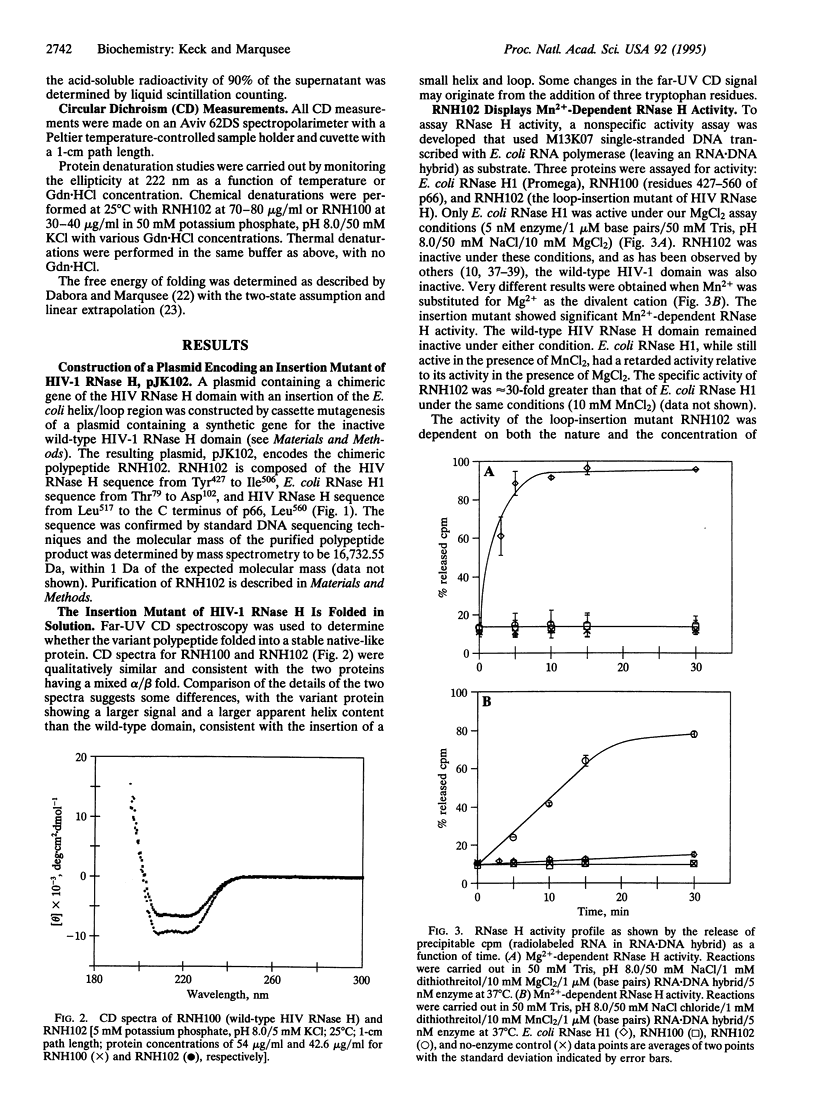
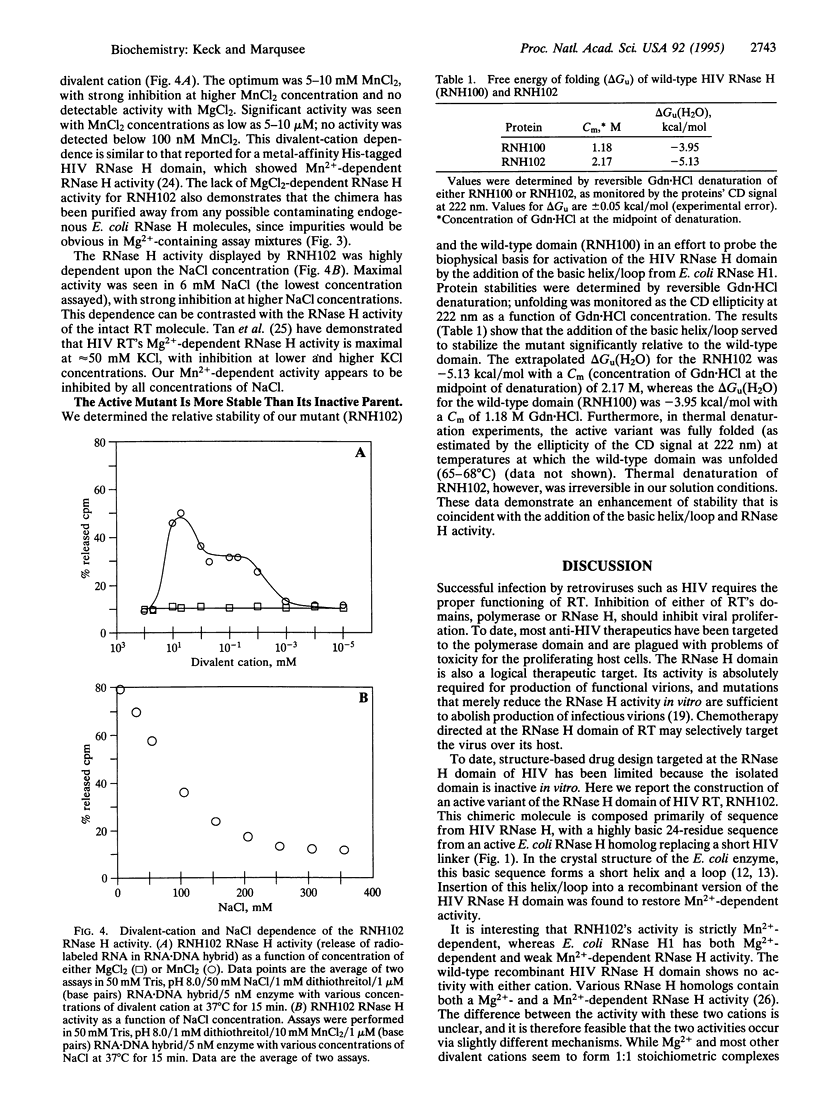
Human immunodeficiency virus (HIV) reverse transcriptase (RT) is a multifunctional protein, containing both DNA polymerase and RNase H activity. The RNase H activity of HIV RT catalyzes the hydrolysis of the RNA strand of RNA.DNA hybrids. While the domain that carries out the RNase H activity in HIV RT can be expressed as an independent, folded polypeptide, it is inactive as an RNase H. Here, we report the overexpression and purification of an active, recombinant HIV RNase H domain in which the sequence corresponding to the Escherichia coli RNase H1 basic helix/loop has been substituted for the corresponding sequence of HIV RNase H. The resulting polypeptide (RNH102) has Mn(2+)-dependent RNase H activity and is more stable than the independently expressed wild-type HIV RNase H domain.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becerra S. P., Clore G. M., Gronenborn A. M., Karlström A. R., Stahl S. J., Wilson S. H., Wingfield P. T. Purification and characterization of the RNase H domain of HIV-1 reverse transcriptase expressed in recombinant Escherichia coli. FEBS Lett. 1990 Sep 17;270(1-2):76–80. doi: 10.1016/0014-5793(90)81238-j. [DOI] [PubMed] [Google Scholar]
- Boyer P. L., Ferris A. L., Hughes S. H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 1992 Feb;66(2):1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay D., Finzel B. C., Munson S. H., Evans D. B., Sharma S. K., Strakalaitus N. A., Brunner D. P., Eckenrode F. M., Dauter Z., Betzel C. Crystallographic analyses of an active HIV-1 ribonuclease H domain show structural features that distinguish it from the inactive form. Acta Crystallogr D Biol Crystallogr. 1993 Jul 1;49(Pt 4):423–427. doi: 10.1107/S0907444993002409. [DOI] [PubMed] [Google Scholar]
- Dabora J. M., Marqusee S. Equilibrium unfolding of Escherichia coli ribonuclease H: characterization of a partially folded state. Protein Sci. 1994 Sep;3(9):1401–1408. doi: 10.1002/pro.5560030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Evans D. B., Brawn K., Deibel M. R., Jr, Tarpley W. G., Sharma S. K. A recombinant ribonuclease H domain of HIV-1 reverse transcriptase that is enzymatically active. J Biol Chem. 1991 Nov 5;266(31):20583–20585. [PubMed] [Google Scholar]
- Ferris A. L., Hizi A., Showalter S. D., Pichuantes S., Babe L., Craik C. S., Hughes S. H. Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure. Virology. 1990 Apr;175(2):456–464. doi: 10.1016/0042-6822(90)90430-y. [DOI] [PubMed] [Google Scholar]
- Hansen J., Schulze T., Mellert W., Moelling K. Identification and characterization of HIV-specific RNase H by monoclonal antibody. EMBO J. 1988 Jan;7(1):239–243. doi: 10.1002/j.1460-2075.1988.tb02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A., Hughes S. H., Shaharabany M. Mutational analysis of the ribonuclease H activity of human immunodeficiency virus 1 reverse transcriptase. Virology. 1990 Apr;175(2):575–580. doi: 10.1016/0042-6822(90)90444-v. [DOI] [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Hudson G. O., Moomaw E. W., Nodes B. R. Reconstitution in vitro of RNase H activity by using purified N-terminal and C-terminal domains of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1148–1152. doi: 10.1073/pnas.88.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. W., Cowan J. A. Metallobiochemistry of the magnesium ion. Characterization of the essential metal-binding site in Escherichia coli ribonuclease H. Eur J Biochem. 1994 Jan 15;219(1-2):253–260. doi: 10.1111/j.1432-1033.1994.tb19936.x. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S., Katsuda-Nakai C., Ikehara M. Importance of the positive charge cluster in Escherichia coli ribonuclease HI for the effective binding of the substrate. J Biol Chem. 1991 Jun 25;266(18):11621–11627. [PubMed] [Google Scholar]
- Kanaya S., Kohara A., Miura Y., Sekiguchi A., Iwai S., Inoue H., Ohtsuka E., Ikehara M. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem. 1990 Mar 15;265(8):4615–4621. [PubMed] [Google Scholar]
- Kane C. M. Renaturase and ribonuclease H: a novel mechanism that influences transcript displacement by RNA polymerase II in vitro. Biochemistry. 1988 May 3;27(9):3187–3196. doi: 10.1021/bi00409a010. [DOI] [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- Katayanagi K., Okumura M., Morikawa K. Crystal structure of Escherichia coli RNase HI in complex with Mg2+ at 2.8 A resolution: proof for a single Mg(2+)-binding site. Proteins. 1993 Dec;17(4):337–346. doi: 10.1002/prot.340170402. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Lightfoote M. M., Coligan J. E., Folks T. M., Fauci A. S., Martin M. A., Venkatesan S. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J Virol. 1986 Nov;60(2):771–775. doi: 10.1128/jvi.60.2.771-775.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Yoshida M., Kanaya S. Role of histidine 124 in the catalytic function of ribonuclease HI from Escherichia coli. J Biol Chem. 1993 Jan 5;268(1):88–92. [PubMed] [Google Scholar]
- Powers R., Clore G. M., Bax A., Garrett D. S., Stahl S. J., Wingfield P. T., Gronenborn A. M. Secondary structure of the ribonuclease H domain of the human immunodeficiency virus reverse transcriptase in solution using three-dimensional double and triple resonance heteronuclear magnetic resonance spectroscopy. J Mol Biol. 1991 Oct 20;221(4):1081–1090. doi: 10.1016/0022-2836(91)90920-2. [DOI] [PubMed] [Google Scholar]
- Powers R., Clore G. M., Stahl S. J., Wingfield P. T., Gronenborn A. Analysis of the backbone dynamics of the ribonuclease H domain of the human immunodeficiency virus reverse transcriptase using 15N relaxation measurements. Biochemistry. 1992 Sep 29;31(38):9150–9157. doi: 10.1021/bi00153a006. [DOI] [PubMed] [Google Scholar]
- Prasad V. R., Goff S. P. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc Natl Acad Sci U S A. 1989 May;86(9):3104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restle T., Müller B., Goody R. S. RNase H activity of HIV reverse transcriptases is confined exclusively to the dimeric forms. FEBS Lett. 1992 Mar 23;300(1):97–100. doi: 10.1016/0014-5793(92)80172-d. [DOI] [PubMed] [Google Scholar]
- Santoro M. M., Bolen D. W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988 Oct 18;27(21):8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- Schatz O., Cromme F. V., Grüninger-Leitch F., Le Grice S. F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989 Nov 6;257(2):311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- Smith J. S., Roth M. J. Purification and characterization of an active human immunodeficiency virus type 1 RNase H domain. J Virol. 1993 Jul;67(7):4037–4049. doi: 10.1128/jvi.67.7.4037-4049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S. J., Kaufman J. D., Vikić-Topić S., Crouch R. J., Wingfield P. T. Construction of an enzymatically active ribonuclease H domain of human immunodeficiency virus type 1 reverse transcriptase. Protein Eng. 1994 Sep;7(9):1103–1108. doi: 10.1093/protein/7.9.1103. [DOI] [PubMed] [Google Scholar]
- Stammers D. K., Tisdale M., Court S., Parmar V., Bradley C., Ross C. K. Rapid purification and characterisation of HIV-1 reverse transcriptase and RNaseH engineered to incorporate a C-terminal tripeptide alpha-tubulin epitope. FEBS Lett. 1991 Jun 3;283(2):298–302. doi: 10.1016/0014-5793(91)80613-8. [DOI] [PubMed] [Google Scholar]
- Tan C. K., Zhang J., Li Z. Y., Tarpley W. G., Downey K. M., So A. G. Functional characterization of RNA-dependent DNA polymerase and RNase H activities of a recombinant HIV reverse transcriptase. Biochemistry. 1991 Mar 12;30(10):2651–2655. doi: 10.1021/bi00224a013. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A., Blain S. W., Goff S. P. Defects in Moloney murine leukemia virus replication caused by a reverse transcriptase mutation modeled on the structure of Escherichia coli RNase H. J Virol. 1992 Feb;66(2):615–622. doi: 10.1128/jvi.66.2.615-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale M., Schulze T., Larder B. A., Moelling K. Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J Gen Virol. 1991 Jan;72(Pt 1):59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- Wöhrl B. M., Volkmann S., Moelling K. Mutations of a conserved residue within HIV-1 ribonuclease H affect its exo- and endonuclease activities. J Mol Biol. 1991 Aug 5;220(3):801–818. doi: 10.1016/0022-2836(91)90119-q. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986 Mar 14;231(4743):1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]




