Abstract
Background
Several major outbreaks in healthcare facilities have occurred with the emergence of multi-resistant bacteria. A possible route for dissemination is the hospital textiles and inadequate laundering of them. The aim of this study was to develop an easy-to-use method for simulating the laundering process of hospital textiles, and thereafter apply the method when evaluating the decontaminating efficacy of two different washing temperatures.
Methods
The laundering process, including tumble drying, took place at two professional laundries. Enterococcus faecium was used as bioindicator.
Results
The results showed that a lowering of the washing temperature from 70°C to 60°C did not affect the decontamination efficacy; the washing cycle alone reduced the number of bacteria with 3–5 log10 CFU, whereas the following tumble drying reduced the bacterial numbers with another 3–4 log10 CFU, yielding the same final result independent of washing temperature. Without tumble drying, there was an obvious risk of adding non-fermenting gram-negative bacteria to the fabric. These bacteria originated from the washing cycle.
Conclusion
A simple method to simulate hospital laundering was developed. To save energy, it is possible to use a washing temperature of 60°C, but the washing cycle should be followed by tumble drying, and the whole laundering process needs to be monitored to maintain sufficient textile hygiene.
Keywords: laundry, high temperature, tumbling, bacterial cleanness, textiles
Several tons of laundry is produced in hospitals every day. A small-sized primary care hospital in Sweden can easily handle 40 tons of laundry per year, whereas the corresponding figure for a county or a region is rather 40 tons per day. The cost for washing these large amounts of textiles is high. In an American study from 1981, it was estimated that hospital laundries accounted for 10–15% of the energy consumed in hospitals (1). Not surprisingly, there has, in late years, been a trend of economizing on these costs by reducing time, energy, water, detergents, and disinfecting agents in the washing machines. The risk of bacteria surviving the laundry process has thereby increased. Since bacteria can survive on fabrics for a month or more (2), clothing, bed linen, towels, etc., used in hospitals can act as sources of infections for patients (2–6) although they are seldom implicated (7, 8).
To ensure a certain level of quality, resulting in clean and microbiologically acceptable products, there are official recommendations for hospital laundering in many countries. The recommendations in Sweden state that hospital laundry should be washed at 70°C or more for at least 10 min. This recommendation is to a large extent followed, but high washing temperatures for longer periods of time result in a significant environmental impact through large energy expenditure and shortened lifetime of the fabrics. A common alternative is to wash at lower temperatures and combine this with a biocide. The latter alternative threatens, however, human health and the environment, and contamination of water with biocides can in the long run be more problematic than high-energy consumption (9). There is, unfortunately, a lack of standardized methods to validate the decontaminating effects of a washing process. It is thereby difficult to evaluate the impact of changes in a full laundering process, including both washing and drying, on microorganisms of clinical importance.
The aims of this study were to develop an easily performed method for evaluating the decontaminating efficacy of a laundering process. Focus was on how the bacterial survival was affected by different washing temperatures followed by tumble drying.
Materials and methods
Settings
During the years 2005–2007, there was a major outbreak at Uppsala University Hospital caused by a multi-resistant Klebsiella pneumoniae strain producing CTX-M-15 (10). While searching for possible sources, it was noticed that the outbreak strain, like many other extended-spectrum β-lactamase (ESBL)-producers (11) had a high ability to survive on different types of fabrics. There was concern that the current laundering process at the hospital and in homes for senior citizens could be insufficient. A study was therefore initiated.
To simulate a true situation as much as possible, all washing and drying processes were carried out at two professional laundries with years of experience of hospital laundering. The laundering was initially planned to take place in a single professional laundry, but due to technical problems related to the lowered washing temperature (from 70°C to 60°C), another laundry was engaged to complete the study. The preparation of the contaminated cloth samples and the control of the level of decontamination after different procedures were performed at the Department of Clinical Microbiology, Uppsala University Hospital, Uppsala, Sweden.
Bacteria and inocula
Two bacterial strains were included in the study: the outbreak strain K. pneumoniae CCUG54718 and Enterococcus faecium NCTC7171/CCUG33573. Both species are common causes of healthcare-associated infections. The relatively thermo tolerant E. faecium was also included since it is frequently used as a bioindicator in laundry studies and in European tests for determining basic bactericidal disinfectant efficiency (12–16).
Prior to the experiments, a single colony from an overnight culture of each strain was inoculated in brain heart infusion broth (BBL™, Becton, Dickinson and Co., Sparks, USA) and incubated at 35°C for 5 h to obtain a bacterial concentration of 108–109 CFU/mL.
Determination of viable bacteria
The bacterial suspension was diluted serial 1/100 with phosphate-buffered saline (PBS), and from each dilution 10 µL and 100 µL were spread in duplicates on blood agar (Neogen, Lansing, MI, USA). After incubating for 20 h at 35°C, viable counts were performed, using three different plates containing 30–300 CFU. Viable counts were performed to verify the bacterial concentration at each experiment.
Control of bacterial tolerance to heat
Tubes containing 1 mL of 5-h broth cultures of E. faecium or K. pneumoniae were exposed in duplicates to two different temperatures (65°C and 85°C) in a thermo-block for up to 30 min. Viable counts were performed after 0, 10, 20, and 30 min. As control, a bacterial culture kept at 35°C was used. The experiment was repeated for the E. faecium strain with a broader temperature range (60°C–90°C).
Preparation of test samples
In each experiment, 14 pieces of sterilized cloth (cotton/polyester 50/50, originating from hospital staff clothes and measuring 10×10 cm) were used. They will hereafter be referred to as test samples. All test samples were contaminated with 1 mL of an E. faecium CCUG33573 suspension. Each test sample was allowed to dry at 35°C in sterile plastic petri dishes (diameter 14 cm).
Processing of test samples
Test samples 1–2 were used for controlling the bacterial number on day 0. The remaining 12 test samples were left in the petri dishes, which were sealed with tape and placed in plastic bags for transport to the laundry. See Fig. 1 for flow chart.
Fig. 1.
Flow chart showing how the test samples were processed at professional laundry I (70°C) and professional laundry II (60°C).
At arrival, test samples 3–4 stayed packaged; they were controls and did not pass through the washing process, consisting of washing, rinsing and spinning phases in a continuous batch (tunnel) washer. In experiments 1–10 out of 13, test samples 5–14 were put into separate laundry bags and washed the same day at 70°C. Water and tumbling temperatures were monitored throughout every process.
After the washing and drying by tumbling, the test samples were placed in separate clean plastic bags. In experiments 11–13, the test pieces were handled in the same way with one important exception: test samples 5–9 were excluded from the drying process. The test samples were thereafter transported back to the laboratory and refrigerated. The temperature inside the refrigerated transport box was measured when it arrived to the laboratory.
Within 20 h after completion of the laundering, test samples 3–14 were examined for the presence of bacteria. The Stomacher plastic bags were filled with 50 mL peptone water (Neogen, Lansing, Michigan, USA) and processed in the Stomacher® 400 (Circulator, Seward medical, UK) for 5 min. The suspension without textile was transferred into sterile bottles containing 25 glass beads. The bottles were shaken at 240 rpm for 10 min on a 4010 Multi-Tube Vortexer, Corning. From each suspension, 10 µL, 100 µL, and 2×200 µL were cultured on blood agar plates for the quantification of growing bacteria and E. faecium in particular. The remaining suspension was filtered through a 0.22-µm cellulose filter (Millipore Corporation, Bedford, USA), which was placed on a blood agar plate. The plate was incubated at 37°C for 48 h, and the total numbers of bacteria and of E. faecium were counted. The cellulose filter was thereafter transferred to a plate selective for enterococci (Enterococcosel™ Agar, Becton, Dickinson and Company, Sparks, USA) and incubated overnight at 37°C for identification of the inoculums strain.
After the first 13 experiments, 10 more followed. In these latter 10 experiments, three changes were made: the laundering process took place at another professional laundry, the temperature was lowered to 60°C, and the tumble drying process was excluded for test samples 5–9 in all experiments. Both laundries followed their current routines and used their own choice of detergent. No biocides were included in any washing process.
Results
Bacterial tolerance to heat
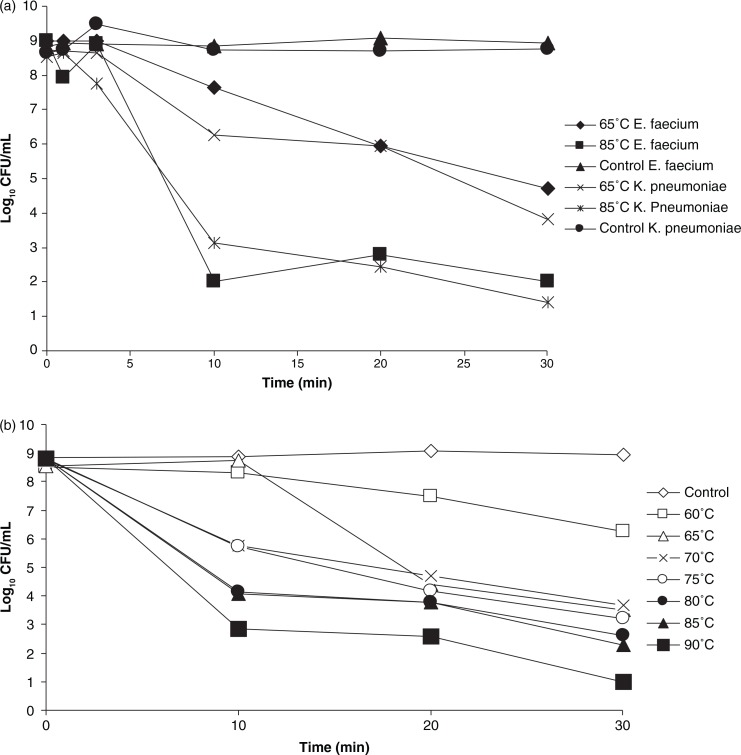
Results of the heat tolerance experiment showed that there was a reduction of K. pneumoniae CCUG 54718 with 4–7 log10 CFU after 30 min exposure to temperatures between 65°C and 85°C. E. faecium NCTC7171 exhibited a thermotolerance that was similar to that of K. pneumoniae CCUG 54718 (see Fig. 2a). To reduce the risks of spreading an epidemic strain, the susceptible enterocccal strain, and not the multi-resistant K. pneumoniae strain, was used in all the following laundering experiments.
Fig. 2.
(a) Tolerance of strains K. pneumoniae CCUG54718 and E. faecium NCTC7171/CCUG33573 to the temperatures 65°C and 85°C. (b) Tolerance of E. faecium strain NCTC7171/CCUG33573 to temperatures 60°C–90°C.
The enterococcal strain showed that the lower the temperature, the longer time of exposure was needed for reducing the bacterial number. At temperatures ≥70°C, the major killing took place during the first 10 min (see Fig. 2b).
Laundering at 70°C
All test samples were impregnated with an average of 108–109 CFU E. faecium (test samples 1–2). Unwashed test samples (3–4) contained 4–9 log10 CFU when returned to the laboratory.
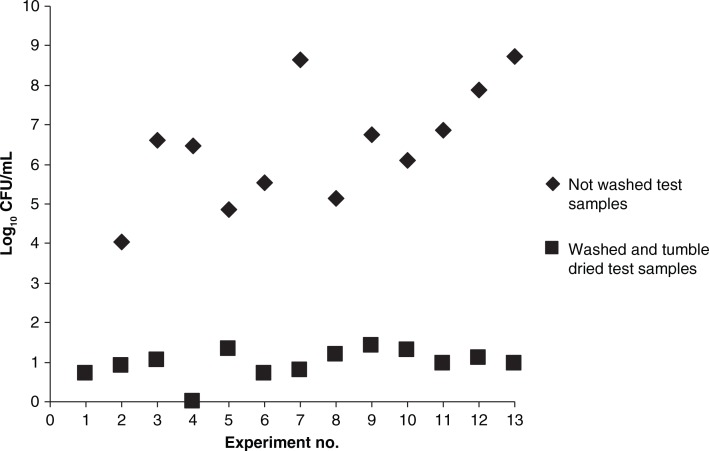
The median washing time at 70°C was 15 min (range 10–20 min) and for the full washing circle 30 min (range 30–55 min). The washing process alone reduced the number of bacteria with about 5 log10 CFU. The washing process followed by tumble drying, reduced the bacterial numbers with up to 9 log10 CFU. The observed killing effect of the tumble drying, was the background to the changes in the study protocol; in the following experiments the washing process and the tumble drying was separated at all times (see Fig. 3). The median drying temperature during tumbling was 78°C (range 63°C–85°C), and the drying process lasted for a median of 22 min (range 17–30 min).
Fig. 3.
Viable counts from test samples before and after a full laundry process, using 70°C wash cycle and tumbling at a median temperature of 78°C. Each point in the graph represents the median number of bacterial cells from two samples before washing and 10 samples after the tumbling.
Laundering at 60°C
The test samples were impregnated with the same level of bacteria as in the first 13 experiments (average of 108–109 CFU), but the unwashed test samples (3–4) contained essentially the same amount of bacteria as test samples 1–2 at the return to the laboratory.
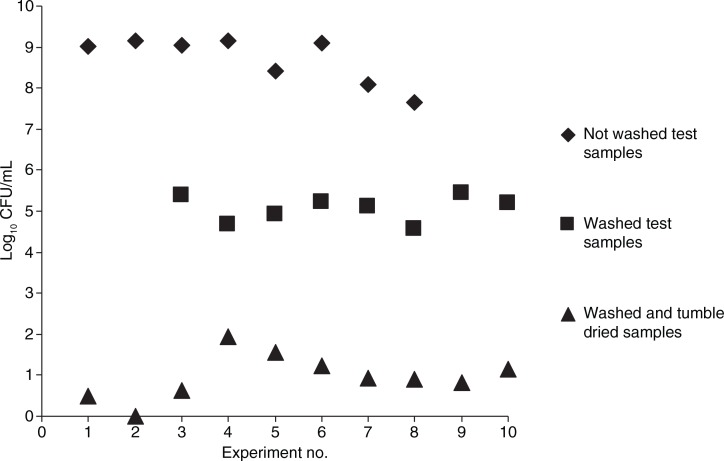
The median washing time at 60°C was 15 min (range 13–17 min) and for the full washing circle 30 min (range 30–33 min). The washing process alone reduced the number of bacteria with 4–5 log10 CFU. The corresponding figure for the tumble drying was 3–4 log10 CFU, yielding a total reduction of bacteria of up to 9 log10 CFU for a full laundering process including the tumble drying (see Fig. 4).
Fig. 4.
Viable counts from test samples before the laundering, after a wash cycle at 60°C, and after a full laundry process (washing at 60°C and tumble drying at a median temperature of 117.5°C). Each point in the graph represents the median number of bacterial cells from two samples before washing and five samples after washing without tumble drying and five samples after washing with tumble drying.
The median temperature during tumble drying was 117.5°C (range 112°C–123°C), whereas the total time for drying was a median of 14.8 min (range 12–17.4 min). The temperature during the tumble drying was thereby higher but lasted for a shorter time period than during the first 13 experiments.
The number of E. faecium cells was not possible to count in the first two experiments due to overgrowth of non-fermenting gram-negative bacteria. This contamination had the washing process as its source.
Discussion
In the present study, a simple method for simulating a full laundering process in a professional laundry was developed to evaluate how different washing temperatures affected the bacterial decontamination. The results showed that the decontamination of the test samples was comparable at 70 and 60°C. Furthermore, it became evident that the following drying process made an important contribution to the reduction of the bacterial number. In fact, without tumble drying there was an obvious risk of contaminating the test samples heavily with non-fermenting gram-negative bacteria during the washing process. Other studies have yielded similar results (17, 18).
In recent years, the interest in laundering processes and their ability to reduce microbial contamination has mounted. The basis for this is to a large extent the increased frequency of outbreaks caused by multi-resistant bacteria in both healthcare facilities and in the community. During outbreak investigations, infection control teams usually check the laundry routines, especially if an outbreak does not seem to have an obvious cause. To give sound advice in these situations is a difficult task. Studies dealing with laundry have yielded a wide range of results, and they are not always easy to interpret or draw conclusions from. This is to a large extent due to the high number of variables that can affect the outcome, that is, the type of microorganisms/washing machine/fabric, inoculum concentration, laundering conditions, added detergents and/or biocides, drying routines, etc. (19).
Some studies have focused on the washing temperature, whereas others have included the drying temperature or the energy costs (13). However, tumble drying the textiles has in several studies, including this, been shown to have significant impact on the decontamination (17). Ironing also seems to be of importance for reducing the bacterial number (18, 20). The role of procedures like tumble drying and ironing, which follow the washing cycle, probably increases when the laundry process cannot be fully monitored. This is usually the case when conventional washing machines are used in old people's homes, other types of healthcare facilities and under home-like conditions. The temperature in these instances is frequently lowered to 30–40°C to not damage the clothes. At these temperatures, the risk of laundry malodor and contamination is obvious (21–23). To let the hospital staff wash their hospital clothing in their private washing machines without tumble drying or ironing is also a risk that needs to be further explored.
Enterococci are notoriously difficult to get rid of and are therefore often used as bioindicators (13–15). Previous studies have shown that they can survive laundering at 60°C for 10 min (12–13) but not 75°C (24). Their high tolerance to heat was also shown in the present study by E. faecium strain NCTC7171. There was, however, a difference between the tolerance at temperatures below 70°C and those at 70°C or more. In the former case, 10 min was not enough to reduce the bacterial number. This difference in temperature tolerance was not noticed when laundering at 70°C and 60°C, indicating that not only the washing temperature but also the centrifugation and rinsing contributes to the decontamination in a washing process.
The multi-resistant K. pneumoniae strain, which caused the major outbreak at the hospital, was surprisingly heat tolerant. Due to the risks of further dissemination, it was not used during the laundering experiments. It can be argued that a susceptible member of the Enterobacteriaceae family should have been included in the laundering experiments instead. They seem, however, to be not as good environmental survivors as their multi-resistant variants (25), and there was a need for rapid answers with a limited budget. A more comprehensive study, including also susceptible K. pneumoniae strains, viruses and fungi, was therefore not possible.
When starting up a new method, there are often problems. One of them was the transportation of samples to and from the professional laundry, which was not optimal during the first part of the study (when washing at 70°C). Furthermore, the laboratory technician who performed the initial experiments was not accustomed to dealing with this type of sample or carrying out viable counts. A combination of these factors is probably the cause of the large variation in bacterial numbers during the laundering at 70°C. After corrections, the reproducibility increased.
In conclusion, a method for simulating a full laundering process in a professional laundry was developed in this study. When applied, the method showed that it is possible to maintain sufficient textile hygiene where bacteria are concerned when washing at 60°C without adding biocides. However, for the best result the washing cycle must be followed by tumble drying. Furthermore, the whole laundering process should be monitored.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Battles DR, Vesley D. Wash water temperature and sanitation in the hospital laundry. J Environ Health. 1981;43:244–50. [PubMed] [Google Scholar]
- 2.Fijan S, Šostar-Turk S. Hospital textiles, are they a possible vehicle for healthcare-associated infections? Int J Environ Res Public Health. 2012;9:330–43. doi: 10.3390/ijerph9093330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weernink A, Severin WPJ, Tjernberg I, Dijkshoorn L. Pillows, an unexpected source of Acinetobacter. J Hosp Infect. 1995;29:189–99. doi: 10.1016/0195-6701(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 4.Balm MND, Jureen R, Teo C, Yeoh AEJ, Lin RTP, Dancer SJ, et al. Hot and steamy: outbreak of Bacillus cereus in Singapore associated with construction work and laundry practices. J Hosp Infect. 2012;81:224–30. doi: 10.1016/j.jhin.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Dohmae S, Okubo T, Higuchi W, Takano T, Isobe H, Baranovich T, et al. Bacillus cereus nosocomial infection from reused towels in Japan. J Hosp Infect. 2008;69:361–7. doi: 10.1016/j.jhin.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Hosein IK, Hoffman PN, Ellam S, Asseez TM, Fakokunde A, Silles J, et al. Summertime Bacillus cereus colonization of hospital newborns traced to contaminated, laundered linen. J Hosp Infect. 2013;85:149–54. doi: 10.1016/j.jhin.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Brunton WAT. Infection and hospital laundry. Lancet. 1995;345:1574–5. doi: 10.1016/s0140-6736(95)91124-3. [DOI] [PubMed] [Google Scholar]
- 8.Brich BR, Perera BS, Hyde WA, Ruehorn V, Gaguli LA, Kramer JM, et al. Bacillus cereus cross-infection in a maternity-unit. J Hosp Infect. 1981;2:349–54. doi: 10.1016/0195-6701(81)90067-0. [DOI] [PubMed] [Google Scholar]
- 9.Fijan S, Škerget M, Knez Z, Šostar-Turk S, Neral B. Determining the disinfection of textiles in compressed carbon dioxide using various indicator microbes. J Appl Microbiol. 2011;112:475–84. doi: 10.1111/j.1365-2672.2011.05216.x. [DOI] [PubMed] [Google Scholar]
- 10.Lytsy B, Sandegren L, Tano E, Torell E, Andersson DI, Melhus Å. The first major extended-spectrum beta-lactamase outbreak in Scandinavia was caused by clonal spread of multiresistant Klebsiella pneumoniae producing CTX-M-15. APMIS. 2008;116:302–8. doi: 10.1111/j.1600-0463.2008.00922.x. [DOI] [PubMed] [Google Scholar]
- 11.Starlander G, Yin H, Edquist P, Melhus Å. Survival in the environment is a possible key factor for the expansion of Escherichia coli strains producing extended-spectrum β-lactamases. APMIS. 2013;122:59–67. doi: 10.1111/apm.12102. [DOI] [PubMed] [Google Scholar]
- 12.Orr KE, Holliday MG, Jones AL, Robson I, Perry JD. Survival of enterococci during hospital laundry processing. J Hosp Infect. 2002;50:133–9. doi: 10.1053/jhin.2001.1137. [DOI] [PubMed] [Google Scholar]
- 13.Wilcox MH, Jones BL. Enterococci and hospital laundry. Lancet. 1995;345:594. doi: 10.1016/s0140-6736(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 14.Bradley CR, Fraise AP. Heat and chemical resistance of enterococci. J Hosp Infect. 1996;34:191–6. doi: 10.1016/s0195-6701(96)90065-1. [DOI] [PubMed] [Google Scholar]
- 15.Fijan S, Šostar-Turk S, Cencič A. Implementing hygiene monitoring systems in hospital laundries in order to reduce microbial contamination of hospital textiles. J Hosp Infect. 2005;61:30–8. doi: 10.1016/j.jhin.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Fijan S, Cencič A, Šostar-Turk S. Hygiene monitoring of textiles used in the food industry. Braz J Microbiol. 2006;37:356–61. [Google Scholar]
- 17.Rutala WA, Weber DJ. Uses of inorganic hypochlorite (bleach) in health-care facilities. Clin Microbiol Rev. 1997;10:597–610. doi: 10.1128/cmr.10.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel SN, Murray-Leonard J, Wilson APR. Laundering of hospital staff uniforms at home. J Hosp Infect. 2006;62:89–93. doi: 10.1016/j.jhin.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Cristian RR, Manchester JT, Mellor MT. Bacteriological quality of fabrics washed at lower-than-standard temperatures in hospital laundry facility. Appl Environ Microbiol. 1983;45:591–7. doi: 10.1128/aem.45.2.591-597.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakdawala N, Pham J, Shah M, Holton J. Effectiveness of low-temperature domestic laundry on the decontamination of healthcare workers' uniforms. Infect Control Hosp Epidemiol. 2011;32:1103–8. doi: 10.1086/662183. [DOI] [PubMed] [Google Scholar]
- 21.Hall TJ, Wren MWD, Jeanes A, Gant VA. Decontamination of laundry at low temperature with CuWB50, a novel copper-based biocidal compound. Am J Infect Control. 2009;37:478–83. doi: 10.1016/j.ajic.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 22.Kubota H, Mitani A, Niwano Y, Takeuchi K, Tanaka A, Yamaguchi N, et al. Moraxella species are primarily responsible for generating malodor in laundry. Appl Environ Microbiol. 2012;78:3317–24. doi: 10.1128/AEM.07816-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stapleton K, Hill K, Day K, Perry JD, Dean JR. The potential impact of washing machines on laundry malodour generation. Lett Appl Microbiol. 2013;56:299–306. doi: 10.1111/lam.12050. [DOI] [PubMed] [Google Scholar]
- 24.Fijan S, Koren S, Cencič A, Šostar-Turk S. Antimicrobial disinfection effect of a laundering procedure for hospital textiles against various indicator bacteria and fungi using different substrates for simulating human excrements. Diagn Microbiol Infect Dis. 2007;57:251–7. doi: 10.1016/j.diagmicrobio.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Starlander G, Melhus Å. Minor outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in an intensive care unit due to a contaminated sink. J Hosp Infect. 2012;82:122–4. doi: 10.1016/j.jhin.2012.07.004. [DOI] [PubMed] [Google Scholar]