Abstract
AIM: To investigate the associations of interleukin-17 (IL-17) genetic polymorphisms and serum levels with ulcerative colitis (UC) risk.
METHODS: Relevant articles were identified through a search of the following electronic databases, excluding language restriction: (1) the Cochrane Library Database (Issue 12, 2013); (2) Web of Science (1945-2013); (3) PubMed (1966-2013); (4) CINAHL (1982-2013); (5) EMBASE (1980-2013); and (6) the Chinese Biomedical Database (1982-2013). Meta-analysis was conducted using STATA 12.0 software. Crude odds ratios and standardized mean differences (SMDs) with corresponding 95% confidence intervals (CIs) were calculated. All of the included studies met all of the following five criteria: (1) the study design must be a clinical cohort or a case-control study; (2) the study must relate to the relationship between IL-17A/F genetic polymorphisms or serum IL-17 levels and the risk of UC; (3) all patients must meet the diagnostic criteria for UC; (4) the study must provide sufficient information about single nucleotide polymorphism frequencies or serum IL-17 levels; and (5) the genotype distribution of healthy controls must conform to the Hardy-Weinberg equilibrium (HWE). The Newcastle-Ottawa Scale (NOS) criteria were used to assess the methodological quality of the studies. The NOS criteria included three aspects: (1) subject selection: 0-4; (2) comparability of subjects: 0-2; and (3) clinical outcome: 0-3. NOS scores ranged from 0 to 9, with a score ≥ 7 indicating good quality.
RESULTS: Of the initial 177 articles, only 16 case-control studies met all of the inclusion criteria. A total of 1614 UC patients and 2863 healthy controls were included in this study. Fourteen studies were performed on Asian populations, and two studies on Caucasian populations. Results of the meta-analysis revealed that IL-17A and IL-17F genetic polymorphisms potentially increased UC risk under both allele and dominant models (P < 0.001 for all). The results also showed that UC patients had higher serum IL-17 levels than healthy controls (SMD = 5.95, 95%CI: 4.25-7.65, P < 0.001). Furthermore, serum IL-17 levels significantly correlated with the severity of UC (moderate vs mild: SMD = 2.59, 95%CI: 0.03-5.16, P < 0.05; severe vs mild: SMD = 7.09, 95%CI: 3.96-10.23, P < 0.001; severe vs moderate: SMD = 5.84, 95%CI: 5.09-6.59, P < 0.001). The NOS score was ≥ 5 for all of the included studies. Based on the sensitivity analysis, no single study influenced the overall pooled estimates. Neither the Begger’s funnel plots nor Egger’s test displayed strong statistical evidence for publication bias (IL-17A/F genetic polymorphisms: t = -2.60, P = 0.019; serum IL-17 levels: t = -1.54, P = 0.141).
CONCLUSION: The findings strongly suggest that IL-17A/F genetic polymorphisms and serum IL-17 levels contribute to the development and progression of UC.
Keywords: Ulcerative colitis, Interleukin-17, Polymorphism, Serum, Meta-analysis
Core tip: This is the first meta-analysis focusing on the associations of interleukin-17 (IL-17) genetic polymorphisms and serum IL-17 levels with the risk of ulcerative colitis. The results of the study indicate that both IL-17A/F genetic polymorphisms and serum IL-17 levels may play a role in the development and progression of ulcerative colitis.
INTRODUCTION
Ulcerative colitis (UC) is a chronic relapsing intestinal inflammatory disorder of the colon. Although the disease has a variable distribution, it is limited to the distal bowel[1]. Clinically, UC affects the colon and rectum, involving the innermost mucosal lining and manifesting as a continuous area of inflammation without unaffected mucosal segments[2]. The most common symptoms of UC are abdominal discomfort and blood or pus present in diarrhea[3]. Although UC can occur in people of any age, it usually develops between the ages of 15 and 30 and is less common between the ages of 60 and 80[3]. It is estimated that the highest annual incidence of UC was 24.3 per 100000 people per year in Europe. Other parts of the world fared better with 6.3 per 100000 person-years in Asia and the Middle East, and 19.2 per 100000 person-years in North America[4]. In general, UC is a disease caused by a complex interaction of environmental, genetic, and immunoregulatory factors[5,6]. Environmental risk factors, such as infectious agents, drugs, diets, and stress, are crucial to UC susceptibility[7]. A dysregulated T helper (Th) cell response has been suggested to play a major role in causing chronic gut inflammation[8,9]. Furthermore, recent studies have also suggested that interleukin-17 (IL-17) genetic polymorphisms and serum levels may be associated with an increased risk of UC[10,11].
The IL-17 family is generally regarded as a bridge system connecting innate and adaptive immunity[12]. Six related ligands (IL-17 A-F) and five receptors have been identified in this family[13]. Among them, IL-17A and IL-17F are the most researched cytokines responsible for the pathogenic activity of Th17 cells[14,15]. The most important role of IL-17 is induction and mediation of proinflammatory responses[16]. Previous studies have demonstrated a strong and independent association between IL-17 and the pathogenesis of multiple T-cell-mediated autoimmune diseases including rheumatoid arthritis, multiple sclerosis, and inflammatory bowel diseases[17,18]. In short, the dynamic equilibrium between pro-inflammatory and anti-inflammatory cytokines may be decisive in normal cells[19]. Although IL-17 stimulates the expression and production of proinflammatory cytokines in human cells, it may disrupt this equilibrium. As a result, proinflammatory cytokines are overexpressed, leading to facilitation of the pathologic lesions of the colonic mucosa, which is inevitably related to the severity of gut inflammation[20,21]. It has been hypothesized that IL-17 is significantly associated with the development and progression of UC[22], with the majority of the studies focusing on the role of IL-17A and IL-17F[23]. Recently, however, some studies have also suggested that elevated serum IL-17 levels are associated with an increased UC risk[14,24]. Moreover, several studies on single nucleotide polymorphisms have suggested that there is a connection between UC susceptibility and the IL-17 gene cluster[10,25]. Genetic polymorphisms in IL-17A and IL-17F may influence the expression of IL-17 via CXC chemokine induction and subsequent neutrophil recruitment, thus affecting UC susceptibility[26]. Recent evidence has proposed that genetic polymorphisms in IL-17A and IL-17F and serum IL-17 levels may be involved in the incidence of UC[14,24]. However, the results of these studies have been contradictory. Therefore, the aim of this meta-analysis was to evaluate the associations of IL-17A/F genetic polymorphisms and serum IL-17 levels with UC risk.
MATERIALS AND METHODS
Literature search
Without language restriction, the following databases were searched through August 1, 2013: PubMed, Web of Science, Google Scholar, Cochrane Library, CISCOM, CINAHL, EBSCO, and Chinese Biomedical Database. The following keywords and Medical Subject Headings terms were used for the search: (“SNP” or “mutation” or “genetic polymorphism” or “variation” or “polymorphism” or “single nucleotide polymorphism” or “variant”) and (“ulcerative colitis” or “ulcer colitis” or “ulcerative colonitis” or “ulcer enterocolitis” or “UC”) and (“interleukin-17” or “interleukin-17A” or “IL-17A” or “interleukin-17F” or “IL-17F”). A manual search was also performed to identify other potential articles.
Selection criteria
Studies had to meet all five of the following criteria in order to be included in the analysis: (1) the study design must be a clinical cohort or a case-control study; (2) the study must relate to the relationship between IL-17A/F genetic polymorphisms or serum IL-17 levels and risk of UC; (3) all patients must meet the UC diagnostic criteria; (4) the study must provide sufficient information about SNP frequencies or serum IL-17 levels; and (5) the genotype distribution of healthy controls must conform to the Hardy-Weinberg equilibrium (HWE). If a study did not meet all of the inclusion criteria, it was excluded. In cases where authors published several studies using the same subjects, either the most recent or the largest sample size publication was included.
Data extraction
Two researchers systematically extracted all relevant data from all included studies using a standardized form, including: (1) publication language; (2) publication year; (3) first author’s surname; (4) geographic location of the study; (5) study design; (6) sample size; (7) source of the subjects; (8) allele frequencies; (9) tissue sample source; (10) SNP genotyping method; (11) evidence of HWE in healthy controls; and (12) serum IL-17 levels.
Study quality assessment
Methodological qualities of the studies were independently assessed by two researchers according to the Newcastle-Ottawa Scale (NOS)[27]. The NOS included three criteria: (1) subject selection: 0-4; (2) comparability of subjects: 0-2; and (3) clinical outcome: 0-3. NOS scores ranged from 0 to 9, with a score ≥ 7 indicating good quality.
Statistical analysis
The STATA version 12.0 software (Stata Corp, College Station, TX, United States) was used for analyses. Crude odds ratios (ORs) or standardized mean differences (SMDs), with their corresponding 95% confidence intervals (CIs), were used to evaluate the specified relationships. The Z test was used to estimate the statistical significance of the pooled data. The Cochran’s Q-statistic and I2 test were used to evaluate inter-study heterogeneity[28]. If the Q-test showed a P < 0.05 or I2 test exhibited > 50%, indicating the presence of significant heterogeneity, the random effects model was used; otherwise, the fixed-effects model was used. Subgroup and meta-regression analyses were performed to investigate potential sources of heterogeneity. Sensitivity analysis was performed to evaluate the influence of a single study on the overall estimate. Begger’s funnel plots and Egger’s linear regression tests were conducted to investigate the possibility of publication bias[29]. A P-value < 0.05 was considered statistically significant.
RESULTS
Characteristics of the included studies
An initial literature search identified 177 potential articles. Subsequent review of the titles and abstracts excluded 78 articles. Examination of the remaining studies’ full texts and data integrity led to the exclusion of an additional 83 articles. Finally, 16 case-control studies, which met all the inclusion criteria, were included in the meta-analysis[10,14,26,30-42]. The study selection process is shown in Figure 1. The distribution of the number of topic-related publications in electronic databases for the last decade is shown in Figure 2. A total of 1614 UC patients and 2863 healthy controls were included in this meta-analysis. Overall, 14 studies were conducted on Asian populations and two studies on Caucasian populations. Genotyping methods included polymerase chain reaction (PCR)-single-strand conformation polymorphism assays, PCR-ligase detection reactions, direct sequencing, and PCR-restriction fragment length polymorphism assays. The control genotype frequencies were all found to be within the HWE. Serum IL-17 levels were determined by ELISA assays in all of the studies. The NOS scores were ≥ 5 for all the included studies. Tables 1 and 2 contain summaries of the study characteristics and methodological quality on IL-17A/F genetic polymorphisms and serum IL-17 levels, respectively.
Figure 1.
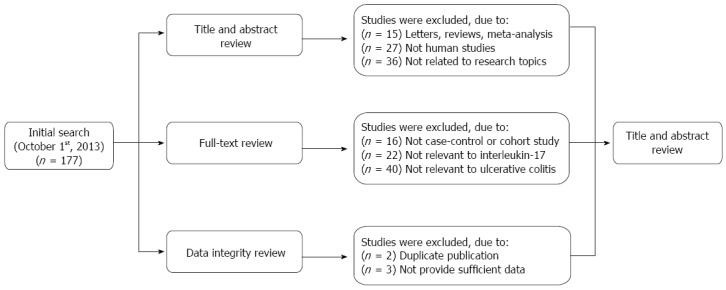
Flow chart of the literature search and study selection.
Figure 2.
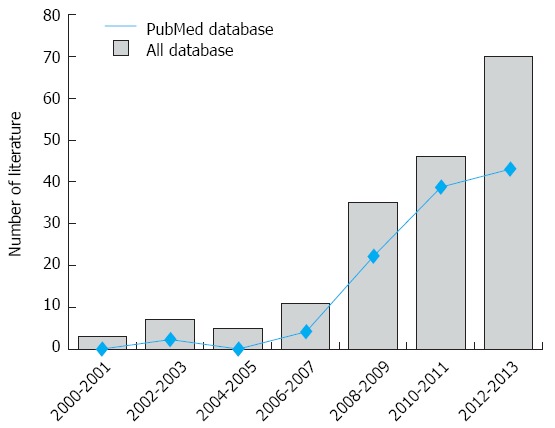
Distribution of the number of topic-related literatures in the electronic databases.
Table 1.
Baseline characteristics and methodological quality of the included studies focused on IL-17A/F genetic polymorphisms
| Ref. | Year | Country | Ethnicity |
Number |
Gender (M/F) |
Age (yr) |
Genotyping method | Genetype | NOS score | |||
| Case | Control | Case | Control | Case | Control | |||||||
| Yu et al[42] | 2012 | China | Asian | 270 | 268 | 130/140 | 136/132 | 47.4 ± 14.1 | 47.1 ± 12.9 | PCR-LDR | IL-17A/F | 8 |
| Hayashi et al[10] | 2012 | Japan | Asian | 202 | 475 | 114/88 | 241/234 | 41.1 ± 14.3 | 59.4 ± 12.7 | PCR-SSCP | IL-17A | 8 |
| Kim et al[41] | 2011 | South Korea | Asian | 268 | 258 | - | - | - | - | Direct sequencing | IL-17A | 6 |
| Chen[40] | 2009 | China | Asian | 148 | 373 | 83/65 | 233/140 | 38.4 ± 12.9 | 37.9 ± 16.1 | PCR-RFLP | IL-17F | 8 |
| Seiderer et al[26] | 2008 | Germany | Caucasian | 216 | 967 | 110/106 | 609/358 | 42.6 ± 14.8 | 47.4 ± 11.3 | PCR-RFLP | IL-17F | 8 |
| Arisawa et al[39] | 2008 | Japan | Asian | 111 | 248 | 56/55 | 133/115 | 39.0 ± 14.3 | 46.6 ± 18.4 | PCR-SSCP | IL-17A/F | 7 |
F: Female; LDR: Ligase detection reaction; M: Male; NOS: Newcastle-Ottawa Scale; PCR: Polymerase chain reaction; RFLP: Restriction fragment length polymorphism; SSCP: Single-strand conformation polymorphism.
Table 2.
Baseline characteristics and methodological quality of the included studies focused on serum interleukin-17 levels
| Ref. | Year | Country | Ethnicity |
Number |
Gender (M/F) |
Age (yr) |
Detection method | NOS score | |||
| Case | Control | Case | Control | Case | Control | ||||||
| Ohman et al[14] | 2013 | China | Asian | 99 | 10 | 70/29 | 4/6 | 32 | 48 | ELISA | 6 |
| Liu[38] | 2013 | China | Asian | 20 | 30 | - | - | - | - | ELISA | 7 |
| Lin et al[37] | 2012 | China | Asian | 50 | 50 | 22/28 | 25/25 | 38.0 ± 6.0 | 37.0 ± 5.0 | ELISA | 7 |
| Chen[36] | 2012 | China | Asian | 24 | 20 | 10/14 | 7/13 | 37.6 ± 9.4 | 38.9 ± 9.9 | ELISA | 7 |
| Zhen et al[35] | 2011 | China | Asian | 54 | 30 | 30/24 | 19/11 | 40.9 ± 8.3 | 50.6 ± 5.3 | ELISA | 7 |
| Luo et al[34] | 2011 | China | Asian | 24 | 30 | 14/10 | 17/13 | - | - | ELISA | 8 |
| Xin et al[33] | 2010 | China | Asian | 29 | 26 | 18/11 | 13/13 | 41.4 ± 9.6 | 32.0 ± 11.2 | ELISA | 6 |
| He et al[32] | 2010 | China | Asian | 25 | 10 | 12/13 | - | - | - | ELISA | 8 |
| Rovedatti et al[31] | 2009 | United Kingdom | Caucasian | 34 | 38 | - | - | 35.6 (14-53) | - | ELISA | 8 |
| Liang et al[30] | 2005 | China | Asian | 40 | 30 | 28/12 | 19/11 | 41.5 ± 12.0 | 41.3 ± 12.0 | ELISA | 8 |
F: Female; M: Male; NOS: Newcastle-Ottawa Scale.
Quantitative data synthesis
Six studies focused on the relationships between the IL-17A/F genetic polymorphisms and susceptibility to UC[10,26,39-42]. The random effects model was used due to the existence of significant heterogeneity among studies. The meta-analysis shows strong correlations between IL-17A (allele model: OR = 1.40, 95%CI: 1.18-1.66, P < 0.001; dominant model: OR = 1.36, 95%CI: 1.11-1.66, P < 0.01) and IL-17F (allele model: OR = 1.47, 95%CI: 1.29-1.67, P < 0.001; dominant model: OR = 1.41, 95%CI: 1.22-1.63, P < 0.001) genetic polymorphisms and an increase in UC risk (Figure 3A and B). A subgroup analysis by country suggests that there are associations between the IL-17A/F genetic polymorphisms and an increased risk of UC in Chinese and Japanese populations (Figure 3C and D), but not in Korean and German populations.
Figure 3.
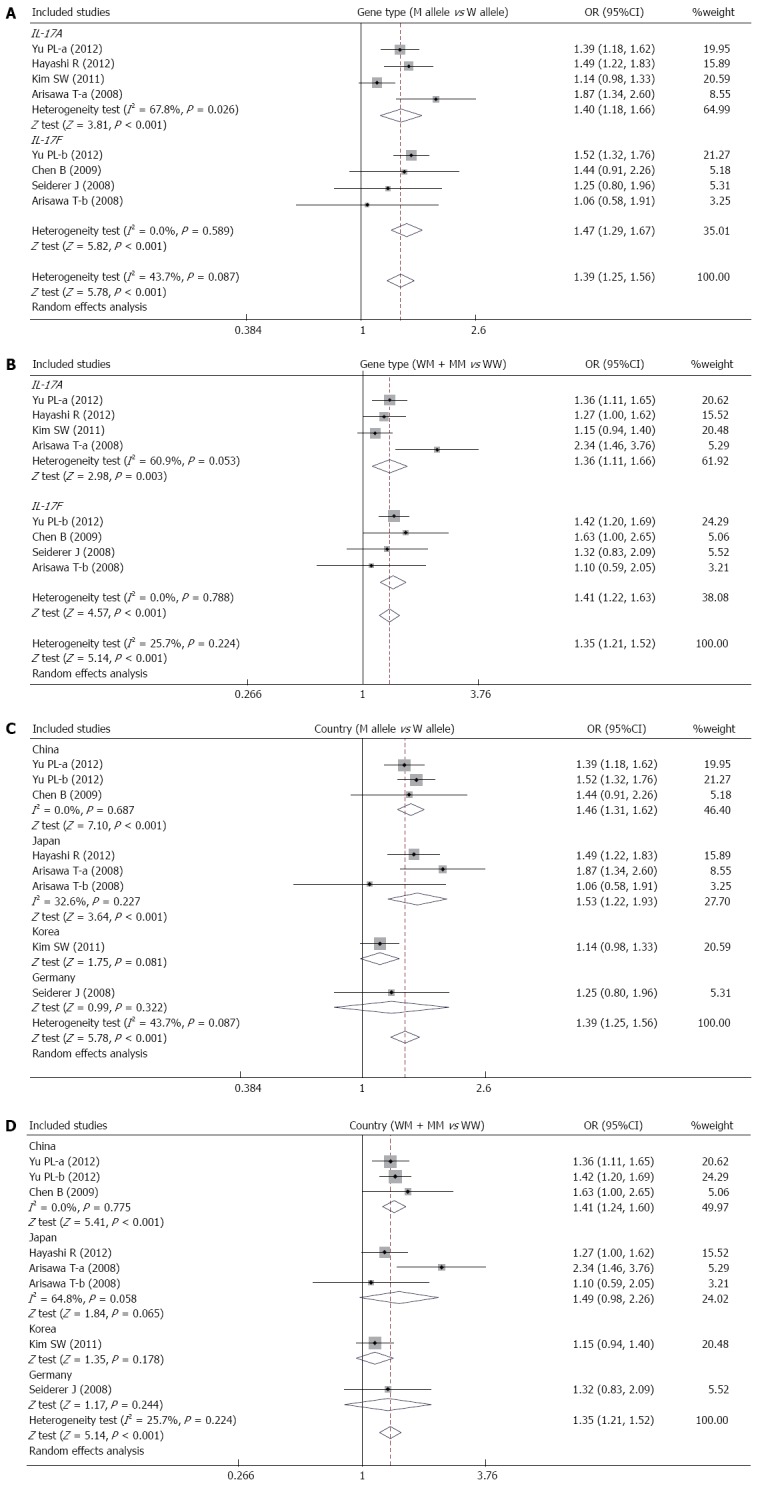
Forest plots of the relationship between IL-17A/F genetic polymorphisms and ulcerative colitis risk under the allele and dominant models.
Ten studies reported differences in serum IL-17 levels between UC patients and healthy controls. Due to obvious heterogeneity, the random effects model was used to analyze the data. Our results demonstrate that UC patients have higher serum IL-17 levels than healthy controls (SMD = 5.95, 95%CI: 4.25-7.65, P < 0.001) (Figure 4A). Furthermore, serum IL-17 levels showed significant correlations with the severity of UC (moderate vs mild: SMD = 2.59, 95%CI: 0.03-5.16, P < 0.05; severe vs mild: SMD = 7.09, 95%CI: 3.96-10.23, P < 0.001; severe vs moderate: SMD = 5.84, 95%CI: 5.09-6.59, P < 0.001) (Figure 4B-D).
Figure 4.
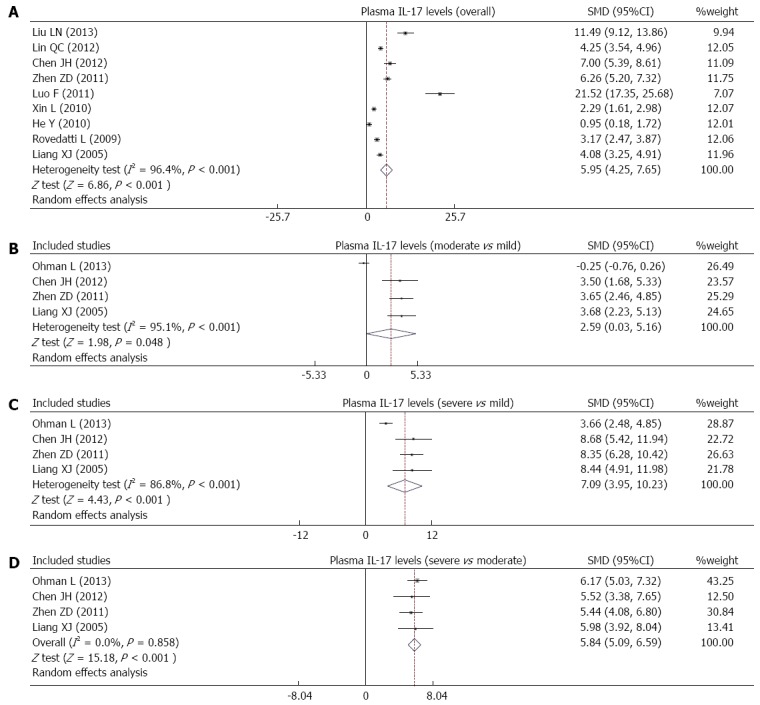
Forest plots of the relationship between serum interleukin-17 levels and ulcerative colitis risk under the allele and dominant models.
The sensitivity analysis results suggest that no single study influenced the overall pooled estimates (Figure 5). There was no evidence of obvious asymmetry in the Begger’s funnel plots (Figure 6). An Egger’s test also did not display strong statistical evidence for publication bias (IL-17A/F genetic polymorphisms: t = -2.60, P = 0.019; IL-17 serum levels: t = -1.54, P = 0.141).
Figure 5.
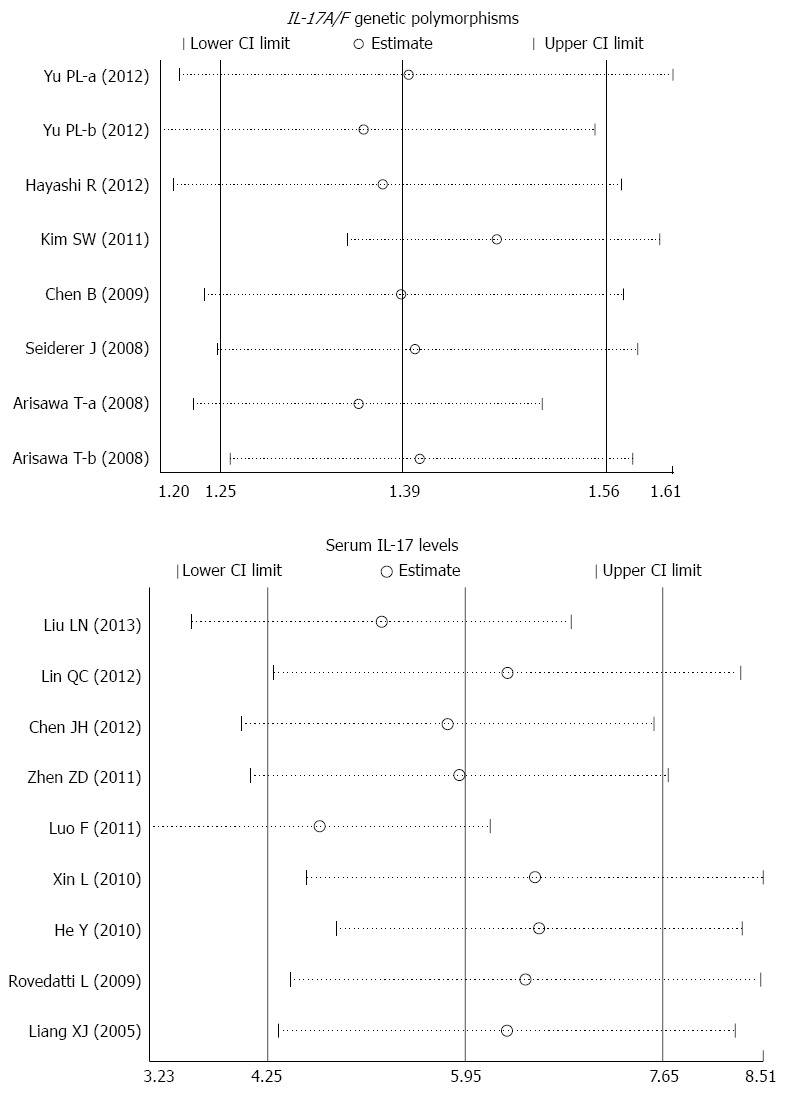
Sensitivity analyses of the summary odds ratio coefficients for the associations of interleukin-17/F polymorphisms and serum interleukin-17 levels with susceptibility to ulcerative colitis. Results were computed by omitting each study in turn. Meta-analysis random effects estimates (exponential form) were used. The two ends of the dotted lines represent the 95%CI.
Figure 6.
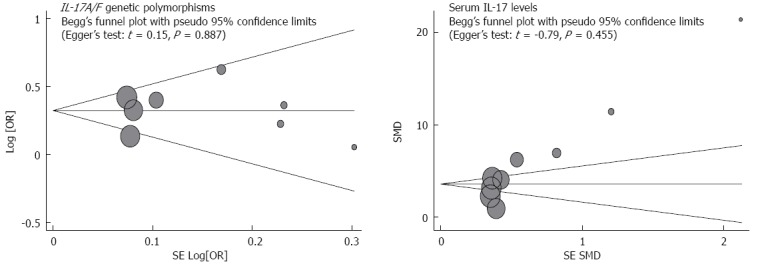
Begger’s funnel plot of publication biases on the associations of interleukin-17/F polymorphisms and serum interleukin-17 levels with susceptibility to ulcerative colitis. Each point represents a separate study for the indicated association. Log (OR) refers to the natural logarithm of the odds ratio; Horizontal line indicates the mean magnitude of the effect. IL-17: Interleukin-17.
DISCUSSION
IL-17, a relatively recently described cytokine, has been shown to act as a bridge between adaptive and innate immune systems[43]. Generally, IL-17 functions as a proinflammatory cytokine, which induces destruction of the pathogen’s cellular matrix upon invasion of the immune system by extracellular pathogens[44]. Six members of the IL-17 family have been identified, including IL-17A, IL-17B, IL-17C, IL-17D, IL-17E (also called IL-25), and IL-17F[45]. Of these, IL-17A and IL-17F, responsible for the pathogenic activity of Th17 cells, have been widely investigated. Recent studies have shown a potentially important role between IL-17 polymorphisms and the risk of UC[39,41]. UC, an inflammatory disorder of the gastrointestinal tract, is a result of the dysregulation of the immunologic response to microbial flora in intestinal lumen[31]. IL-17 may stimulate cytokine and chemokine production in endothelial cells via macrophage activation, thereby leading to neutrophil recruitment and inflammation[46]. Consequently, IL-17 has been presumed to be a pleiotropic cytokine in an autocrine loop of the mucosal immune system during UC pathogenesis[14]. Several investigations have shown that IL-17 increased intercellular adhesion molecule-1 cell surface expression and upregulated the production of nitric oxide. Proinflammatory activities and effects of IL-17 on adhesion molecules suggest its importance in UC[47,48]. As members of the IL-17 family, IL-17A and IL-17F share similar functions, especially in terms of their ability to induce chemokine expression, which is important in neutrophil recruitment and activation. Consequently, IL-17A and IL-17F may also significantly correlate with the risk of UC[44,49].
The human IL-17 gene was mapped to chromosome 2q31[50]. Several studies have indicated that the IL-17A G197A (rs2275913)[39] and IL-17F 7488 A>G (rs763780)[40] alleles, as well as serum IL-17 levels[24,51], are significantly linked to the development of UC. However, contradictory conclusions on the exact role of IL-17 gene polymorphisms and expression in UC exist[52,53]. Potential explanations for the inconsistent results include ethnic differences, the various gene areas examined, and small sample sizes used in the studies. Thus, this meta-analysis aimed to provide a more comprehensive and reliable conclusion regarding the associations of IL-17 gene polymorphisms and serum IL-17 levels with the risk of UC.
The results of this meta-analysis show that polymorphisms in both the IL-17A (G197A) and IL-17F (7488T/C) genes are associated with an increased incidence of UC and suggest that these polymorphisms may be significantly involved in the development of UC. Although the mechanism has yet to be identified, mutations in IL-17A and IL-17F may affect their activation and pro-inflammatory functions, which are necessary for the response to immune system invasion, resulting in cellular matrix destruction, colonic injuries, and thereby increasing susceptibility to UC[25,41]. Consistent with our study, Hayashi et al[10] have reported a significantly higher G197A (IL-17A) allele frequency in UC patients than in healthy controls, suggesting an increased risk of developing UC with homozygosity. A study conducted by Arisawa et al[39] revealed that both G197A (IL-17A) and 7488 A>G (IL-17F) alleles were independently and significantly associated with susceptibility to and pathophysiologic features of UC.
Additionally, IL-17 serum levels were significantly correlated with the severity of UC, suggesting serum IL-17 might be closely related to the inflammatory progress of this disease. A reasonable explanation for this correlation may be that IL-17 mutations alter the stability of a larger number of related pro-inflammatory cytokines secreted into the serum of UC patients[54]. Consistent with these findings, Ajduković et al[47] revealed that the median serum IL-17 concentration in patients with UC was significantly higher than that in controls. Ohman et al[14] also found that serum IL-17 levels in treatment-naive UC patients, measured at the onset of the disease, reflected the clinical disease severity, and thus, might be a valuable tool in the clinical management of newly diagnosed UC patients. Despite the relatively small sample sizes in the present study, a subgroup analysis by country suggests that IL-17A/F genetic polymorphisms are associated with an increased risk of UC in Chinese and Japanese populations, but not in Korean and German populations. Taken together, the findings are partially consistent with previous studies, which suggested that IL-17A and IL-17F gene polymorphisms, as well as serum IL-17 levels, contribute to an individual’s susceptibility to UC and may be useful biomarkers in the detection and clinical management of UC.
As the first meta-analysis on the association of IL-17 genetic polymorphisms and serum IL-17 levels with the risk of UC, our study has some limitations. First, the results lacked sufficient statistical power to assess associations between IL-17 and UC risk due to a relatively small sample size. In addition, a meta-analysis is a retrospective study that may lead to subject selection bias. Third, this meta-analysis failed to obtain the original data of included studies, potentially limiting further clinical assessment of importance of IL-17 genetic polymorphisms and serum IL-17 levels in UC. Importantly, the inclusion criteria of cases and controls were not always well defined in the included studies, thus potentially influencing the results.
In conclusion, the meta-analysis suggests a potential role of IL-17A/F polymorphisms and serum IL-17 levels in the development and progression of UC and they can thus potentially be used as biomarkers for early UC detection. However, due to limitations mentioned above, additional detailed studies are still necessary to confirm these findings.
ACKNOWLEDGMENTS
We would like to thank all of our colleagues working in the Department of Gastroenterology and Department of Infectious Diseases at The Third Affiliated Hospital of Liaoning Medical University, Jinzhou, China.
COMMENTS
Background
Ulcerative colitis (UC), a chronic relapsing intestinal inflammatory disorder of the colon, has a variable distribution, but is limited to the distal bowel. In short, UC is a disease caused by a complex interaction of environmental, genetic, and immunoregulatory factors.
Research frontiers
This is the first meta-analysis focused on the association between interleukin-17 (IL-17) genetic polymorphisms and serum IL-17 levels with respect to UC risk.
Innovations and breakthroughs
As the first reported meta-analysis, the results of this study suggest a potentially important role of IL-17A/F polymorphisms and serum IL-17 levels in the development and progression of UC.
Applications
IL-17A/F genetic polymorphisms and serum IL-17 levels could be useful biomarkers for early detection of UC.
Terminology
Crude odds ratios or standardized mean difference, with their 95% confidence intervals, were used to evaluate specified relationships. The Z test was used to estimate the statistical significance of pooled statistics. The Cochran’s Q-statistic and I2 test were used to evaluate potential heterogeneity among studies.
Peer review
This manuscript describes a meta-analysis of the association of IL-17 genetic polymorphisms and serum levels with UC risk. A significant association between IL-17A/F gene polymorphisms and serum IL-17 levels with the risk of UC has been found. This study selected appropriate methods for the literature search, data extraction, and quality assessment of the literature.
Footnotes
P- Reviewer: Vavricka SR, Yoshino J S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Zhang DN
References
- 1.Kornbluth A, Sachar DB. Ulcerative colitis practice guidelines in adults: American College Of Gastroenterology, Practice Parameters Committee. Am J Gastroenterol. 2010;105:501–523; quiz 524. doi: 10.1038/ajg.2009.727. [DOI] [PubMed] [Google Scholar]
- 2.Tahara T, Shibata T, Nakamura M, Okubo M, Yamashita H, Yoshioka D, Yonemura J, Kmiya Y, Ishizuka T, Fujita H, et al. Host genetic factors, related to inflammatory response, influence the CpG island methylation status in colonic mucosa in ulcerative colitis. Anticancer Res. 2011;31:933–938. [PubMed] [Google Scholar]
- 3.Rönnblom A, Samuelsson SM, Ekbom A. Ulcerative colitis in the county of Uppsala 1945-2007: incidence and clinical characteristics. J Crohns Colitis. 2010;4:532–536. doi: 10.1016/j.crohns.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42; quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Molodecky NA, Kaplan GG. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (N Y) 2010;6:339–346. [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SC, Woodrow S, Patel N, Subhani J, Harbord M. Role of genetic and environmental factors in British twins with inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:725–736. doi: 10.1002/ibd.21747. [DOI] [PubMed] [Google Scholar]
- 7.Jantchou P, Monnet E, Carbonnel F. [Environmental risk factors in Crohn’s disease and ulcerative colitis (excluding tobacco and appendicectomy)] Gastroenterol Clin Biol. 2006;30:859–867. doi: 10.1016/s0399-8320(06)73333-4. [DOI] [PubMed] [Google Scholar]
- 8.Thompson AI, Lees CW. Genetics of ulcerative colitis. Inflamm Bowel Dis. 2011;17:831–848. doi: 10.1002/ibd.21375. [DOI] [PubMed] [Google Scholar]
- 9.Pearl DS, Shah K, Whittaker MA, Nitch-Smith H, Brown JF, Shute JK, Trebble TM. Cytokine mucosal expression in ulcerative colitis, the relationship between cytokine release and disease activity. J Crohns Colitis. 2013;7:481–489. doi: 10.1016/j.crohns.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi R, Tahara T, Shiroeda H, Saito T, Nakamura M, Tsutsumi M, Shibata T, Arisawa T. Influence of IL17A polymorphisms (rs2275913 and rs3748067) on the susceptibility to ulcerative colitis. Clin Exp Med. 2013;13:239–244. doi: 10.1007/s10238-012-0206-5. [DOI] [PubMed] [Google Scholar]
- 11.Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidlak D, Kielian T. Differential effects of interleukin-17 receptor signaling on innate and adaptive immunity during central nervous system bacterial infection. J Neuroinflammation. 2012;9:128. doi: 10.1186/1742-2094-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolls JK, Lindén A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Ohman L, Dahlén R, Isaksson S, Sjöling A, Wick MJ, Sjövall H, Van Oudenhove L, Simrén M, Strid H. Serum IL-17A in newly diagnosed treatment-naive patients with ulcerative colitis reflects clinical disease severity and predicts the course of disease. Inflamm Bowel Dis. 2013;19:2433–2439. doi: 10.1097/MIB.0b013e3182a563cb. [DOI] [PubMed] [Google Scholar]
- 15.Afzali B, Lombardi G, Lechler RI, Lord GM. The role of T helper 17 (Th17) and regulatory T cells (Treg) in human organ transplantation and autoimmune disease. Clin Exp Immunol. 2007;148:32–46. doi: 10.1111/j.1365-2249.2007.03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryzhakov G, Lai CC, Blazek K, To KW, Hussell T, Udalova I. IL-17 boosts proinflammatory outcome of antiviral response in human cells. J Immunol. 2011;187:5357–5362. doi: 10.4049/jimmunol.1100917. [DOI] [PubMed] [Google Scholar]
- 17.Hundorfean G, Neurath MF, Mudter J. Functional relevance of T helper 17 (Th17) cells and the IL-17 cytokine family in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:180–186. doi: 10.1002/ibd.21677. [DOI] [PubMed] [Google Scholar]
- 18.Wendling D, Cedoz JP, Racadot E, Dumoulin G. Serum IL-17, BMP-7, and bone turnover markers in patients with ankylosing spondylitis. Joint Bone Spine. 2007;74:304–305. doi: 10.1016/j.jbspin.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Szkaradkiewicz A, Marciniak R, Chudzicka-Strugała I, Wasilewska A, Drews M, Majewski P, Karpiński T, Zwoździak B. Proinflammatory cytokines and IL-10 in inflammatory bowel disease and colorectal cancer patients. Arch Immunol Ther Exp (Warsz) 2009;57:291–294. doi: 10.1007/s00005-009-0031-z. [DOI] [PubMed] [Google Scholar]
- 20.Qu N, Xu M, Mizoguchi I, Furusawa J, Kaneko K, Watanabe K, Mizuguchi J, Itoh M, Kawakami Y, Yoshimoto T. Pivotal roles of T-helper 17-related cytokines, IL-17, IL-22, and IL-23, in inflammatory diseases. Clin Dev Immunol. 2013;2013:968549. doi: 10.1155/2013/968549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuett H, Schieffer B. Targeting cytokine signaling as an innovative therapeutic approach for the prevention of atherosclerotic plaque development. Curr Atheroscler Rep. 2012;14:187–189. doi: 10.1007/s11883-012-0246-z. [DOI] [PubMed] [Google Scholar]
- 22.Feng T, Qin H, Wang L, Benveniste EN, Elson CO, Cong Y. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. J Immunol. 2011;186:6313–6318. doi: 10.4049/jimmunol.1001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean LP, Cross RK, Shea-Donohue T. Combined blockade of IL-17A and IL-17F may prevent the development of experimental colitis. Immunotherapy. 2013;5:923–925. doi: 10.2217/imt.13.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Perálvarez ML, García-Sánchez V, Villar-Pastor CM, González R, Iglesias-Flores E, Muntane J, Gómez-Camacho F. Role of serum cytokine profile in ulcerative colitis assessment. Inflamm Bowel Dis. 2012;18:1864–1871. doi: 10.1002/ibd.22865. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Yu P, Wang Y, Jiang W, Shen F, Wang Y, Tu H, Yang X, Shi R, Zhang H. Genetic polymorphisms of interleukin 17A and interleukin 17F and their association with inflammatory bowel disease in a Chinese Han population. Inflamm Res. 2013;62:743–750. doi: 10.1007/s00011-013-0629-9. [DOI] [PubMed] [Google Scholar]
- 26.Seiderer J, Elben I, Diegelmann J, Glas J, Stallhofer J, Tillack C, Pfennig S, Jürgens M, Schmechel S, Konrad A, et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm Bowel Dis. 2008;14:437–445. doi: 10.1002/ibd.20339. [DOI] [PubMed] [Google Scholar]
- 27.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 28.Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 29.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 30.Liang XJ, Ma ZL, Zeng YJ. The clinical significance of interleukin 4 & interleukin 17’s blood plasma level in patients with ulcerative colitis. Youjiang Yixue Zazhi. 2005;33:6–8. [Google Scholar]
- 31.Rovedatti L, Kudo T, Biancheri P, Sarra M, Knowles CH, Rampton DS, Corazza GR, Monteleone G, Di Sabatino A, Macdonald TT. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut. 2009;58:1629–1636. doi: 10.1136/gut.2009.182170. [DOI] [PubMed] [Google Scholar]
- 32.He Y, Li CM, Hou FX, Liu YJ. Expression of IL-17 type cytokin and IFN-γ type cytokin in ulcerative clitis patients. Zhongguo Xiandai Yixue. 2010;17:36–37. [Google Scholar]
- 33.Xin L, Han Y, Wang ZH, Wang JH. Role of IL-17 and IL-23/IL-17 Immune-regulatory Pathway in the Pathogenesis of Ulcerative Colitis. Xinyixue Zazhi. 2010;20:538–541, 534. [Google Scholar]
- 34.Luo F, Wang C. Influence of olsalazine on TNF-α and IL-17 in patients with ulcerative colitis. Yixue Luntan Zazhi. 2011;32:48–49. [Google Scholar]
- 35.Zhen ZD, Wan XQ, Liu LY. Serum contents of IL-23 and IL-17 in the patients with ulcerative colitis and the clinical significance. Xibao Yu Fenzimianyixue Zazhi. 2011;27:203–204. [PubMed] [Google Scholar]
- 36.Chen JH. Clinical significance and serum contents of IL-23, IL-17 and IL25in the patients with inflammatory bowel disease. Zhongguo Linchang Shiyong Zazhi (Dianziban) 2012;6:698–699. [Google Scholar]
- 37.Lin QC, Liao BX, Huang FN. Expression of interleukin IL-4, IL-17 in ulcerative colitis. Zhongguo Yixue Gongcheng Zazhi. 2012;2:44–45. [Google Scholar]
- 38.Liu LN. Serum level and significance of IL-17 in the children with infectious diarrhea and ulcerative colitis. Shiyong Yixue Zazhi. 2013;17:141–142. [Google Scholar]
- 39.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Nakamura M, Yoshioka D, Arima Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol. 2008;28:44–49. doi: 10.1007/s10875-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 40.Chen B, Zeng Z, Hou J, Chen M, Gao X, Hu P. Association of interleukin-17F 7488 single nucleotide polymorphism and inflammatory bowel disease in the Chinese population. Scand J Gastroenterol. 2009;44:720–726. doi: 10.1080/00365520902795430. [DOI] [PubMed] [Google Scholar]
- 41.Kim SW, Kim ES, Moon CM, Park JJ, Kim TI, Kim WH, Cheon JH. Genetic polymorphisms of IL-23R and IL-17A and novel insights into their associations with inflammatory bowel disease. Gut. 2011;60:1527–1536. doi: 10.1136/gut.2011.238477. [DOI] [PubMed] [Google Scholar]
- 42.Yu PL, Zhang XF, Shen FC, Zhang HJ. Association between interleukin gene polymorphisms and iflammatory bowel diseases in Chinese patients. Shijie Huaren Xiaohua Zazhi. 2012;20:875–882. [Google Scholar]
- 43.Gaffen SL. An overview of IL-17 function and signaling. Cytokine. 2008;43:402–407. doi: 10.1016/j.cyto.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds JM, Angkasekwinai P, Dong C. IL-17 family member cytokines: regulation and function in innate immunity. Cytokine Growth Factor Rev. 2010;21:413–423. doi: 10.1016/j.cytogfr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kono T, Korenaga H, Sakai M. Genomics of fish IL-17 ligand and receptors: a review. Fish Shellfish Immunol. 2011;31:635–643. doi: 10.1016/j.fsi.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 46.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 47.Ajduković J, Tonkić A, Salamunić I, Hozo I, Simunić M, Bonacin D. Interleukins IL-33 and IL-17/IL-17A in patients with ulcerative colitis. Hepatogastroenterology. 2010;57:1442–1444. [PubMed] [Google Scholar]
- 48.Yu P, Shen F, Zhang X, Cao R, Zhao X, Liu P, Tu H, Yang X, Shi R, Zhang H. Association of single nucleotide polymorphisms of IL23R and IL17 with ulcerative colitis risk in a Chinese Han population. PLoS One. 2012;7:e44380. doi: 10.1371/journal.pone.0044380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyun YS, Han DS, Lee AR, Eun CS, Youn J, Kim HY. Role of IL-17A in the development of colitis-associated cancer. Carcinogenesis. 2012;33:931–936. doi: 10.1093/carcin/bgs106. [DOI] [PubMed] [Google Scholar]
- 50.Rouvier E, Luciani MF, Mattéi MG, Denizot F, Golstein P. CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J Immunol. 1993;150:5445–5456. [PubMed] [Google Scholar]
- 51.Peluso I, Raguzzini A, Villano DV, Cesqui E, Toti E, Catasta G, Serafini M. High fat meal increase of IL-17 is prevented by ingestion of fruit juice drink in healthy overweight subjects. Curr Pharm Des. 2012;18:85–90. doi: 10.2174/138161212798919020. [DOI] [PubMed] [Google Scholar]
- 52.Fonseca-Camarillo G, Mendivil-Rangel E, Furuzawa-Carballeda J, Yamamoto-Furusho JK. Interleukin 17 gene and protein expression are increased in patients with ulcerative colitis. Inflamm Bowel Dis. 2011;17:E135–E136. doi: 10.1002/ibd.21816. [DOI] [PubMed] [Google Scholar]
- 53.Hölttä V, Klemetti P, Salo HM, Koivusalo A, Pakarinen M, Westerholm-Ormio M, Kolho KL, Vaarala O. Interleukin-17 immunity in pediatric Crohn disease and ulcerative colitis. J Pediatr Gastroenterol Nutr. 2013;57:287–292. doi: 10.1097/MPG.0b013e3182979252. [DOI] [PubMed] [Google Scholar]
- 54.Zheng ZD, Wan XQ, Liu LY. Serum contents of IL-23 and IL-17 in the patients with ulcerative colitis and the clinical significance. Xibao Yu Fenzimianyixue Zazhi. 2011;27:203–206. [PubMed] [Google Scholar]


