Abstract
Background:
Currently, none of the available colorectal adenocarcinoma (CAC) testing has been established as a well-accepted diagnosis tool, particularly for the early stage of CAC. The recent discovery of serum microRNA (miRNA) profile has provided a new auxiliary approach for tumour diagnosis. Our study is involved in the global analysis of serum miRNAs during the normal–colorectal adenoma (CA)–CAC sequence.
Methods:
Serum samples were collected from 307 CAC patients, 164 CA patients and 226 healthy controls. Differentially expressed serum miRNAs were screened with Miseq sequencing followed by the reverse transcription PCR (RT–qPCR) validation. The miRNA panel was developed with a logistic regression model and validated using an independent cohort. The miRNA levels in CAC patients of different clinical stages and CA patients of different grades were compared. Receiver operating characteristic curves were constructed to evaluate the diagnostic accuracy of the panel.
Results:
The Miseq sequencing results revealed 15 differentially expressed miRNAs in the intersection of CAC vs CA and CA vs healthy controls according to our criteria. After the selection and validation process via RT–qPCR, we identified a four-miRNA panel (miR-19a-3p, miR-223-3p, miR-92a-3p and miR-422a) with a high diagnostic accuracy of CAC. Even in the low-carcinoembryonic antigen level group, the diagnostic accuracy of this miRNA panel was still acceptable (AUC=0.810). Surprisingly, our results indicated that the miRNA panel could differentiate stage I/II CAC from controls. In addition, this panel could also differentiate CA from CAC (AUC=0.886) and healthy controls (AUC=0.765).
Conclusions:
We established a serum four-miRNA panel with considerable clinical value in the early-stage diagnosis of CAC.
Keywords: microRNA, colorectal adenocarcinoma, colorectal adenoma, miseq sequencing, RT–qPCR
Colorectal adenocarcinoma (CAC) is one of the leading causes of cancer-related death with increasing incidence and mortality in the past several decades. Approximately 1.23 million people worldwide develop CAC annually, leading to 0.6 million deaths (Jemal et al, 2009; Ferlay et al, 2010). However, its current high mortality and most CAC-related deaths would be significantly decreased if the early-stage CAC and precancerous lesions could be detected by more suitable diagnostic tools (Lieberman, 2009). Among the available CAC testing, colonoscopy is recommended as the most reliable tool for high-risk people. However, its wide application in large-scale screening is hampered owing to its invasive nature and high cost (Kim et al, 2007). In addition, the faecal occult blood test (FOBT) and carcinoembryonic antigen (CEA) detection have relatively poor sensitivity (Collins et al, 2005; Levin et al, 2008; Mandel, 2008). None of these methods have been established as a well-accepted diagnostic tool, particularly for the early stage of CAC. Therefore, there is an urgent need to identify new biomarkers with high sensitivity and specificity for the early-stage CAC and precancerous lesions.
MicroRNAs (miRNAs) are a class of small non-coding RNAs with 19–24 nucleotides, and they have attracted a great deal of attention in cancer research. Functional studies have indicated that deregulation of miRNAs is involved in the initiation and progression of human cancer (Garzon et al, 2006; Cho, 2007). Recently, serum miRNAs have been shown to be stable, and they could be excellent candidates for the early diagnosis of cancer (Zhang et al, 2010; Liu et al, 2011; Chen et al, 2012). Several studies have examined the alteration of serum miRNA expression in colorectal cancer patients compared with healthy controls (Chen et al, 2008; Ng et al, 2009; Huang et al, 2010; Pu et al, 2010). Additional studies have examined the expression of certain selected miRNAs in colorectal adenoma (CA), a usual precursor to CAC (Asaga et al, 2011; Oberg et al, 2011). However, those studies were limited by one or more of the following factors: limited numbers of screened miRNAs, small sample size, lack of independent validation and failure to differentiate CA from CAC and healthy controls.
In the present study, we characterised the genome-wide miRNA expression profile in the serum of patients with CAC and CA and of healthy controls using a high-throughput Miseq sequencing screening followed by two phases of reverse transcription PCR (RT–qPCR) assay. Finally, we developed a 4-miRNA panel using logistic regression model and provided useful serum biomarkers with high diagnostic accuracy for the early diagnosis of CAC.
Materials and methods
Study design
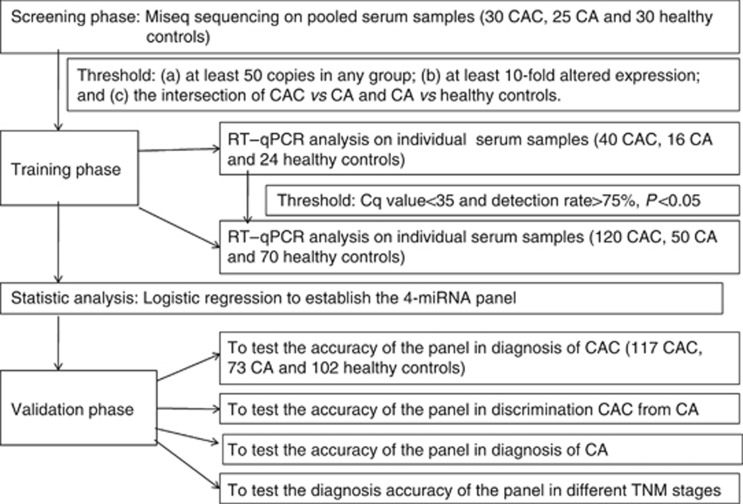
In the present investigation, our experimental process was separated into three phases (Figure 1). In the initial screening phase, serum samples were collected from 30 CAC patients, 25 CA patients and 30 healthy controls, and differentially expressed miRNAs were assessed by Miseq sequencing using the intersection of two pairwise comparisons: CAC vs CA and CA vs healthy controls. In the training phase, miRNAs identified via Miseq sequencing were first evaluated by RT–qPCR in an independent cohort of serum samples from 80 participants, including 40 CAC patients, 16 CA patients and 24 healthy controls. Subsequently, the validated miRNAs, which were differentially expressed between CAC and control group (CA and healthy controls), were further examined in additional 240 participants, including 120 CAC patients, 50 CA patients and 70 healthy controls. The diagnostic miRNA panel was then constructed using the above-mentioned 320 serum samples based on the logistic regression model. In the validation phase, parameters of the logistic model obtained from the training phase were applied to another independent cohort of 292 participants (117 CAC patients, 73 CA patients and 102 healthy controls) to validate the diagnostic performance of the miRNA panel. This study was approved by the Clinical Research Ethics Committee of Qilu Hospital of Shandong University.
Figure 1.
Overview of the design strategy.
Patients and control subjects
Written informed consent was obtained from every participant for the use of venous blood sample. All the CAC and CA patients were recruited from the Department of General Surgery and Gastroenterology, Qilu Hospital of Shandong University between 2010 and 2013. The healthy controls were enrolled from the Department of Physical Examination Center, Qilu Hospital of Shandong University.
All blood samples were collected before any therapeutic procedures, such as surgery, chemotherapy and radiotherapy, were performed. CAC and CA diagnosis was confirmed by histopathology or biopsy. Tumours were staged according to the tumour–node–metastasis (TNM) staging system of the Union for International Cancer Control (UICC). Age- and sex-matched healthy controls were recruited from a large pool of healthy individuals seeking a routine health check-up (Table 1).
Table 1. Clinicopathological data for patients and healthy controls.
| Characteristics | Screening phase | Training phase | Validation phase |
|---|---|---|---|
|
Healthy control | |||
| N | 30 | 94 | 102 |
| Age (mean±s.d.), years |
54.37±16.24 |
52.33±13.16 |
52.76±11.36 |
|
Sex, n (%) | |||
| Male | 18 (60.0) | 51 (54.3) | 59 (57.8) |
| Female |
12 (40.0) |
43 (45.7) |
43 (42.2) |
|
CA | |||
| Number | 25 | 66 | 73 |
| Age (mean±s.d.), years |
54.60±17.36 |
51.31±10.80 |
52.43±12.38 |
|
Sex, n (%) | |||
| Male | 16 (64.0) | 39 (59.1) | 42 (57.5) |
| Female |
9 (36.0) |
27 (40.9) |
31 (42.5) |
|
Location of adenoma, n (%) | |||
| Colon | 18 (72.0) | 46 (69.7) | 53 (72.6) |
| Rectum |
7 (28.0) |
20 (30.3) |
20 (27.4) |
|
Adenoma grade, n (%) | |||
| Low-grade intraepithelial neoplasia | 14 (56) | 42 (63.6) | 39 (53.4) |
| High-grade intraepithelial neoplasia |
11 (44) |
24 (36.4) |
34 (46.6) |
|
CAC | |||
| Number | 30 | 160 | 117 |
| Age (mean±s.d.), years |
56.56±19.67 |
60.19±13.54 |
56.28±14.72 |
|
Sex, n (%) | |||
| Male | 19 (63.3) | 93 (58.1) | 68 (58.2) |
| Female |
11 (36.7) |
67 (41.9) |
49 (41.8) |
|
Location of tumours, n (%) | |||
| Colon | 22 (73.3) | 112 (70.0) | 83 (70.9) |
| Rectum |
8 (26.7) |
48 (30.0) |
34 (29.1) |
|
Tumour size, n (%) | |||
| ⩽3 cm | 4 (13.3) | 26 (16.3) | 18 (15.4) |
| >3 cm |
26 (86.7) |
134 (83.7) |
99 (84.6) |
|
Differentiation, n (%) | |||
| Well | 4 (13.3) | 34 (21.3) | 18 (15.4) |
| Moderate | 19 (63.3) | 91 (56.9) | 76 (65.0) |
| Poor | 5 (16.7) | 35 (21.8) | 23 (19.6) |
| N/A |
2 (6.7) |
|
|
|
TNM stage, n (%) | |||
| Stage I | 11 (36.7) | 23 (14.4) | 19 (16.2) |
| Stage II | 8 (26.6) | 49 (30.6) | 31 (26.5) |
| Stage III | 5 (16.7) | 71 (44.4) | 46 (39.3) |
| Stage IV |
4 (13.3) |
17 (10.6) |
21 (18.0) |
| N/A | 2 (6.7) | ||
Abbreviations: CA=colorectal adenoma; CAC=colorectal adenocarcinoma; TNM=tumour–node–metastasis.
Serum preparation
Briefly, 5 ml of venous blood was collected from each subject. The whole blood was separated into serum and cellular fractions within 2 h by centrifugation at 4000 r.p.m for 10 min. The supernatant (serum) was collected and further centrifuged at 12 000 r.p.m for 15 min to completely remove the cell debris. The whole process was strictly controlled to avoid haemolysis. The obtained serum was stored at −80 °C before further analysis. The CEA level of each sample was determined using electrochemiluminescence immunoassay.
Miseq sequencing
For Miseq sequencing, equal volumes of serum from age- and sex-matched subjects, including 30 CAC patients, 25 CA patients and 30 healthy controls, were pooled. The sequencing procedure was conducted as described in our previous study (Zheng et al, 2013).
Quantification of miRNAs by RT–qPCR analysis
RT–qPCR was performed on an ABI PRISM 7500 Sequence Detection System (Applied biosystems, Carlsbad, CA, USA) using the SYBR PrimeScript miRNA qPCR Kit (Takara Bio, Otsu, Japan). The reverse transcription was conducted in a 20-μl reaction system containing 10 μl of 2 × miRNA reaction buffer mix, 2 μl of miRNA Primescript RT Enzyme Mix (Takara Bio), 2 μl of 0.1% BSA and 3 μl of serum pre-mixed with 3 μl of serum buffer (2.5% Tween-20, 50 mmol l−1 Tris and 1 mmol l−1 EDTA). The reaction was performed at 37 °C for 60 min, followed by 85 °C for 5 s. The synthesised cDNA was centrifuged at 10 000 r.p.m for 10 min. The PCR was conducted in a 25-μl reaction system containing 12.5 μl of SYBR Premix Ex Taq (Takara Bio), 0.5 μl of dye, 2 μl of 5 μM forward primer, 1 μl of 10 μM Uni-miR qPCR Primer (Takara Bio), 7 μl of ddH2O and 2.0 μl of cDNA template. Briefly, after a denaturation step at 95 °C for 30 s, the amplification was carried out for 45 cycles at a melting temperature of 95 °C for 5 s and an annealing temperature of 57 °C for 34 s. A dissociation curve was analysed for each PCR experiment to assess the primer–dimer formation or contamination. All reactions were performed in triplicate, and the Cq values were determined using the default threshold setting. The combination of miR-191-5p and U6 snRNA was used as reference genes for serum miRNAs RT–qPCR detection as described in our previous study (Zheng et al, 2013).
Statistical analysis
All statistical analyses were performed with SPSS 17.0, and non-parametric Mann–Whitney U-test was used to compare the difference in serum miRNA concentrations between the cancer group and the control group. A P-value of <0.05 was considered as statistically significant. Multiple logistic regression analysis was used to establish the miRNA panel. Receiver operating characteristic (ROC) curves was constructed, and area under the ROC curve (AUC) was used to evaluate the diagnostic performance of the selected miRNA panel. MedCalc software version 9.3.9.0 (MedCalc, Mariakerke, Belgium) was used to perform the ROC curve.
Results
Discovery of candidate biomarkers by Miseq sequencing
Of the 740 sequenced serum miRNAs, 348, 303 and 283 miRNAs were detectable (>10 copies) in CAC patients, CA patients and healthy controls, respectively. During the Miseq sequencing, the final reads of each miRNA in each pooled group were determined through the normalisation with total reads of all called miRNAs. The miRNAs were selected as candidate biomarkers based on the following the criteria: (a) having at least 50 copies in any group; (b) exhibiting at least 10-fold altered expression; and (c) the intersection of CAC vs CA and CA vs healthy controls. Levels of 12 miRNAs, including miR-195-5p, miR-92a-3p, miR-1290, miR-582-5p, miR-223-3p and miR-136-5p, miR-3074-5p, miR-29a-3p, miR-221-3p, miR-148a-3p, miR-19a-3p and miR-17-3p were significantly higher in CAC than those in CA and healthy controls (fold change=10.78–399.02; P<0.05). In contrast, levels of 3 miRNAs, including miR-422a, miR-1260a and miR-4502, in the CAC group were significantly lower (fold change=0.002–0.09; P<0.05). In summary, a total of 15 differentially expressed miRNAs were identified as candidate biomarkers, which should be further tested via RT–qPCR.
Confirmation of miRNAs by RT–qPCR analysis
We first tested the 15 candidate miRNAs using an independent cohort of 80 serum samples with RT–qPCR. Four miRNAs (miR-136-5p, miR-195-5p, miR-221-3p and miR-582-5p) with detection rate of <75% and two miRNAs (miR-148-3p and miR-3074-5p) with Cq value of >35 were excluded from further analysis, resulting in nine qualified candidate miRNAs. In addition, five miRNAs (miR-17-3p, miR-29a-3p, miR-1260a, miR-1290 and miR-4502) with a P-value of >0.05 were excluded. Consequently, only four miRNAs (miR-19a-3p, miR-223-3p, miR-92a-3p and miR-422a) with a differential expression between the CAC vs CA and CA vs healthy control group were selected as candidate biomarkers.
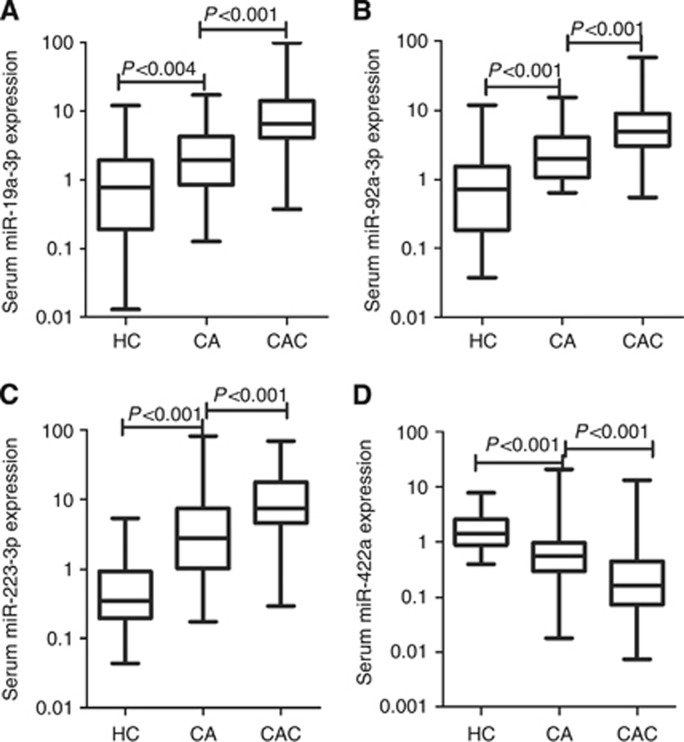
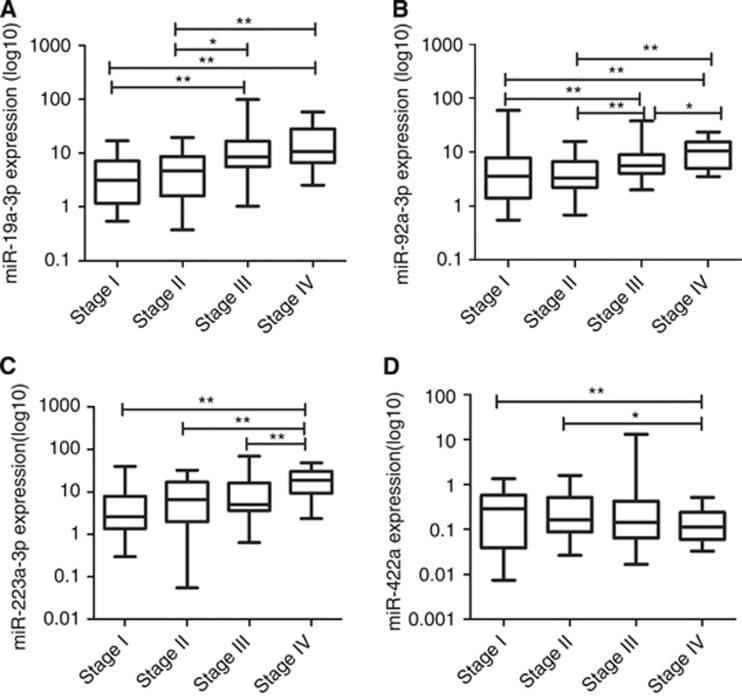
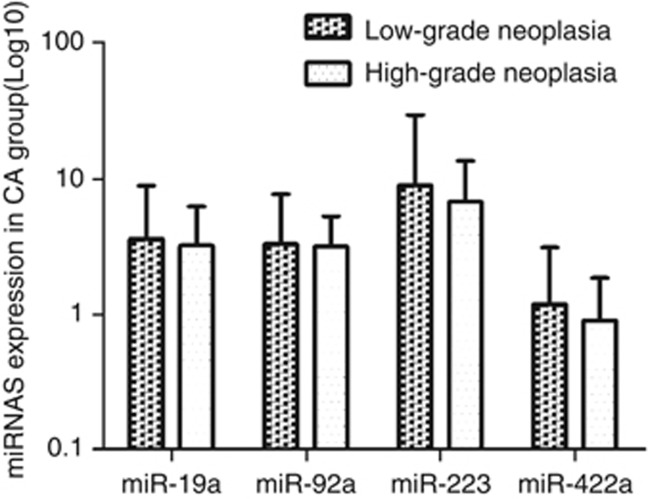
These four miRNAs were further evaluated by RT–qPCR using additional independent 240 serum samples. Figure 2 shows that high expression levels of miR-19a-3p, miR-92a-3p and miR-223-3p as well as the low expression level of miR-422a were detected in CAC patients compared with CA and healthy control group. Moreover, the levels of these four miRNAs in CAC of different clinical stages and CA of different grades were compared as shown in Figure 3 and 4.
Figure 2.
Large-scale validation of miR-19a-3p (A), miR-92a-3p (B), miR-223-3p (C) and miR-422a (D) in 94 healthy controls, 66 colorectal adenoma (CA) patients and 160 CAC patients. Expression levels of the microRNAs (miRNAs) (Log10 scale at Y-axis) are normalised to the combination of miR-191-5p and U6. Mann–Whitney U-test was used to determine the statistical differentiation.
Figure 3.
The expression of miR-19a-3p (A), miR-92a-3p (B), miR-223-3p (C) and miR-422a (D) in CAC of different clinical stages. *P<0.05, **P<0.01.
Figure 4.
The expression of four miRNAs in the CA group of different grades. There was no significant statistical difference for miR-19a-3p, miR-92a-3p, miR-223-3p and miR-422a expression between the low-grade and high-grade intraepithelial neoplasia groups.
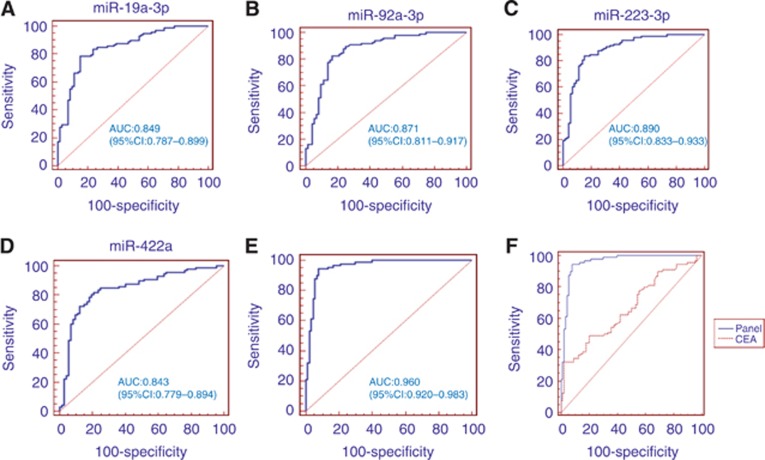
The diagnostic accuracy of these four miRNAs was measured by ROC and their corresponding AUCs were 0.849, 0.871, 0.890 and 0.843, respectively (Figure 5A–D).
Figure 5.
Receiver operating characteristic (ROC) curves for the ability of the four individual miRNAs (A–D) and the 4-miRNA panel (E) to differentiate the CAC patients from the control group in the training phase. Comparison of ROC curves for the ability of the 4-miRNA panel and carcinoembryonic antigen (CEA) (F) to differentiate the CAC patients from the control group in the validation phase.
Establishing the predictive miRNAs panel
In the present study, we constructed the miRNA panel for CAC diagnosis using 320 serum samples as the training data. The predicted probability of diagnosis with CAC from the stepwise logistic regression model was calculated using the equation as follows: logit(P)=0.3313-0.0081 × miR-19a-3p-0.0257 × miR-92a-3p-0.0406 × miR-223-3p+0.1328 × miR-422a.
The diagnostic performance of the established miRNA panel was evaluated by the ROC analysis. Figure 5E shows that the AUC of the established 4-miRNA panel was 0.960.
Validation of the miRNAs panel
We further validated the diagnostic performance of the established 4-miRNA panel in another independent validation phase. The AUC of the 4- miRNA panel was 0.951 (95% CI: 0.907–0.978; sensitivity=84.3%, specificity=91.6%), which was >CEA detection (AUC: 0.667, 95% CI: 0.593–0.735, P<0.001, Figure 5F).
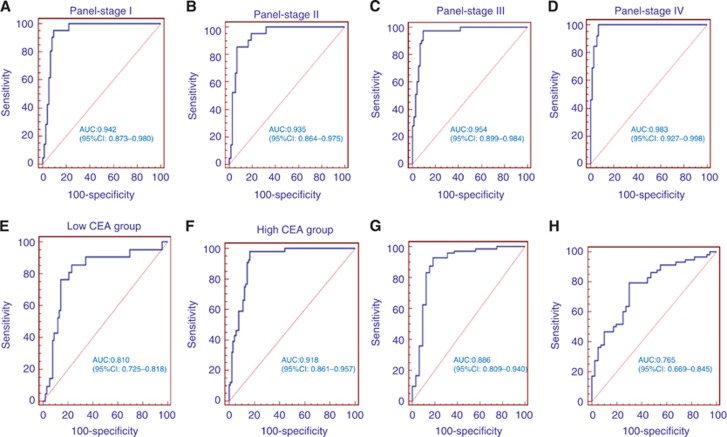
Moreover, we further evaluated the diagnostic performance of the established miRNA panel at different TNM stages. The corresponding AUCs for patients with TNM stages I, II, III and IV were 0.942, 0.935, 0.954 and 0.983, respectively (Figure 6A–D).
Figure 6.
ROC curves for the ability of the miRNA panel to differentiate the stages I (A), II (B), III (C), IV (D), low-CEA level group (E) and high-CEA level group (F) of CAC from the control group, CAC group from CA group (G), CA group from healthy control group (H) in the validation phase.
In addition, we then evaluated the diagnostic accuracy of the established miRNA panel according to the CEA level. In the low-CEA level (<5 ng ml−1) group, the AUC of the established miRNA panel was 0.810 (95% CI, 0.725–0.818; sensitivity=57.14%, specificity=86.67% Figure 6E). In the elevated CEA level (>5 ng ml−1) group, the AUC of the established miRNA panel was 0.918 (95% CI, 0.861–0.957; sensitivity=76.79% and specificity=85.56% Figure 6F).
Finally, we also evaluated the diagnostic performance of the 4-miRNA panel in discriminating the CA from CAC and healthy controls group. The analysis demonstrated that the miRNA panel possessed a high accuracy in discriminating CA from CAC (AUC=0.886; 95% CI: 0.809–0.940; Figure 6G) and CA from healthy controls (AUC=0.765; 95% CI: 0.669–0.845; Figure 6H).
Discussion
Current methods for the CAC diagnosis have been used for many years, including FOBT, CEA detection and colonoscopy. However, the diagnostic performance of these methods is unsatisfactory, especially for the diagnosis of early-stage CAC (Collins et al, 2005; Kim et al, 2007; Lieberman, 2009). The lack of effective detection measures in the early stage at least partly contributes to the high mortality of CAC. The recent discovery of serum miRNA profile in human cancer has provided a new auxiliary approach for the tumour diagnosis (Zhang et al, 2010; Zhou et al, 2011; Liu et al, 2012). Our study is the genome-wide analysis of serum miRNAs during the normal–CA–CAC sequence.
In this study, we established a proof of principle approach to identify a serum miRNA profile of CAC patients. As an initial screening stage, we performed a high-throughput Miseq sequencing assay and excluded the possible contaminations by other small RNAs. However, the sequencing results from pooled samples might not be consistent with the RT–qPCR results performed on individual serum samples. Therefore, the screening stage was followed by two phases of RT–qPCR validation in our study. Through this approach, four significantly altered miRNAs (miR-19a-3p, miR-223-3p, miR-92a-3p and miR-422a) in the serum were identified. Our results demonstrated that the 4-miRNA panel possessed a high accuracy in the CAC diagnosis. Surprisingly, this panel had the potential to separate stage I/II CAC patients from controls and could predict CAC at a relative early stage. Furthermore, we also demonstrated that the 4-miRNA panel was a more sensitive indicator of CAC than the conventional CEA biomarker. Even in the low-CEA level group, the diagnostic accuracy of the miRNA panel was still acceptable. In addition, we also evaluated the diagnostic performance of the 4-miRNA panel in discriminating the CA group from the CAC and healthy control group, and satisfactory results were obtained.
Previous studies have identified a spectrum of dysregulated miRNAs associated with the tumorigenesis and development of CAC. However, these studies mainly focused on the miRNAs in tissues or cells (Slaby et al, 2007; Schetter et al, 2008; Chen et al, 2009; Diosdado et al, 2009; Motoyama et al, 2009; Earle et al, 2010; Liu et al, 2010; Chiang et al, 2011). The reliance on surgical section and invasive procedure for tissue sample collection limits the application of tissue miRNAs in cancer diagnosis (Liu et al, 2012). It becomes possible to comprehensively analyse cancers through a serum miRNA-based biomarker without the need for biopsy, surgery or other invasive methods. In fact, the differential expression of several circulating miRNAs has been reported, including miR-92a, miR-29a, miR-17-3p and miR-221 (Ng et al, 2009; Huang et al, 2010; Pu et al, 2010; Faltejskova et al, 2012; Liu et al, 2013). Previous studies showed that miR-92a is significantly increased in colorectal cancer plasma compared with healthy controls, suggesting its possibility of serving as a potential CAC biomarker. However, its expression levels are not correlated with TNM stages. MiR-92a can also discriminate advanced adenoma from normal controls but fails to distinguish CAC from CA (Ng et al, 2009; Liu et al, 2013). However, one study reported that no significant difference was observed in miR-92a levels in the sera of CRC patients and controls, which provided an evidence against the usage of serum miR-92a as a new biomarker for early detection of CRC (Faltejskova et al, 2012). In the present study, we found that miR-92a-3p was upregulated in both Miseq sequencing and RT–qPCR results and correlated with TNM stages. It could distinguish not only CAC from healthy controls (AUC: 0.918, see ), CA from healthy controls (AUC: 0.826, See Supplementary Figure 1B) but also CAC from CA (AUC: 0.762, see Supplementary Figure 1C). We did not further validate miR-92a-5p by RT–qPCR because it was <50 copies in all the three groups according to our sequencing results. Moreover, some studies showed that plasma levels of miR-29a and miR-17-3p were significantly increased in CAC patients compared with healthy controls (Ng et al, 2009; Huang et al, 2010; Pu et al, 2010). One study reported that no significant differences were observed in miR-17-3p and miR-29a levels in the sera of CRC patients and controls, which was contradictory to previous studies (Faltejskova et al, 2012). In the present study, we found miR-17-3p and miR-29a were not gradually upregulated in the healthy control–CA–CAC sequence in the selection phase. Although it has been found that miR-19a (Wang et al, 2010) and miR-223 (Fu et al, 2012; Wu et al, 2012) are upregulated, and that miR-422a (Faltejskova et al, 2012) is downregulated in CAC tissues compared with normal mucosa, our study was the first report, to our knowledge, showing the diagnostic value of serum expression of miR-19a-3p, miR-223-3p and miR-422a in CAC patients.
Compared with those studies of circulating miRNAs in CAC diagnosis, our study was unique owing to reasons as follows. First, instead of measuring several miRNAs selected from literatures, we screened the genome-wide serum miRNA profiles of CAC via Miseq sequencing. Miseq sequencing is a recent introduction of next-generation sequencing technology, and it can measure the absolute abundance of miRNAs, resulting in a better chance to identify potential diagnostic markers. Furthermore, we included not only the CAC and healthy controls, but also the CA group. It is well known that CA is the usual benign precursor lesion in the transformation to CAC (Oberg et al, 2011). The clinical pathway of most CAC may follow the healthy, CA and CAC states. Therefore, the intersection of differentially expressed miRNAs from CAC vs CA and CA vs healthy control should be considered. Failure to do so might be the reason of the unable discrimination of CA from CAC and healthy controls in previous studies. In addition, we validated the newly developed serum miRNA panel by a large independent cohort using a combination of miR-191-5p and U6 snRNA as reference genes. Currently, there is no standard reference gene for the circulating miRNA studies. In our previous studies, we profiled pooled serum of CAC, CA and healthy controls (same samples used in this study) followed by two phases of validation. We found that there is no differential expression of the combination of miR-191-5p and U6 snRNA among the three groups. Therefore, they could be used as reference genes for serum miRNA RT–qPCR study in CAC (Zheng et al, 2013).
Taken together, we established a serum 4-miRNA panel using a large number of individuals, and this serum miRNA panel could differentiate CAC from CA and healthy controls with a high accuracy. Our study also demonstrated that the 4-miRNA panel had important clinical value for the diagnosis of stage I/II CAC and CA patients. In conclusion, our finding provided a relatively accurate CAC diagnosis at the early stage, and more patients, who would have otherwise missed the curative treatment window, might benefit from the early diagnosis.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81271916, 81301506), the Research Fund for the Doctoral Program of Higher Education of China (No. 20120131110055, 20130131120067), Independent Innovation Foundation of Shandong University (IIFSDU, 2012TS174), the Shandong Technological Development Project (STDP, 2013GSF11859) and the Natural Science Foundation of Shandong (Grant No. ZR2013HM104).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, Zhang CY. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, Zhuang R, Zhang Y, Hu H, Liu C, Xu L, Wang J, Shen H, Zhang J, Zen K, Zhang CY. Identification of ten serum microRN as from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for non small cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- Chiang Y, Song Y, Wang Z, Chen Y, Yue Z, Xu H, Xing C, Liu Z. Aberrant expression of miR-203 and its clinical significance in gastric and colorectal cancers. J Gastrointest Surg. 2011;15:63–70. doi: 10.1007/s11605-010-1367-8. [DOI] [PubMed] [Google Scholar]
- Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JF, Lieberman DA, Durbin TE, Weiss DG. Accuracy of screening for fecal occult blood on a single stool sample obtained by digital rectal examination: a comparison with recommended sampling practice. Ann Intern Med. 2005;142:81–85. doi: 10.7326/0003-4819-142-2-200501180-00006. [DOI] [PubMed] [Google Scholar]
- Diosdado B, VandeWiel MA, TerhaarSiveDroste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B, Meijer GA. MiR-17–92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707–714. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle JS, Luthra R, Romans A, Abraham R, Ensor J, Yao H, Hamiton SR. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433–440. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faltejskova P, Bocanek O, Sachlova M, Svoboda M, Kiss L, Vyzula R, Slaby O. Circulating miR-17-3p, miR-29a, miR-92a and miR-135b in serum: evidence against their usage as biomarkers in colorectal cancer. Cancer Biomark. 2012;4:199–204. doi: 10.3233/CBM-130308. [DOI] [PubMed] [Google Scholar]
- Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss L, Vyzula R, Slaby O. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16:2655–2666. doi: 10.1111/j.1582-4934.2012.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Fu J, Tang W, Du P, Wang G, Chen W, Li J, Zhu Y, Gao J, Cui L. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol. 2012;6:68. doi: 10.1186/1752-0509-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- Kim DH, Pickhardt PJ, Taylor AJ, Leung WK, Winter TC, Hinshaw JL, Gopal DV, Reichelderfer M, Hsu RH, Pfau PR. CT colonography versus colonoscopy for the detection of advanced neoplasia. N Engl J Med. 2007;357:1403–1412. doi: 10.1056/NEJMoa070543. [DOI] [PubMed] [Google Scholar]
- Levin B, Lieberman DA, McFarland B, Smith RA, Brooks D, Andrews KS, Dash C, Giardiello FM, Glick S, Levin TR, Pickhardt P, Rex DK, Thorson A, Winawer SJ. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer. J Clin. 2008;58:130–160. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- Lieberman DA. Clinical practice. Screening for colorectal cancer. N Engl J Med. 2009;361:1179–1187. doi: 10.1056/NEJMcp0902176. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhou Z, Chen R, Wang M, Zhou B, Li Y, Sun XF. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis ofcolorectal cancer. Tumour Biol. 2013;34:2175–2181. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, Yuan Y, Li Z, Zen K, Ba Y, Zhang CY. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R, Deng T, Liu H, Yin J, Wang S, Zen K, Ba Y, Zhang CY. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Mandel JS. Screening for colorectal cancer. Gastroenterol Clin North Am. 2008;37:97–115. doi: 10.1016/j.gtc.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Motoyama K, Inoue H, Takatsuno Y, Anaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- Oberg AL, French AJ, Sarver AL, Subramanian S, Morlan BW, Riska SM, Borralho PM, Cunningham JM, Boardman LA, Wang L, Smyrk TC, Asmann Y, Steer CJ, Thibodeau SN. miRNA expression in colon polyps provides evidence for a multihitmodel of colon cancer. PLoS One. 2011;6:e20465. doi: 10.1371/journal.pone.0020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu X, Huang G, Guo H, Guo C, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Neutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang X, Zhang B, Yang C, Chen X, Gao H. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- Wu L, Li H, Jia C, Cheng W, Yu M, Peng M, Zhu Y, Zhao Q, Dong YW, Shao K, Wu A, Wu XZ. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012;586:1038–1043. doi: 10.1016/j.febslet.2012.02.050. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, Xing J, Liang Z, Ren B, Yang C, Zen K, Zhang CY. Expression profile of microRNAs in serum: a fingerpr int for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- Zheng G, Wang H, Zhang X, Yang Y, Wang L, Du L, Li W, Li J, Qu A, Liu Y, Wang C. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One. 2013;12:e83025. doi: 10.1371/journal.pone.0083025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X, Wang Z, Qiu S, Wang X, Yang G, Sun H, Tang Z, Wu Y, Zhu H, Fan J. Plasma microRNA panel to diagnose hepatitis B virus–related hepatocellular carcinoma. J of Clin Oncol. 2011;29:4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.