Bacterial cell-to-cell signaling
In order to regulate energy expenditure, microbes rely on a variety of mechanisms to control gene expression in response to changing environmental conditions. One mechanism used by microorganisms was originally described as quorum sensing (QS). During QS, a bacterial cell produces and secretes a signaling molecule, called an autoinducer. As the density of the bacterial population increases, so does the concentration of secreted autoinducer molecules. When the concentration of the autoinducer reaches a critical threshold, it diffuses back into the cell and activates or represses certain target genes. This type of signaling enables bacteria to regulate genes in a manner that reflects population density. Bacteria are also able to detect signal molecules produced by other species of bacteria as well as hormones produced by their mammalian hosts. Therefore, cell-to-cell signaling involves more than just taking a bacterial census, but is also involved in communicating about the local environment and growth potential of a population of cells (6, 70).
As currently understood, E. coli and Salmonella utilize three main types of cell-to-cell signaling processes. In the LuxR process, E. coli and Salmonella detect an autoinducer synthesized by other types of bacteria. During the LuxS/AI-2 signaling system, E. coli and Salmonella participate in intra- and interspecies signaling. Finally, during the AI-3/epinephrine/norepinephrine system, E. coli and Salmonella recognize self-produced autoinducer, signal produced by other microbes, or the human stress hormones epinephrine or norepinephrine.
Overview: LuxR-I
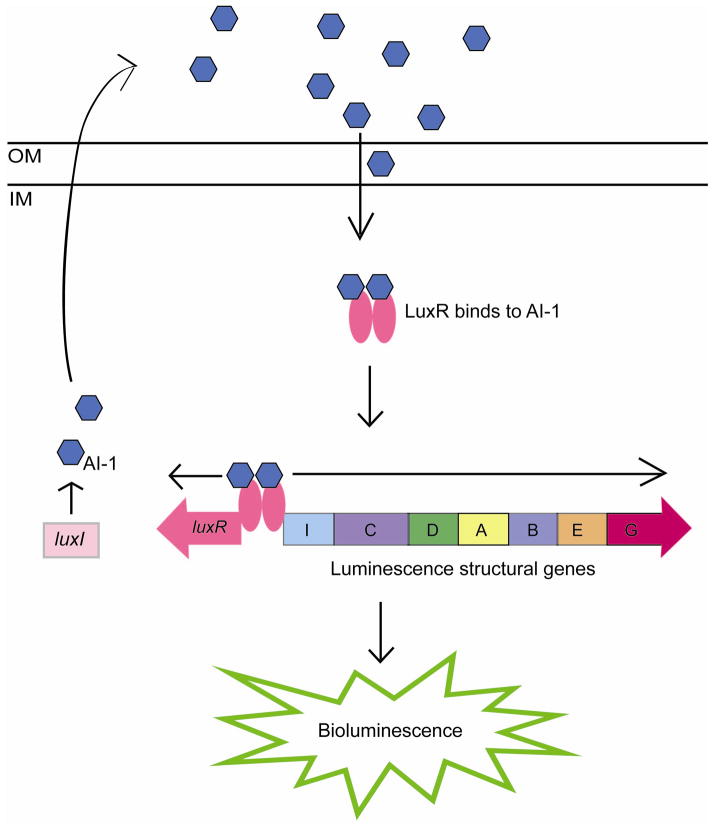
Quorum sensing using the LuxR-I system was initially described as regulating the bioluminescence in Vibrio fischeri (47). Two proteins regulate the luciferase operon in V. fischeri, LuxI and LuxR. LuxI is responsible for the synthesis of the autoinducer molecule N-acyl-homoserine-lactone (AHL), called autoinducer 1 (AI-1). After synthesis, AI-1 freely diffuses across the bacterial membrane into the surrounding environment (33). As the number of bacteria in a population increases, so does the concentration of AI-1 molecules. At a particular threshold concentration, AI-1 diffuses back into the bacterial cell and binds to its specific receptor protein LuxR. LuxR complexed with AI-1 activates transcription of itself as well as transcription of the luciferase operon (16, 17, 33) (Fig. 1). In all of the LuxR-LuxI systems, bacteria produce the AHL molecule, autoinducer-1 (AI-1), which then binds to the LuxR protein to regulate transcription of different genes involved in a variety of phenotypes (12, 49).
Figure 1.
Model of LuxI/ LuxR quorum sensing in Vibrio fischeri. LuxI synthesizes AI-1 and diffuses outside of the cell. When cell density, and therefore, AI-1 concentration, is high, AI-1 diffuses back into the cell where it binds to LuxR. LuxR complexed with AI-1 subsequently activates transcription of itself as well as the luciferase operon.
IM, inner membrane; OM, outer membrane.
AI-1
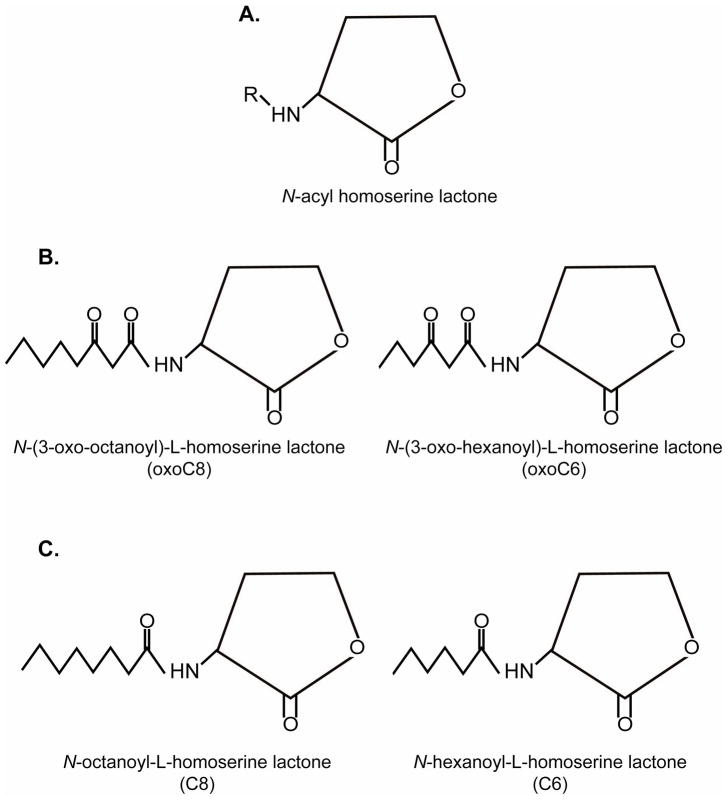
LuxI and its homologues synthesize autoinducer molecules by transferring a fatty acid chain from an acylated acyl carrier protein to S-adenosylmethionine, releasing the AHL and methylthioadenosine (58). AHLs consist of a homoserine lactone ring joined to a fatty acid side chain (Fig. 2A). Although each species of bacteria produces a distinct autoinducer molecule, variations in AHLs are found among different species of bacteria. For example, the length of acyl chain may contain four to 18 carbons, and acyl chains differ in the degree of saturation. Additionally, the AHL may be modified at the third carbon of the acyl chain and contain a hydrogen, hyrdroxyl, carbonyl, or oxo group (20, 42, 81).
Figure 2.
Structures of AI-1. A. AI-1 is composed of a homoserine lactone ring bound to a fatty acid side chain; B. SdiA detects oxoC8 and oxoC6 with greatest sensitivities; C. SdiA can also detect other forms of AI-1 such as C8 and C6, although with less sensitivity.
Similar to the AHL, the receptor molecule (LuxR and its homologues) is species specific; however, some LuxR homologs can detect other related AHLs produced by other species of bacteria, but with lower specificity (68). The AHL receptor molecules detect nanomolar concentrations of its corresponding AHL molecule and are stabilized by this interaction. In the absence of its specific AHL, AHL receptor proteins are targeted to degradation (89, 90).
Overview of AHL/SdiA in E. coli & Salmonella
E. coli and Salmonella are unique in this cell-signaling process in that these bacteria rely on AHL detection for interspecies communication as opposed to intra-species communication that was the paradigm of this mechanism for many years (44). E. coli and Salmonella lack LuxI, and thus do not synthesize AHLs; however, both encode the protein SdiA that apparently recognizes and binds to AHLs produced by other species of bacteria. SdiA requires these AHL compounds to fold properly (86, 87).
SdiA detects a much broader range of AHLs than other LuxR homologs (68). SdiA is most strongly activated by 3O-AHLs with chains between six and eight carbons long (Fig. 2B), sensing concentrations as low as 1 nm to 5 nm of these AHLs. However, SdiA can also recognize oxoC10, 6, and 8 AHLs at approximately 50 nm (1, 30, 44) (Fig 2C). When a sulfur atom replaces the 3′-oxygen molecule in a laboratory-synthesized derivative, SdiA is also strongly activated (30); however, it is not know whether this molecule naturally exists in nature.
AI-1 signaling in E. coli
Initial experiments suggested that SdiA (suppressor of cell division inhibition) played a role in the regulation of cell division genes ftsQAZ in E. coli (79). These results were based on sdiA cloned into a multi-copy plasmid, yet the sdiA mutant has no apparent cell division defects (79). Additional experiments demonstrated that SdiA repressed the LEE and motility genes in enterohemorrhagic E. coli (EHEC) (31); however, these effects were observed only by overexpression of SdiA, and no sdiA mutant was examined (31).
The precise role of SdiA was elusive for many years until the discovery that SdiA did not sense self-produced AHLs, but AHLs produced by other bacterial species. Many LuxR-type proteins rely on the AHL autoinducer as a co-factor for proper folding, and that in the absence of AHLs, the protein is targeted for degradation (89, 90). Indeed, the NMR structure of the SdiA protein indicates that AHL-binding allows proper protein folding (86), and the phenotypes associated with SdiA expression are only observed in the presence of AHLs (44).
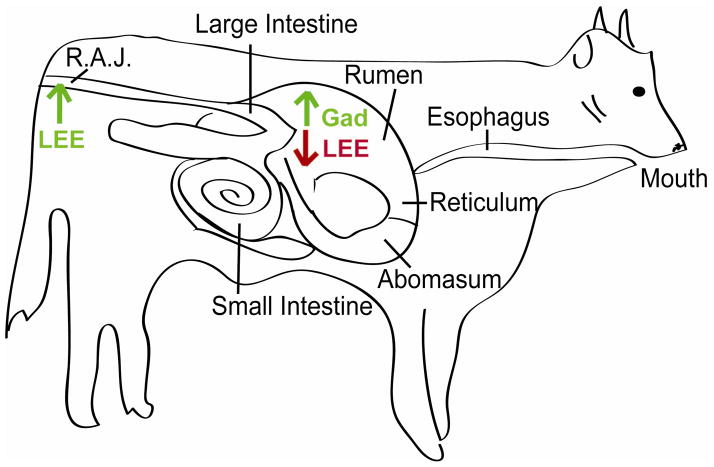
SdiA seems to integrate external stimuli such as temperature and pH (28, 73), which may allow enterohemorrhagic E. coli (EHEC) O157:H7 to colonize the gastrointestinal (GI) tract of cattle (15, 28, 40), the main reservoir for this bacterium (32). During passage through the cattle GI tract, EHEC encounters broad ranges in pH, and thus, must regulate gene expression to ensure survival and colonization (Fig. 3). Upon entering the rumen, EHEC is subjected to a neutral pH and AHLs (18). Here the AHLs activate SdiA which in turn increases expression of the gad-encoded acid resistance genes (28, 51). After passage through the rumen, EHEC traverses through the low pH (2.0 to 2.5) environment of the abomasum en route to the colon. Potentially, the up-regulation of the gad acid resistance genes in the rumen primes EHEC for entry into the acidic environment of the abomasum (pH 2.0 to 2.5) (51), the next stop for EHEC en route to the colon.
Figure 3.
Model of SdiA-AHL dependent EHEC gene expression in the GI of cattle. Once EHEC enters the rumen, it encounters AHLs. In the presence of AHLs, SdiA is functionally stable and acts to increase expression of acid-tolerance genes in the gad operon and represses expression of the LEE genes. Up-regulation of the gad genes allows EHEC to survive passage through the acidic abomasum. AHLs have not been detected in the colon, thus SdiA is unstable, and EHEC can activate the LEE and colonize the renal anal junction of the colon.
Additionally, SdiA directly regulates expression of the LEE genes. The LEE genes (and corresponding AE lesion formation) are necessary for EHEC colonization of the renal-anal-junction (RAJ) site in the colon (40). Five major operons (LEE1-5) comprise the LEE. The LEE encoded regulator (Ler) that is encoded in LEE1 activates the other LEE operons (22, 57, 63). In the presence of AHLs, SdiA directly binds to ler, acting as a repressor of this gene, and consequently, the other LEE genes expression of the LEE (28). AHLs have not been detected in the RAJ, thus in the absence of AHLs, SdiA will be degraded, relieving repression of the LEE and allowing EHEC to successfully colonize the cattle. Accordingly, competition experiments demonstrated that an sdiA mutant does not colonize cattle as efficiently as WT EHEC (28).
In nonpathogenic E. coli, SdiA, in conjunction with indole, may regulate biofilm formation, motility formation, and indole production (36–38). However, more research is needed to elucidate this signaling pathway as differential regulation by SdiA could be observed in the presence or absence of AHLs (37), the compounds required by SdiA for proper folding and function (86, 87).
AI-1 signaling in Salmonella
Upon detecting AHLs produced by other species of bacteria, SdiA in Salmonella activates expression of two srg (SdiA-regulated gene) loci, the rck operon that encodes seven genes and the srgE gene (1, 61). The rck operon is located on the Salmonella virulence plasmid pSLT and contains six genes: pefI, srgD, srgA, srgB, rck, and srgC (1, 19, 44, 61) (Fig. 4). The pefI and srgA genes regulate expression and folding of the upstream pef operon (5, 41, 48). The pef operon encodes frimbriae that function in adhesion to the small intestine of mice (3, 19). SrgD and SrgC are putative transcription factors whose target genes have not been identified (19, 44). The lipoprotein SrgB has no known function (19). Rck, an 8-stranded β-barrel protein localized to the outer membrane, (8) functions in resistance to complement killing (23, 24) by preventing the polymerization of complement component C9 on the bacterial cell envelope (25) as well as acting as an adhesin to host tissues (11). The second SdiA-regulated locus is encoded in the chromosome and consists of the gene srgE that is predicted to encode a protein containing a coiled-coil domain (2, 61). The function of SrgE has not yet been elucidated (61). Neither the rck operon nor the srgE gene are found in E. coli (1).
Figure 4.
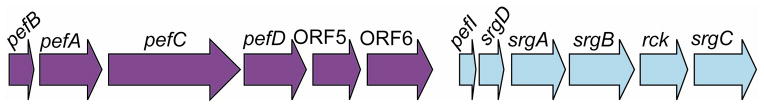
SdiA in Salmonella activates expression of the rck operon. The pefI and the srgA genes encoded in this operon affect the transcription and folding, respectively, of plasmid-encoded fimbriae that are encoded by the upstream pef genes. SrgA plays a role in protein folding in the pef-encoded fimbriae by catalyzing disulfide bond formation (5, 41).
Similar to E. coli, SdiA in Salmonella appears to play a role in adapting and recognizing environmental conditions. In Salmonella, SdiA is induced at low pH (pH 4) under aerobic conditions (56), and the rck operon is expressed at 37°C, but not at 30°C or at 22°C (1). Contrary to E. coli, SdiA in Salmonella does not appear to play a role in cow colonization, as it is not activated during passage through the GI tract of cattle (62). SdiA is activated, however, during passage through the GI tract of the turtle, a reptile that is commonly associated with Salmonella (62). Potentially, the age or diet of cattle influences AHL production by its commensal microbiota (62), and therefore, colonization by Salmonella.
LuxS/AI2
Overview
Since the discovery of cell-to-cell signaling using AHLs, another cell-to-cell signaling system has been discovered. The luxS QS system is present in roughly half of all sequenced bacterial genomes (80) and was first characterized as the regulator of bioluminescence in Vibrio harveyi (70).
LuxS synthesizes AI-2
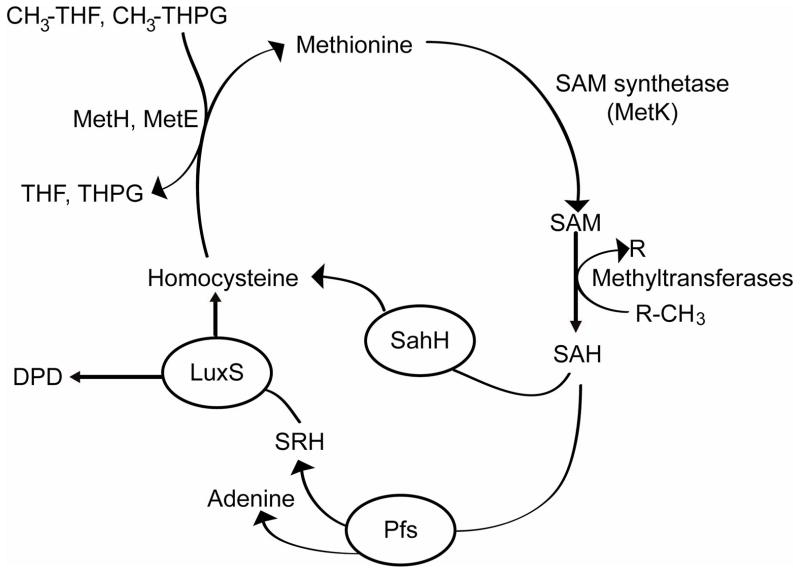
LuxS is a small metalloenzyme that catalyzes the terminal step in the essential activated methyl cycle in bacterial cells (Fig. 5). In this pathway, S-adenosylmethionine (SAM) is recycled ultimately to homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD) (59, 60, 82–84). SAM is a major cellular methyl donor in bacterial cells, and transfer of its methyl group to various substrates produces the toxic by-product S-adenosylhomocysteine (SAH) (60). The nucleoside Pfs subsequently detoxifies SAH, yielding adenine and S-ribosylhomocysteine (SRH). In the final step of the pathway, LuxS coverts SRH to homocysteine and DPD. DPD is an extremely unstable compound that reacts with water and cyclizes to form several different furanones, one of which is thought to be the precursor of AI-2 (39, 60, 66, 82).
Figure 5.
Synthesis of AI-2. In the activated methyl cycle, SAM is converted ultimately to homocysteine and DPD. LuxS catalyzes the last step in the pathway, resulting in DPD. AI-2 is derived from the unstable compound DPD.
Crystal structures of LuxS have revealed that this protein is a homodimer with two identical active sites that are formed at the dimmer interface by residues from both subunits (27, 39, 52). Each active site contains a divalent metal ion, Fe2+, that catalyzes an internal redox reaction (50, 88).
AI-2
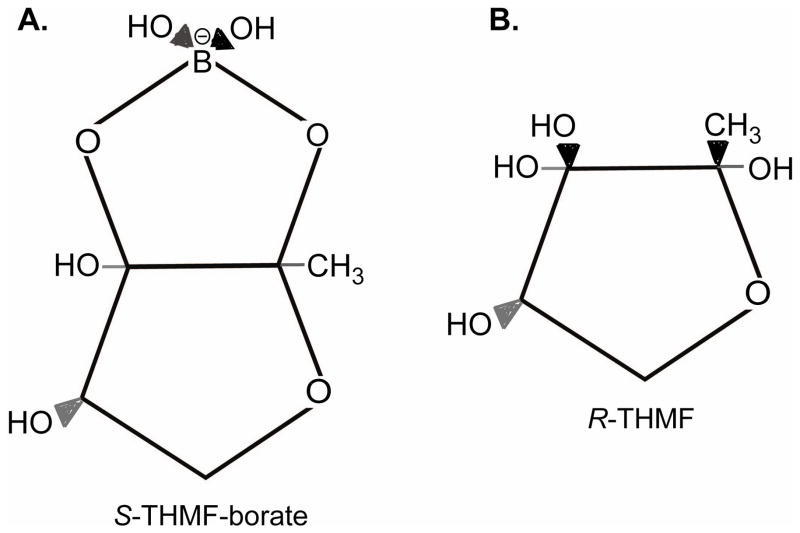
The structure of AI-2 has been solved by co-crystallization of the ligand with its receptor LuxP in Vibrio harveyi (7). In V. harveyi, AI-2 is a furanosyl-borate diester, (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran-borate (S-THMF borate) (7). LuxP homologues and homologues from the V. harveyi signaling cascade appear to be unique to this genera, suggesting that AI-2 recognition varies among bacterial species. Indeed, co-crystallization of AI-2 and its receptor, the periplasmic protein LsrB, in Salmonella, revealed that Salmonella recognizes a chemically distinct form of AI-2, (2R,4S)-2-methyl-2,3,3,4-tetrahydroxyte-tetrahydrofuran (R-THMF) (45) (Fig. 6).
Figure 6.
Structure of AI-2 signaling molecules. A. AI-2 produced by Vibrio harveyi is (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran-borate (S-THMF borate); B. AI-2 produced by Salmonella typhimurium is (2R,4S)-2-methyl-2,3,3,4-tetrahydroxyte-tetrahydrofuran (R-THMF).
AI-2 production is dependent on growth conditions, such as nutrient availability, pH, osmolarity, oxygen availability, growth rate and stress factors- both intracellular and from the environment (heat shock, amino acid limitation) (13, 70).
AI-2 signaling in Salmonella and E. coli
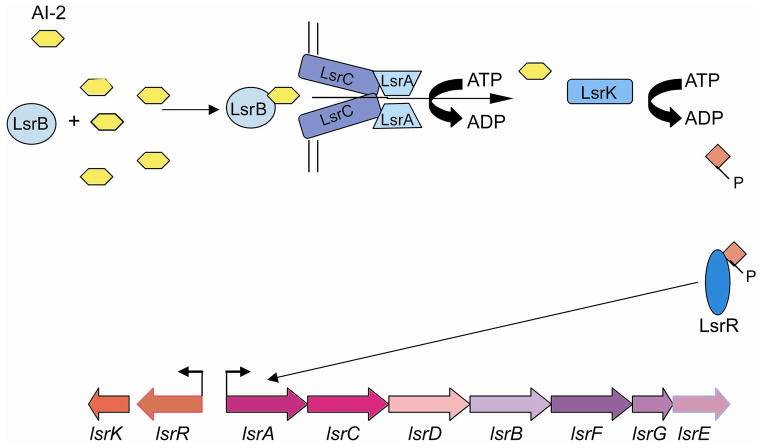
In Salmonella enterica serovar Typhimurium, AI-2 regulation involves genes that encode an ABC transporter named Lsr (LuxS-regulated) (Fig. 7). Seven genes comprise the lsr operon, lsrACDBFGE, and AI-2 activates its transcription (72). This transporter is also present in E. coli, and in both species, the Lsr transporter is homologous to sugar transporters. As extracellular AI-2 concentrations increase, AI-2 binds the periplasmic protein LsrB and is subsequently imported inside the cell by the Lsr ABC transport system. Once it is inside the cell, AI-2 is modified by phosphorylation. Phosphorylated AI-2 is thought to interact with LsrR, a SorC-like transcription factor. Upon binding AI-2, LsrR represses expression of the lsr operon (71, 72).
Figure 7.
The LuxS/AI-2 signaling pathway. The lsrACDBFGE genes are transcribed in an operon, whereas lsrK and lsrR are divergently transcribed. LsrB binds AI-2 which is then imported by the Lsr ABC transport system. Inside the cell, AI-2 is phosphorylated by LsrK and is thought to interact with LsrR, relieving the repression of the lsr operon.
AI-2 and virulence
Many phenotypes have been attributed to cell signaling via AI-2; however, the precise role of AI-2 in bacterial cell-to-cell signaling is debated. Some studies have suggested that AI-2 is involved in biofilm formation, motility (14, 21, 26); however, another report examining a luxS mutant in W3110 strain did not see any effects on growth, motility, or biofilm formation (78). Additional experiments compared luxS mutants with wild-type strains and genetically complemented mutants or added preconditioned media to bacterial cultures and found that AI-2 signaling affected expression of the LEE-encoded type three secretion system and motility in EHEC (64, 65). However, using purified and in vitro- synthesized AI-2, it has been revealed that the signaling molecule affecting TTSS and motility in EHEC is not AI-2, but a distinct autoinducer, AI-3, that is not dependent upon luxS for synthesis (66, 76) (see below). Therefore, the role of AI-2 in pathogenesis needs to be examined further.
AI-2 and cell metabolism
In contrast, some researchers have suggested that the role of AI-2 in Salmonella and E. coli functions primarily in metabolism because of the similarities in function and sequence homology of the lsr operon to other sugar transporters (74, 78, 82, 83). Similar to other carbon sources, the synthesis and import of AI-2 is strictly controlled. In both E. coli and Salmonella typhimurium, AI-2 production is dependent on the amount of carbohydrates in the medium and is growth phase dependent. Cells in exponential phase produce and secrete AI-2, whereas those entering stationary phase no longer produce AI-2 (6, 69). Furthermore, AI-2 is not imported in the presence of glucose because the lsr operon is not transcribed due to cAMP-CAP-mediated repression; however, when glucose becomes limiting, cells import AI-2 (77, 85).
Gene expression profiles also seem to suggest that the function of AI-2 is primarily metabolic. For instance, the lsr operon was not induced when E. coli was grown in medium containing glucose, whereas, in glucose-free medium, the operon was expression (78). Moreover, the lsr operon was induced only in the absence of glucose, and that the luxS mutation in E. coli primarily affected genes related to AI-2 production and transport (78). When a luxS mutant was compared to WT EHEC as well as the luxS mutant with DPD added, the majority of genes with an altered profile are associated with central metabolism and core biological processes (35). Finally, a study using phenotype microarrays observed that the luxS mutation resulted in numerous metabolic changes, especially in those processes that involve nitrogen and carbon metabolism (76).
AI-3/epinephrine (epi)/norepinephrine (NE) signaling system
Overview
The third major type of cell-to-cell signaling involves inter-kingdom signaling between prokaryotic and eukaryotic cells. In this system, the autoinducer-3 (AI-3) produced by the commensal GI microflora and/ or epinephrine (epi) and norepinephrine (NE) produced by the host (66) interact with a two-component regulatory system to activate transcription of genes involved in pathogenesis (64, 67). This signaling system was initially characterized in EHEC (66), but it is not unique to this strain (53, 76).
Signaling molecules AI-3, epi, NE
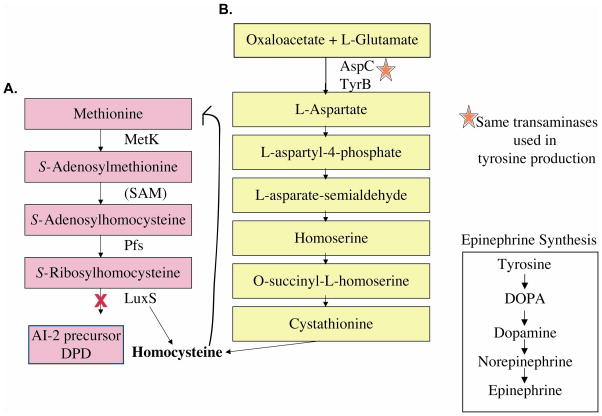
AI-3 quorum sensing system was discovered through its association with the luxS system. Although AI-3 does not directly depend upon luxS for synthesis, a luxS mutation has several effects on AI-3 production. A mutation in the luxS gene affects AI-3 synthesis by altering cellular metabolism (Fig. 8). More specifically, the luxS mutation requires that the cell utilize oxaloacetate, instead of SAM, for de novo synthesis of methionine. Exclusive use of this pathway may alter cellular metabolism and cellular concentrations of amino acids, potentially lead to reduced tyrosine levels and, consequently, diminished AI-3 concentrations (76).
Figure 8.
Pathways for synthesis of homocysteine in E. coli. A. Methionine is important in the cell for production of the vital metabolic enzyme SAM (involved in the methylation of lipids, RNA, DNA, and protein) and de novo synthesis of methionine requires homocysteine. B. The luxS mutant cannot produce homocysteine through SRH hydrolysis; therefore, the oxaloacetate/L-glutamate pathway must be used. Oxaloacetate, L-glutamate, and the AspC and TyrB transaminases are used to produce aspartate, which than can proceed through a series of reactions, that result in the synthesis of homocysteine. Exclusive use of this pathway may lead to altered metabolism and amino acid content in the luxS mutant, leading to reduced AI-3 synthesis.
Additional studies have demonstrated that AI-3 is a chemically distinct molecule from AI-2. AI-2 is a polar furanone that does not bind to C-18 columns, whereas AI-3 binds to C-18 columns and can only be eluted with methanol (66). Moreover, electrospray mass spectrometry also revealed differences between the structures of AI-3 and AI-2 (7, 66). AI-2 and AI-3 activity can be differentiated using two different assays. The AI-2 assay is based on the production of bioluminescence in V. harveyi, and AI-3 does not show any activity for this assay. Conversely, the AI-3 activates transcription of the EHEC virulence genes, and AI-2 has no effect on this assay.
AI-3 is an aromatic aminated signal, but its final structure has not yet been elucidated (29). Because Epi/NE can replace AI-3 in the regulation of EHEC virulence gene regulation and because the regulatory effects of epi/NE and AI-3 can be inhibited by adrenergic receptor antagonists, it has been hypothesized that AI-3 may be structurally similar to epi/NE (66) (Fig. 9). In addition to EHEC, Salmonella and several species of pathogenic and nonpathogenic bacteria produce AI-3 (76).
Figure 9.
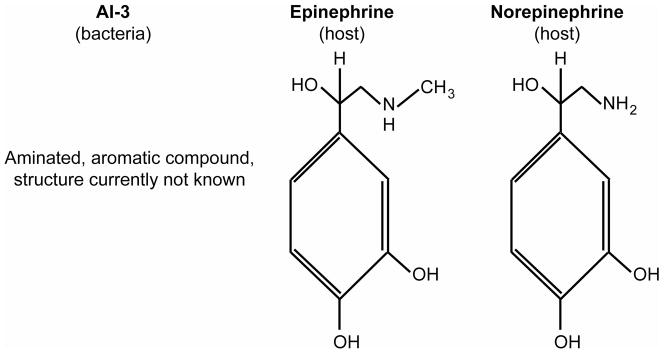
The AI-3, epinephrine, and norepinephrine signaling molecules. The structure of AI-3 is not known, but may resemble the aromatic compounds epinephrine and norepinephrine.
AI-3/ epi/ NE receptor, QseC
The membrane-bound protein QseC (quorum sensing in E. coli) is a bacterial adrenergic receptor that directly interacts with AI-3 and epi/NE (10). QseC has two transmembrane domains, a histidine sensor kinase (HK) domain, and an ATPase domain. The HK domain allows autophosphorylation upon sensing AI-3 and, especially, epi/NE. QseC also contains an ATPase domain that enables it to phosphorylate QseB. The action of QseC can be blocked by phentolamine, an antagonist of α-adrenergic receptors (10). The QseC sensor is conserved among other enteric bacteria of the genera including Shigella, Salmonella, and Yersinia (10).
AI-3/epi/NE signaling in E. coli
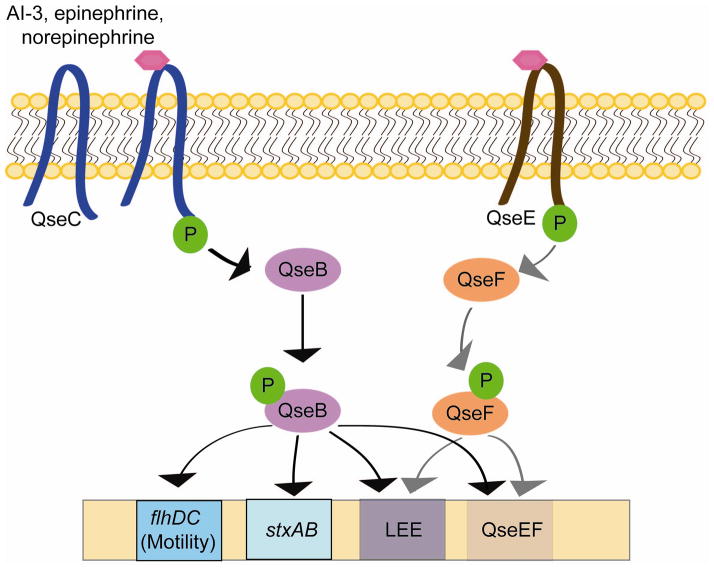
QseBC plays an important role in EHEC pathogenesis and inter-kingdom signaling (29). Upon binding AI-3 and epi/NE, QseC augments its phosphorylation state and then activates a complex regulatory cascade that includes its response regulator QseB (9, 10, 65–67) (Fig. 10). Rabbit and bovine infection models have shown that recognition of these three signals is essential for in vivo virulence expression (10, 53, 75). QseBC activates transcription of the flagella regulon responsible for swimming motility in EHEC (9), production of Shiga toxin, expression of the LEE genes, as well as another two-component system, QseEF that is involved in AE lesion formation (29, 54, 55).
Figure 10.
The AI-3/epi/NE signaling pathway. AI-3, epinephrine, and norepinephrine bind the membrane receptor protein QseC, resulting in autophosphorylation. Subsequently, QseC phosphorylates its response regulator QseB, initiating a complex phosphorelay signaling cascade that results in expression of the flagellar biosynthesis and motility genes (flhDC), Shiga toxin (stxAB), the locus of enterocyte effacement (LEE), and a second two-component system, QseEF that also promotes expression of the LEE.
Recently, the AI-3/epi/NE quorum sensing has been implicated in biofilm formation in enteropathogenic E. coli (EPEC) (46). Microcolony formation in an initial step in biofilm development, and in EPEC, it is mediated by several adhesins including the bundle-forming pilus and the EspA (46). Expression of espA is controlled by the AI-3/epi/NE quorum sensing system, suggesting that biofilm formation is regulated, at least in part, by AI-3.
AI-3/ epi/NE signaling in Salmonella
The AI-3/epi/NE signaling system S. typhimurium appears very similar to that of EHEC. S. typhimurium encodes a functionally interchangeable homologue of the EHEC QseC (87% similarity) that similarly regulates virulence gene expression (43). A S. typhimurium qseC mutant is defective for colonization of the swine GI tract (4) and attenuated for systemic disease in mice (53). Moreover, microarray and real-time RT-PCR data indicated that in response to norepinephrine, motility genes as well as early-, mid-, and late-genes involved in flagellar synthesis were up-regulated in S. Typhimurium (4). As in EHEC, the QseBC two-component system in S. Typhimurium seems necessary for optimal induction of motility in response to norepinephrine (4). Norepinephrine also regulates genes encoding a lipid A modification system, iron transport, and type three secretion (34). The phenotypes observed in response can be blocked by propranolol (34).
Acknowledgments
This work was supported by NIH Grant AI053067 and a Burroughs Wellcome Fund. MMK was supported by a NIH NIAID fellowship 1F32AI080115-01
References
- 1.Ahmer BM. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol. 2004;52:933–945. doi: 10.1111/j.1365-2958.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahmer BM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumler AJ, Tsolis RM, Bowe FA, Kusters JG, Hoffmann S, Heffron F. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect Immun. 1996;64:61–68. doi: 10.1128/iai.64.1.61-68.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearson BL, Bearson SMD. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microbiol Path. 2008;44:271–278. doi: 10.1016/j.micpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Bouwman CW, Koli M, Killoran A, Touchie GA, Kadner RJ, Martin ML. Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J Bacteriol. 2003;185:991–1000. doi: 10.1128/JB.185.3.991-1000.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cappuyns AM, Bernaerts K, Vanderleyden J, Van Impe JF. A dynamic model for diauxic growth, overflow metabolism, and AI-2 mediated cell-cell communication of Salmonella Typhimurium based on systems biology concepts. Biotechnol Bioeng. 2008;102:280–293. doi: 10.1002/bit.22044. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Schauder S, Potier N, VanDorssealaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo DM, Heffernan EJ, Wu L, Harwood J, Fierer J, Guiney DG. Identification of a domain in Rck, a product of the Salmonella typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect Immun. 1996;64:2019–2023. doi: 10.1128/iai.64.6.2019-2023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke M, Sperandio V. Transcriptional regulation of flhDC by QseBC and sigma 28 (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2005;57:1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- 10.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crago AM, Koronakis V. Binding of extracellular matrix laminin to Escherichia coli expressing the Salmonella outer membrane proteins Rck and PagC. FEMS Microbiol Lett. 1999;176:495–501. doi: 10.1111/j.1574-6968.1999.tb13703.x. [DOI] [PubMed] [Google Scholar]
- 12.de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLisa MP, Valdes JJ, Bentley WE. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J Bacteriol. 2001;183:2918–2928. doi: 10.1128/JB.183.9.2918-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domka J, Lee J, Wood TK. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol. 2006;72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dziva F, vanDiemen PM, Stevens MP, Smith AJ, Wallis TS. Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology. 2004;150:3631–3645. doi: 10.1099/mic.0.27448-0. [DOI] [PubMed] [Google Scholar]
- 16.Engebrecht J, Nealson KH, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 17.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson DL, V, Nsereko L, Morgavi DP, Selinger LB, Rode LM, Beauchemin KA. Evidence of quorum sensing in the rumen ecosystem: detection of N-acyl homoserine lactone autoinducers in ruminal contents. Can J Microbiol. 2002;48:374–378. doi: 10.1139/w02-022. [DOI] [PubMed] [Google Scholar]
- 19.Friedrich MJ, Kinsey NE, Vila J, Kadner RJ. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 20.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Esherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haack KR, Robinson CL, Miller KJ, Fowlkes JW, Mellies JL. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect Immun. 2003;71:384–392. doi: 10.1128/IAI.71.1.384-392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackett J, Wyk P, Reeves P, Mathan V. Mediation of serum resistance in Salmonella typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect Immun. 1987;64:2019–2023. [Google Scholar]
- 24.Heffernan EJ, Harwood J, Fierer J, Guiney DG. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol. 1992;174:84–91. doi: 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heffernan EJ, Reed S, Hackett J, Fierer J, Roudier C, Guiney DG. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J Clin Invest. 1992;90:953–964. doi: 10.1172/JCI115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzberg M, I, Kaye K, Petri W, Wood TK. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higers MT, Ludwig ML. Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site. Proc Natl Acad Sci USA. 2001;98:11169–11174. doi: 10.1073/pnas.191223098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes DT, Hovde CJ, Gonzales JE, Edrington TS, Rasko DA, Sperandio V. SdiA: a bacterial sensor for the rumen environment in cattle. 2009 submitted. [Google Scholar]
- 29.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nature Microbio Rev. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janssens JCA, Metzger K, Daniels R, Ptacek D, Verhoeven T, Habel LW, Vanderleyden J, DeVos DE, DeKeersmaecker SCJ. Synthesis of N-acyl homoserine lactone analogues reveals strong activators of SdiA, the Salmonella enterica serovar Typhimurium LuxR homologue. Appl Environ Microbiol. 2007;73:535–544. doi: 10.1128/AEM.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. SdiA, an Escherichia coli homologue of quorum sensing-regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2000;38:805–816. doi: 10.1046/j.1365-2958.2000.02171.x. [DOI] [PubMed] [Google Scholar]
- 32.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nature Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karavolos MH, Spencer H, Bulmer DM, Thompson A, Winzer K, Williams P, Hinton JCD, Khan CMA. Adrenaline modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC Genomics. 2008;9:548. doi: 10.1186/1471-2164-9-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kendall MM, Rasko DA, Sperandio V. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect Immun. 2007;75:4875–4884. doi: 10.1128/IAI.00550-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Maeda T, Hong SH, Wood TK. Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactone. Appl Environ Microbiol. 2009;75:1703–1716. doi: 10.1128/AEM.02081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Zhang XS, Hedge M, Bentley WE, Jayaraman A, Wood TK. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- 39.Lewis HA, Furlong EV, Laubert B, Eroshkina GA, Batiyenko Y, Adams JM, Bergseid MG, Marsh CD, Peat TS, Sanderson WE, Sauder JM, Buchanan SG. A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs. Structure. 2001;9:527–537. doi: 10.1016/s0969-2126(01)00613-x. [DOI] [PubMed] [Google Scholar]
- 40.Lim JY, Sheng H, Seo KS, Park YH, Hovde CJ. Characterization of an Escherichia coli O157:H7 plasmid O157 deletion mutant and its survival and persistence in cattle. Appl Environ Microbiol. 2007;73:2037–2047. doi: 10.1128/AEM.02643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin D, Rao CV, Slauch JM. The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J Bacteriol. 2008;180:87–97. doi: 10.1128/JB.01323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marketon MM, Gronquist MR, Eberhard A, González JE. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J Bacteriol. 2002;184:5685–5695. doi: 10.1128/JB.184.20.5686-5695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merighi M, Carroll-Portillo A, Septer AN, Bhatiya A, Gunn JS. Role of Salmonella enterica serovar Typhimurium two-component system PreA/PreB in modulating PmrA-regulated gene transcription. J Bacteriol. 2006;188:141–149. doi: 10.1128/JB.188.1.141-149.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson RM. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 46.Moreira CG, Palmer K, Whiteley M, Sircili MP, Trabulsi LR, Castro AFP, Sperandio V. Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J Bacteriol. 2006;188:3952–3961. doi: 10.1128/JB.00177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminsescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholson B, Low D. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol Microbiol. 2000;35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- 49.Parsek MR, Greenberg EP. Acyl-homoserine lactone quorum sensing in Gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci USA. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pei D, Zhu J. Mechanism of action of S-ribosylhomocysteinase (LuxS) Curr Opion Chem Biol. 2004;8:492–497. doi: 10.1016/j.cbpa.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Price SB, Wright JC, DeGraves FJ, Castanie-Cornet MP, Foster JW. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl Environ Microbiol. 2004;70:4792–4799. doi: 10.1128/AEM.70.8.4792-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajan R, Zhu J, Hu X, Pei D, Bell CE. Crystal structure of S-ribosylhomoscysteinase (LuxS) in complex with a catalytic 2-ketone intermediate. Biochemistry. 2005;44:3745–3753. doi: 10.1021/bi0477384. [DOI] [PubMed] [Google Scholar]
- 53.Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reading NC, Rasko D, Torres AAG, Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci USA. 2009;106:5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reading NC, Torres AG, Kendall MM, Hughes DT, Yamamoto K, Sperandio V. A novel two-component signaling system that activates transcription of an enterohemorrhagic E. coli (EHEC) effector involved in remodeling of host actin. J Bacteriol. 2007;189:2468–2476. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rychlik I, Barrow PA. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev. 2005;29:1021–1040. doi: 10.1016/j.femsre.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Sánchez-SanMartín C, Bustamante VH, Calva E, Puente JL. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J Bacteriol. 2001;183:2823–2833. doi: 10.1128/JB.183.9.2823-2833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaefer AL, Val DL, Hanzelka BL, Cronan JE, Greenberg EP. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schauder S, Bassler BL. The languages of bacteria. Gen Dev. 2001;15:468–1480. doi: 10.1101/gad.899601. [DOI] [PubMed] [Google Scholar]
- 60.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 61.Smith JN, Ahmer BMM. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J Bacteriol. 2003;185:1357–1366. doi: 10.1128/JB.185.4.1357-1366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith JN, Dyszel JL, Soares JA, Ellermeier CD, Altier C, Lawhon SD, Adams LG, Konjufca V, Curtis R, III, Slauch JM, Ahmer BM. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS ONE. 2008;3:e2826. doi: 10.1371/journal.pone.0002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sperandio V, Mellies JL, Delahay RM, Frankel G, Crawford JA, Nguyen W, Kaper JB. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol Microbiol. 2000;38:781–793. doi: 10.1046/j.1365-2958.2000.02168.x. [DOI] [PubMed] [Google Scholar]
- 64.Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sperandio V, Torres AG, Girón JA, Kaper JB. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 68.Steindler L, Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett. 2007;266:1–9. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 69.Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50:1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- 72.Taga ME, Semmelhack JL, Bassler BL. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonellas typhimurium. Mol Microbiol. 2001;50:1411–1427. doi: 10.1046/j.1365-2958.2001.02669.x. [DOI] [PubMed] [Google Scholar]
- 73.VanHoudt R, Aertsen A, Moons P, Vanoirbeek K, Michiels CW. N-acyl-l-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol Lett. 2006;256:83–89. doi: 10.1111/j.1574-6968.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- 74.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nature Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 75.Vlisidou I, Lyte M, van Diemen PM, Hawes P, Monaghan P, Wallis TS, Stevens MP. The neuroendocrine stress hormone norepiniphrine augments Escherichia coli O157:H7-induced enteritis and adherence in a bovine ligated ileal loop model of infection. Infect Immun. 2004;72:5446–5451. doi: 10.1128/IAI.72.9.5446-5451.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. doi: 10.1128/JB.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J Bacteriol. 2005;187:2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Li J, March JC, Valdes JJ, Bentley WE. LuxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J Bacteriol. 2005;187:8350–8360. doi: 10.1128/JB.187.24.8350-8360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang XD, deBoer PA, Rothfield LI. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waters CM, Bonnie BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 81.Whitehead NA, Barnard AML, Slater H, Simpson NJ, Salmond GPC. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 82.Winzer K, Hardie KR, Burgess N, Doherty N, Kirke DF, Holden MT, Linforth R, Cornell KA, Taylor AJ, Hill PJ, Williams P. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology. 2002;148:909–922. doi: 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- 83.Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can’t talk now- gone to lunch! Curr Opin Microbiol. 2002;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- 84.Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 85.Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol. 2006;355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 87.Yao Y, Martinez-Yamout MA, Dyson HJ. Backbone and side chain 1H, 13C and 15N assignments for Escherichia coli SdiA1-171, the autoinducer-binding domain of a quorum sensing protein. J Biomol NMR. 2005;31:373–374. doi: 10.1007/s10858-005-2470-0. [DOI] [PubMed] [Google Scholar]
- 88.Zhu J, Dizin E, Hu X, Wavreille AS, Park J, Pei D. S-Ribosylhomocysteinase (LuxS) is a mononuclear iron protein. Biochemistry. 2003;42:4717–4726. doi: 10.1021/bi034289j. [DOI] [PubMed] [Google Scholar]
- 89.Zhu J, Winans SC. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc Natl Acad Sci USA. 1999;98:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc Natl Acad Sci USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]