Abstract
Epithelioid hemangioma (EH) is a benign neoplasm with distinctive vasoformative features, which occasionally shows increased cellularity, cytologic atypia, and/or loco-regional aggressive growth, resulting in challenging differential diagnosis from malignant vascular neoplasms. Based on two intra-osseous EH index cases with worrisome histologic features, such as the presence of necrosis, RNA sequencing was applied for possible fusion gene discovery and potential subclassification of a novel atypical EH subset. A ZFP36-FOSB fusion was detected in one case, while a WWTR1-FOSB chimeric transcript in the other, both were further validated by FISH and RT-PCR. These abnormalities were then screened by FISH in 44 EH from different locations with 7 additional EH revealing FOSB gene rearrangements, all except one being fused to ZFP36. Interestingly, 4/6 penile EH studied showed FOSB abnormalities. Although certain atypical histologic features were observed in the FOSB-rearranged EH, including solid growth, increased cellularity, mild to moderate nuclear pleomorphism, and necrosis in 3/9 cases, no overt sarcomatous areas were discerned to objectively separate the lesions from the fusion-negative EH. No patient has developed recurrence to date, but the follow-up was relatively limited and short to draw definitive conclusions regarding behavior. Although FOSB-rearranged EH do not show significant morphologic overlap with SERPINE1-FOSB fusion-positive pseudomyogenic hemangioendothelioma, FOSB oncogenic activation is emerging as an important event in these benign and intermediate groups of vascular tumors.
Keywords: epithelioid hemangioma, epithelioid hemangioendothelioma, FOSB, ZFP36, translocation
INTRODUCTION
Epithelioid vascular tumors encompass a wide histologic spectrum, including benign epithelioid hemangioma (EH); low grade malignant epithelioid hemangioendothelioma (EHE); and high grade epithelioid angiosarcoma (Fletcher et al., 2013). Recurrent chromosomal translocations have been described recently in both conventional EHE, t(1;3) resulting in WWTR1-CAMTA1 fusion (Errani et al., 2011;Tanas et al., 2011) and in the less common variant of EHE with vasoformative features, showing YAP1-TFE3 fusion (Antonescu et al., 2013). However, these genetic abnormalities were not identified in EH, which can be used as adjunct molecular tests in difficult cases to exclude a malignant epithelioid vascular tumor (Errani et al., 2011;Errani et al., 2012;Antonescu et al., 2013). Furthermore, a recurrent SERPINE1-FOSB fusion has been identified in pseudomyogenic hemangioendothelioma (PHE, a.k.a. epithelioid sarcoma-like hemangioendothelioma) (Walther et al., 2014), an unusual vascular lesion of intermediate malignancy, which rarely metastasizes (Hornick and Fletcher, 2011;Fletcher et al., 2013).
EH are ubiquitously located and have been described at most anatomic sites, including skin, soft tissue, bone and viscera (Fletcher et al., 2013). Its diagnostic challenge, particularly in the osseous locations, stems not only from its overlapping cytologic features with malignant lesions, but also from its somewhat aggressive clinical characteristics, with occasional destructive growth and multifocality (Evans et al., 2003;Nielsen et al., 2009;Errani et al., 2012). The importance of distinguishing EH from other malignant epithelioid vascular tumors is paramount as a result of differences in their management and clinical outcome. Based on two index cases of EH that showed worrisome histologic features, namely areas of necrosis, next generation paired-end RNA sequencing was applied in an attempt to achieve genetic characterization and potential identification of a novel/atypical subset of EH. The validated molecular abnormalities were then screened in a large extended group of EH with available archival material.
MATERIAL AND METHODS
The Pathology files of MSKCC and the personal consultations of the corresponding authors (CRA, CDF) were searched for cases of epithelioid hemangioma (EH). Based on the two index cases, a particular emphasis for selection for the extended study cohort was given to lesions with increased cellularity and evidence of atypical histologic features, such as presence of necrosis and/or mild-moderate nuclear pleomorphism. However, all cases selected had otherwise characteristic features of EH, including predominance of epithelioid morphology, with moderate to abundant eosinophilic cytoplasm, intra-cytoplasmic vacuoles and clearly vasoformative features. Furthermore, overtly malignant lesions, best classified as angiosarcoma or EHE, were not included. Pathologic diagnosis and immunohistochemical stains were re-reviewed by the corresponding authors in all cases. In addition, 6 cases of PHE were included as a control group and studied separately by FISH to exclude overlapping genetic abnormalities.
RNA Sequencing
Total RNA was prepared for RNA sequencing in accordance with the standard Illumina mRNA sample preparation protocol (Illumina). Briefly, mRNA was isolated with oligo(dT) magnetic beads from total RNA (10 μg) extracted from the case. The mRNA was fragmented by incubation at 94°C for 2.5 min in fragmentation buffer (Illumina). To reduce the inclusion of artifactual chimeric transcripts due to random priming of transcript fragments into the sequencing library, because of inefficient A-tailing reactions that lead to self ligation of blunt-ended template molecules (Quail et al., 2008), an additional size-selection step (capturing 350–400 bp) was introduced prior to the adapter ligation step. The adaptor-ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA-sequencing at read lengths of 50 or 51 bp was performed with the HiSeq 2500 (Illumina). Across the two samples a total of about 141 million paired-end reads were generated, corresponding to about 21 billion bases.
Analysis of RNA Sequencing Results with FusionSeq
All reads were independently aligned with STAR alignment software against the human genome reference sequence (hg19) and a splice junction library, simultaneously (Dobin et al., 2013). The mapped reads were converted into Mapped Read Format (Habegger et al., 2011) and analyzed with FusionSeq (Sboner et al., 2010) to identify potential fusion transcripts. FusionSeq is a computational method successfully applied to paired-end RNA-seq experiments for the identification of chimeric transcripts (Tanas et al., 2011;Pierron et al., 2012) (Mosquera et al., 2013). Briefly, paired-end reads mapped to different genes are first used to identify potential chimeric candidates. A cascade of filters, each taking into account different sources of noise in RNA-sequencing experiments, was then applied to remove spurious fusion transcript candidates. Once a confident list of fusion candidates was generated, they were ranked with several statistics to prioritize the experimental validation. In these cases, we used the DASPER score (Difference between the observed and Analytically calculated expected SPER): a higher DASPER score indicated a greater likelihood that the fusion candidate was authentic and did not occur randomly. See (Sboner et al., 2010) for further details about FusionSeq.
Fluorescence In Situ Hybridization (FISH)
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking genes that were identified as potential fusion partners in the RNA-seq experiment. BAC clones were chosen according to USCS genome browser (http://genome.uscs.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
An aliquot of the RNA extracted above from frozen tissue (Trizol Reagent; Invitrogen; Grand Island, NY) was used to confirm the novel fusion transcripts identified by FusionSeq. RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product). Three micrograms of total RNA was used for cDNA synthesis by SuperScript® III First-Strand Synthesis Kit (Invitrogen, Grand Island, NY). RT-PCR was performed using the Advantage-2 PCR kit (Clontech, Mountain View, CA) for 33 cycles at a 64.5°C annealing temperature, using the following primers: ZFP36 Fwd Exon 1: 5’–GGATCTGACTGCCATCTACGAG–3’; WWTR1 Fwd Exon3: 5’– CCTGATTCGGGCTCGCACTC–3’; and FOSB Rev Exon 2: 5’– GAGACCCCGAGAGGAGACG – 3’. For the reciprocal FOSB-ZFP36 RT-PCR the following primers were used: FOSB Exon 1: 5’ – CTCCTCACCCTCTGCCGAGTC – 3’and ZFP36 Exon 2: 5’– CTGAGCCCCTCCGACTCCAG – 3’.
DNA PCR
Genomic DNA was extracted from frozen tissue using the Phenol/Chloroform assay and quality was confirmed by electrophoresis. 0.5 μg genomic DNA was amplified with the Advantage 2 PCR kit (Clontech, Mountain View, CA) in order to assess the intronic breaks of the ZFP36-FOSB, using the same primer set as for the RT-PCR above. Additionally for the WWTR1-FOSB intronic break Long-Range DNA was carried out by using QIAGEN LongRange PCR Kit (QIAGEN, Germantown, MD), from 1 μg of genomic DNA. The primers used were: WWTR1-Intron 3 Fwd: 5’ –CCACTCTGGATTAGATGCTGCTG– 3’ and FOSB-Intron 1 Rev: 5’ –CAAACTGCAAAGAGTCGGGAGATG – 3’.
RESULTS
RNA sequencing identifies ZFP36-FOSB and WWTR1-FOSB novel fusion candidates
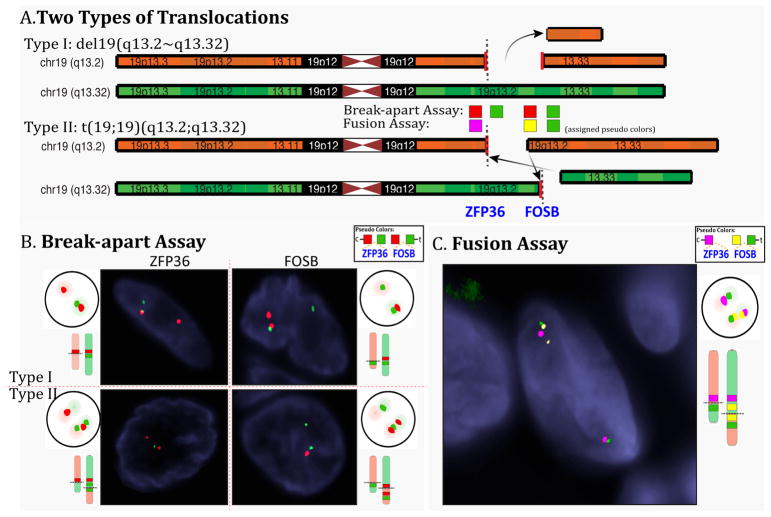
In EH1 the FusionSeq analysis identified a ZFP36-FOSB fusion candidate (Fig 1). ZFP36 and FOSB are 6 Mb apart on chromosome 19 and show similar directions of transcription. A potential fusion can thus occur either as a result of an interstitial deletion or from a balanced t(19;19) translocation. Experimental validation by RT-PCR confirmed a 192 bp product composed of ZFP36 exon 1 fused to FOSB exon 2 (Fig. 1). FISH studies performed confirmed a break-apart in both ZFP36 and FOSB genes, with a typical split signal lacking evidence of genetic loss (Fig. 2). This result was further confirmed by a FISH fusion assay. DNA PCR identified the intronic break within the first 554 bp of intron 1 of ZFP36 fused to the last 54 bp of FOSB intron 1 (Fig. 1).
Figure 1. ZFP36-FOSB gene fusion in an atypical epithelioid hemangioma of bone.
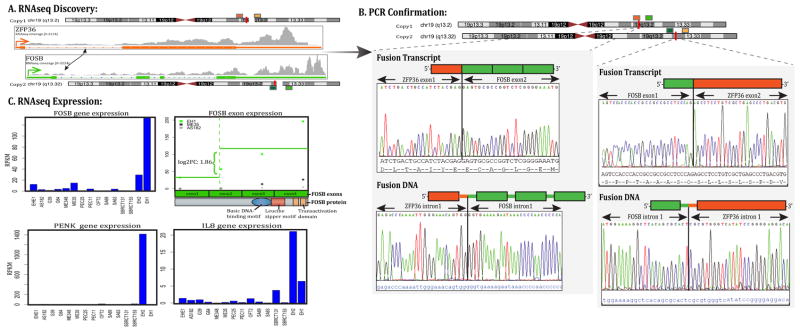
(A) Schematic representation indicating the ZFP36 locus located on one of the chromosome 19 is joined to FOSB, situated on the other chromosome 19 copy, resulting in a t(19;19)(q13.2; q13.2) translocation. (B) Fusion candidates were validated first by RT-PCR showing ZFP36 exon 1 being fused to FOSB exon 2 (top left image), followed by the DNA PCR confirming the intronic break within intron 1 of ZFP36 being fused to intron 1 of FOSB (bottom left image). In addition RT-PCR and DNA-PCR depict the FOSB-ZFP36 reciprocal transcripts (bottom images). (C) Normalized RNA sequencing (RPKM) showed significant FOSB mRNA upregulation in the two FOSB-rearranged index cases (EH1 and EH2), compared to other sarcoma types (top left image), with specific FOSB overexpression of exons 2–4 which participate in the fusion (top right image). PENK and IL8 overexpression in FOSB-positive EH cases compared to other sarcoma types (bottom panels)
Figure 2. Schematic depiction of two possible ZFP36-FOSB translocation types and fluorescence in situ hybridization (FISH) results.
A. The Type I translocation occurs through a simple interstitial deletion event involving 6 Mb region on 19q13.2, while type II fusion results from a balanced t(19;19)(q13.2;q13.32). Assigned pseudocolors are used for the different BAC probes tested either in the break-apart or fusion FISH assays to better illustrate the two fusion patterns. B. Schematic and actual FISH showing the break-apart signal patterns for both type I and type II translocations for each ZFP36 and FOSB assays (red, centromeric, green telomeric) C. Diagrammatic and actual FISH images for the three-color fusion assay for the type II balanced translocation (magenta, ZFP36 centromeric; yellow, FOSB centromeric; green FOSB telomeric).
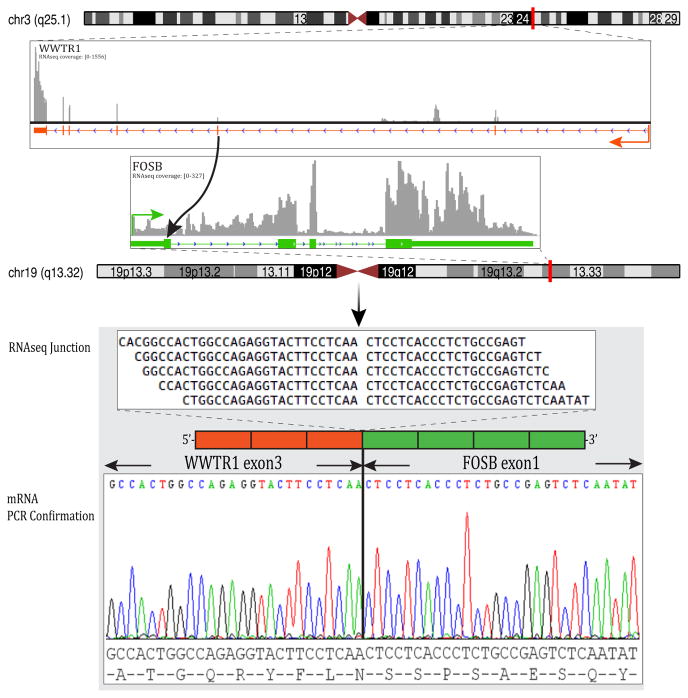
In EH2 the FusionSeq analysis identified a WWTR1-FOSB fusion candidate (Fig. 3). RT-PCR confirmed the presence of exon 3 of WWTR1 fused to exon 1 of FOSB (Fig. 3). FISH studies confirmed a break-apart in FOSB genes, but did not identify the presence of a WWTR1 rearrangement, possible due to an unbalanced or more complex translocation. However, long-range DNA PCR confirmed the intronic break within the first 11,088 bp of WWTR1 intron 3 and the last 635 bp of FOSB exon 1.
Figure 3. WWTR1-FOSB gene fusion in a skeletal EH with necrosis.
(A) Diagrammatic representation of WWTR1 locus on 3q25 joined together with the FOSB on 19q12 detected by RNAseq (top image). RNA reads covering the fusion junction were isolated independent to the FusionSeq analysis work flow, supporting the WWTR1-FOSB fusion candidate (middle image); RT-PCR experimental validation of the fusion shows the junction sequence between WWTR1 exon 3 and FOSB exon 1 (bottom image).
Recurrent ZFP36-FOSB fusions are identified by FISH in a subset of cellular EH
As FOSB gene rearrangement emerged as a recurrent event upon RNAseq of the 2 index cases, the entire cohort of 46 EH cases was subjected to FISH analysis for detecting FOSB gene abnormalities. A total of 9 (20%) EH cases were found to have FOSB break-apart alterations, including 2 in the bone (index cases: EH1 and EH2), 2 in soft tissue , 4 penile and one involving an axillary lymph node (Table 1). In all except one case, FISH also showed ZFP36 gene rearrangements. Only one FOSB-rearranged case (EH7) showed no FISH abnormalities in ZFP36, WWTR1, or SERPINE1 genes. FISH analysis revealed two distinct patterns of ZFP36-FOSB fusions, one as described above in keeping with a balanced t(19;19) translocation seen in 4 cases and the other consistent with a simple interstitial deletion seen in 2 cases (EH4, EH8), with the other copy of chromosome 19 being unaffected (Fig. 2). One tumor showed a mixed FISH pattern (EH9).
Table 1.
Clinical and Pathologic features of FOSB-rearranged EH
| EH# | Age/Sex | Location | Morphologic Features | Nuclear Pleomorphism | Necrosis | Follow-up | ZFP36 FISH |
|---|---|---|---|---|---|---|---|
| 1* | 52/M | Bone, tibia | Vasoformative, eosinophils | mild | Yes | 1yr NED |
+ |
| 2* | 11/M | Bone, calcaneus (multifocal, foot and ankle) | Vasoformative | mild | Yes | 1yr AWD |
− |
| 3 | 44/M | ST, iliac crest | Mostly solid, focal vasoformative, rare blister cells | moderate nuclear enlargement macronucleoli | No | NA | + |
| 4 | 41/M | ST, knee | Mostly solid, intra-vascular growth, eosinophils | mild | No | NA | + |
| 5 | 28/F | Lymph node, axilla | Mostly vasoformative, focally solid, scattered blister cells & eosinophils | mild | No | 3 yrs NED |
+ |
| 6 | 44/M | Penis | Mostly solid, with prominent blister cells, eosinophils | mild | No | NA | + |
| 7 | 39/M | Penis | vasoformative, blister cells | mild to moderate, nuclear enlargement, macronucleoli | No | NA | − |
| 8 | 27/M | Penis | vasoformative, blister cells rare eosinophils | mild to moderate, nuclear indentations macronucleoli | Yes | NA | + |
| 9 | 51/M | Penis | vasoformative | mild to moderate nuclear indentations multinucleated GC | No | 11 yrs NED |
+ |
Index cases studied by RNA sequencing and confirmed by RT-PCR to have a ZFP36-FOSB fusion in EH1 and a WWTR1-FOSB fusion in EH2;
M, male; F, female; ST, soft tissue; GC giant cell; yrs, years; NA, not available; NED, no evidence of disease; AWD, alive with disease.
Pathologic Findings of FOSB-rearranged EH
All lesions were marginally excised or curetted and displayed a lobulated growth with a pushing rather than infiltrative pattern. In EH1 the radiographic appearance showed a lytic lesion with hazy peripheral sclerosis in proximal tibial diametaphysis, associated with irregular endosteal destruction (Supplem Fig 1). One patient (EH2) presented with multifocal lesions in his foot and ankle, with only the dominant lesion being curetted and he is being followed for residual gross disease. Tumors showed a variable degree of solid growth, characterized by diffuse increased cellularity and nuclear crowding. Tumor cells often displayed a moderate amount of densely eosinophilic cytoplasm (Fig. 4). All cases showed at least focal vasoformative features in addition to varying number of cells with distinctive intra-cytoplasmic vacuoles, frequently as an obvious finding at low power (Fig. 4, Table 1). Eosinophils were present in 5 cases, often as a focal feature. Most cases showed some degree of atypical nuclear features, including nuclear enlargement, nuclear grooves and indentations, reminiscent of a histiocytic phenotype (Fig. 4). Most tumors showed cells with vesicular and open chromatin, with prominent nucleoli, but nuclear hyperchromasia was typically not seen. Rare cases showed multinucleated giant cells. Mitotic figures were rare, with most cases showing no more than 1 mitosis/10HPFs and a low Ki67 proliferative index of <5% nuclear labeling. However, 3 (33%) cases showed areas of necrosis despite otherwise classic EH morphologic features, which has triggered our initial RNA sequencing investigation.
Figure 4. Morphologic spectrum of FOSB-rearranged EH tumors.
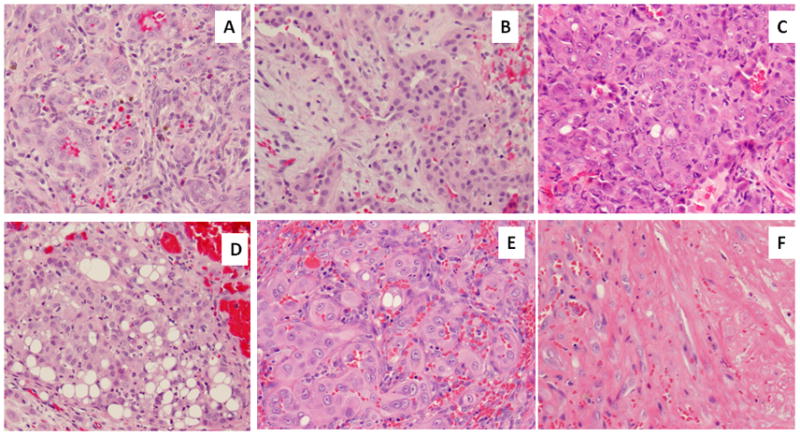
A. Index EH1 case of a tibial metaphyseal lytic lesion showing well-formed vasoformative features; B. WWTR1-FOSB positive of index EH2 showing typical tombstone pattern; C. a cellular example of EH with predominant solid component and atypical cytomorphology, with enlarged vesicular nuclei with prominent nucleoli (EH3); D. solid growth with prominent blister cells (EH6); E. EH with nuclear enlargements and prominent nucleoli, abundant glassy eosinophilic cytoplasm and F. areas of necrosis (EH8).
Morphologic Features of FOSB-Fusion Negative EH
The FOSB-negative EH group was composed of 37 cases, which were retrospectively re-reviewed after the FISH results became available. These lesions were overall less cellular and had less solid components (Fig 4). Furthermore, the presence of atypical cytologic features, such as nuclear enlargement and mild nuclear pleomorphism, were only occasionally present as a focal finding. Only one of the 37 (3%) cases showed the presence of necrosis. The cases were located in the soft tissue (n=23), bone (n=12) and penis (n=2). The most common anatomic distribution was in the extremities, 22 cases, with 12 tumors being diagnosed in acral locations, followed by head and neck, 7 cases and trunk, 6 cases. At least 3 patients with available clinical information presented with multifocal disease in the same anatomic location.
DISCUSSION
Epithelioid vascular tumors remain somewhat controversial because of their rarity, unusual morphology, and unpredictable biologic behavior (Antonescu, 2014). Specifically, the differential diagnosis at the benign/low grade end of the spectrum, between EH and EHE has been challenging. The recurrent t(1;3)(p36;q25) chromosomal translocation, resulting in WWTR1-CAMTA1 fusion, identified in EHE of various anatomic sites, but not in other epithelioid vascular neoplasms, suggests distinct pathogeneses and provides an objective and powerful diagnostic tool to distinguish EH from EHE in limited biopsy material (Errani et al., 2011;Tanas et al., 2011). Furthermore, the recent characterization of YAP1-TFE3 fusion in the less common variant of EHE displaying conspicuous vasoformative features and nested/alveolar growth, has highlighted the significant overlap at morphologic level with EH (Antonescu et al., 2010). Particularly in skeletal locations the diagnosis of EH has been often debated, some authors’ challenging its fully benign biologic potential and favoring a morphologic spectrum with EHE, rather than sharply defined pathologic entities (O'Connell et al., 2001;Evans et al., 2003). For example, Evans et al argued that EH is not a distinct clinicopathologic entity but rather a misdiagnosed hemangioendothelioma, a tumor that, unlike hemangioma, has malignant potential (Evans et al., 2003). This illustrates the lack of objective diagnostic criteria and the confusion surrounding the classification of this rare subset of vascular tumors. Our study highlights this controversy by genetically confirming a distinct subset of EH, that is associated with certain atypical histologic traits. However, this chromosomal translocation is distinct from the fusions so far reported in EHE and the morphologic appearance of FOSB-rearranged atypical EH still fits better within the conventional EH category than any other malignant epithelioid vascular neoplasm. Obviously, due to the benign diagnoses rendered, the follow-up information is rather limited and brief to allow drawing definitive conclusions on their clinical behavior.
A novel SERPINE1-FOSB fusion was recently described in pseudomyogenic hemangioendothelioma (PHE) (a.k.a. epithelioid-sarcoma-like hemangioendothelioma) (Walther et al., 2014). Despite the shared FOSB gene rearrangements resulting in FOSB up-regulation, no significant morphologic overlap is discerned between EH and PHE. PHE typically lacks vasoformative features, often resembling a spindle myoid tumor or less frequently showing prominent epithelioid morphology, reminiscent of an epithelioid sarcoma (Billings et al., 2003;Hornick and Fletcher, 2011). However, in order to rule out a histologic spectrum between PHE and the newly identified subset of FOSB-rearranged atypical EH, we screened 6 PHE cases by FISH for possible overlapping molecular abnormalities. In 3/6 cases, only the FOSB gene showed rearrangement, while the SERPINE1 gene was uninvolved. These 3 SERPINE1-negative PHE were then screened for both ZFP36 and WWTR1 gene abnormalities by FISH but no break-apart changes were noted, in keeping with a distinct pathogenesis. Similar to SERPINE1-FOSB positive PHE tumors (Walther et al., 2014), both our ZFP36-FOSB and WWTR1-FOSB fusion-positive EH index cases showed high levels of FOSB mRNA expression. Nevertheless, the FOSB break in both PHE and this EH subset was similar, including the C-terminal starting FOSB exon 2 (Walther et al., 2014), preserving the critical domains of FOSB protein, including the basic DNA binding motif, leucine zipper domain and the transactivating domain (Fig. 1C).
FOSB members dimerise with Jun proteins to form the AP-1 transcription factor complex, regulating the expression of various genes involved in cell proliferation, differentiation, and cell death (Nakabeppu et al., 1988;Shaulian and Karin, 2001). The expression of fosB is induced in neural progenitor cells following transient forebrain ischemia in the rat brain (Yutsudo et al., 2013) fosB-null mice exhibit impaired neurogenesis and spontaneous epilepsy, in addition to depressive-like behaviors. Gene expression profiling of fosB-null mice suggests that expression of Vgf, Gal, Trh, Penk, and Srxn1 is likely to be regulated by FosB. FosB dramatically enhances Jun transcription regulation of AP-1–dependent promoters, whereas, ΔFosB, lacking the C-terminal transactivation domain and the TATA-binding protein (TBP)-binding domain (Ohnishi et al., 2008), suppresses Jun function in a dominant-negative manner (Nakabeppu and Nathans, 1991). FosB also plays a major role in IL8 induction in response to stress hormones (Shahzad et al., 2010). IL8 has been shown to modulate matrix metalloproteinase expression in tumor and endothelial cells, thereby regulating angiogenic activity (Li et al., 2003). IL8 and FosB silencing reduced tumor growth and microvessel density in an orthotopic xenograft model of ovarian carcinoma (Shahzad et al., 2010). As FOSB-fusion positive EH and PHE result in upregulation of FOSB mRNA, we investigated the expression of genes reportedly regulated by FOSB. By transcriptome sequencing, IL8 and PENK mRNA expression were significantly up-regulated in one or both FOSB-positive EH index cases compared to other sarcomas types (Fig. 1C).
Zinc finger protein 36 (ZFP36), a.k.a. Tristetraprolin (TTP) is a tandem CCCH zinc-finger RNA-binding protein that regulates the stability of certain adenosine/uridine AU-rich element (ARE) mRNAs, through removal of the poly(A) tail and increased mRNA turnover (Lai et al., 2003;Al-Souhibani et al., 2010). Nearly 15% of the human transcriptome have an ARE in their 3′-untranslated region (UTR), encoding for cytokines, chemokines, growth factors, and pro-inflammatory enzymes involved in inflammation and immune responses. Many target cytokine and chemokine mRNAs for ZFP36 have been identified, including TNF-α, GM-CSF, IFN-γ, IL-2, IL-3, COX-2, VEGF, IL-6, and IL-8 (Carrick et al., 2004). Mice lacking the ZFP36 gene suffer from severe chronic inflammation, induced predominantly due to an excess of circulating tumor necrosis factor-α (TNF-α) (Carballo and Blackshear, 2001). The expression and function of ZFP36 are regulated by the p38 mitogen activated protein kinase (p38MAPK) signaling pathway (Taylor et al., 1995;Mahtani et al., 2001).
In summary, we are describing a novel recurrent ZFP36-FOSB fusion in a subset (20%) of EH showing increased cellularity and occasional areas of necrosis and mild nuclear pleomorphism. Compared to the more common fusion negative EH cohort, a disproportionate number of FOSB-rearranged EH were identified in a penile location. Although some atypical histologic features were noted in this genetic subgroup, the overall morphologic findings and the short follow-up available are not definitive to suggest either a more aggressive behavior or classification into a borderline malignancy group. At least temporarily, until larger series are examined, we propose designating these lesions descriptively as EH with atypical features.
Supplementary Material
(A) Anteroposterior and (B) lateral radiographs of proximal tibia show a vague lytic lesion with hazy peripheral sclerosis (arrows) in proximal tibial diametaphysis. Lesion has resulted in irregular endosteal destruction, most pronounced medially and posteriorly. Lesion contains no evident calcified matrix. An incidental sclerotic lesion (black oval), oriented along long axis of tibia, is consistent with a bone island. (C) Anteroposterior image from whole-body bone scan demonstrates intense uptake in the right proximal tibial lesion.
Acknowledgments
The authors would like to thank Milagros Soto for editorial assistance and Dr G Taylor (Hospital for Sick Children, Toronto, Canada) for sharing material in consultation. Also we would like to thank the following Pathologists for providing cases in consultation to Dr Christopher Fletcher: G. Amaan & M. Kolz, Vienna, Austria, J.H. Choi, Daegu City, South Korea, B.G. Kulander, Seattle, WA, C. Richards, Leicester, UK, M. Smolkin, Exeter, NH
Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA), Cycle for Survival and Angiosarcoma Awareness.
Footnotes
Conflict of interest: none
References
- Al-Souhibani N, Al-Ahmadi W, Hesketh JE, Blackshear PJ, Khabar KS. The RNA-binding zinc-finger protein tristetraprolin regulates AU-rich mRNAs involved in breast cancer-related processes. Oncogene. 2010;29:4205–4215. doi: 10.1038/onc.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu C. Malignant vascular tumors--an update. Mod Pathol. 2014;27(Suppl 1):S30–38. doi: 10.1038/modpathol.2013.176. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, Chen HW, Pathan N, Krausz T, Dickson BC, Weinreb I, Rubin MA, Hameed M, Fletcher CD. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–784. doi: 10.1002/gcc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings SD, Folpe AL, Weiss SW. Epithelioid sarcoma-like hemangioendothelioma. Am J Surg Pathol. 2003;27:48–57. doi: 10.1097/00000478-200301000-00006. [DOI] [PubMed] [Google Scholar]
- Carballo E, Blackshear PJ. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98:2389–2395. doi: 10.1182/blood.v98.8.2389. [DOI] [PubMed] [Google Scholar]
- Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errani C, Zhang L, Panicek DM, Healey JH, Antonescu CR. Epithelioid hemangioma of bone and soft tissue: a reappraisal of a controversial entity. Clin Orthop Relat Res. 2012;470:1498–1506. doi: 10.1007/s11999-011-2070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, Healey JH, Antonescu CR. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans HL, Raymond AK, Ayala AG. Vascular tumors of bone: A study of 17 cases other than ordinary hemangioma, with an evaluation of the relationship of hemangioendothelioma of bone to epithelioid hemangioma, epithelioid hemangioendothelioma, and high-grade angiosarcoma. Hum Pathol. 2003;34:680–689. doi: 10.1016/s0046-8177(03)00249-1. [DOI] [PubMed] [Google Scholar]
- Fletcher C, Bridge JA, Hogendoorn PC, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. IARC; Lyon: 2013. [Google Scholar]
- Fletcher CD, Bridge JA, Hogendoorn PC, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. Lyon: IARC Press; 2013. [Google Scholar]
- Habegger L, Sboner A, Gianoulis TA, Rozowsky J, Agarwal A, Snyder M, Gerstein M. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick JL, Fletcher CD. Pseudomyogenic hemangioendothelioma: a distinctive, often multicentric tumor with indolent behavior. Am J Surg Pathol. 2011;35:190–201. doi: 10.1097/PAS.0b013e3181ff0901. [DOI] [PubMed] [Google Scholar]
- Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Dubey S, Varney ML, Dave BJ, Singh RK. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol. 2003;170:3369–3376. doi: 10.4049/jimmunol.170.6.3369. [DOI] [PubMed] [Google Scholar]
- Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol Cell Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera JM, Sboner A, Zhang L, Kitabayashi N, Chen CL, Sung YS, Wexler LH, LaQuaglia MP, Edelman M, Sreekantaiah C, Rubin MA, Antonescu CR. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y, Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991;64:751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Ryder K, Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988;55:907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- Nielsen GP, Srivastava A, Kattapuram S, Deshpande V, O'Connell JX, Mangham CD, Rosenberg AE. Epithelioid hemangioma of bone revisited: a study of 50 cases. Am J Surg Pathol. 2009;33:270–277. doi: 10.1097/PAS.0b013e31817f6d51. [DOI] [PubMed] [Google Scholar]
- O'Connell JX, Nielsen GP, Rosenberg AE. Epithelioid vascular tumors of bone: a review and proposal of a classification scheme. Adv Anat Pathol. 2001;8:74–82. doi: 10.1097/00125480-200103000-00003. [DOI] [PubMed] [Google Scholar]
- Ohnishi YN, Sakumi K, Yamazaki K, Ohnishi YH, Miura T, Tominaga Y, Nakabeppu Y. Antagonistic regulation of cell-matrix adhesion by FosB and DeltaFosB/Delta2DeltaFosB encoded by alternatively spliced forms of fosB transcripts. Mol Biol Cell. 2008;19:4717–4729. doi: 10.1091/mbc.E07-08-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad MM, Arevalo JM, Armaiz-Pena GN, Lu C, Stone RL, Moreno-Smith M, Nishimura M, Lee JW, Jennings NB, Bottsford-Miller J, Vivas-Mejia P, Lutgendorf SK, Lopez-Berestein G, Bar-Eli M, Cole SW, Sood AK. Stress effects on FosB- and interleukin-8 (IL8)-driven ovarian cancer growth and metastasis. J Biol Chem. 2010;285:35462–35470. doi: 10.1074/jbc.M110.109579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, Mitsopoulos C, Zvelebil M, Hoch BL, Weiss SW, Debiec-Rychter M, Sciot R, West RB, Lazar AJ, Ashworth A, Reis-Filho JS, Lord CJ, Gerstein MB, Rubin MA, Rubin BP. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J Biol Chem. 1995;270:13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- Walther C, Tayebwa J, Lilljebjorn H, Magnusson L, Nilsson J, von Steyern FV, Ora I, Domanski HA, Fioretos T, Nord KH, Fletcher CD, Mertens F. A novel SERPINE1-FOSB fusion gene results in transcriptional up-regulation of FOSB in pseudomyogenic haemangioendothelioma. J Pathol. 2014;232:534–540. doi: 10.1002/path.4322. [DOI] [PubMed] [Google Scholar]
- Yutsudo N, Kamada T, Kajitani K, Nomaru H, Katogi A, Ohnishi YH, Ohnishi YN, Takase K, Sakumi K, Shigeto H, Nakabeppu Y. fosB-null mice display impaired adult hippocampal neurogenesis and spontaneous epilepsy with depressive behavior. Neuropsychopharmacology. 2013;38:895–906. doi: 10.1038/npp.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Anteroposterior and (B) lateral radiographs of proximal tibia show a vague lytic lesion with hazy peripheral sclerosis (arrows) in proximal tibial diametaphysis. Lesion has resulted in irregular endosteal destruction, most pronounced medially and posteriorly. Lesion contains no evident calcified matrix. An incidental sclerotic lesion (black oval), oriented along long axis of tibia, is consistent with a bone island. (C) Anteroposterior image from whole-body bone scan demonstrates intense uptake in the right proximal tibial lesion.