Abstract
Animals locate and track chemoattractive gradients in the environment to find food. With its small nervous system, Caenorhabditis elegans is a good model system1,2 in which to understand how the dynamics of neural activity control this search behavior. Extensive work on the nematode has identified the neurons that are necessary for the different locomotory behaviors underlying chemotaxis through laser ablation3–7, activity recording in immobilized animals and the study of mutants4,5. However, we do not know the neural activity patterns in C. elegans that are sufficient to control its complex chemotactic behavior. To understand how the activity in its interneurons coordinate different motor programs to lead the animal to food, we used optogenetics and new optical tools to directly manipulate neural activity in freely moving animals to evoke chemotactic behavior. By deducing the classes of activity patterns triggered during chemotaxis and exciting individual neurons with these patterns, we identified interneurons that control the essential locomotory programs for this behavior. Surprisingly, we discovered that controlling the dynamics of activity in just one interneuron pair (AIY) was sufficient to force the animal to locate, turn towards and track virtual light gradients. Two distinct activity patterns triggered in AIY as the animal moved through the gradient, controlled reversals and gradual turns to drive chemotactic behavior. Since AIY are post-synaptic to most chemosensory and thermosensory neurons8, these activity patterns in AIY are likely to play an important role in controlling and coordinating different taxis behaviors of the animal.
Organisms, from bacteria to multicellular eukaryotes, have to search for food to survive. Complex internal circuits process external signals to evoke and coordinate multiple motor programs, leading the animal to track attractive odors and find food. Are there master nodes in the circuits that control and coordinate search behavior? Here we ask if the neural circuits generating chemotactic behavior in C. elegans can be controlled through such key nodes.
The nematode C. elegans uses reversals (backward movement), sharp and gradual turns to locate, and track gradients of chemo-attractive signals4,5,9,10. Previous work on C. elegans has identified about 14 pairs of inter and motor neurons including the interneurons AIY, AIZ and AIB that are necessary for the locomotory behaviors underlying chemotaxis (Supplementary Table 1)3–7. The neuro-anatomy of the animal shows that a majority of the amphid chemosensory and thermosensory neurons synapse onto one or more of the first layer of interneuron pairs AIY, AIZ, and AIB11, which are further connected to a dense network of interneurons8. The activity dynamics in this network must process sensory signals to produce and coordinate the different locomotory behaviors underlying chemotaxis through the downstream motor neurons. Despite the experiments in the literature involving ablation, genetics, and calcium imaging, we do not know if chemotaxis is driven by key interneurons or if the generation of this complex behavior is achieved by the dynamics of a more diffuse neural network.
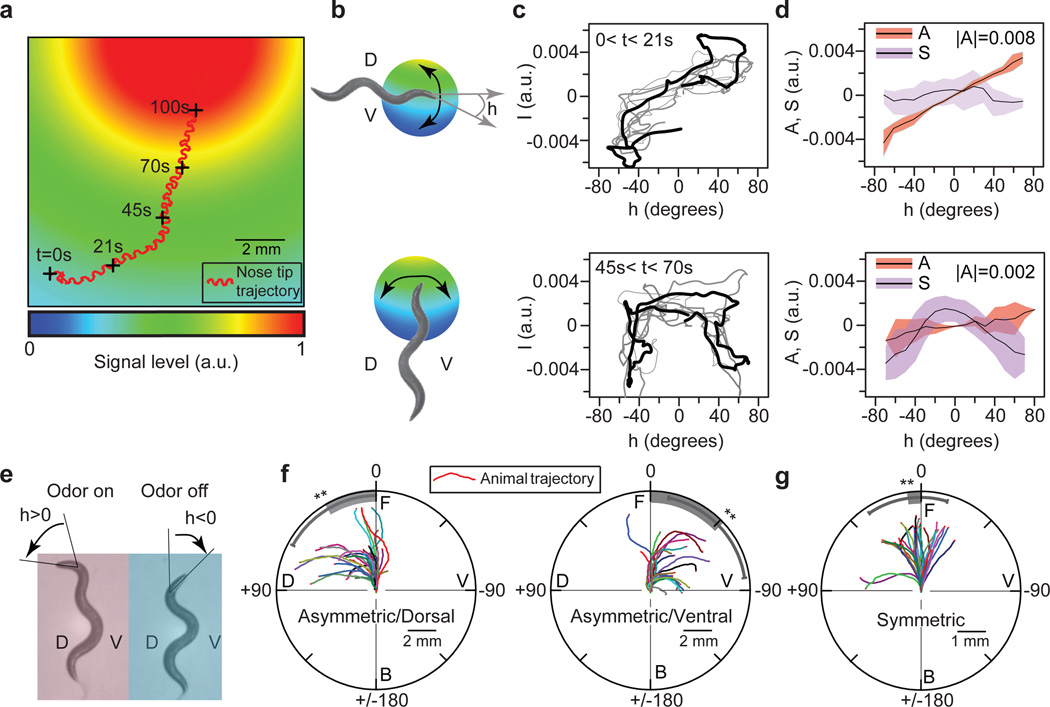
To evoke chemotactic behavior by directly controlling interneuron activity, we have to answer two intricately linked questions: which sets of interneurons do we control, and what activity patterns do we stimulate in them? To deduce the classes of activity patterns triggered in the nervous system during chemotaxis, we followed animals as they crawled towards a bacterial lawn (Fig. 1a). The undulatory head swings (from dorsal to ventral, since the animal crawls on its side) caused the angle at which the head bends relative to the locomotory direction, h(t) (Fig. 1b), to oscillate between positive and negativevalues. Due to this changing head-bending angle and the movement of the animal, sensory cilia at the nose tip experienced the spatial profile of the chemoattractants (Fig. 1a, b) as a temporally fluctuating odor signal, I(h,t). In general, this signal can be written as a sum of two terms I(h,t) = SI (h,t) + AI(h,t), where SI is a symmetric function of h : S(h,t) = S(−h,t) and AI is an asymmetric function of h : A(h,t) = A(−h,t). When the animal moves perpendicular to the gradient direction I is dominated by AI (Fig. 1b–d, top). As the animal turns and tracks the gradient, the magnitude of AI, |AI| decreases and I is dominated by SI (Fig. 1b–d, bottom, Supplementary Fig. 1a).
Figure 1. Asymmetric component of the odor signal controls gradual turning.
a, Trajectory of a nematode’s nose tip overlaid on modeled exponential profile (decay length 1 cm) of the gradient of chemoattractants (pseudocolor) from the bacterial lawn. Plus signs: positions of animal at indicated times. a.u.: arbitrary units. b, Illustration of an animal crawling perpendicular to (above) and along (below) the odor gradient. h: head-bending angle; D: dorsal (h > 0); V: ventral (h < 0). c, Odor signal at nose tip, I(t), vs. head-bending angle, h(t), over the last (black) and sequentially prior (grey) head swings for the nose-tip trajectory in (a). d, Plot of the mean (black line) and s.d. (colored bar) of the asymmetric (A(t)) and symmetric (S(t)) components of I(t) in (c). |A|: magnitude of A(t). e, Dorsal asymmetric odor stimulation (see supplementary method). f–g, Sample trajectories of center of mass of the animals upon (f) dorsal and ventral asymmetric, (g) symmetric odor stimulation. Grey bar: mean turning angle; D: dorsal; V: ventral; F: front; B: back; angles (0, 90, −90, 180) define the turning angles with respect to initial orientation of animal. Mean (grey bar), error bar: s.d. over n=10; **, p<0.05, two-sample t-test.
To determine how these asymmetric and symmetric components of I (t) controlled locomotory behavior, we built a microscopy system that delivered odors on a freely crawling animal in precise temporal patterns determined by h(t) (Supplementary Fig. 1b). To mimic AI, we exposed the animal to asymmetric odor stimulation: air with chemoattractant vapors (10−3 M isoamyl alcohol) blown on the entire animal when the head was bent in one direction (for example, dorsally, h > 0) and odor-free air blown when the head was bent in the other (ventrally, h < 0) (Fig. 1e). The animal turned gradually in the direction in which its head was bent when the odor was delivered (Fig. 1f, Supplementary Fig. 1c–d, Supplementary Movie 1). To mimic SI we delivered vapors of isoamyl alcohol constantly. independent of h. The animal reduced its reversal frequency11 and did not turn (Fig. 1g). The most parsimonious hypothesis based on these results is that asymmetric and symmetric odor components generate activity patterns in the nervous system with corresponding symmetries to separately control turning and reversal frequency during chemotaxis.
We therefore identified interneurons that triggered the different locomotory behaviors necessary for chemotaxis by directly stimulating individual neurons in a freely moving animal in asymmetric and symmetric patterns. To do so, we expressed channelrhodopsin-2 (ChR2)12,13 or archaerhodopsin-3 (Arch)14,15 in different neurons. Light activation of ChR2 (by 480 nm light) and Arch (540 nm) leads to neural excitation and inhibition, respectively.
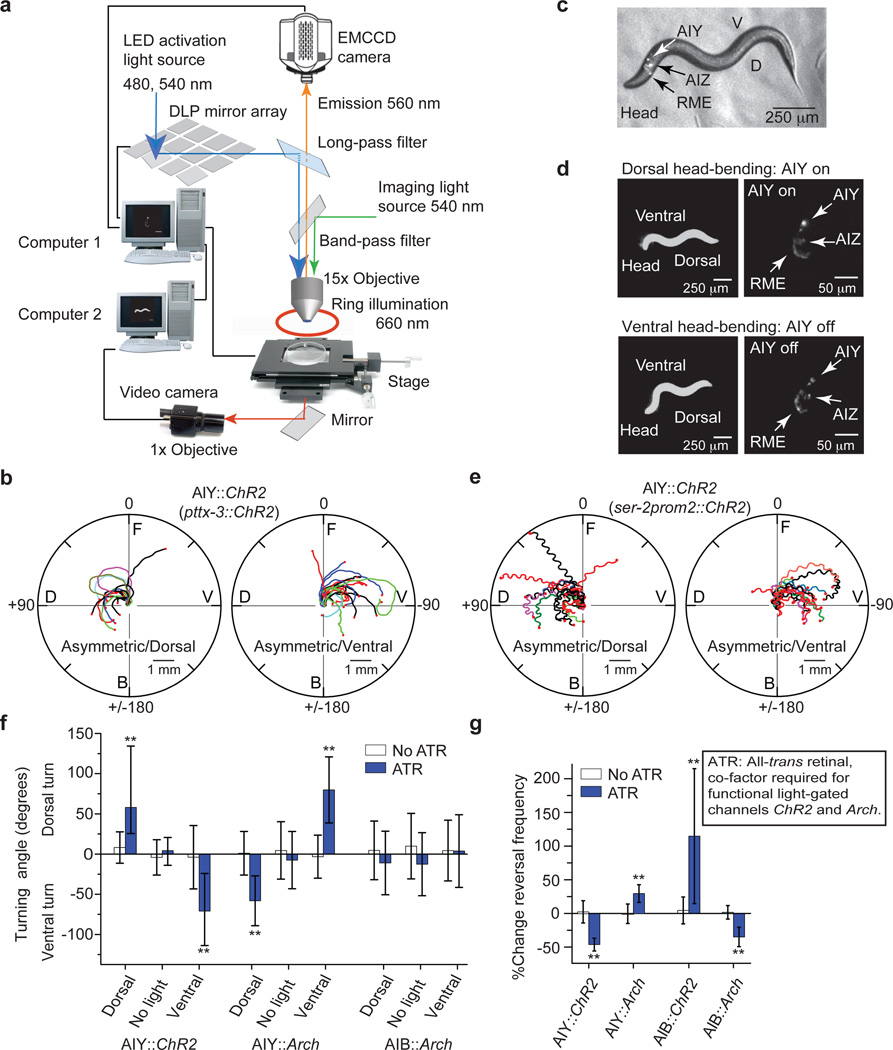
Targeted illumination of specific neurons in motionless animals16 or of body segments in freely moving animals17,18 shave been developed to excite neurons for which specific promoters are not known. Since the neurons in the nerve ring are as close as 5–10 µm to each other and their relative positions change quickly as the animal moves (Supplementary Fig. 2a), we could not use these techniques. To optically stimulate one of many neurons (each with a diameter of 5–10 µm) expressing light-gated ion channels in the nerve ring of an animal that typically moves at 150±50 µm/s (Supplementary Fig. 2b), our setup tracks, identifies, and specifically illuminates the neuron(s) of interest, all within 25 milliseconds to achieve a 5 µm spatial resolution of excitation (Fig. 2a).
Figure 2. Asymmetric and symmetric excitation of AIY control gradual turning and reversal frequency.
a, Setup for closed-loop single neuron stimulation (see supplementary method). b, Sample trajectories of center of mass of animals upon dorsal or ventral asymmetric excitation of AIY (n=10, pttx-3::ChR2). c, Fluorescence image overlaid on the bright field image of a nematode co-expressing ChR2 and mKO in AIY, AIZ and RME (ser-2prom2::mKO and ser-2prom2::ChR2). d, Asymmetric dorsal stimulation of the animal in (c). Upon dorsal head-bending (top, left), ChR2 and mKO were excited in AIY using setup in (a) (top, right, higher fluorescence in AIY) but not upon ventral head-bending (bottom, right, decreased fluorescence in AIY). Left: 1× dark-field image of animal; right: 15× fluorescent images of neurons in that animal. e, Sample trajectories of nose tip of animals upon dorsal (left) or ventral (right) asymmetric excitation of AIY (n=10, ser-2prom2::ChR2). f, Turning angle of animals upon asymmetric stimulation of AIY::ChR2 (n=10), AIY::Arch (n=10) and AIB::Arch (n=7). Dorsal: asymmetric dorsal stimulation. No light: unstimulated. Ventral: asymmetric ventral stimulation. No ATR: Control without all-trans retinal (ATR). g, Reversal frequencies upon symmetric stimulation of AIY::ChR2 (n=10), AIY::Arch (n=19), AIB::ChR2 (n=14), and AIB::Arch (n=11) (f–g mean: white and blue bars, error bars: 1 s.d over n animals, white bars: no ATR; **, p<0.05, two-sample t-test).
Using this setup, we first tested how stimulating the interneurons AIY and AIB affected locomotory behavior. Both neuron pairs receive chemical synapses from the AWC sensory neurons that detect isoamyl alcohol8 and showed calcium activity when animals were stimulated with this chemoattractant11. Asymmetric excitation of AIY with light in animals that expressed ChR2 only in AIY (under the promoter ttx-3) caused the animal to turn in the direction in which the head was bent when AIY were excited (Fig. 2b, Supplementary Movie 2). We validated our setup by reproducing these results in animals that expressed ChR2 in AIY alone and fluorescence protein monomeric Kusabira-Orange (mKO) in neurons AIY, AIZ and RME (ser-2prom2 promoter, Supplementary Fig. 2c–f). Asymmetric stimulation of AIY in animals expressing ChR2 in AIY, AIZ and RME (ser-2prom2) showed the same results (Fig. 2e–f, Supplementary Fig. 2g, Supplementary Movie 3). Consistently, inhibiting the activity in AIY asymmetrically (pttx-3::Arch) caused the animal turn in the opposite direction in which the head was bent when AIY were inhibited (Fig. 2f, Supplementary Fig. 3a, Movie 4). Symmetric excitation and inhibition of AIY decreased and increased the reversal frequency respectively, but did not cause turning (Fig. 2f–g, Supplementary Fig. 3, Supplementary Movie 5).
Both asymmetric and symmetric excitation or inhibition of AIB in AIB::ChR2 and AIB::Arch animals (npr-9 promoter) affected the reversal frequency of the animal but did not produce any gradual turning (Fig. 2f–g, Supplementary Fig. 4). We could thus control the two locomotory behaviors crucial for chemotaxis, gradual turns and reversal frequency, by driving different patterns of activity in AIY alone.
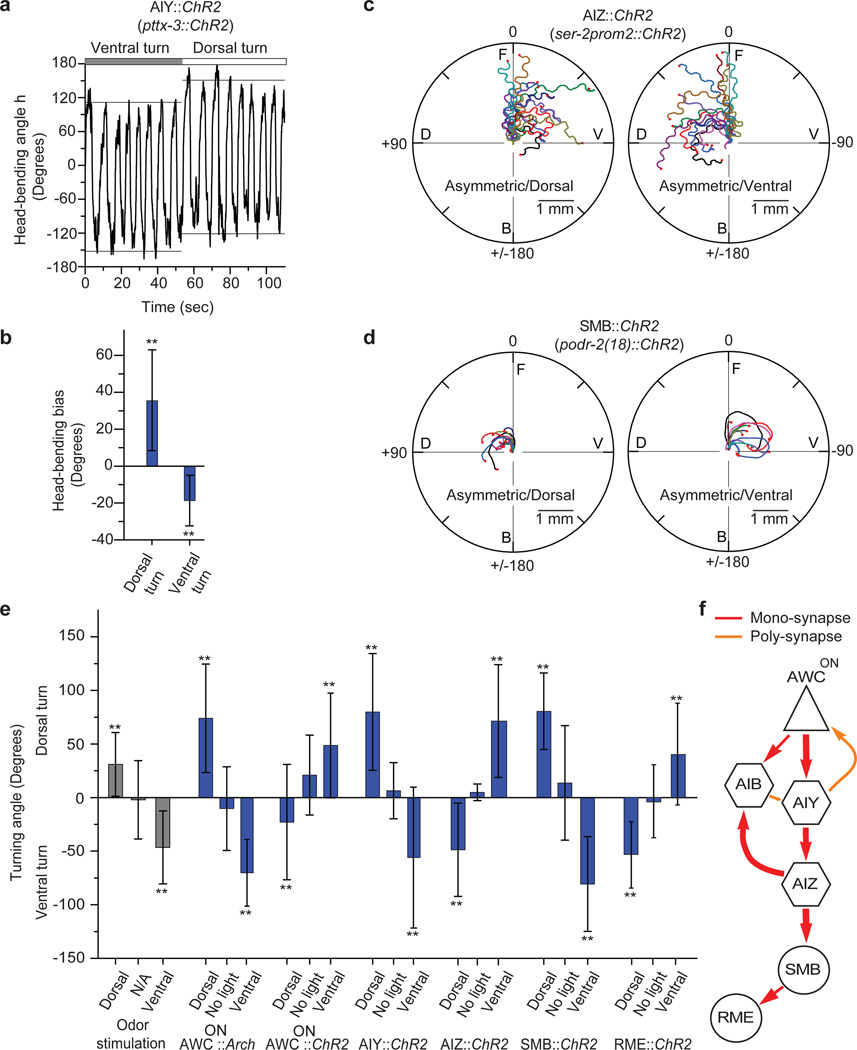
Turning is initiated by a larger head-bending angle in one direction19,20. When we forced the animal to turn by asymmetrically stimulating AIY (pttx-3::ChR2), the head-bending angle in the direction of the turn increased (Fig. 3a, b), suggesting that asymmetric activation of AIY controlled head bending through head motor neurons to cause gradual turning. AIY are most directly connected to the head muscles through the interneurons AIZ which synapse onto the head motor neurons SMB and RME8. AIZ neurons have been implied to play a role in gradual turns by laser ablation4. Ablations of SMB and RME change the head-bending angle during crawling and exhibit loopy behavior7,21.
Figure 3. Asymmetric AIY excitation modulates head-bending angle to cause turning.
a, Representative head-bending angle (horizontal lines: means of maximum and minimum h during each full head swing) of an AIY::ChR2 animal forced to first turn ventrally and then dorsally (positive: dorsal, negative: ventral). b, Histogram of head-bending angle bias (max(h)+min(h)) during dorsal and ventral turn caused by asymmetric AIY excitation (error bar, s.d. over n=5). c–d, Sample tracks of animals upon dorsal (left) or ventral (right) asymmetric excitation (c) of AIZ::ChR2 (ser-2prom2::ChR2), (d) of SMB::ChR2 (podr-2(18)::ChR2) animals. e, Turning angle upon asymmetric stimulation with odor (n=10) and optically of AWCON::Arch (n=10), AWCON::ChR2 (n=9), AIY::ChR2 (n=10), AIZ::ChR2 (n=5), SMB::ChR2 (n=5), and RME::ChR2 (n=5). Dorsal: asymmetric dorsal stimulation; no light: unstimulated; ventral: asymmetric ventral stimulation (error bar: s.d. over n, **, p<0.05, two-sample t-test). f, The monadic (red) and polyadic (orange) synapses between AWCON, AIY, AIB, AIZ, SMB and RME (thickness: proportional to synapse number). Triangle: sensory neuron; hexagon: interneuron; circle: motor neuron.
When we specifically excited AIZ::ChR2 (ser-2prom2::ChR2), SMB::ChR2 (podr-2(18)::ChR2), or RME::ChR2 (ser-2prom2::ChR2) asymmetrically using our setup, the animals turned (Fig. 3c–e, Supplementary Fig. 5, Supplementary Movie 6–8). These results in conjunction with those from asymmetric optical stimulation of the isoamyl alcohol sensing neuron AWCON (Supplementary Fig. 6, Supplementary Movie 9), show that asymmetric stimulation of the sequence of anatomically connected neurons from AWCON, through the interneurons AIY, AIZ to the head motor neurons SMB and RME8 (Fig. 3f) all cause turning. These set of neurons thus sense and respond to the component of the sensory signal that oscillates asymmetrically and in synchrony with head movement to control head bending and turning.
Since different patterns of activity in AIY are sufficient to control both the frequency of reversals and turning, we tested if controlling AIY activity alone was sufficient to coordinate reversal frequency and turning to evoke chemotactic behavior. To do so, we measured the locomotory behavior of animals in a fixed spatial light gradient that directly excited AIY::ChR2 (Supplementary Fig. 7a, Supplementary Methods). The animals were unable to track the gradient direction (Supplementary Fig. 7c, Supplementary Movie 10).
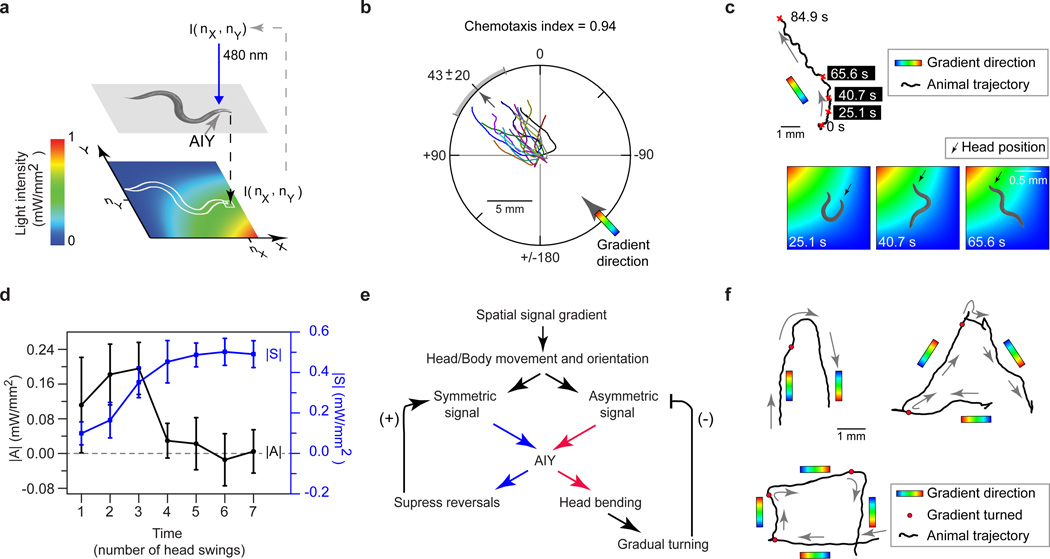
AIY's somas and processes are 150±25 µm behind the nose tip of an adult animal (Supplementary Fig. 7b). Since the animal’s speed is 150±50 µm/sec, AIY follow the position of the nose tip with approximately a one second delay. We argued that the dynamics of AIY excitation caused by the animal’s movement through the gradient were not in synchrony with head bending due to this delay, preventing the animals from tracking the gradient. Therefore, we designed a virtual light gradient where the excitation light intensity on AIY depended not on the positions of AIY in space but on the position of the nose tip (Fig. 4a, Supplementary Fig. 7a, Supplementary Methods).
Figure 4. Controlling AIY activity is sufficient to evoke chemotactic behavior.
a, Virtual light gradient algorithm (see supplementary method). At each time t, AIY::ChR2 animals (pttx-3::ChR2) are stimulated with 480 nm blue light with an intensity (I(nx,ny)) of the virtual gradient at the nose-tip position (nx,ny). b, Trajectories of AIY::ChR2 animals moving in a virtual light gradient (as in (a)) with a gradient direction at 45 degrees (black tick: mean direction of trajectory, grey bar: s.d., n=10). c, Top: A sample trajectory of an animal in (b). Bottom: Snapshots of the animal making a gradual turn to reorient itself to the gradient direction (pseudocolor: same as (a)). d, Magnitude of AI (black), and SI (blue, right axis), over a head swing, as a function of numbers of head swings during a gradual turn (n=5, from trajectories in (b), error bar: 1 s.d.). e, Model for chemotaxis in the virtual light gradient. f, Trajectories of center of mass of animals when the gradient direction was suddenly rotated (at times when the animal reached the red dots) by 180, 120 or 90 degrees.
In this setup, animals changed their locomotory direction using reversals and gradual turns to stably track the gradient direction (the fraction of the animals moving up the gradient and hence to the correct quadrant, defined as chemotaxis index22 =0.94, Fig. 4b). As with the odor profile at the nose tip (Fig. 1d), the temporal light intensity pattern that excited AIY could be written as a sum of an asymmetric (AI(h, t)) and a symmetric (SI(h,t)) component. When the animal's locomotory direction was not along the direction of the light gradient, the magnitude of AI(h,t) over each head swing, , was larger, exciting AIY asymmetrically to make the animal turn. As the animal oriented itself along the direction of the gradient, the magnitude of AI(h,t) continuously diminished, suppressing turns while the magnitude of SI(h,t), |S|, increased and suppressed reversals (Fig. 4c–d). Thus, manipulating the dynamics of activity in just the AIY interneuron pair is sufficient to evoke chemotactic behavior. This is because the head bending and locomotion of the animal through the virtual light gradient together generate and modulate the levels of symmetric and asymmetric excitation of AIY which in turn control future locomotory behavior (Fig. 4e).
This model would predict robust chemotactic behavior in the light gradient stimulating AIY, with the symmetric and asymmetric components modulating their relative magnitudes to stably guide the animal in the correct direction. To test for robust tracking, we suddenly rotated the virtual light gradient direction by different angles and measured the animal’s response. The animals followed the gradient direction as this direction was suddenly and repeatedly rotated by 180 (Supplementary Movie S11), 120 or 90 degrees (Fig. 4f).
Previous studies have identified neurons involved in chemotaxis by showing that defects in these neurons compromise locomotory programs and sensory modalities necessary for this behavior. Through our approach we can identify key neurons in the neural network whose dynamics are sufficient to drive chemotactic behavior and hence act as control nodes in the network. Our study leads to questions of how activity patterns stimulated in the AIY neurons during chemotaxis are disentangled by the downstream neurons to drive the different motor programs. Since many chemosensory and thermosensory neurons synapse onto AIY, the modulation of symmetric and asymmetric activity patterns in these interneurons are likely to play a central role in the different taxis behaviors of C. elegans. Our techniques provide avenues to eventually identify and generate neural activity patterns to control all the behaviors of this nematode.
Methods
Strains were grown and maintained under standard conditions2 unless indicated otherwise. Transgenic lines with pha-1 selection marker were grown under 24°C22. All optical stimulation experiments were done in lite-1 mutants to minimize the animal's sensitivity to blue light23. A complete strain list andinformation on transgenes are included in the Supplementary Table 2.
Chemotaxis analysis
The animals' chemotaxis to a bacterial lawn were assayed on an open-lid, 10 cm nematode growth medium (NGM) plate incubated in room temperature overnight with 10 µL Escherichia coli OP50 at the center. N2 young adults were placed on the plate, 1.5 cm away from the center of the bacterial lawn. The animals' behaviors were recorded under 6× magnification by an EMCCD camera at 20Hz and analyzed by customized LabView scripts.
Odor stimulation setup
An N2 young adult was placed on an open-lid, food-free, 10 cm NGM plate for at least 1 minute. An electric valve (Supplementary Fig. 1b) was used to determine whether the air (100 sccm) was bubbled through water or 10−3 M isoamyl alcohol based on the posture of the freely moving animal. The images were recorded under 6× magnification by an EMCCD camera at 20 Hz and analyzed by customized LabView scripts.
Single neuron stimulation setup
L4 animals were transferred to a new NGM plate with a thin layer of E. coli OP50, containing 100 µM ATR (all-trans retinal, the co-factor required for functional light-gated ion channels)16 if required, 24 hours before experiments in room temperature. An animal then was placed on an open-lid, food-free, 6 cm NGM plate for at least 1 minute. Dark-field 660 nm illumination was used to visualize the posture of the animal under 1× magnification by a video camera at 20 Hz (Fig. 2a, computer 2). Low-power 540 nm (0.1 mW/mm2) epifluorescent illumination was used to visualize the neurons co-expressing light-gated ion channels and monomeric Kusabira-Orange (mKO)24 at 15× by an EMCCD camera at 40 Hz (computer 1). Image thresholding and particle detection were used as the image processing algorithms to identify the mKO-tagged neurons. As the animal swung its head, the neurons in the field rotated and changed their positions. To offset the rotation effect, positions of the neurons were measured using the principle axis and the distance from the center of mass of the processed image. The processed images were then used to track the animal and to position the DLP mirrors to deliver light (4 mW/mm2, 480 or 540 nm) on the neurons of interest with any desired temporal patterns to excite or inhibit the activity of the neurons. Feedback between motorized stage, DLP mirrors and image processing software was operated at 40 frames/sec to achieve a 5 µm spatial resolution of excitation on a freely moving animal. The images were processed, recorded, and analyzed by customized LabView scripts.
Reversal frequency
L4 animals were transferred to a new NGM plate with a thin layer of E. coli OP50, containing 100 µM ATR if required, 24 hours before experiments in room temperature. A 6 cm copper ring was placed in an open-lid, food-free, 10 cm NGM plate immediately before experiments to keep the animals in the field of view. Young adults then were transferred to the assay plate for 1 minute before the experiment started. The desired wavelength of light (1 mW/mm2, 480 nm or 540 nm) was delivered in alternate 3-minute intervals for 1 hour using customized LabView scripts. The experiments were recorded by a video camera at 20 Hz. Reversal frequency was calculated from pirouettes determined by an automated worm tracker (http://wormsense.stanford.edu/tracker)25.
Virtual light gradient setup
An AIY::ChR2 animal was placed on an open-lid, food-free, 10 cm NGM plate for at least 1 minute before starting the experiment. A virtual-gradient of light, I(r) = exp (−r2/r02), from 0 to 1 mW/mm2 over 1.3 mm, where r0 =0.8 mm, was defined in an x–y coordinate system tied to the center of mass of the animal which was always at the center of the gradient profile (0.65 mm, 0.65 mm) (Supplementary Fig. 7a). The virtual light gradient moved with the center of mass of the animal but at a fixed orientation. At each time t, the coordinates of the nose tip were identified to calculate the corresponding intensity of light I(nx,ny) (Fig. 4a). The animal was then illuminated with blue (480 nm) light at intensity I(nx,ny), and thus, AIY instantaneously experienced the intensity of light at the nose tip.
Molecular biology
Arch::EGFP14 was cloned into Fire lab vector kit plasmid pPD96.52 (ligation number L2534, Addgene plasmid 1608). Chop-2(H134R)::TagRFP was obtained by swapping TagRFP with YFP in the previously cloned chop-2(H134R)::YFP16. Arch::TagRFP and Arch::mKO were obtained respectively by swapping TagRFP26 ormKO24 with EGFP in Arch::EGFP. TagRFP and GCaMP-327 were codon-optimized for C. elegans (de novo synthesized by GenScript) whereas others were optimized for mammalian cells. We amplified 2kb str-2p28, 1kb ttx-3p29, 3kb npr-9p30, 4.5kb ser-2prom231, and 2.4kb odr-2(18)p32 by PCR from C. elegans genomic DNA. All the promoters were then fused with desired light-gated ion channels by PCR fusion33.
Supplementary Material
Acknowledgements
We thank John Dowling, Shawn Lockery, Jeff Lichtman, Kathryn McCormick, Andrew Murray, Erin O’Shea, Alex Schier, Bodo Stern and members of the Ramanathan lab for discussions and comments, Human Frontier Science Program (HFSP) Postdoctoral Fellowship (A.K.), NSF Graduate Fellowship (CH.S.), NSF Career Award, Pew Scholar, Klingenstein and the NIH Pioneer Awards (S.R.) for support.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions AK, CHS, ZG and SR designed the experiments. AK, CHS and ZG performed the experiments. AK, CHS and SR wrote the manuscript.
The authors have no competing financial interests.
References
- 1.Brenner S. The genetics of behaviour. Br Med Bull. 1973;29:269–271. doi: 10.1093/oxfordjournals.bmb.a071019. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior inCaenorhabditis elegans. Journal of Neurobiology. 2003;56:178–197. doi: 10.1002/neu.10245. [DOI] [PubMed] [Google Scholar]
- 4.Iino Y, Yoshida K. Parallel Use of Two Behavioral Mechanisms for Chemotaxis in Caenorhabditis elegans. Journal of Neuroscience. 2009;29:5370–5380. doi: 10.1523/JNEUROSCI.3633-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19:9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakabayashi T, Kitagawa I, Shingai R. Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neuroscience Research. 2004;50:103–111. doi: 10.1016/j.neures.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Gray JM. Inaugural Article: A circuit for navigation in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White JG, Southgate E, Thomson JN, S B. The structure of the nervous system fo the nematode c elegans. Phil. Trans. R. Soc. Lond. B. 1986:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 9.Ward S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci U S A. 1973;70:817–821. doi: 10.1073/pnas.70.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izquierdo EJ, Lockery SR. Evolution and Analysis of Minimal Neural Circuits for Klinotaxis in Caenorhabditis elegans. Journal of Neuroscience. 2010;30:12908–12917. doi: 10.1523/JNEUROSCI.2606-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalasani SH. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 12.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 13.Nagel G, et al. Light Activation of Channelrhodopsin-2 in Excitable Cells of Caenorhabditis elegans Triggers Rapid Behavioral Responses. Current Biology. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki A, Sudo Y, Takagi S. Optical Silencing of C. elegans Cells with Arch Proton Pump. PLoS ONE. 2012;7:e35370. doi: 10.1371/journal.pone.0035370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nature Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel ADT. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nature Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stirman JN, et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Methods. 2011;8:153–158. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockery SR. The computational worm: spatial orientation and its neuronal basis in C. elegans. Current Opinion in Neurobiology. 2011;21:782–790. doi: 10.1016/j.conb.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D, Park S, Mahadevan L, Shin JH. The shallow turn of a worm. Journal of Experimental Biology. 2011;214:1554–1559. doi: 10.1242/jeb.052092. [DOI] [PubMed] [Google Scholar]
- 21.McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature. 1993;364:337–341. doi: 10.1038/364337a0. [DOI] [PubMed] [Google Scholar]
- 22.Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bono M, et al. A Novel Molecular Solution for Ultraviolet Light Detection in Caenorhabditis elegans. PLoS Biology. 2008;6:e198. doi: 10.1371/journal.pbio.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
References associated with Full Methods Section only
- 24.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aramayo R et al. The Parallel Worm Tracker: A Platform for Measuring Average Speed and Drug-Induced Paralysis in Nematodes. PLoS ONE. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merzlyak EM, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 27.Hires SA, Tian L, Looger LL. Reporting neural activity with genetically encoded calcium indicators. Brain Cell Biology. 2008;36:69–86. doi: 10.1007/s11068-008-9029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- 29.Hobert O, et al. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 30.Bendena WG, et al. A Caenorhabditis elegans allatostatin/galanin-like receptor NPR-9 inhibits local search behavior in response to feeding cues. Proc Natl Acad Sci U S A. 2008;105:1339–1342. doi: 10.1073/pnas.0709492105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsalik E. LIM homeobox gene-dependent expression of biogenic amine receptors in restricted regions of the C. elegans nervous system. Developmental Biology. 2003;263:81–102. doi: 10.1016/s0012-1606(03)00447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou JH, Bargmann CI, Sengupta P. The Caenorhabditis elegans odr-2 gene encodes a novel Ly-6-related protein required for olfaction. Genetics. 2001;157:211–224. doi: 10.1093/genetics/157.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulin T. Reporter gene fusions. WormBook. 2006 doi: 10.1895/wormbook.1.106.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.