Abstract
Although the discrete Fourier transform played an enabling role in the development of modern NMR spectroscopy, it suffers from a well-known difficulty providing high-resolution spectra from short data records. In multidimensional NMR experiments, so-called indirect time dimensions are sampled parametrically, with each instance of evolution times along the indirect dimensions sampled via separate one-dimensional experiments. The time required to conduct multidimensional experiments is directly proportional to the number of indirect evolution times sampled. Despite remarkable advances in resolution with increasing magnetic field strength, multiple dimensions remain essential for resolving individual resonances in NMR spectra of biological macromolecues. Conventional Fourier-based methods of spectrum analysis limit the resolution that can be practically achieved in the indirect dimensions. Nonuniform or sparse data collection strategies, together with suitable non-Fourier methods of spectrum analysis, enable high-resolution multidimensional spectra to be obtained. Although some of these approaches were first employed in NMR more than two decades ago, it is only relatively recently that they have been widely adopted. Here we describe the current practice of sparse sampling methods and prospects for further development of the approach to improve resolution and sensitivity and shorten experiment time in multidimensional NMR. While sparse sampling is particularly promising for multidimensional NMR, the basic principles could apply to other forms of multidimensional spectroscopy.
Introduction
Since the introduction of Fourier Transform NMR by Richard Ernst and Weston Anderson1, NMR spectra have mainly been determined by measuring the response of the spin system to a broad-band RF pulse. The response (the free induction decay, FID) is measured at regular intervals, and the spectrum is obtained by computing the discrete Fourier transform (DFT). To correctly determine frequencies, the sampling interval must be at least as short as the reciprocal of the spectral width spanned by frequency components in the signal, the famous Nyquist condition. Conversely, the resolution of the spectrum is related to the longest time sampled. Together, the twin requirements of sufficiently rapid uniform sampling and long evolution times means that high resolution spectra require large data sets.
The problem becomes more acute with multidimensional experiments that are based on the concept first introduced by Jean Jeener2. Jeener proposed the introduction of additional time domains through parametric sampling, for example by repeating a two-pulse experiment while incrementing a time delay separating the pulses. The time domains that are sampled parametrically are referred to as indirect dimensions, whereas the real-time evolution of the signal is said to occur in the direct or acquisition dimension. Three- and four-dimensional NMR experiments have become routine, and indeed indispensible for resolving individual resonances of biological macromolecules. A problem attending these experiments, however, is that conventional methods of spectrum analysis employing the DFT mandate impractically lengthy experiments in order to obtain high frequency resolution in the indirect dimensions.
An approach employing linear prediction (LP) to improve the resolution attainable from short data records emerged in the 1980s3. LP extrapolation posits that the time domain data for a given evolution time can be described as a linear combination of past values. Coefficients fit to the measured interval are used to extrapolate the data beyond the measured interval. The assumption underlying LP extrapolation is equivalent to assuming the data can be described as a sum of exponentially decaying sinusoids. While LP extrapolation represents a significant advance, it is prone to generation of false peaks when applied to noisy data, and can lead to frequency bias when applied too aggressively or to signals that deviate from exponential decay4. Like the DFT, LP extrapolation requires data sampled at uniform intervals.
The development of non-Fourier methods of spectrum analysis that do not require data sampled at uniform intervals has enabled an alternative approach to the problem of high resolution from short data records5. Using nonuniform sampling (NUS), these methods permit the collection of data at long evolution times – required for high resolution – while avoiding the requirement of measuring every intervening time separated by multiples of the Nyquist interval. NUS methods are not a panacea, however, because they introduce sampling artifacts or “sampling noise”. Some methods of spectrum analysis and various post-processing schemes are able to diminish the artifacts, but limits on the ability to eliminate sampling artifacts are ultimately imposed by the presence of signal noise.
The combination of non-Fourier methods and NUS applied to multidimensional NMR is beginning to enable high frequency resolution along indirect dimensions, within practical constraints on measuring time. The methods are likely to become routine for experiments in three or more dimensions simply as a means to improve the efficiency of very expensive high-field spectrometers. Additional improvement in data collection strategies and spectral estimation methods promises to expand the applicability of multidimensional NMR to more challenging systems by improving sensitivity and resolution, and shortening experiment time.
Evolution
The provenance of NUS in multidimensional NMR can be traced back 30 years. Bodenhausen and Ernst introduced a means of avoiding the sampling constraints associated with uniform parametric sampling of two indirect dimensions of three-dimensional experiments, by coupling the two indirect evolution times6. By incrementing the evolution times in concert, sampling occurs along a radial vector in t1-t2, with a slope given by the ratio of the increments applied along each dimension. This effectively creates an aggregate evolution time t = t1 + α*t2 that is sampled uniformly, and thus the DFT can be applied to determine the frequency spectrum. According to the projection-cross-section theorem, the resulting spectrum is the projection of the full t1-t2 spectrum onto a vector with angle tan−1(α) in the f1-f2 plane. Bodenhausen and Ernst referred to this as an “accordion” experiment. Although they did not propose reconstruction of the full f1-f2 spectrum from multiple projections, they did discuss the use of multiple projections for characterizing the corresponding f1-f2 spectrum, and thus the accordion experiment is the precursor to more recent radial sampling methods (discussed below). Because the coupling of evolution times effectively combines dimensions, the accordion experiment is an example of a reduced dimensionality (RD) experiment7.
The 3D accordion experiment has much lower sampling requirements than conventional 3D approaches because it avoids sampling the uniform grid of indirect time (t1,t2) values that must be sampled in order to utilize the DFT to compute the spectrum along both t1 and t2. A more general approach is to eschew regular sampling altogether and employ non-Fourier reconstruction techniques that are capable of utilizing nonuniformly sampled data. Laue and colleagues were the first to adopt this approach in multidimensional NMR, by using maximum entropy (MaxEnt) reconstruction to compute the frequency spectrum from NUS data8. While the combination of NUS and MaxEnt reconstruction provided high resolution spectra with dramatic reductions in experiment time compared to conventional uniform sampling, the approach initially was not widely adopted, no doubt because neither MaxEnt reconstruction nor NUS was highly intuitive. Nevertheless a small cadre of investigators continued to explore novel NUS schemes in conjunction with MaxEnt reconstruction throughout the 1990’s9,10. A broader appreciation for NUS was stimulated by a series of papers by Kupče and Freeman, in which they utilized back-projection reconstruction (BPR) from a series of experiments employing radial sampling in t1-t2 to reconstruct the fully-dimensional f1-f2-f3 spectrum11–15. While the radial data sampling employed by the accordion and BPR is also employed by other approaches (see below), it was the use of back-projection (by analogy to computerized tomography) that demonstrated the connection with the 3D spectrum conventionally obtained by uniform sampling and DFT. Despite some drawbacks to radial sampling (discussed below), the BPR approach was important because it provided a powerful heuristic for more general NUS approaches. More recently, however, the promulgation of non-Fourier methods capable of treating NUS data and significant advances in computer speed have led to a renewed interest in the design of more efficient NUS schemes.
Beyond Fourier
An important aspect of NUS is the use of non-Fourier methods of spectrum analysis that properly account for “missing” data. Although the DFT can be applied to NUS data that is collected on-grid, the resulting spectrum is the one that would be obtained if the missing data values had the value zero. Thus applying the DFT to NUS data is not, strictly speaking, the DFT of NUS data, it is instead the DFT of zero-augmented NUS data, and the distinction is important. The orthogonality of the Fourier basis functions (the complex exponentials) breaks down when the complete grid is not sampled, and frequencies in the spectrum thus interfere, giving rise to aliases (sampling artifacts). To paraphrase the French director of a modern silent film, zero is very different from nothing16.
A host of non-Fourier methods has been developed that properly treat the missing data (rather than assuming the value zero). These include nonparametric regularization methods (e.g. maximum entropy or minimum l1-norm), parametric methods that model the signal (e.g. maximum likelihood (MLM17, Bayesian18, and “CLEAN”19), decomposition methods that exploit symmetry properties (e.g. multiway decomposition, MWD), and methods specific for radially-sampled data (e.g. G-matrix Fourier transform (GFT20) BPR, and polar FT21).While these methods differ significantly in their approach, typically they yield very similar results when used with the same NUS data, indicating that the most important determinant of spectral quality using NUS is the sampling scheme employed, and not the method of spectrum analysis.
Tower of Babel
The decades-long gestation of NUS methods in multidimensional NMR is in part responsible for a surfeit of different terminology, which can obscure similarities among, and occasionally the equivalence of, various approaches. This applies both to sampling schemes as well as methods used to obtain spectral estimates from NUS data. The connection among accordion, reduced dimensionality, back projection, and GFT methods, which all utilize radial sampling, was slow to be recognized. Iterative ad hoc approaches involving thresholding (iterative soft threshholding, IST22, spectroscopy by integration of frequency and time domain information (SIFT23) have been shown to be closely related to more formal regularization methods22, and a host of different approaches that assume the signal can be modeled as a sum of exponentially decaying sinusoids (MLM, Bayesian, CLEAN, filter diagonalization method (FDM24), linear prediction – singular value decomposition (LPSVD25), while adopting very different approaches to fitting the model parameters, yield very similar results. The recent introduction of “compressed sensing” (CS26–29) is viewed in some quarters as a novel approach, but amounts to NUS with l1-norm regularization, and thus has a history that precedes the new terminology. Also confusing is the widely-used term “nonuniform DFT” (nuDFT), which lacks many important properties of the DFT. “Sparse recovery” is another common synonym for NUS.
Fundamentals of NUS
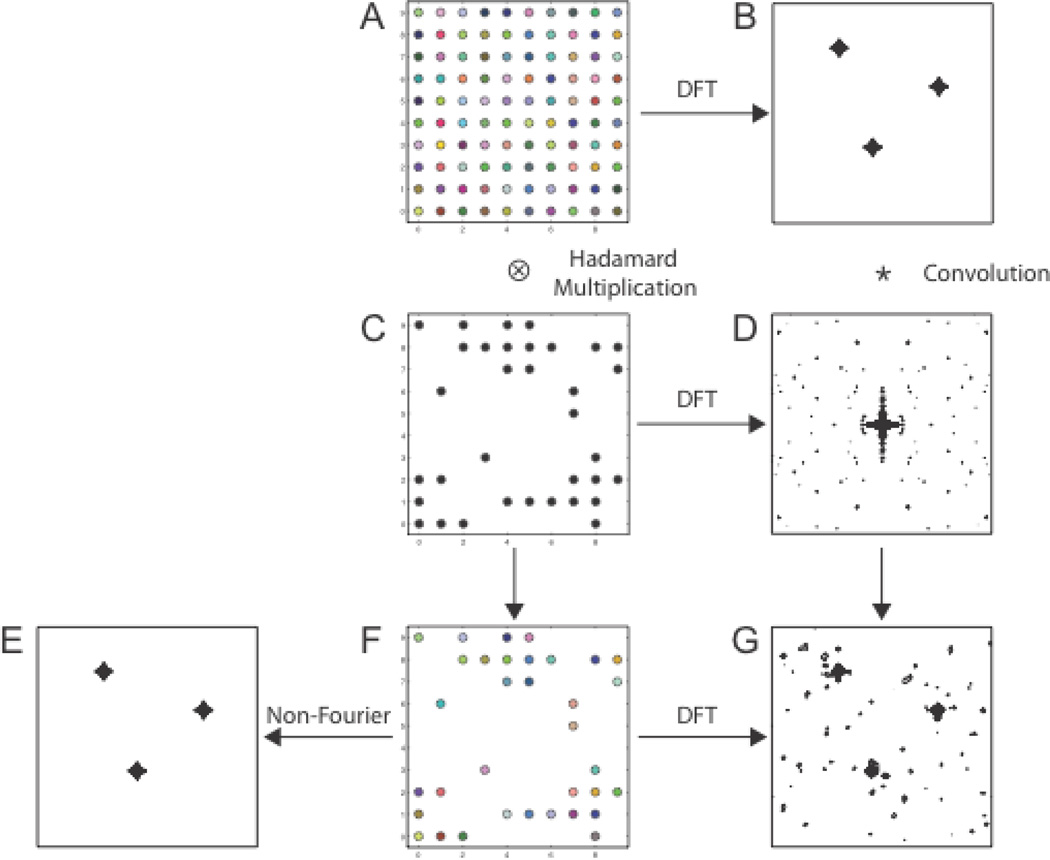
The aim of NUS is to approximate the spectrum that would be obtained using uniform sampling while collecting substantially fewer samples. The relationship between the DFT of zero-augmented NUS data and the DFT spectrum for uniformly sampled data has a particularly simple form when NUS is restricted to a regular or Cartesian grid (Figure 1). If there exists a real-valued sampling function with the property that when it multiplies a uniformly sampled data vector, element-wise (the Hadamard product), the result is the zero-augmented NUS data matrix, then the DFT of the zero-augmented NUS data is the convolution of the DFT spectrum of the uniformly sampled data with the DFT of the sampling function. The sampling function has the value 1 for times that are sampled and zero for times that are not sampled. The DFT of the sampling function is variously called the point-spread function (PSF), the impulse response, or the sampling spectrum.
Figure 1.
The role of the sampling function and PSF in NUS. The uniformly sampled data matrix is shown in A, with color used to indicate different data values. The sampling function (C) has the value 1 for sampled evolution times and 0 for those not sampled. The zero-augmented NUS data set (F) is the Hadamard product of the uniformly sampled data and the sampling function. In the matrices (C) and (F), entries with the value zero appear blank. The two-dimensional DFT of A is the uniformly sampled spectrum (B). The two-dimensional DFT of the sampling function (C) is the PSF (D). The two-dimensional DFT (G) of the zero-augmented NUS data (F) is equivalent to the convolution of the uniformly sampled spectrum with the PSF. Non-Fourier methods of spectrum analysis deconvolve the PSF to approximate (E) the spectrum that would have been obtained using uniform sampling, but from NUS data.
Computing the DFT of zero-augmented NUS data results in a spectrum that reflects coherences in the sampling scheme as well as those in the signal. In contrast, non-Fourier spectral reconstruction methods attempt to diminish sampling artifacts in the reconstructed spectrum. Their ability to suppress sampling artifacts is invariably limited by the presence of noise. While they reduce the magnitude of sampling artifacts, the artifacts nevertheless appear at the same locations as found in the zero-augmented DFT spectrum.
Point spread function
The PSF provides insight into the locations and magnitudes of sampling artifacts that result from NUS, and it can have an arbitrary number of dimensions, corresponding to the number of dimensions in which NUS is applied. The PSF typically consists of a main central component at zero frequency, with smaller non-zero frequency components. Because the PSF enters into the zero-augmented DFT spectrum through convolution, each non-zero frequency component in the PSF will give rise to a sampling artifact for each frequency component in the signal, with positions relative to the true signal components that are the same as the relationship of the satellite peaks to the zero-frequency peak in the PSF. The amplitudes of the sampling artifacts will be proportional to the amplitude of the signal component and the relative height of the satellite peaks in the PSF. Thus the largest sampling artifacts will arise from the largest-amplitude components of the signal spectrum. The effective dynamic range (the ratio between the magnitude of the largest and smallest signal components) of the zero-augmented DFT spectrum can be directly estimated from the ratio between the amplitude of the zero-frequency component and the amplitude of the largest non-zero-frequency component (the peak-to-sidelobe ratio, discussed below) in the PSF.
How long?
The principal rationale for NUS is the ability to sample at long evolution times, necessary for high resolution, while reducing the overall amount of sampling associated with conventional uniform sampling. Using the DFT, resolving spectral features to the natural linewidth requires sampling at evolution times of πT2 or longer. Of course methods such as linear prediction extrapolation have long been used to approximate the data beyond the measured interval without introducing truncation artifacts associated with zero-filling. Methods of spectrum analysis that yield resolution smaller than 1/tmax (tmax is the longest evolution time sampled) are said to provide “super-resolution”. Sampling beyond πT2 is of little benefit unless one employs a parametric method that models the signal (such as MLM, Bayesian, and FDM methods) or takes steps to deconvolve the natural lineshape. Sampling beyond 1.26T2 results in diminishing returns on sensitivity30. When combined with methods of spectrum analysis capable of super-resolution, sampling to 1.26T2 usually provides more than ample resolution to resolve spectral components to the level of the natural linewidth, and thus represents a reasonable compromise between sensitivity and resolution for decaying signals. For experiments in which the evolution period is constant-time or semi-constant-time, the signal decay is determined mainly by field inhomogeneity (RF and B0), and so practical limits on tmax are imposed by the inhomogeneity or length of the constant time period, rather than T2.
In general, NUS that involves sampling a subset of the Cartesian Nyquist matrix yield lower sensitivity than the full uniformly-sampled dataset. However if constructed properly, the sensitivity per unit measurement time is usually higher. The sensitivity of a sampling scheme relative to collecting the full matrix can be estimated based on prior knowledge of the signal decay rate. For an exponentially decaying signal, the relative sensitivity of a scheme with sampling function K spanning a two-dimensional grid with size n1 by n2 is given by
| (1) |
where the elements of p are given by
| (2) |
and R2(1) and R2(2) are the signal envelope decay rates, and SW1 and SW2 are the spectral widths in the two dimensions. Eq. (2) readily generalizes to other signal envelopes.
Artifacts: Bandwidth and aliasing
NUS introduces sampling artifacts, regardless of the method used to estimate the spectrum. Some spectral estimation methods are better than others at reducing the artifacts, but completely eliminating them is difficult, if not impossible, because of the presence of noise. In order to design an efficient sample schedule that minimizes artifacts, it helps to understand how features of a sample schedule translate into the PSF.
The Nyquist sampling theorem, defining the bandwidth in relation to the (uniform) sampling interval, does not hold for NUS; there is no well-defined bandwidth for NUS. NUS artifacts are thus a form of aliasing. They can be diminished by decreasing the greatest common divisor (GCD) of the sampled times, as pointed out by Bretthorst31. The GCD need not correspond to the spacing of the underlying grid. Introducing irregularity is one way to ensure that the GCD is equivalent to the grid spacing, and this helps to explain the usefulness of randomness for reducing artifacts from nonuniform sampling schemes32. The incorporation of randomness can suppress artifacts in otherwise regular sampling schemes, such as radial sampling. Another way to increase the effective bandwidth is to sample from an oversampled grid. When employed with NUS, oversampling has the effect of shifting sampling artifacts out of the original spectral window33.
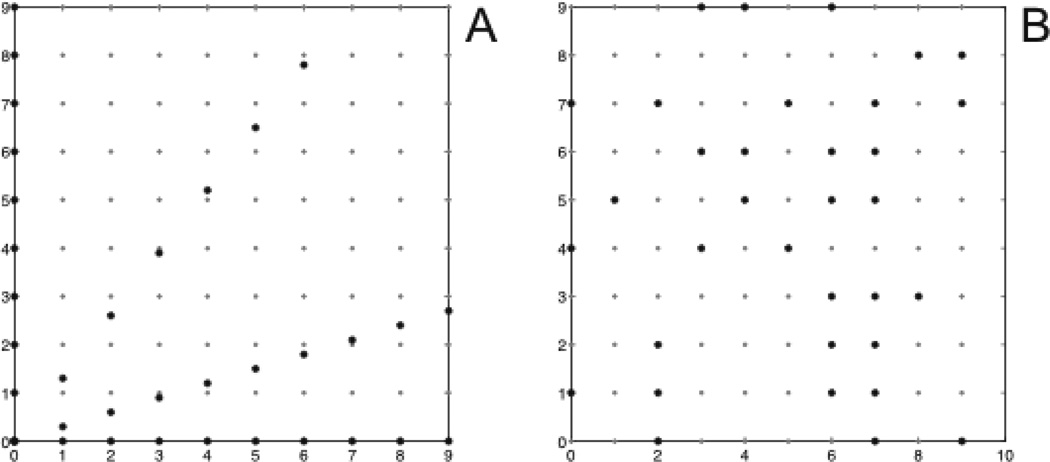
Sampling strategies
Broadly, NUS strategies are described as on-grid or off-grid (Fig. 2). On-grid sampling schemes employ uniform sampling intervals along a given time dimension, and thus amount to selecting a subset from a regular or Cartesian multidimensional grid of sampling intervals. Collecting all the samples of this grid corresponds to uniform or linear sampling, as employed by conventional Fourier-based approaches. The fraction of samples from the full grid employed by a NUS scheme is called the coverage. Radial sampling schemes, which sample along radial vectors in time emanating from the time origin, are usually described as being off-grid, but they can be approximated by on-grid schemes34. Bretthorst has pointed out that in practice all sampling schemes, including radial, are inherently on-grid because of the finite precision of timings used in NMR spectrometers31.
Figure 2.
Examples of off-grid (A) and on-grid (B) sampling schemes. Panel A depicts radial sampling, employed by accordion, RD, GFT, BPR, HIFI, and APSY experiments. Random sampling schemes are typically restricted to a subset of the uniform grid defined by the Nyquist sampling interval in each dimension.
The most widely-used NUS strategies in multidimensional NMR employ radial sampling or some variant of random sampling (Fig. 1). Approaches that utilize radial sampling include BPR, reduced dimensionality (RD), GFT, high-resolution iterative frequency identification (HIFI, (35)), and automated projection spectroscopy (APSY, (36)). Among this group only BPR reconstructs the fully-dimensional spectrum, whereas methods such as APSY and HIFI analyze projected sub-spectra (which can be computed using conventional Fourier-based methods) and use knowledge of the projection angle (and the associated frequency shifts) to extract the true frequencies. Random methods, in contrast, typically utilize on-grid sampling and non-Fourier methods of spectrum analysis to reconstruct the fully-dimensional spectrum. An advantage of the later approach is the results are isomorphic with spectra obtained by conventional methods, and conventional tools can be used to analyze the spectra. APSY and HIFI utilize special analysis tools.
In the first general application of NUS in multidimensional NMR, Laue et al. used a biased random sampling scheme8. By analogy with matched filter apodization (which was first applied in NMR by Ernst, and maximizes the S/N of the uniformly-sampled DFT spectrum), Laue and colleagues reasoned that tailoring NUS so that the signal is sampled more frequently at short times, where the signal is strong, and less frequently when the signal is weak, would similarly improve S/N. They applied an exponential bias to match the decay rate of the signal envelope; we refer to this as envelope-matched sampling (EMS). Generalizations of the approach to sine-modulated signals, where the signal is small at the beginning, and constant-time experiments, where the signal envelope does not decay, were described by Schmieder et al.35, 36.
Hyberts and Wagner37 noted empirically that the distribution of the gaps in a sampling schedule are also important. Long gaps near the beginning or end of a sample schedule were particularly detrimental. They adapted an idea employed in computer graphics, Poisson gap sampling, to generate sampling schedules that avoid long gaps while ensuring the samples are randomly distributed. Similar distributions can be generated using other approaches, for example quasi-random (e.g. Sobolev38) sequences. In addition to being robust, Poisson gap sampling schedules show less variation with the random deviate than other sampling schemes. A potential weakness of Poisson gap sampling, however, is that the minimum distance between samples must not be too small, otherwise aliasing can become significant.
Methods such as HIFI and APSY employ knowledge about chemical shift distributions in proteins to select radial vectors to minimize the likelihood of overlap in the projected cross-section. The idea of using prior knowledge about expected frequencies can be applied to random sampling by extending the concept behind EMS to consider finer details of the signal39. For example, a signal containing two strong frequency components will exhibit beats in the time domain signal separated by the reciprocal of the frequency difference between the components. As the signal becomes more complex, with more frequency components, more beats will occur corresponding to frequency differences between the various components. If one knows a priori the expected frequencies of the signal components, one can predict the location of the beats (and nulls, or zero-crossings), and tailor sampling accordingly. The procedure is entirely analogous to EMS, except that the sampling density is matched to the fine detail of predicted time-domain data, not just the signal envelope. We refer to this approach as beat-matched sampling (BMS). Possible applications where the frequencies are known a priori include relaxation experiments or multidimensional experiments in which scout scans or complementary experiments provide knowledge of the frequencies. In practice, BMS sampling schedules appear similar to EMS (e.g. exponentially biased) schedules, however they tend not to be as robust, as small difference in noise level or small frequency shifts can have pronounced effects on the location of beats or nulls in the signal.
Other deterministic sampling schemes have been explored. Delsuc and colleagues employed triangular sampling in two time dimensions to capture the strongest part of a two-dimensional signal40. Coggins and Zhou introduced the concept of concentric ring sampling (CRS), and showed that radial sampling is a special case of CRS41. They showed that the DFT could be adapted to CRS (and radial sampling) by changing to polar coordinates from Cartesian coordinates (essentially by introducing the Jacobian for the coordinate transformation as weighting factors). Optimized CRS that linearly increases the number of samples in a ring as the radius increases and incorporates randomness were shown to provide resolution comparable to uniform sampling for the same measurement time, but with fewer sampling artifacts than radial sampling.
Random phase detection
In most applications of NUS, quadrature detection (either States-Haberkorn-Ruben42 or time-proportional phase incrementation (TPPI43) is used to determine the sign of frequency of signal components. Either approach incurs a factor of two sampling burden, relative to single-phase detection, just to disambiguate the sign. However single-phase detection using uniform sampling in time but with random phase (random phase detection, RPD44) is able to resolve the frequency sign ambiguity without oversampling. This results in a factor of two reduction in the number of samples required, compared to quadrature or TPPI detection methods, for each indirect dimension of a multidimensional experiment. For experiments not employing quadrature or TPPI detection, it provides a factor of two increase in resolution for each dimension. The relationship among the PSF, the uniformly sampled spectrum, and the zero-augmented DFT is more complicated than the depiction given in Fig. 1, because a separate real-valued sampling function is required for each quadrature component. Combination of conventional NUS approaches with RPD is feasible, and affords additional flexibility in the design of efficient NUS schemes.
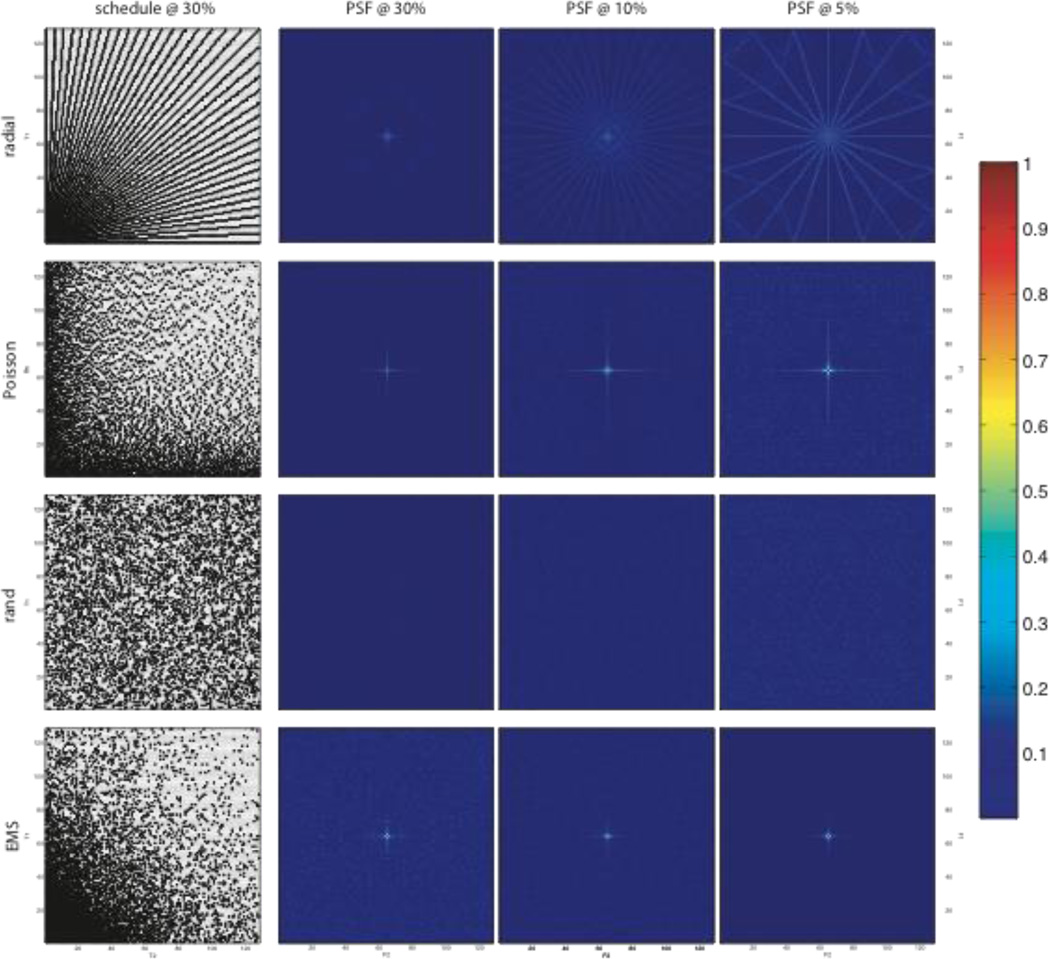
Performance
While the efficacy of a particular sampling scheme depends on a host of factors, including the nature of the signal being sampled, the PSF provides a useful first-order tool for comparing sampling schedules. PSFs that exhibit the largest peak-to-sidelobe ratio (PSR, which can be measured peak-to-peak or peak-to-rms) will give rise to the smallest sampling artifacts. Figure 3 illustrates examples of several common two-dimensional NUS schemes, together with PSFs computed for varying levels of coverage (30%, 10%, and 5%) of the underlying uniform grid. Some of the schemes are off-grid schemes, but they are approximated here by mapping onto a uniform grid. (As noted previously, on-grid approximation of off-grid sampling schemes coupled with reconstruction methods such as MaxEnt gives results that are very similar of off-grid sampling.) For a given sampling scheme, the PSR increases with sampling coverage.
Figure 3.
Examples of NUS sampling functions and PSFs in two nonuniformly sampled dimensions. Metrics reflecting the relative performance of these sampling schemes are given in Table 1. Purely random sampling (third row) yields the smallest sampling artifacts for a given level of coverage.
The PSF alone does not tell the whole story, because it does not address relative sensitivity. For example, while the random schedule has a PSF with very weak side-lobes, and gives rise to fewer artifacts than a radial sampling scheme for the same level of coverage, it has lower sensitivity for exponentially decaying sinusoids than a radial scheme (which concentrates more samples at short evolution times where the signal is strongest). NUS approaches always reduce sensitivity compared to uniform sampling that encompass the same maximum evolution times, both because sampling artifacts behave like additional noise, and because fewer samples are collected with NUS. Sensitivity per unit measurement time can be greater for NUS than with uniform sampling, however, when the sampling density is biased to capture more of the signal energy, e.g. at short evolution times for decaying signals.
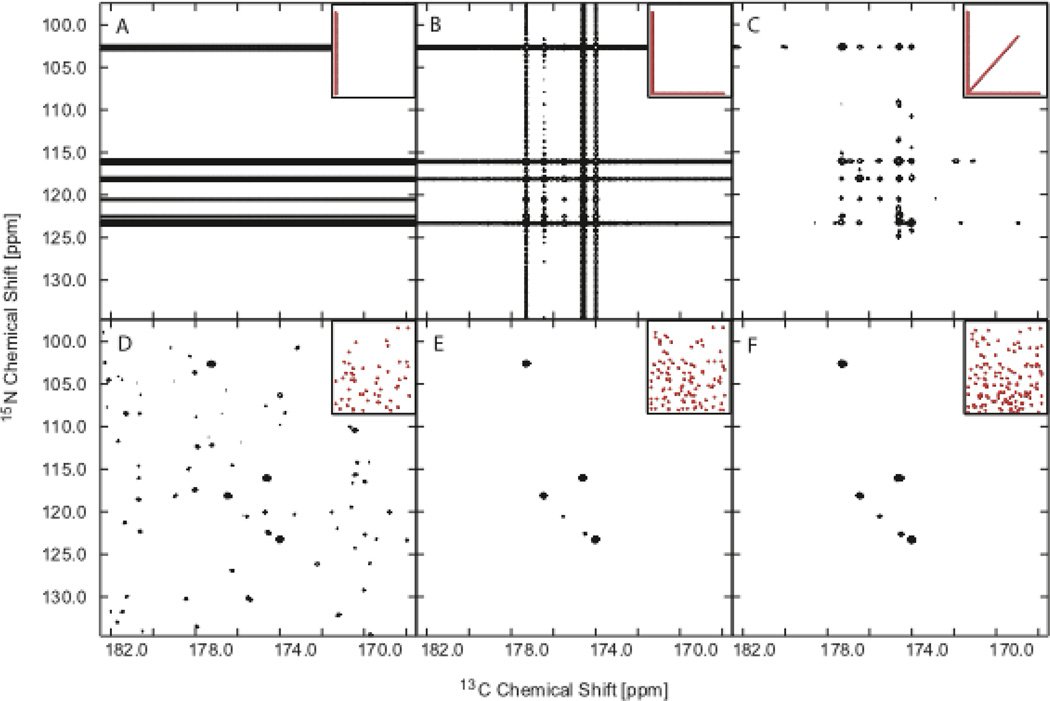
A fair test of the relative performance of two NUS schemes is their application to the same sample, keeping the number of samples fixed (total measuring time). Figure 4 shows results using radial sampling (top row of Fig. 3) compared with EMS (bottom row of Fig. 3 in two indirect dimensions of a 3D HNCA experiment for the protein Ubiquitin. The sampling distributions are shown in the insets in Fig. 4. The results for increasing sample size corresponding to one, two, and three radial sampling vectors (left to right, top row) and the matching sample size using EMS (bottom row) vividly illustrate the general principle that less regular sampling strategies more efficiently suppress sampling artifacts (consistent with the PSFs shown in Fig. 3).
Figure 4.
Influence of coverage and sampling scheme on NUS HNCO spectra of ubiquitin. All six panels depict the f3 (1H) plane of the spectrum corresponding to 8.14 ppm. For each panel the sampling scheme in t1-t2 is depicted in the red inset at the upper right. Top panels (A, B, C) show the addition of 0°, 90° and 30° projections of the two jointly sampled indirect dimensions, reconstructed using BPR. Each projection contains 52 complex points, thus the total number of complex points sampled in panels A, B, and C is 52, 104, and 156, respectively. The lower panels show MaxEnt reconstructions using the same sampling coverage, but distributed differently: randomly along the 15N dimension (constant time) and with an exponentially decreasing sampling density corresponding to a 15 Hz decay rate in the 13C dimension. The MaxEnt reconstruction parameters were selected using an automated protocol49, 50.
An important characteristic of the non-Fourier methods of spectrum analysis that are used with NUS is that they are all nonlinear. The nonlinearities manifest to different degrees and in different ways, depending on the method. A general consequence of nonlinearity is signal-to-noise ratio (SNR) is no longer a reliable proxy for sensitivity45, and more robust measures of sensitivity such as weak peak detection and false peak rejection – sometimes categorized as the false discovery rate – need to be employed. Sensitivity, after all, is the ability to distinguish signals from noise or artifact.
Similarly, resolution using NUS depends on a host of factors, including the method used to estimate the spectrum, but it correlates strongly with the average evolution time for the sampling scheme46.
Future prospects
The use of nonuniform sampling in all its guises is transforming the practice of multidimensional NMR, most importantly by lifting the sampling limited obstacle to obtaining high resolution along the indirect dimensions. Nonuniform sampling is also beginning to have tremendous impact in magnetic resonance imaging, where even small reductions in the time required to collect an image can have tremendous clinical impact. For all of the successes using NUS, our understanding of how to design optimal sampling schemes remains incomplete. A major limitation is that we lack a comprehensive theory able to predict the performance of a given NUS scheme a priori. This in turn is related to the absence of a consensus on performance metrics, i.e., measures of spectral quality. Further advances in NUS will be enabled by the development of robust, shared metrics. An additional hurdle has been the absence of a common set of test or reference data, which is necessary for critical comparison of competing approaches. Once shared metrics and reference data are established, we anticipate an acceleration of the pace of improvements in the design and application of NUS to multidimensional NMR spectroscopy. This development will help to expand the range of systems amenable to investigation by multidimensional NMR spectroscopy. As many multidimensional spectroscopic techniques first applied in NMR have migrated to other forms of spectroscopy (e.g. EPR47 and IR48), we anticipate that NUS similarly will find wider application.
Acknowledgements
We thank Gerhard Wagner and Sven Hyberts for providing the Poisson gap sampling schedules used in Figure 3, and for helpful discussions. JCH gratefully acknowledges support from the US National Institutes of Health (grants GM047467 and RR020125).
References
- 1.Ernst RR, Anderson WA. Rev. Sci. Instrum. 1966;37:93–102. [Google Scholar]
- 2.Jeener J. Oral Presentation. Basko Polje Yugoslavia: Ampere International Summer School; 1971. [Google Scholar]
- 3.Ni F, Scheraga HA. J. Magn. Reson. 1986;70:506–511. [Google Scholar]
- 4.Stern AS, Li K-B, Hoch JC. J. Am. Chem. Soc. 2002;124:1982–1993. doi: 10.1021/ja011669o. [DOI] [PubMed] [Google Scholar]
- 5.Mobli M, Hoch JC, King GF. In: Advances in Biomedical spectroscopy. Dingley AJ, Pascal SM, editors. Amsterdam: IOS Press; 2011. [Google Scholar]
- 6.Bodenhausen G, Ernst RR. J Magn Reson. 1981;45:367–373. doi: 10.1016/j.jmr.2011.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Ding K, Gronenborn AM. J. Magn. Reson. 2002;156:262–268. doi: 10.1006/jmre.2002.2537. [DOI] [PubMed] [Google Scholar]
- 8.Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP. J. Magn. Reson. 1987;73:69–77. [Google Scholar]
- 9.Aggarwal K, Delsuc M-A. Magn. Reson. Chem. 1997;35:593–596. [Google Scholar]
- 10.Barna JCJ, Tan SM, Laue ED. J. Magn. Reson. 1988;78:327. [Google Scholar]
- 11.Kupče E, Freeman R. J. Biomol. NMR. 2003;27:101–113. doi: 10.1023/a:1024960302926. [DOI] [PubMed] [Google Scholar]
- 12.Kupče E, Freeman R. J. Am. Chem. Soc. 2003;125:13958–13959. doi: 10.1021/ja038297z. [DOI] [PubMed] [Google Scholar]
- 13.Kupče E, Freeman R. J. Biomol. NMR. 2003;25:349–354. doi: 10.1023/a:1023066715071. [DOI] [PubMed] [Google Scholar]
- 14.Kupče E, Freeman R. J. Amer. Chem. Soc. 2004;126:6429–6440. doi: 10.1021/ja049432q. [DOI] [PubMed] [Google Scholar]
- 15.Kupče E, Freeman R. J Magn Reson. 2005;173:317–321. doi: 10.1016/j.jmr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Dowd M. New York: New York Times; 2011. [Google Scholar]
- 17.Chylla RA, Markley JL. J. Biomol. NMR. 1995;5:245–258. doi: 10.1007/BF00211752. [DOI] [PubMed] [Google Scholar]
- 18.Bretthorst GL. J. Magn. Reson. 1990;88:533–551. [Google Scholar]
- 19.Barna JCJ, Tan SM, Laue ED. J. Magn. Reson. 1988;78:327–332. [Google Scholar]
- 20.Kim S, Szyperski T. J. Amer. Chem. Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- 21.Coggins BE, Zhou P. J Magn Reson. 2006;182:84–95. doi: 10.1016/j.jmr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 22.Stern AS, Donoho DL, Hoch JC. J Magn Reson. 2007;188:295–300. doi: 10.1016/j.jmr.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuki Y, Eddy MT, Herzfeld J. J Am Chem Soc. 2009;131:4648–4656. doi: 10.1021/ja807893k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandelshtam VA, Taylor HS, Shaka AJ. J. Magn.Reson. 1998;133:304–312. doi: 10.1006/jmre.1998.1476. [DOI] [PubMed] [Google Scholar]
- 25.Barkhuijsen H, Beer Rd, Bovee WM, Creyghton JH, Ormondt Dv. Magn. Reson. Med. 1985;2:86–89. doi: 10.1002/mrm.1910020111. [DOI] [PubMed] [Google Scholar]
- 26.Holland DJ, Bostock MJ, Gladden LF, Nietlispach D. Angew Chem Int Ed Engl. 2011;50:6548–6551. doi: 10.1002/anie.201100440. [DOI] [PubMed] [Google Scholar]
- 27.Kazimierczuk K, Orekhov VY. Angew Chem Int Ed Engl. 2011;50:5556–5559. doi: 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- 28.Lustig M, Donoho D, Pauly JM. Magn Reson Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 29.Donoho DL. IEEE Trans. Inf. Theory. 2006;52:1289–1306. [Google Scholar]
- 30.Rovnyak D, Hoch JC, Stern AS, Wagner G. J Biomol NMR. 2004;30:1–10. doi: 10.1023/B:JNMR.0000042946.04002.19. [DOI] [PubMed] [Google Scholar]
- 31.Bretthorst GL. Concepts Magn. Reson. 2008;32A:417–435. [Google Scholar]
- 32.Hoch JC, Maciejewski MW, Filipovic B. J Magn Reson. 2008;193:317–320. doi: 10.1016/j.jmr.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maciejewski MW, Qui HZ, Rujan I, Mobli M, Hoch JC. J Magn Reson. 2009 doi: 10.1016/j.jmr.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mobli M, Stern AS, Hoch JC. J Magn Reson. 2006;182:96–105. doi: 10.1016/j.jmr.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Schmieder P, Stern AS, Wagner G, Hoch JC. J. Biomol. NMR. 1993;3:569–576. doi: 10.1007/BF00174610. [DOI] [PubMed] [Google Scholar]
- 36.Schmieder P, Stern AS, Wagner G, Hoch JC. J. Biomol. NMR. 1994;4:483–490. doi: 10.1007/BF00156615. [DOI] [PubMed] [Google Scholar]
- 37.Hyberts SG, Takeuchi K, Wagner G. J Am Chem Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in Fortran: The Art of Scientific Programming. Cambridge, England: Cambridge University Press; 1992. pp. 299–306. [Google Scholar]
- 39.Schuyler AD, Maciejewski MW, Arthanari H, Hoch JC. J Biomol NMR. 50:247–262. doi: 10.1007/s10858-011-9512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aggarwal K, Delsuc MA. Magn. Reson. Chem. 1997;35:593–596. [Google Scholar]
- 41.Coggins BE, Zhou P. J Magn Reson. 2007;184:207–221. doi: 10.1016/j.jmr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.States DJ, Haberkorn RA, Ruben DJ. J. Magn.Reson. 1982;48:286–292. [Google Scholar]
- 43.Marion D, Wüthrich K. Biochem. Biophys. Res. Comm. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 44.Maciejewski MW, Fenwick M, Schuyler AD, Stern AS, Gorbatyuk V, Hoch JC. Proc Natl Acad Sci U S A. 108:16640–16644. doi: 10.1073/pnas.1103723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donoho DL, Johnstone IM, Stern AS, Hoch JC. Proc Natl Acad Sci U S A. 1990;87:5066–5068. doi: 10.1073/pnas.87.13.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuyler AD, Maciejewski MW, Arthanari H, Hoch JC. J Biomol NMR. 2011;50:247–262. doi: 10.1007/s10858-011-9512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweiger A, Gunnar J. Principles of pulse electron paramagnetic resonance. New York: Oxford University Press; 2001. [Google Scholar]
- 48.Wright JC. Int. Rev. Phys. Chem. 2002;21:185–255. [Google Scholar]
- 49.Mobli M, Maciejewski MW, Gryk MR, Hoch JC. Nat Methods. 2007;4:467–468. doi: 10.1038/nmeth0607-467. [DOI] [PubMed] [Google Scholar]
- 50.Mobli M, Maciejewski MW, Gryk MR, Hoch JC. J Biomol NMR. 2007;39:133–139. doi: 10.1007/s10858-007-9180-8. [DOI] [PubMed] [Google Scholar]