Abstract
Chronic periodontitis (CP) is an inflammatory disease of the teeth-supporting tissues in which genetic predisposition, dental plaque bacteria, and immune mechanisms all play important roles. The aim of this study was to evaluate the occurrence of IL-4 gene polymorphisms in chronic periodontitis and to investigate the association between polymorphisms and cytokines production after bacterial stimulation. Sixty-two subjects (47 CP patients and 15 healthy controls) with detected two polymorphisms in the IL-4 gene (-590C/T and intron 3 VNTR) were examined. Production of cytokines (IL-1α, IL-1β, IL-4, IL-5, IL-6, IL-10, IL-17, TNFα, INFγ, and VEGF) was studied after in vitro stimulation of isolated peripheral blood by mitogens (Pokeweed mitogen, Concanavalin A), dental plaque bacteria (Aggregatibacter actinomycetemcomitans, Tannerella forsythia, Porphyromonas gingivalis, and Prevotella intermedia), and Heat Shock Protein (HSP) 70 by the Luminex multiplex cytokine analysis system. The results were correlated with IL-4 genotypes in patients with CP and healthy controls. The mononuclear cells isolated from peripheral blood of CP patients with selected IL-4 polymorphisms significantly altered the production of IFNγ, IL-10, IL-1β, IL-1α, TNFα, and IL-6 after stimulation by HSP 70 or selected bacteria (from P < 0.001 to P < 0.05). IL-4 gene polymorphisms may influence the function of mononuclear cells to produce not only interleukin-4 but also other cytokines, especially in patients with CP.
1. Introduction
Chronic periodontitis (CP) is an inflammatory disease of the teeth-supporting tissues in which genetic predisposition, dental plaque bacteria, and immune mechanisms all play important roles. Bacteria which are involved in the pathogenesis of CP include Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, Tannerella forsythia, Prevotella intermedia, and Fusobacterium nucleatum. The participation of these bacteria in the pathogenesis of CP correlates with an increased level of specific antibodies in sera [1]. Suspect pathogens isolated from dental plaque continually stimulate cells in the periodontal tissue to produce cytokines as well as other biologically active molecules. The development and regulation of the immune response depend on the local production and quantities of cytokines and other mediators which influence disease progression or resolution. The disease progression depends on the increased production of proinflammatory cytokines (IL-1α, IL-1β, IL-6, IL-8, and TNFα), metalloproteinases, and prostaglandins or decreased production of anti-inflammatory cytokines (IL-10, TGFβ) and inhibitors of metalloproteinases [2].
Based on the concept of different functions of TH1 and TH2 lymphocytes [3], Seymour et al. [4] proposed the hypothesis that, in patients with progressing CP, clones of TH2 are generated upon activation with bacteria, while, in the nonprogressing disease, stable lesions are regulated by TH1 clones. We confirmed these findings in our previous longitudinal studies in which we followed, for a period of 10 years, patients with early onset periodontitis (EOP, now known as aggressive periodontitis) with changes in periodontal tissues diagnosed between the ages of 17 and 25. Such patients repeatedly exhibited increased production of IL-4 (TH2) upon stimulation of peripheral blood lymphocytes with dental plaque bacteria while their healthy siblings and other genetically unrelated controls with intact periodontal tissues produced a significantly increased level of IFNγ (TH1) [5]. As mentioned above, both the onset and progression of periodontal diseases are strongly influenced by the genetic predisposition of the individual. The heritability of the disease varies from a nearly 100% share in “Mendelian/syndromological” forms to states with a low proportion of heritability and a high effect of environmental factors. Rateitschak et al. [6] described the genetic predisposition for early onset periodontitis; according to Michalowicz [7], genes are responsible for more than 50% of the risk of chronic periodontitis. The 1990s witnessed the beginning of the phase of genetic analysis at the level of so-called “candidate” genes, that is, genes mostly coding proinflammatory cytokines, chemokines, metalloproteinases, and other factors associated with the production of these mediators and their presumed role in the pathogenesis of the disease [8, 9]. The current development of methodological options underlies the present era of genome-wide studies [10]. Polymorphisms in certain alleles of cytokines have been associated with susceptibility to a wide range of infectious or immune diseases, including periodontitis [11–16] and others. The human interleukin-4 (IL-4) gene is mapped within the cytokine gene cluster on chromosome 5q31-33 and contains several polymorphisms; some of them are implicated in the regulation of IL-4 production. Recently, these polymorphisms have attracted widespread attention, especially the IL-4 gene promoter -590C/T (rs2243250) polymorphism and a 70 bp variable number of tandem repeat (VNTR) polymorphism in its third intron. It was proposed that an increased responsiveness of the -590C/T allele and the (70 base pairs (bp)) 2 repeat allele to transcriptional activation might lead to overexpression of IL-4 [17].
However, the role of these polymorphisms in cytokine genes in the production of cytokines in response to stimulation by dental plaque bacteria in periodontal disease has not yet been studied. The aim of this study was to assess the influence of IL-4 (-590C/T) and IL-4 VNTR (variable number of tandem repetitions in intron 3 gene) polymorphisms on cytokine production after stimulation of isolated peripheral blood mononuclear cells by dental plaque bacteria, mitogens, and HSP70 in patients with periodontitis and in healthy controls.
2. Materials and Methods
2.1. Study Population
All patients with chronic periodontitis (CP, N = 47) were recruited from the patient pool of the Periodontology Department, Clinic of Stomatology, St. Anne's Faculty Hospital Brno, from 2010 to 2012. Inclusion criteria were good general health, diagnosis of generalized chronic periodontitis according to the International Workshop for a Classification of Periodontal Diseases and Conditions for Chronic Periodontitis [18], and agreement with sample collection for genetic/immunological examinations. Exclusion criteria included history of cardiovascular disorders (such as coronary artery diseases or hypertension), diabetes mellitus, malignant diseases, immunodeficiency, current pregnancy or lactation, and smoking. The control group (healthy/nonperiodontitis subjects, N = 15) were selected randomly during the same period as patients and matched for age, gender, and nonsmoking status. All controls had at least 20 remaining teeth, were general healthy, and agree with genetic/immunological examinations. Exclusion criteria were the same as those applied with patients with periodontitis.
Diagnosis of nonperiodontitis/periodontitis was based on a detailed clinical examination, medical and dental history, tooth mobility, and radiographic assessment. Probing depth (PD) and clinical attachment loss (CAL) were collected by UNC-15 periodontal probe from six sites on every tooth present. All patients had to have at least three teeth with bleeding on probing (BOP), PD of >4 mm, and CAL of ≥3 mm in all quadrants (excluding the third molars); however, they were treated patients without active phase of disease. The loss of alveolar bone was determined radiographically. We used the Mühlemann index to evaluate decreases in alveolar bone levels [19]. The control group (healthy periodontium) consisted of subjects with no history or clinical signs of gingivitis and/or periodontitis (no PPD of >4 mm, no loss of clinical attachment around any tooth, and no radiographic sign of bone resorption).
The study was performed with the approval of the Committees for Ethics of the Medical Faculty, Masaryk University Brno and St. Anne's Faculty Hospital. Written informed consent was obtained from all participants in line with the Helsinki declaration before inclusion in the study.
2.2. Isolation of Genomic DNA
DNA for genetic analysis was extracted from the peripheral blood leukocytes (5 mL) using standard phenol/chloroform procedures with proteinase K according to Sambrook et al. [20].
2.3. Genetic Analysis
The GenBank accession number was AF395008 for the reference genomic sequence used for IL-4 alignments. The IL-4 SNP polymorphism at position -590C/T (rs2243250) was genotyped by PCR-RFLP analysis according to previously published methods [21] with slight modification. DNA was amplified with the following primers: [5′-ACT AGG CCT CAC CTG ATA CG-3′ (sense), 5′-AGG TGT CGA TTT GCA GTG AC-3′ (antisense)] as a product with 646 bp length. PCR was carried out in a volume of 15 μL containing 50 ng of genomic DNA, 0.3 μM of each primer, 0.5 U of DNA polymerase (Biotools B&M Labs S. A., Madrid, Spain), 5 mM of MgCl2, 10x reaction buffer MgCl2 free (Biotools B&M Labs S. A., Madrid, Spain), and 0.5 mM of deoxyribonucleoside triphosphate mix (Roche, Basel, Switzerland). Denaturation for 5 min at 95°C was followed by 35 cycles of 95°C for 45 s, 56°C for 45 s, and 72°C for 1 min. The last synthesis step was extended to 10 min at 72°C. The PCR products were then digested with BsmFI restriction enzyme and separated on 3% agarose gel. The restriction was performed in a volume of 12 μL containing 7 μL of the PCR product, 10x NeBuffer 4, and 4 U of BsmFI enzyme (New England Biolabs, Hitchin, UK) and incubated for 4 hours at 65°C. Genotypes were described as CC (601 + 45 bp), CT (646 + 601 + 45 bp), and TT (646 bp).
VNTR (IL-4 70-bp repeat) polymorphism in intron 3 of the IL-4 gene was detected by a modification of the PCR method described by Mout et al. [22]. DNA was amplified with the following primers: [5′-TAG GCT GAA AGG GGG AAA GC-3′ (sense), 5′-CTG TTC ACC TCA ACT GCT CC-3′ (antisense)]. PCR was carried out in a volume of 15 μL containing 100 ng of genomic DNA, 0.3 μM of each primer, 0.5 U of DNA polymerase (Biotools B&M Labs S.A., Madrid, Spain), 3.5 mM of MgCl2, 10x reaction buffer MgCl2 free (Biotools B&M Labs S.A., Madrid, Spain), and 0.5 mM of deoxyribonucleoside triphosphate mix (Roche, Basel, Switzerland). Denaturation for 5 min at 95°C was followed by 35 cycles of 95°C for 45 s, 56°C for 45 s, and 72°C for 1 min. The last synthesis step was extended to 10 min at 72°C. Amplified products were separated on 2% agarose gel; the size of the product corresponds to the number of the repeats present (allele 1 = 3 repeats (254 bp); allele 2 = 2 repeats (184 bp)). The rarely occurring allele type 3 with only a single repeat (114 bp) was not present in our set.
The genotyping was performed by P. B. L. unaware of the phenotype. DNA samples with known genotypes, which were identified by sequencing of these samples in the previous study by Holla et al. [14], were used as positive controls for both polymorphisms.
2.4. Cultivation of Dental Plaque Bacteria
Samples of the sulcular fluid for the cultivation of bacteria (A. actinomycetemcomitans, P. gingivalis, T. forsythia, and P. intermedia) were collected from sites of periodontium with the worst affected areas of periodontium. A volume of 50 μL of a sample diluted by 2 mL Brain Heart medium was inoculated on the surface of microbiological diagnostic media in Petri dishes. Selective cultivation of bacteria was performed according to the method of Slots [23] on TSBV agar incubated at 37°C in 10% CO2 atmosphere created by GasPak CO2 bags (Oxoid) in an anaerobic pot. Bacteria colonies were harvested after 5 to 7 days and bacterial suspension containing 1010 cells/mL was inactivated by incubation at 120°C for 20 minutes. Samples were centrifuged for 10 minutes at 600 g. Sediments were diluted with PBS and centrifuged for 10 minutes at 600 g and then washed 3 times. The washed sediments were diluted by X-Vivo medium to optical density corresponding to 1010 cells/mL.
2.5. Immunological Examination
20 mL of peripheral blood was collected from the subjects under examination to BD Vacutainer with sodium heparin as anticoagulant (BD, Plymouth, UK). Peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood by Histopaque 1.077 (Sigma, St. Louis, MO, USA) gradient centrifugation for 30 min at 600 g. The white layer in the interphase containing mononuclear cells was collected and washed with X-vivo 10 medium at 400 g and 200 g and then diluted with X-Vivo 10 serum-free tissue medium (Cambrex) to the concentration of 107 cells/mL. 100 μL cells were cultivated for 3 days at 37°C in 5% CO2 atmosphere and stimulated with 100 μL of the bacteria with concentration of 109/mL (A. actinomycetemcomitans, P. gingivalis, T. forsythia, and P. intermedia), HSP 70 (Sigma, 5 μg/mL), Pokeweed mitogen (PWM, Sigma, 2.5 μg/mL), and Pokeweed mitogen in costimulation with Concanavalin A (PWM, Sigma, 2.5 μg/mL + ConA, Sigma, 10 μg/mL) and supplemented with X-vivo 10 medium to l mL (Table 2). After the 3 days of cultivation, the medium was collected and stored at −20°C for the determination of cytokines (IL-1α, IL-1β, IL-4, IL-5, IL-6, IL-10, IL-17, IFNγ, TNFα, and VEGF) by the Luminex multiplex method (LUMINEX 100TM analyzer, R&D Systems, USA). Cytokine production in subjects with different genotypes of IL-4 polymorphism was then compared.
Table 2.
Scheme of in vitro stimulation of mononuclear cells.
| Cells (107 cells/mL) |
Stimulation | Cultivation X-Vivo medium |
|
|---|---|---|---|
| PWM | 100 µL | 100 µL PWM | 800 µL |
| PWM + ConA | 100 µL | 100 µL PWM + 100 µL ConA | 700 µL |
| HSP70 | 100 µL | 100 µL | 800 µL |
| A.a. | 100 µL | 100 µL A.a. | 800 µL |
| P.g. | 100 µL | 100 µL P.g. | 800 µL |
| T.f. | 100 µL | 100 µL T.f. | 800 µL |
| Unstim. | 100 µL | 900 µL |
PWM: Pokeweed mitogen, ConA: Concanavalin A, HSP70: Heat Shock Protein 70, A.a.: Aggregatibacter actinomycetemcomitans, P.g.: Porphyromonas gingivalis, T.f.: Tannerella forsythia, and unstim.: without stimulation.
2.6. Statistical Analysis
Statistically significant results (for P < 0.05, P < 0.01, and P < 0.001) are indicated in Tables 3(a) and 3(b) representing medians for dichotomous variants, separately for patients with periodontitis (N = 47) and for healthy controls (N = 15). Calculations were performed in SPSS (version 17.0).
Table 3.
Sample medians for cytokine levels expressed by mitogens (PWM, P + C), by HSP70, and by selected bacteria in comparison to patients with periodontitis (N = 47) and healthy controls (N = 15) in division to groups by IL-4 genotypes (polymorphisms).
(a).
| Locus | Periodontitis patients | Healthy controls | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-4-590 | IL-4VNTR | IL-4-590 | IL-4VNTR | |||||||||||||
| Polymorphism | CC | CT + TT | 11 | 12 + 22 | CC | CT + TT | 11 | 12 + 22 | ||||||||
| (Number of patients) | (30) | (16 + 1) | (28) | (17 + 1) | (8) | (5 + 2) | (10) | (5 + 0) | ||||||||
| Cytokines [pg/mL] | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | median | (Min/max) | Median | (Min/max) | Median | (Min/max) |
| IFNg_PWM | 116.7 | (0/7816.5) | 180.6 | (0.8/3898.5) | 116.8 | (0.2/7816.5) | 147.6 | (0/3898.5) | 4.5 | (0.5/983.9) | 178.6 | (2.2/1073.7) | 4.5 | (0.5/983.9) | 178.6 | (2.2/1073.7) |
| IFNg_P + C | 92.8 | (0.3/7215) | 71.2 | (0.3/1280.4) | 113.6 | (0.6/7215) | 56.0 | (0.3/1280.4) | 126.9 | (1.1/1009.9) | 81.6 | (2/1081) | 126.9 | (1.1/1009.9) | 81.6 | (2/1081) |
| IFNg_HSP70 | 1.5 | (0/7282) | 0.3 | (0/781.9) | 1.1 | (0/7282) | 0.3 | (0/781.9) | 1.1 | (0.1/550.5) | 0.3 | (0.2/552.4) | 1.1 | (0.1/550.5) | 0.3 | (0.2/552.4) |
| IFNg_Pg | 0.9 | (0/633.3) | 1.6 | (0/9.2) | 0.7 | (0/633.3) | 2.1 | (0/9.2) | 0.6 | (0/4.8) | 0.4 | (0.2/1) | 0.5 | (0/4.8) | 0.4 | (0.2/1) |
| IFNg_Aa | 2.9 | (0/230.1) | 1.1 | (0/405.2) | 2.9 | (0/230.1) | 1.1 | (0/405.2) | 1.0 | (0/119.1) | 2.1 | (0.2/240) | 1.0 | (0/119.1) | 3.2 | (0.2/240) |
| IFNg_Tf | 2.6 # | (0.1/633.3) # | 0.4 | (0/9.9) | 2.1 * | (0.1/633.3) * | 0.4 | (0/9.9) | 1.6 | (0/89.4) | 0.3 | (0.1/99.5) | 0.4 | (0/89.4) | 0.4 | (0.1/99.5) |
| IFNg_PI | 0.8 | (0/238.7) | 0.2 | (0/1.9) | 0.8 | (0/238.7) | 0.3 | (0/2.2) | 0.7 | (0/61.1) | 0.2 | (0.1/809.9) | 0.3 | (0/61.1) | 0.2 | (0.1/809.9) |
| IFNg_unstim. | 0.7 | (0/21.9) | 0.1 | (0/8.6) | 0.6 | (0/21.9) | 0.1 | (0/8.6) | 0.0 | (0/91) | 0.1 | (0/7) | 0.0 | (0/91) | 0.1 | (0.1/0.4) |
|
| ||||||||||||||||
| IL-10_PWM | 63.5 | (0/3447) | 60.3 | (0/518.6) | 90.3 | (0/3447) | 55.1 | (0/518.6) | 76.0 | (0/1017) | 85.7 | (37.4/326) | 76.0 | (0/1017) | 180.7 | (37.4/326) |
| IL-10_P + C | 37.2 | (0/866.7) | 30.4 | (0/1732.9) | 41.0 | (0/866.7) | 27.6 | (0/1732.9) | 125.9 | (8.8/497.2) | 105.1 | (26.3/537.7) | 125.9 | (8.8/497.2) | 105.1 | (26.3/537.7) |
| IL-10_HSP70 | 5.1 | (0/581.3) | 1.7 | (0/271.7) | 5.6 | (0/581.3) | 3.2 | (0/271.7) | 9.7 | (1.3/250.3) | 10.3 | (3.9/215.4) | 10.4 | (1.3/250.3) | 4.8 | (3.9/215.4) |
| IL-10_Pg | 2.5 | (0/164.5) | 2.3 | (0/81.9) | 1.8 | (0/164.5) | 3.6 | (0/81.9) | 0.5 | (0/45.7) | 0.3 | (0.1/9) | 0.5 | (0/45.7) | 0.3 | (0.1/6.2) |
| IL-10_Aa | 28.8 | (0.8/823.6) | 25.2 | (0.1/589.9) | 36.8 | (0.8/823.6) | 24.0 | (0.1/589.9) | 26.6 | (11.2/303.1) | 58.2 | (4.4/663.1) | 26.6 | (4.4/303.1) | 92.5 | (10.7/663.1) |
| IL-10_Tf | 68.8† | (3.6/1510.4)† | 18.2 | (0.1/374.8) | 106.9† | (9.4/1510.4)† | 20.6 | (0.1/374.8) | 134.2 * | (4.3/320.1) * | 52.9 | (0.6/151.6) | 92.6 | (0.6/320.1) | 63.8 | (0.7/151.6) |
| IL-10_PI | 25.0 # | (0/207.2) # | 2.7 | (0/27.6) | 22.7 * | (0/207.2) * | 3.0 | (0/81.9) | 9.2 | (0/173.6) | 3.7 | (0.4/29.1) | 5.7 | (0/173.6) | 5.5 | (1.1/29.1) |
| IL-10_unstim. | 2.7 | (0/14.7) | 0.4 | (0/7.2) | 2.0 | (0/14.7) | 0.6 | (0/7.2) | 0.4 | (0/11) | 2.1 | (0.7/6.2) | 0.7 | (0/11) | 2.8 | (0.7/6.2) |
|
| ||||||||||||||||
| IL-17_PWM | 8.0 | (0/2200) | 15.0 | (5.1/1248.5) | 8.5 | (0.5/2200) | 13.5 | (0/1248.5) | 47.2 | (0.8/2064.1) | 127.0 | (2.3/947.3) | 47.2 | (0.8/2064.1) | 127.0 | (3/947.3) |
| IL-17_P + C | 34.4 | (1.1/2200) | 62.1 | (2.8/2200) | 50.2 | (1.1/2200) | 42.2 | (1.5/2200) | 239.2 | (2.8/2200) | 140.4 | (0.7/2376.8) | 239.2 | (0.7/2200) | 140.4 | (1.1/2376.8) |
| IL-17_HSP70 | 6.3 | (0/1107.5) | 1.2 | (0/968.6) | 4.0 | (0/1107.5) | 1.4 | (0/968.6) | 7.2 | (0.1/349.5) | 0.3 | (0.1/737.9) | 7.2 | (0.1/349.5) | 0.3 | (0.1/737.9) |
| IL-17_Pg | 1.1 | (0/543.1) | 0.4 | (0/12.9) | 1.0 | (0/543.1) | 1.2 | (0/12.9) | 0.1 | (0/6.9) | 0.3 | (0.1/0.6) | 0.1 | (0/6.9) | 0.3 | (0.1/0.4) |
| IL-17_Aa | 2.6 | (0.8/88.3) | 1.1 | (0.1/45.1) | 2.4 | (0.8/88.3) | 1.9 | (0.1/45.1) | 0.3 | (0/10.2) | 1.2 | (0/72.2) | 0.4 | (0/10.2) | 1.2 | (0/72.2) |
| IL-17_Tf | 1.6 | (0/2064.1) | 1.5 | (0/22.6) | 1.5 | (0/2064.1) | 2.1 | (0/22.6) | 0.7 | (0/11.6) | 0.4 | (0/168.9) | 0.3 | (0/11.6) | 1.4 | (0/168.9) |
| IL-17_PI | 1.5 | (0/62.1) | 0.0 | (0/6.3) | 1.5 | (0/62.1) | 0.1 | (0/6.3) | 0.4 | (0/10.2) | 0.4 | (0/2.2) | 0.3 | (0/10.2) | 1.5 | (0/2.2) |
| IL-17_unstim. | 1.2 | (0/5.1) | 0.5 | (0/6.3) | 1.0 | (0/5.1) | 0.8 | (0/6.3) | 0.2 | (0/2.8) | 0.3 | (0/22.7) | 0.2 | (0/22.7) | 0.3 | (0/2.5) |
|
| ||||||||||||||||
| IL-1 alfa_PWM | 141.3 | (0/2400) | 160.7 | (22/7780) | 152.0 | (0/2400) | 158.7 | (22/7780) | 231.3 | (0/967) | 258.7 | (112/774.1) | 197.1 | (0/967) | 412.2 | (212.2/774.1) |
| IL-1 alfa_P + C | 140.0 | (2.3/4701.5) | 124.6 | (3.6/1643.5) | 140.6 | (4.3/4701.5) | 124.2 | (2.3/1643.5) | 159.9 | (15.4/1153.3) | 247.1 | (121.9/436.8) | 159.9 | (15.4/1153.3) | 261.1 | (242.5/436.8) |
| IL-1 alfa_HSP70 | 32.8 | (0/1139.8) | 13.0 | (0/187.4) | 52.3 | (0/1139.8) | 13.0 | (0/187.4) | 24.1 | (6.4/398.9) | 124.0 * | (43.3/496.4) * | 31.1 | (6.4/398.9) | 124.0 | (43.3/496.4) |
| IL-1 alfa_Pg | 47.5 | (4.3/635.4) | 44.0 | (1.9/339) | 47.5 | (4.4/635.4) | 44.7 | (1.9/339) | 38.4 | (6.8/372.5) | 69.9 | (53.8/263.5) | 44.2 | (6.8/372.5) | 63.8 | (53.8/263.5) |
| IL-1 alfa_Aa | 97.2 | (11.2/724.2) | 99.3 | (2.8/1745.6) | 97.2 | (13.3/724.2) | 110.5 | (2.8/1745.6) | 129.7 | (13/238.7) | 350.3 * | (108.3/821.5) * | 133.5 | (13/238.7) | 405.3 * | (167.8/821.5) * |
| IL-1 alfa_Tf | 106.3 | (0/450.7) | 37.7 | (3.3/188.4) | 106.3 | (0/450.7) | 46.7 | (3.3/194.9) | 121.4 | (18.6/720.9) | 236.9 | (0.2/465.6) | 111.5 | (18.6/720.9) | 355.3 | (0.2/465.6) |
| IL-1 alfa_PI | 134.2 | (15.9/248.2) | 17.0 | (1.9/238.7) | 133.3 | (15.9/248.2) | 34.4 | (1.9/238.7) | 84.4 | (11.9/218.5) | 242.3 | (11.9/346.4) | 84.6 | (11.9/346.4) | 242.3 | (11.9/287) |
| IL-1 alfa_unstim. | 17.4 | (0/185.5) | 6.8 | (0/397.3) | 18.8 * | (0/185.5) * | 9.0 | (0/397.3) | 20.8 | (2.3/120.6) | 140.2 * | (5.9/308.8) * | 22.5 | (2.3/178.1) | 140.2 | (5.9/308.8) |
|
| ||||||||||||||||
| IL-1 beta_PWM | 583.3 | (0/2586) | 267.1 | (0/1597) | 655.9 * | (0/2586) * | 214.2 | (0/1597) | 676.4 | (2.6/1600) | 839.7 | (100.9/1700) | 676.4 | (2.6/1600) | 839.7 | (285.3/1700) |
| IL-1 beta_P + C | 533.3 | (1.3/2650) | 278.3 | (1.6/1600) | 600.2 * | (2.6/2650) * | 266.5 | (1.3/1600) | 695.2 | (47.2/1600) | 610.5 | (168.2/1657) | 571.9 | (47.2/1600) | 733.8 | (324.8/1657) |
| IL-1 beta_HSP70 | 141.5 | (0/1600) | 34.7 | (0/616.1) | 228.9 * | (0/1600) * | 34.7 | (0/616.1) | 93.6 | (44.4/1600) | 335.9 | (183.2/1338.1) | 143.4 | (44.4/1600) | 335.9 | (198.5/1338.1) |
| IL-1 beta_Pg | 33.2 | (0/851.4) | 13.8 | (0/600.4) | 33.2 | (0/851.4) | 16.3 | (0/600.4) | 22.9 | (3.7/106) | 8.2 | (2.4/289.7) | 22.9 | (2.9/289.7) | 8.2 | (2.4/189.5) |
| IL-1 beta_Aa | 533.3 | (0/4216.5) | 482.3 | (0/1600) | 533.3 * | (0/4216.5) * | 405.2 | (0/1600) | 439.5 | (47.7/1271.5) | 483.6 | (167.5/1531.1) | 439.5 | (47.7/1271.5) | 748.0 | (344.3/1531.1) |
| IL-1 beta_Tf | 516.8 * | (0/2952) * | 109.6 | (0/952.7) | 533.3 # | (0/2952) # | 101.8 | (0/952.7) | 521.6 | (47.7/1600) | 448.5 | (0/1700) | 479.4 | (47.7/1600) | 558.8 | (0/1700) |
| IL-1 beta_PI | 251.0 | (0/552.8) | 63.8 | (0/578.6) | 244.7 | (0/552.8) | 68.7 | (0/578.6) | 302.9 | (145.5/985) | 342.0 | (44.4/748.2) | 302.9 | (145.5/985) | 390.8 | (44.4/748.2) |
| IL-1 beta_unstim. | 83.5 * | (0/968.5) * | 21.9 | (0/533.3) | 91.8 * | (0/968.5) * | 16.6 | (0/533.3) | 66.0 | (9.5/621.8) | 195.2 | (21.5/415.6) | 66.0 | (9.5/621.8) | 247.7 | (21.5/415.6) |
(b).
| Locus | Periodontitis patients | Healthy controls | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-4-590 | IL-4VNTR | IL-4-590 | IL-4VNTR | |||||||||||||
| Polymorphism | CC | CT + TT | 11 | 12 + 22 | CC | CT + TT | 11 | 12 + 22 | ||||||||
| Number of patients | (30) | (16 + 1) | (28) | (17 + 1) | (8) | (5 + 2) | (10) | (5 + 0) | ||||||||
| Cytokines [pg/mL] | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) | Median | (Min/max) |
| IL-4_PWM | 3.6 | (0/163.6) | 3.9 | (0/17.8) | 3.7 | (0/163.6) | 3.4 | (0/17.8) | 4.3 | (0.6/163.6) | 3.6 | (0.7/9.9) | 4.3 | (0.6/163.6) | 3.6 | (0.7/9.9) |
| IL-4_P + C | 3.7 | (0/40.7) | 4.4 | (0/25.7) | 4.0 | (0/40.7) | 4.0 | (0/25.7) | 0.9 | (0/20.1) | 2.2 | (0.7/19.9) | 0.9 | (0/20.1) | 2.2 | (0.7/11.9) |
| IL-4_HSP70 | 0.7 | (0/6.5) | 0.6 | (0/5.5) | 0.0 | (0/6.5) | 1.2 | (0/5.5) | 1.2 | (0/3.1) | 1.6 | (0.7/2.3) | 1.2 | (0/3.1) | 1.6 | (0.7/2.3) |
| IL-4_Pg | 0.5 | (0/12.9) | 0.1 | (0/7.7) | 0.1 | (0/12.9) | 0.1 | (0/7.7) | 0.7 | (0/96.6) | 0.7 | (0/1.2) | 0.7 | (0/96.6) | 0.7 | (0.6/1.2) |
| IL-4_Aa | 0.4 | (0/11.2) | 0.1 | (0/3.7) | 0.1 | (0/4.7) | 0.1 | (0/11.2) | 1.9 | (0.7/113.8) | 0.7 | (0/4.5) | 1.9 | (0.7/113.8) | 0.7 | (0/4.5) |
| IL-4_Tf | 1.8 | (0/113.8) | 0.8 | (0/3.7) | 0.1 | (0/113.8) | 1.6 | (0/96.6) | 1.2 | (0/5.7) | 1.2 | (0.7/2.6) | 1.2 | (0/5.7) | 1.2 | (0.7/2.6) |
| IL-4_PI | 0.7 | (0/4) | 0.0 | (0/3.7) | 0.3 | (0/4) | 0.6 | (0/3.7) | 0.7 | (0/4.1) | 0.7 | (0/1.9) | 0.7 | (0/4.1) | 0.7 | (0/1.9) |
| IL-4_unstim. | 0.0 | (0/3.7) | 0.0 | (0/3.3) | 0.0 | (0/3.7) | 0.0 | (0/3.3) | 0.9 | (0/2.8) | 0.7 | (0/4.5) | 0.9 | (0/4.5) | 0.7 | (0/2.6) |
|
| ||||||||||||||||
| IL-5_PWM | 5.7 | (0/1882) | 3.7 | (0/98.9) | 8.4 | (0/1882) | 1.4 | (0/98.9) | 7.3 | (0.6/212.4) | 10.6 | (1.8/143) | 7.3 | (0.6/212.4) | 10.6 | (1.8/112.4) |
| IL-5_P + C | 8.7 | (0/230.1) | 9.5 | (0/375.5) | 9.0 | (0/230.1) | 6.9 | (0/375.5) | 20.0 | (2.3/226.2) | 9.2 | (1.2/403.2) | 20.0 | (2.3/403.2) | 9.2 | (1.2/169.3) |
| IL-5_HSP70 | 1.5 | (0/64.8) | 0.3 | (0/61) | 1.0 | (0/64.8) | 0.6 | (0/61) | 3.8 | (0.6/158) | 6.0 | (1.8/16.9) | 4.3 | (0.6/158) | 5.2 | (1.8/16.9) |
| IL-5_Pg | 1.0 | (0/322.7) | 1.2 | (0/8.1) | 0.9 | (0/322.7) | 1.2 | (0/8.1) | 3.0 | (0.6/6) | 3.8 | (3.2/7.7) | 3.5 | (0.6/7.7) | 3.7 | (3.2/6) |
| IL-5_Aa | 1.7 | (0/12.5) | 1.0 | (0/7.9) | 1.5 | (0/12.5) | 1.2 | (0/7.9) | 3.3 | (2.2/5.2) | 5.2 | (0/6) | 3.3 | (2.2/6) | 5.2 | (0/6) |
| IL-5_Tf | 1.9 | (0/212.4) | 0.6 | (0/3.7) | 1.8 | (0/212.4) | 1.2 | (0/3.9) | 2.0 | (1.2/5.2) | 2.9 | (1.2/6) | 2.4 | (1.2/6) | 2.9 | (1.2/6) |
| IL-5_PI | 1.5 | (0/28.6) | 0.6 | (0/3.7) | 1.7 | (0/28.6) | 0.6 | (0/3.7) | 2.4 | (0.6/4.2) | 3.4 | (0.6/6.9) | 2.9 | (0.6/4.7) | 3.4 | (0.6/6.9) |
| IL-5_unstim. | 0.7 | (0/8.1) | 0.7 | (0/7.9) | 0.6 | (0/8.1) | 0.9 | (0/7.9) | 2.5 | (0/6) | 3.4 | (0/6.9) | 2.5 | (0/6) | 3.4 | (1.8/6.9) |
|
| ||||||||||||||||
| IL-6_PWM | 3500.0 | (0/5960) | 3500.0 | (0/4699) | 3500.0 | (5.4/5960) | 3500.0 | (0/4699) | 2143.1 | (0/4824.3) | 3281.8 | (130.5/5200) | 2143.1 | (0/5200) | 3281.8 | (1186.1/4272.5) |
| IL-6_P + C | 3196.5 | (0/6457) | 2280.7 | (44.7/5528.5) | 3500.0 | (9.3/6457) | 2267.5 | (0/5528.5) | 2876.5 | (211.1/4900) | 1593.1 | (123.2/5193.4) | 2876.5 | (211.1/5193.4) | 1593.1 | (123.2/4931.7) |
| IL-6_HSP70 | 773.9 | (3.3/4207) | 141.4 | (2.7/5833.5) | 1143.1 | (3.3/4207) | 51.9 | (2.7/5833.5) | 888.5 | (165.6/3500) | 2040.7 | (167.5/4673.4) | 888.5 | (165.6/4673.4) | 2040.7 | (167.5/3111.3) |
| IL-6_Pg | 19.8 | (0/4191.6) | 10.6 | (0.4/3577.2) | 19.8 | (0/4191.6) | 11.5 | (0.4/3577.2) | 13.8 | (0/791) | 6.5 | (0.3/2438.4) | 13.8 | (0/2438.4) | 6.5 | (0.3/1675.2) |
| IL-6_Aa | 3500.0 | (27.5/6667) | 3500.0 | (71.9/7362.5) | 3500.0 | (839/6667) | 3500.0 | (27.5/7362.5) | 3500.0 | (108/4686) | 2076.7 | (258.5/3600) | 3366.9 | (108/4686) | 2076.7 | (1802.6/3600) |
| IL-6_Tf | 3500.0 | (25.1/6760) | 1035.3 | (1.4/4916) | 3500.0 * | (108/6760) * | 913.9 | (1.4/4916) | 3457.2 | (33.9/4203.3) | 2037.2 | (0.2/3500) | 3449.9 | (33.9/4203.3) | 2037.2 | (0.2/3500) |
| IL-6_PI | 1052.6 * | (336.1/4032) * | 350.9 | (0.7/3521.6) | 1102.1 * | (336.1/4032) * | 585.5 | (0.7/3521.6) | 3011.5 | (6.2/4686) | 324.3 | (87.5/1937.7) | 2387.8 | (6.2/4686) | 324.3 | (87.5/1337) |
| IL-6_unstim. | 214.8 | (2/3200.5) | 20.0 | (1/2051.9) | 227.2* | (2/3200.5)* | 8.0 | (1/2051.9) | 468.4 | (33.9/1796.2) | 558.5 | (14.2/2046.3) | 418.9 | (14.2/1796.2) | 601.9 | (25.2/2046.3) |
|
| ||||||||||||||||
| TNF alfa_PWM | 1085.9 | (2.8/3701) | 1002.3 | (0/3880) | 1250.1 | (3.5/3701) | 963.4 | (0/3880) | 1506.6 | (3.5/2800) | 1209.1 | (85.1/2800) | 1494.2 | (3.5/2800) | 1209.1 | (85.1/2800) |
| TNF alfa_P + C | 1071.5 | (0/3871) | 450.2 | (74/2800) | 1127.2 | (7.3/3871) | 442.1 | (0/2800) | 1579.4 | (35.6/2800) | 1531.3 | (22.9/2800) | 1596.6 | (35.6/2800) | 1531.3 | (22.9/2800) |
| TNF alfa_HSP70 | 32.1 | (2.6/3494.6) | 11.6 | (2.3/2800) | 92.6 | (2.6/3494.6) | 10.6 | (2.3/2800) | 63.4 | (9.6/1249.1) | 152.2 | (11.8/1656.7) | 87.6 | (9.6/1656.7) | 152.2 | (11.8/432) |
| TNF alfa_Pg | 20.6 | (0.5/612.2) | 13.6 | (1.9/576.6) | 21.1 | (0.5/612.2) | 13.3 | (1.9/576.6) | 2.1 | (0.3/54.6) | 0.8 | (0.2/146.3) | 2.1 | (0.2/146.3) | 0.8 | (0.4/51.8) |
| TNF alfa_Aa | 407.1 | (10.7/4760) | 462.2 | (3.7/3880) | 510.8 | (10.7/4760) | 441.8 | (3.7/3880) | 933.0 | (104.4/3569.7) | 527.6 | (51.9/4347.3) | 933.0 | (51.9/3569.7) | 527.6 | (235.7/4347.3) |
| TNF alfa_Tf | 650.9 | (4.2/2845.4) | 179.7 | (2.1/2393.9) | 687.5 * | (104.4/2845.4) * | 147.2 | (2.1/2393.9) | 899.8 | (8.5/1813.5) | 387.6 | (0.1/965.3) | 886.2 | (8.5/1813.5) | 387.6 | (0.1/965.3) |
| TNF alfa_PI | 74.0 | (8.3/1448.7) | 39.1 | (1.2/182.6) | 73.4 | (8.3/1448.7) | 55.0 | (1.2/297.7) | 80.5 | (22.2/130.7) | 38.2 | (9.5/223) | 73.4 | (10.4/130.7) | 38.2 | (9.5/223) |
| TNF alfa_unstim. | 14.0 | (1.8/2336.7) | 6.6 | (1.7/91.8) | 16.7 | (1.8/2336.7) | 4.8 | (1.7/91.8) | 7.0 | (2.1/52.1) | 21.1 | (3.5/112.1) | 10.5 | (2.1/52.1) | 18.7 | (3.5/112.1) |
|
| ||||||||||||||||
| VEGF_PWM | 13.7 | (3.4/66.9) | 5.5 | (1.5/94.4) | 14.4 | (3.4/66.9) | 5.1 | (1.5/94.4) | 17.1 | (0.2/83.4) | 4.9 | (0.1/33.1) | 11.0 | (0.2/83.4) | 11.9 | (0.1/33.1) |
| VEGF_P + C | 21.7 | (4.7/96.1) | 9.4 | (1.6/52.6) | 27.2 | (4.7/96.1) | 9.3 | (1.6/52.6) | 12.3 | (0/20.6) | 5.2 | (0.2/17.2) | 9.6 | (0/20.6) | 5.2 | (0.2/17.2) |
| VEGF_HSP70 | 32.8 | (5.8/101.1) | 10.6 | (1.1/45.3) | 35.5 | (5.8/101.1) | 16.0 | (1.1/45.3) | 54.0 | (3/121.7) | 14.4 | (1.5/91.9) | 42.7 | (3/121.7) | 15.7 | (1.5/91.9) |
| VEGF_Pg | 17.9 | (0.5/48.1) | 9.4 | (0.8/63.6) | 17.4 | (0.5/48.1) | 15.3 | (0.8/63.6) | 2.9 | (0.3/33.7) | 5.5 | (0.1/30) | 5.1 | (0.3/33.7) | 2.1 | (0.1/30) |
| VEGF_Aa | 27.4 | (8.8/119.1) | 27.7 | (3.5/57.8) | 27.0 | (8.8/119.1) | 29.5 | (3.5/57.8) | 47.8 | (4.1/73.7) | 27.8 | (7.6/36.8) | 23.9 | (4.1/73.7) | 30.4 | (7.6/36.8) |
| VEGF_Tf | 18.4 | (4.1/89.4) | 17.0 | (4/90.1) | 17.2 | (4.1/89.4) | 19.0 | (4/90.1) | 18.9 | (3.6/126.8) | 11.1 | (0.1/27.3) | 10.6 | (3/126.8) | 13.9 | (0.1/27.3) |
| VEGF_PI | 30.2 | (7.1/94.4) | 15.2 | (4.6/72.8) | 29.1 | (7.1/94.4) | 22.6 | (4.6/72.8) | 30.2 | (20/97.8) | 15.2 | (0.1/49.9) | 29.4 | (10/97.8) | 15.2 | (0.1/49.9) |
| VEGF_unstim. | 24.9 | (0.9/91) | 22.8 | (4.5/135.5) | 29.6 | (0.9/91) | 21.9 | (4.5/135.5) | 39.8 | (15.3/79.4) | 10.2 | (6.4/42.4) | 35.6 | (9.5/79.4) | 10.2 | (6.4/42.4) |
* , #, and † denote significantly higher levels of cytokines for P < 0.05, P < 0.01, and P < 0.001 (resp.), how they were found by Kruskal-Wallis tests in application to couple locus in table header (the minor homozygous locus was connected to a heterogeneous locus for the formal increase the statistical significance of results).
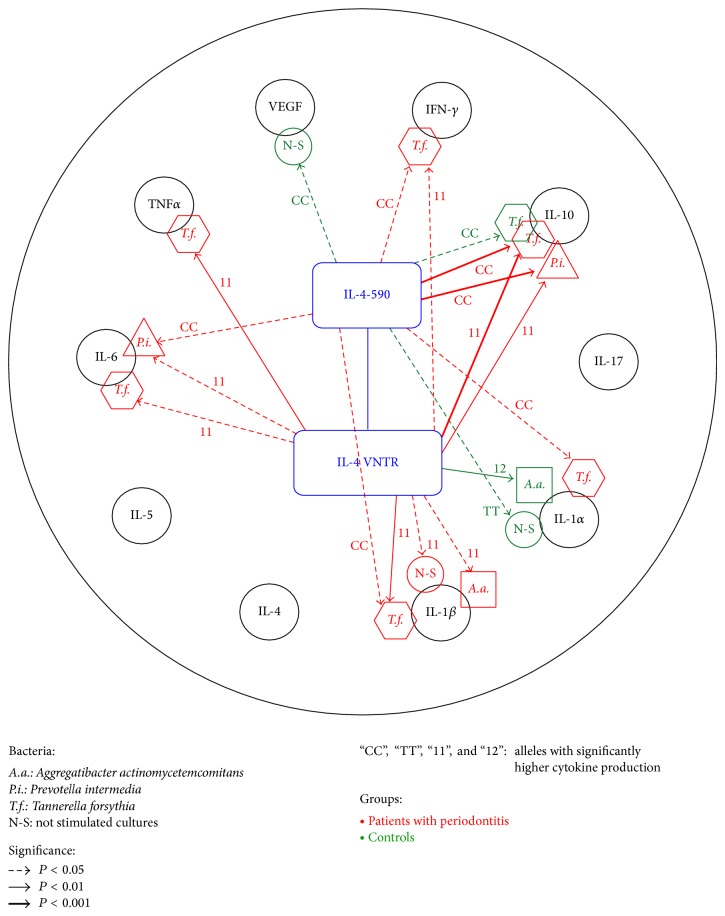
The differences in production of cytokines inside the two groups of patients (with periodontitis and controls) in combination with investigated IL-4 polymorphisms were preliminarily analyzed by multiple use of the Kruskal-Wallis test [24] with two adjacent alleles with minority representations in sample populations being always tied into one (see Tables 3(a) and 3(b) where alleles with significantly higher levels of cytokines are marked right over sample medians). Alternatively, we computed two matrices of Spearman rank correlation coefficients [25] (one for patients with periodontitis and another one for controls) between observed levels of cytokines and IL-4 polymorphisms and compared the observed levels of significance for the two methods (see Figure 1, where only statistically significant relations are highlighted by arrows.) Statistically significant results (for P < 0.05, P < 0.01, and P < 0.001) are indicated in Tables 3(a) and 3(b) and Figure 1.
Figure 1.
Statistically significant (higher) production of cytokines (unstimulated and stimulated by bacteria) on alleles of IL-4 SNP genes for patients with periodontitis (N = 47) and healthy controls (N = 15), evaluation by Spearman rank correlations. Remark: our application of Spearman correlation analysis gives us the very similar results in comparison with levels of significance observed by K-W method with preliminary aggregation of two adjacent minority alleles (see legend under Table 3); application of Spearman rank correlations does not reduce any data and slightly more supports the hypotheses related to expression of cytokines on homozygous allele.
3. Results
The mean ages for periodontitis patients (51.53 ± 6.52 years, mean ± SD) and healthy subjects (46.13 ± 9.65) did not differ between the two groups. There were no significant differences (P > 0.05) between subjects with periodontitis and controls regarding the average age, nonsmoking status, and ratio of males/females (4/11 in controls, 21/26 in patients with periodontitis) (Table 1).
Table 1.
Demographic and clinical data in patients with chronic periodontitis (CP) and nonperiodontitis controls.
| Patients with CP | Controls | |
|
| ||
| Number of subjects | 47 | 15 |
| Mean age (yrs ± SD) | 51.53 ± 6.52 | 46.13 ± 9.65 |
| Age range (yrs) | From 35 to 65 | From 35 to 66 |
| Sex (M/F) | 21/26 | 4/11 |
| PD (mm; mean ± SD) | 4.9 ± 1.5* | 1.9 ± 0.6 |
| CAL (mm; mean ± SD) | 5.4 ± 2.1* | 2.1 ± 0.7 |
* P < 0.05.
Both studied polymorphisms (IL-4 -590C/T and VNTR in intron 3) were in the Hardy-Weinberg equilibrium in the control group. Allele and genotype frequencies of IL-4 polymorphisms were not significantly different between the subjects with periodontitis and controls (P > 0.05; data not shown). Among 45 patients with CP, there were 2 patients with slight (1 or 2 mm CAL), 11 patients with moderate (3 or 4 mm CAL), and 32 patients with severe form of CP (≥5 mm CAL). When comparing genotypes (IL-4 -590CC versus CT + TT and IL-4 VNTR 11 versus 12 + 22) between patients with the slight/moderate (N = 13) and severe (N = 32) form of periodontitis, we found no statistically significant differences between both groups (P = 0.50 for IL-4 -590C/T and P = 0.61 for IL-4 VNTR polymorphisms).
Both patients and healthy controls were divided into groups according to the polymorphisms in IL-4 gene (homozygote for CC versus CT + TT genotypes for -590C/T variant and homozygote 11 versus 12 + 22 genotypes for VNTR). Tables 3(a) and 3(b) illustrate medians of the cytokine levels measured after a 3-day in vitro stimulation of lymphocytes isolated from peripheral blood of patients and healthy controls by dental plaque bacteria, mitogens, and HSP70.
The results in Table 3(b) show that the measured values of IL-4 and IL-5 (in pg/mL) were very low and no significant differences between groups in the levels of these cytokines were detected. Conversely, the level of IL-6 was relatively high in both patients with periodontitis and healthy controls. The levels of IL-6 after stimulation of lymphocytes by the bacterium P. intermedia in patients carrying the IL-4 -590CC and IL-4 VNTR 11 genotype were significantly higher than in the carriers of -590CT genotypes and IL-4 VNTR 12 + 22 (P < 0.05). A similar statistically significant difference in the production of IL-6 and also TNFα after stimulation by T. forsythia was detected in the carriers of IL-4 VNTR 11 genotype when compared with patients carrying genotypes 12 or 22 (P < 0.05). The results in Table 3(a) show that lymphocytes from patients with periodontitis carrying CC genotype of the IL-4 -590 polymorphism or 11 genotype of the IL-4 VNTR produced more IFNγ (P < 0.01 and P < 0.05, resp.) after stimulation with T. forsythia and also more IL-10 after stimulation with T. forsythia and P. intermedia (from P < 0.001 to P < 0.05). IL-10 was significantly increased in group of healthy persons carrying genotypes CC IL-4 -590 after stimulation with T. forsythia. On the other hand, no significant differences in the production of IL-17 and IL-1α after stimulation by dental plaque bacteria were determined in periodontitis patients carrying the IL-4 gene polymorphisms in this study. Production of IL-1β was significantly higher in patients with periodontitis carrying the CC genotype IL-4 -590C/T variant after stimulation with T. forsythia and in the unstimulated culture (P < 0.05). Subjects carrying genotype IL-4 VNTR 11 were found to have a higher production of IL-1β after stimulation with nearly all agents (from P < 0.01 to P < 0.05) except for the stimulation with P. gingivalis and P. intermedia (P > 0.05).
Within the group of healthy controls, statistically significant differences were observed only between subjects with CC versus CT and TT genotypes in IL-4 -590 polymorphism after stimulation with T. forsythia (P < 0.05). The highest difference in the production of cytokine IL-1α was observed between controls carrying genotypes CT and TT versus CC after stimulation with HSP70 and A. actinomycetemcomitans and in unstimulated culture (P < 0.05). A significantly higher production of IL-1α was found only in the unstimulated culture in patients with genotypes IL-4 VNTR 11. We did not find any significant differences in cytokines production after stimulation by PWM, PWM + Con A, and HSP70 in both examined groups.
Figure 1 shows the results of a parallel evaluation of these relationships by Spearman's correlation analysis; they are identical with the data in Tables 3(a) and 3(b) and clearly show the relationship of both IL-4 polymorphisms to cytokine production after the stimulation by the studied bacteria. Figure 1 shows that, in patients with periodontitis, homozygotes “CC” of IL-4 -590C/T polymorphism and “11” of IL-4 VNTR play an important role in the response of peripheral blood lymphocytes to stimulation with bacteria. T. forsythia is an important stimulator of production of cytokines TNFα, IL-6, IL-10, IFNγ, IL-10, and IL-1β while P. intermedia affects the in vitro production of IL-6 and IL-10 in patients with periodontitis. Production of IL-17, IL-4, and IL-5 was not affected by the short-term stimulation by any of the above factors (Figure 1).
4. Discussion
The interactions between periodontal pathogens and the immune response of the organism require knowledge of the pathogenesis of periodontal disease. The activation of dental plaque bacteria involves the development of local and systemic humoral and cellular immunity. The illness is characterized by accumulation of lymphocytes in the periodontal tissues; the inflammatory infiltrate is dominated at the beginning by T lymphocytes while the later chronic phase shows the shift to B lymphocytes and plasma cells [26].
After in vitro stimulation with P. gingivalis and F. nucleatum, most lymphocytes obtained from biopsies of periodontitis-affected tissues were of the TH2 phenotype [4]. Inactivation of IFN-γ and IL-2 by gingipains from P. gingivalis at inflammatory sites could also downregulate TH1 responses (associated with nonaggressive periodontal lesions) and promote TH2 pathways and polyclonal B-cell activation in advanced periodontitis [27]. Based on our previous experience and discussed publications, we have selected periopathogens and mitogens to be tested in this study. PWM was used as T and B cells stimulator and it was used in combination with Concanavalin A that activates T regulatory mechanisms; HSP70 was used as an indicator of shock response of inflamed tissue.
We suppose low levels of investigated cytokines after stimulation with P. gingivalis were found as a consequence of short time stimulation or by inactivation of investigated cytokines by gingipains.
The results of the present study point to the importance of single nucleotide IL-4 gene polymorphisms in periodontal disease. One of the most important findings is the proof that the -590C/T polymorphism in the promoter region and VNTR in intron 3 of the gene encoding IL-4 affect not only the production of this interleukin, as described earlier [28–30], but also the production of other cytokines in response to the action of dental plaque bacteria or selected mitogens, this effect being much more conspicuous in patients with chronic periodontitis when compared with healthy controls.
The greatest effect of stimulation with T. forsythia on production of IL-10 was observed in patients with periodontitis carrying the genotypes CC (-590C/T) and 11 (VNTR) of the gene encoding IL-4 while the increase in the levels of IFNγ, IL-1 β, IL-6, and TNFα in patients with the same genotypes was lower (P < 0.05). The levels of regulatory cytokine IL-10 in patients with periodontitis carrying the appropriate genotypes of IL-4 polymorphisms were also significantly increased, after stimulation with P. intermedia. In contrast to patients that exhibited increased production of the abovementioned cytokines in the presence of genotypes CC or 11, higher production of IL-1α after stimulation with HSP70 and the bacterium A. actinomycetemcomitans was found in control subjects carrying the combination CT + TT (IL-4 -590C/T) and 12 (IL-4 VNTR).
Based on our previous results [5, 15], we assumed that, after stimulation with dental plaque bacteria, IL-4 gene polymorphisms in periodontitis patients would in particular affect the production of cytokine IL-4. Contrary to this assumption, no significant increase was found in the level of IL-4, while stimulation with T. forsythia and P. intermedia resulted in significantly higher levels of other cytokines. These results are in keeping with the data in a number of recent studies which documented that polymorphisms in genes encoding some cytokines or their receptors can affect the production of not only their own but also other mediators. The results of the study by Wallis et al. [31] indicate that I50V SNP in part IL-4R controls production of IL-17 by TH17 cells cultured from healthy individuals. Similarly, the minor allele of the IL-4R SNP (rs1805010), which confers impaired IL-4 signaling and has been associated with an aggressive destructive course of rheumatoid arthritis (RA), contributes to increased TH17 cell frequency, enhanced clinical activity, and accelerated radiographic progression in RA by rendering CD4 T cells from RA [32].
Cytokines play a key role in modulation of immune reaction. All cytokines are pleiotropic. They have the ability to interact with specific receptors on the cell surface and can modulate receptor expression for other cytokines. The same biological function of cytokines is influenced by several distinct cytokines (cytokine cross talk). This is a biologically important feature: if one cytokine is absent or its level limited, substitution by another cytokine exists and may be regulated by IL-4 gene polymorphisms in patients with CP [33]. Our results presented in Table 3(b) show that IL-4 gene polymorphisms influence the production of another TH2 cytokine IL-6. IL-6 is an interleukin that acts as both a proinflammatory and anti-inflammatory cytokine. The role of IL-6 is anti-inflammatory: it has an inhibitory effect on TNFα and IL-1 production and it activates production of IL-1 ra and regulatory IL-10. We suppose that the higher median detected in the healthy group as compared to periodontitis patients group after stimulation of their lymphocytes by P. intermedia may be explained by the anti-inflammatory properties of IL-6.
There were several limitations to this study. For instance, the short cultivation time of lymphocytes isolated from peripheral blood with bacteria and mitogens. Also, the healthy control and periodontitis groups were not precisely matched by sex. Lastly, the small sample size precludes any robust claims and points to the need for additional large longitudinal studies of cohorts of patients with periodontitis. For this reason, two statistical methods for evaluation of results were used.
It appears that the knowledge of the genetic background of chronic periodontitis will remain for quite some time on the level of knowledge of individual “potential” risk or protective factors whose effect on the disease will depend on a broader, still unexplored genetic background and a number of epigenetic factors. The differences in the findings published in individual studies are hindered by the fact that individual populations in the world can differ in the frequencies of the alleles of appropriate polymorphisms. Other factors that may play a role are different approaches to data evaluation and differences in clinical evaluation and selection of patients. For example, the periodontitis of young subjects, formerly presented as early onset periodontitis or juvenile periodontitis, is currently classified as aggressive periodontitis. In a longitudinal (10-year) study, we explored the progression of early onset periodontitis in patients aged 17–25 years at the beginning of the study. Over the years, the disease in some patients progressed while in others it did not [15, 34]. According to current criteria, all patients would be classified at the beginning of the study as suffering from aggressive periodontitis. This fact points to the importance of preventive examinations focused on the initial changes of the periodontium of young subjects and on early preventive therapy.
5. Conclusion
Our study suggests that polymorphisms in the IL-4 gene not only affect the production of the IL-4 cytokine but can influence production of several other cytokines, such as IL-10, IFNγ, IL-1β, IL-6, and TNFα, which in turn can affect periodontal disease. Further studies are needed to clarify the association of these polymorphisms with cytokine production in other populations.
Acknowledgments
The study was supported by PRVOUK-P28/LF1/6 (Charles University of Prague, Czech Republic), by the project of MUNI/A/0951/2013 (Masaryk University), and by the Grants NT 11405-6, NT 14164-3, and NT 13087-3 (Ministry of Health, Czech Republic).
Conflict of Interests
The authors declare that they have no conflict of interests associated with this work.
References
- 1.Mergenhagen S. E., de Araujo W. C., Varah E. Antibody to Leptotrichia buccalis in human sera. Archives of Oral Biology. 1965;10(1):29–33. doi: 10.1016/0003-9969(65)90054-3. [DOI] [PubMed] [Google Scholar]
- 2.Gemmell E., Seymour G. J. Immunoregulatory control of Th1/Th2 cytokine profiles in periodontal disease. Periodontology 2000. 2004;35:21–41. doi: 10.1111/j.0906-6713.2004.003557.x. [DOI] [PubMed] [Google Scholar]
- 3.Mosmann T. R., Coffman R. L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annual Review of Immunology. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 4.Seymour G. J., Gemmell E., Reinhardt R. A., Eastcott J., Taubman M. A. Immunopathogenesis of chronic inflammatory periodontal disease: cellular and molecular mechanisms. Journal of Periodontal Research. 1993;28(6):478–486. doi: 10.1111/j.1600-0765.1993.tb02108.x. [DOI] [PubMed] [Google Scholar]
- 5.Bártová J., Krátká-Opatrná Z., Procházková J., Krejsa O., Dušková J., Mrklas L., Tlaskalová H., Cukrowská B. Th1 and Th2 cytokine profile in patiens with early onset periodontitis and their healthy siblings. Mediators of Inflammation. 2000;9(2):115–120. doi: 10.1080/096293500411587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rateitschak K. H., Rateitschak E. M., Wolf H. F., Hessl M. Periodontology (Color Atlas of Dental Medicine 1) 2nd. G. Thieme Verlag Stuttgart; 1989. [Google Scholar]
- 7.Michalowicz B. S. Genetic and heritable risk factors in periodontal disease. Journal of Periodontology. 1994;65(5):479–488. doi: 10.1902/jop.1994.65.5s.479. [DOI] [PubMed] [Google Scholar]
- 8.Page R. C. The role of inflammatory mediators in the pathogenesis of periodontal disease. Journal of Periodontal Research. 1991;26(3):230–242. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 9.Kornman K. S., di Giovine F. S. Genetic variations in cytokine expression: a risk factor for severity of adult periodontitis. Annals of Periodontology. 1998;3(1):327–338. doi: 10.1902/annals.1998.3.1.327. [DOI] [PubMed] [Google Scholar]
- 10.Vaithilingham R. D., Safii S. H., Baharuddin N. A., Ng C. C., Cheong S. C., Barold P. M., Schaefer A. S., Loos B. G. Moving into a new era of periodontal genetic studies: relevance of large case-control samples using severe phenotypes for genome-wide association studies. Journal of Periodontal Research. 2014 doi: 10.1111/jre.12167. [DOI] [PubMed] [Google Scholar]
- 11.Kornman K. S., Crane A., Wang H. Y., di Giovine F. S., Newman M. G., Pirk F. W., Wilson T. G. J., Higginbottom F. L., Surf W. G. The interleukin -1 genotype as a severity factor in adult periodontal disease. Periodontology 2000. 1997;14:33–53. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 12.Kinane D. F., Hodge P., Eskdale J., Ellis R., Gallagher G. Analysis of genetic polymorphisms at the interleukin-10 and tumour necrosis factor loci in early-onset periodontitis. Journal of Periodontal Research. 1999;34(7):379–386. doi: 10.1111/j.1600-0765.1999.tb02270.x. [DOI] [PubMed] [Google Scholar]
- 13.Kinane D. F., Hart T. C. Genes and gene polymorphisms associated with periodontal disease. Critical Reviews in Oral Biology and Medicine. 2003;14(6):430–449. doi: 10.1177/154411130301400605. [DOI] [PubMed] [Google Scholar]
- 14.Holla L. I., Fassmann A., Augustin P., Halabala T., Znojil V., Vanek J. The association of interleukin-4 haplotypes with chronic periodontitis in a Czech population. Journal of Periodontology. 2008;79(10):1927–1933. doi: 10.1902/jop.2008.080035. [DOI] [PubMed] [Google Scholar]
- 15.Krátká Z., Bártová J., Krejsa O., Otčenášková M., Janatová T., Dušková J. Interleukin-1 gene polymorphisms as assessed in a 10-year study of patiens with early onset periodontitis. Folia Microbiologica. 2007;52(2):183–188. doi: 10.1007/BF02932157. [DOI] [PubMed] [Google Scholar]
- 16.Keen L. J. The extent and analysis of cytokine and cytokine receptor gene polymorphism. Transplant Immunology. 2002;10(2-3):143–146. doi: 10.1016/S0966-3274(02)00061-8. [DOI] [PubMed] [Google Scholar]
- 17.Cantagrel A., Navaux F., Loubet-Lescoulié P., et al. Interleukin-1beta, interleukin-1 receptor antagonist, interleukin-4, and interleukin-10 gene polymorphisms: relationship to occurrence and severity of rheumatoid arthritis. Arthritis & Rheumatology. 1999;42(6):1093–1100. doi: 10.1002/1529-0131(199906)42:6<1093::AID-ANR5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 18.Armitage G. C. Development of a classification system for periodontal diseases and conditions. Annals of periodontology. 1999;4(1):1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Mühlemann H. R., Mazor Z. S. Röntgendiagnostik. Schweiz Monatsschr Zahnheilkd. 1955;65:1005–1013. [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. New York, NY, USA: Cold Spring Laboratory Press; 1989. [Google Scholar]
- 21.Hegab A. E., Sakamoto T., Saitoh W., Massoud H. H., Massoud H. M., Hassanein K. M., Sekizawa K. Polymorphisms of IL4, IL13, and ADRB2 genes in COPD. Chest. 2004;126(6):1832–1839. doi: 10.1378/chest.126.6.1832. [DOI] [PubMed] [Google Scholar]
- 22.Mout R., Willemze R., Landegent J. E. Repeat polymorphisms in the interleukin-4 gene (IL4) Nucleic Acids Research. 1991;19(13):3763. doi: 10.1093/nar/19.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slots J. Selective medium for isolation of Actinobacillus actinomycetemcomitans. Journal of Clinical Microbiology. 1982;15(4):606–609. doi: 10.1128/jcm.15.4.606-609.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruskal J. B., Wallis W. A. Use of ranks in one-criterion variance analysis. Journal of the American Statistical Association. 1952;47(260):583–662. [Google Scholar]
- 25.Spearman C. The proof and measurement of association between two things. The American journal of psychology. 1987;100(3-4):441–471. doi: 10.2307/1422689. [DOI] [PubMed] [Google Scholar]
- 26.Okada H., Shimabukuro Y., Kassai Y., Ito H., Matsuo T., Ebisu S., Harada Y. The function of gingival lymphocytes on the establishment of human periodontitis. Advances of Dental Research. 1988;2(2):364–367. doi: 10.1177/08959374880020022801. [DOI] [PubMed] [Google Scholar]
- 27.Yun L. W. P., Decarlo A. A., Collyer C., Hunter N. Enhancement of Th2 pathways and direct activation of B cells by the gingipains of Porphyromonas gingivalis. Clinical and Experimental Immunology. 2003;134(2):295–302. doi: 10.1046/j.1365-2249.2003.02287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smolnikova M. V., Smirnova S. V., Freidin M. B., Tyutina O. S. Immunological parameters and gene polymorphisms (C-590T IL4, C-597A IL10) in severe bronchial asthma in children from the Krasnoyarsk region, West Siberia. International Journal of Circumpolar Health. 2013;72(1) doi: 10.3402/ijch.v72i0.21159.21159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenwasser L. J., Borish L. Genetics of atopy and asthma: The rationale behind promoter-based candidate gene studies (IL-4 and IL-10) American Journal of Respiratory and Critical Care Medicine. 1997;156(4):S152–S155. doi: 10.1164/ajrccm.156.4.12tac-14. [DOI] [PubMed] [Google Scholar]
- 30.Nakashima H., Miyake K., Inoue Y., Shimizu S., Akahoshi M., Tanaka Y., Otsuka T., Harada M. Association between IL-4 genotype and IL-4 production in the Japanese population. Genes and Immunity. 2002;3(2):107–109. doi: 10.1038/sj.gene.6363830. [DOI] [PubMed] [Google Scholar]
- 31.Wallis S. K., Cooney L. A., Endres J. L., Lee M. J., Ryu J., Somers E. C., Fox D. A. A polymorphism in the interleukin-4 receptor affects the ability of interleukin-4 to regulate Th17 cells: a possible immunoregulatory mechanism for genetic control of the severity of rheumatoid arthritis. Arthritis Research & Therapy. 2011;13(1, article R15) doi: 10.1186/ar3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leipe J., Schramm M. A., Prots I., Schulze-Koops H., Skapenko A. Increased TH17 cell frequency and poor clinical outcome in rheumatoid arthritis are associated with genetic variant in the IL-4R gene, rs 1805010. Arthritis and Rheumatology. 2014;66(5):1165–1175. doi: 10.1002/art.38343. [DOI] [PubMed] [Google Scholar]
- 33.Whiteside Theresa L. Introduction to cytokines as targets for immunomodulation. In: House R. V., Descotes J., editors. Methods in Pharmacology and Toxikology: Cytokines in Human Health: Immnotoxicology, Patology and Therapeutic Applications. 2007. pp. 1–12. [Google Scholar]
- 34.Bartová J., Kratka Z., Svobodova K., Janatova T., Podzimek S., Prochazkova J., Krejsa O. Periodontitis—Risk Factors of Atherosclerosis. Medimond s.r.l., Moduzi Editore International Proceedings Division, ECI; 2009. [Google Scholar]