Abstract
Nitrogen assimilation plays a vital role in plant metabolism. Assimilation of nitrate, the primary source of nitrogen in soil, is linked to generation of the redox signal nitric oxide (NO). An important mechanism by which NO regulates plant development and stress responses is through S-nitrosylation, i.e. covalent attachment of NO to cysteines to form S-nitrosothiols (SNO). Despite the importance of nitrogen assimilation and NO signalling, it remains largely unknown how these pathways are interconnected. Here we show that SNO signalling suppresses both nitrate uptake and reduction by transporters and reductases, respectively, to fine-tune nitrate homeostasis. Moreover, NO derived from nitrate assimilation suppresses the redox enzyme S-nitrosoglutathione Reductase 1 (GSNOR1) by S-nitrosylation, preventing scavenging of S-nitrosoglutathione, a major cellular bio-reservoir of NO. Hence, our data demonstrates that (S)NO controls its own generation and scavenging by modulating nitrate assimilation and GSNOR1 activity.
Introduction
Nitrogen is a conspicuous building block of many central biomolecules, such as nucleic acids, amino acids, and cofactors. The primary source of nitrogen available to land plants is inorganic nitrate (NO3−), the concentration of which can vary from micromolar to millimolar amounts in soils1. To cope with such large fluctuations in nitrate availability, higher plants have evolved sophisticated high-affinity (HATS) and low-affinity (LATS) transport systems2,3. These systems rely mainly on two families of membrane-bound nitrate transporters (NRT) of which NRT2 members are high-affinity, while most members of NRT1 are low-affinity nitrate transporters2,3. NRT2.1 and NRT1.1 (firstly identified as CHL1, for chlorate resistant 1) are particularly important for nitrate uptake by roots of Arabidopsis thaliana plants4. AtNRT2.1 is a pure high-affinity nitrate transporter that is repressed by high nitrate levels and activated under low-nitrate conditions5. AtNRT1.1 is an exception in the NRT1 family being a dual-affinity nitrate transporter: it normally has low affinity uptake but can change to the high-affinity mode under low nitrate levels6,7. The switch from low- to high-affinity transport is mediated by phosphorylation at Thr101 residue of NRT1.1, which enhances its affinity to nitrate8,9, as well as by transcriptional down-regulation of NRT1.1 and up-regulation of NRT2.1 allowing scavenging of available nitrate2,7.
Once taken up by roots nitrate is mainly transported to shoots for further assimilation and in leaves it is reduced to nitrite (NO2−) by the activity of NAD(P)H-dependent cytosolic Nitrate Reductases (NR). Nitrite, in turn, is promptly removed from cells or transported to chloroplasts where it is reduced by Nitrite Reductase (NiR) into ammonium (NH4+) for further assimilation into organic compounds by the glutamine synthetase/glutamine-2-oxoglutarate aminotransferase (GS-GOGAT) system1,10,11.
In Arabidopsis, the catalytic activity of NR, which is considered limiting to nitrogen assimilatory pathways12,13 is conferred by the genes NIA1 and NIA2. Double mutant nia1 nia2 plants display poor growth on media with nitrate as the sole nitrogen source, which is in part due to lack of nitrogen incorporation into amino acids14. Curiously, gene expression of NRT1.1 and NRT2.1 transporters is constitutively upregulated in roots of nia1 nia2 plants, suggesting that NR activity or a nitrogen-containing metabolite derived from nitrate reduction feedback regulate uptake systems15. However, the identity of this regulatory metabolite remains obscure15,16.
In addition to its reduction to NH4+, nitrite can be reduced to nitric oxide (NO) via non-enzymatic as well as various enzymatic pathways17,18. High levels of nitrite allow NR to reduce this assimilate into NO19,20, although genetic evidence suggests that the main role of NR in NO biosynthesis is the production of nitrite21,22. L-arginine, polyamines and hydroxylamines are also potential sources for NO synthesis in higher plants, however the molecular mechanisms responsible for these activities have not been identified so far17,18,23.
NO is a free radical with a wide range of important signalling functions in all eukaryotes. Accordingly, Arabidopsis NO overproducing nox1 (also known as cue1-6) mutants accumulate elevated levels of NO and exhibit defects in floral transition, root apical meristems, and pathogen-induced programmed cell death24-26. The underlying mechanisms of NO action rely on its physicochemical properties that allow reactivity with different kinds of biomolecules, thereby altering the redox state of their active groups. NO and its derivatives can react with thiols, tyrosine residues, metal centers and reactive oxygen species17,27. Particularly, addition of NO to cysteine thiols results in the formation of S-nitrosothiols (SNO), which have been shown to alter the activity, localization or conformation of target proteins27,28.
NO may also react with glutathione to form S-nitrosoglutathione (GSNO), which is thought to be a major cellular reservoir of NO capable of generating protein-SNO. Cellular GSNO levels are controlled by the evolutionary conserved, cytosolic enzyme GSNO Reductase 1 (GSNOR1), which catalyses the NADH-dependent reduction of GSNO to oxidized glutathione and ammonium29-31. Arabidopsis plants with impaired GSNOR1 function display elevated levels of protein-SNO and exhibit deficiencies in development, immunity, and thermotolerance, indicating that GSNOR1 indirectly controls the level of biologically active protein-SNO31-36. Taken together, these studies clearly indicate that the generation and scavenging of NO is connected to molecular pathways of nitrogen assimilation. However, it remains unclear if nitrate or other nitrate-derived metabolites directly affect NO signalling, and vice versa, if NO signalling influences nitrogen homeostasis37. Here we provide genetic and biochemical evidence for intimate interplay between nitrate assimilation and NO signalling. We identified novel NO-mediated feedback pathways that regulate the transcription of nitrate transporters and enzymatic activities of NR and GSNOR1 by redox-based post-translational modification. Our data reveal that nitrate assimilation and NO signalling are connected in unexpected ways, allowing plants to fine-tune NO generation and scavenging.
Results
NO signalling feedback regulates nitrogen assimilation
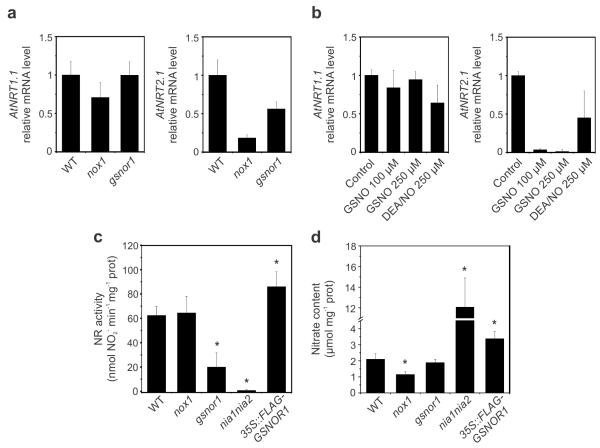
Because significant amounts of NO and derived SNO result from the nitrogen assimilation pathway, we considered that NO may feedback regulate this pathway. Nitrogen assimilation commences by the uptake of nitrate by low- and high-affinity transport systems, in which the NRT1.1 and NRT2.1 transporter genes play key roles2,3. We assessed the expression of these genes in roots of wild-type (WT) plants as well as the NO and SNO signalling mutants, nox124 and gsnor134 (also known as par2-1). While nox1 plants overproduce free NO, gsnor1 plants accumulate high levels of GSNO28, a more stable redox form of NO. Compared to wild type plants grown under moderate nitrate availability, expression of NRT2.1 was strongly suppressed in both nox1 and gsnor1 mutants, whereas expression of NRT1.1 remained unchanged (Fig. 1a). Accordingly, exogenous treatment of WT plants with GSNO or the alternative NO donor, DEA/NO, also led to inhibition of NRT2.1 expression but left NRT1.1 expression unaltered (Fig. 1b).These findings suggest that elevated NO and SNO levels induce a switch from high- to low-affinity nitrate transport.
Figure 1. Nitrate uptake and reduction in plants with altered NO signalling.
(a) Expression of the nitrate transporter marker genes NRT1.1 and NRT2.1 in roots of WT, nox1 and par2-1 plants was determined by qRT-PCR and normalized to expression of ACT2. Error bars represent SD (n = 3). (b) Effect of GSNO on nitrate-induced expression of NRT genes in roots. WT seedlings grown in half-strength MS medium (9.4 mM KNO3 and 10.3 mM NH4NO3) were incubated for 3 hours in water with 1 mM nitrate (KNO3), in the absence or presence of GSNO or DEA/NO. NRT expression was determined by qRT-PCR and normalized to expression of ACT2. Error bars represent SD (n = 3). (c) NR activity and (d) nitrate (NO3−) content determined in leaf extracts of WT plants and genotypes with enhanced (nox1 and par2-1) or impaired (nia1nia2 and 35S::FLAG-GSNOR1) NO homeostasis, after six hours of light. Data points represent means ± SD of three independent experiments. Asterisks indicate statistical differences from the WT (Student’s t test, P < 0.05).
Once taken up into the root, nitrate is mainly transported to the shoots where it is assimilated at the expense of photosynthetic reducing power2. In leaves nitrate is reduced to nitrite by the cytosolic enzyme NR. To examine if NO also regulates this rate-limiting step in nitrogen assimilation, we measured NR activity in leaves of the genotypes with altered (S)NO homeostasis (Fig. 1c). Compared to WT, gsnor1 mutant plants exhibited strongly reduced NR activity, while GSNOR1 overexpressing plants (35S::FLAG-GSNOR1, Supplementary Fig. 1a and 1b) displayed enhanced NR activity. Surprisingly, however, mutant nox1 plants, that only accumulate 30-40% more SNO than WT plants under basal conditions26, did not exhibit altered NR activity (Fig. 1c). Together with the fact that expression of the NR gene NIA2 was indifferent in all mutants (Supplementary Fig. 1c and 1d), these data suggest that GSNO and free NO differentially affect NR activity. To understand the cumulative consequences of (S)NO-modulated nitrate transport and reduction, we also measured nitrate contents of mutant leaves and compared them to WT and NR double mutant nia1 nia2 leaves, the latter of which is known to accumulate high levels of nitrate due to lack of NR activity15. Figure 1d shows that nox1 plants accumulated significantly less nitrate than the WT, likely caused by partial switching to the low-affinity transport system in these mutants (Fig. 1a). However, in gsnor1 plants activation of the low-affinity transport system in conjunction with reduced NR activity apparently resulted in relatively normal nitrate levels. Instead, a regulatory role for GSNO only became apparent in 35S::FLAG-GSNOR1 plants, which accumulated elevated levels of nitrate (Fig. 1d). Taken together, these findings indicate that NO and SNO modulate nitrogen assimilation by differentially inhibiting nitrate uptake and reduction.
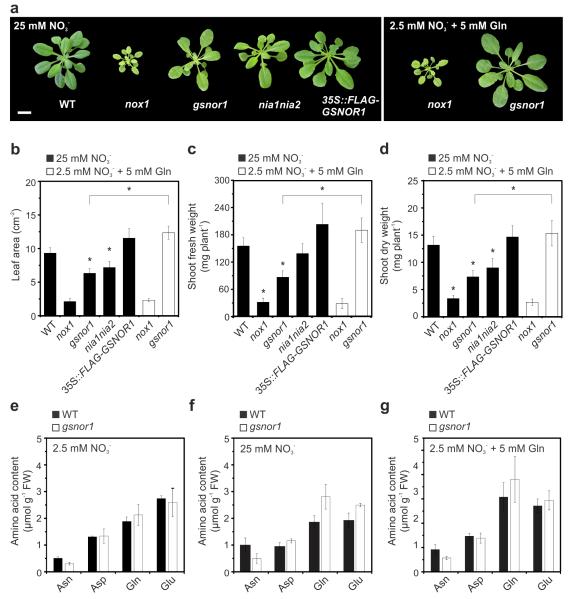
To assess the biological impact of (S)NO on nitrate assimilation, we analysed the vigour of (S)NO signalling mutants by measuring growth and biomass accumulation parameters (Fig. 2a-d). As expected, the inability of nia1 nia2 plants to reduce nitrate led to reduced leaf area and a decrease in dry shoot weight compared to WT. Like nia1 nia2, mutant nox1 and gsnor1 plants also displayed strongly decreased growth vigour. Conversely, leaf area and biomass growth tended to increase, albeit not always statistically significant, in GSNOR1 overexpressing 35S::FLAG-GSNOR1 plants (Fig. 2a-d). These findings suggest (S)NO-mediated suppression of nitrate assimilation may have dramatic effects on plant growth. To confirm the poor growth vigour phenotypes of (S)NO mutants were due to decreased nitrate assimilation, we sought to bypass this pathway by exogenous addition of glutamine (Gln), the main end product of nitrate assimilation. Addition of Gln to WT plants did not further improve growth compared to nitrate-replete conditions (Supplementary Fig. 2). Importantly, however, irrigation of nox1 and gsnor1 mutants in the presence of Gln recovered growth vigour of gsnor1, but not that of nox1, to levels comparable to those of WT and 35S::FLAG-GSNOR1 plants (Fig. 2a-d). Thus, bypassing both nitrate uptake and reduction by feeding Gln rescued the gsnor1 phenotype, while suppression of N assimilation may not be the only cause for lack of growth vigour in nox1 plants. In contrast to GSNOR that is directly involved in NO homeostasis, the metabolic changes in nox1 that lead to an increase in NO production are indirect. The nox1 mutant is defective in a phosphoenolpyruvate/phosphate translocator that imports phosphoenolpyruvate for the synthesis of aromatic amino acids and related compounds through the shikimate pathway39. Consequently, nox1 mutants display an overall marked increase in the levels of free amino acids, including L-arginine, a precursor of NO (Supplementary Fig. 3 and ref 24). Additionally, this mutant exhibits an imbalance of aromatic versus non-aromatic amino acids and a marked reduction in secondary phenolic compounds that are dependent on the shikimate pathway for precursors, severely compromising the establishment of photoautotrophic growth.39 Thus, alterations of amino acid levels as a direct consequence of the nox1 mutation are not expected to be complemented by simply adding Gln, as other imbalances are not corrected by this treatment. Accordingly, the effect of the nox1 mutation on biomass is much more severe than the nia1 nia2 knock out mutation (Fig. 2a-d), indicating that mechanisms unrelated to nitrate assimilation underpin the nox1 phenotype. In contrast, our data indicate that mutation of GSNOR1 affected plant growth by inhibiting nitrate uptake and assimilation (Fig. 1), and accordingly, this phenotype can be rescued by addition of Gln (Fig. 2a-d). To further establish that suppressed nitrate assimilation underpins the poor primary productivity phenotype of gsnor1 plants, we assessed global accumulation of amino acids in this mutant (Supplementary Fig. 3). Particularly the accumulation of glutamine (Gln), glutamate (Glu), asparagine (Asn), and aspartate (Asp) are informative for nitrogen homeostasis, because they represent the primary transported amino acids derived from ammonium11. Several observations made in gsnor1 mutants support the notion that (S)NO-mediated suppression of nitrate assimilation affects primary productivity. Firstly, low nitrogen conditions stimulate the formation of Gln and Glu because of their comparatively lower nitrogen-to-carbon ratios (2N:5C for Gln and 1N:5C for Glu). Importantly, even in presence of high nitrate, gsnor1 plants accumulated more Gln and Glu compared to the WT (Fig. 2e, 2f), indicating these mutants were suffering from nitrogen shortage. Secondly, compared to Gln and Glu, the amino acid Asn is rich in nitrogen (2N:4C ratio) and its production is therefore avoided under low nitrogen availability. Despite the presence of 25 mM nitrate, mutant gsnor1 plants contained decreased levels of Asn compared to WT (Fig. 2e, 2f), further indicating these mutants experience a shortage in nitrogen. Finally, in both WT and gsnor1 plants exogenous addition of 5 mM Gln led to an expected rise in endogenous Gln and also increased Asn content, while the levels of Glu, and Asp remained largely unchanged compared to the low nitrate regime (Fig. 2e-g, Supplementary Fig. 3). Taken together, these data indicate that (S)NO are important regulators of nitrate assimilation and thus, plant growth and development.
Figure 2. Growth vigour and amino acid content of genotypes with impaired and enhanced (S)NO signalling.
(a) Phenotype, (b) leaf area, (c) shoot fresh weight and (d) shoot dry weight of four-week-old plants grown on perlite:vermiculite (1:1) under 12h/12h light/dark and irrigated three times a week with a MS nutrient solution containing 25 mM nitrate (half KNO3 and half NH4NO3) (black bars) or with 2.5 mM nitrate and 5 mM glutamine. Scale bar, 1 cm. Data points represent means ± SD (n = 15 plants). (e-g) Contents of primary transported amino acids in WT and gsnor1 leaves. Asn, asparagine; Asp, aspartate; Gln, glutamine; Glu, glutamate. Data represent means ± SD of three independent analyses. Asterisks indicate statistically significant difference from WT (P < 0.05, Student’s t test).
Nitrogen metabolism regulates GSNOR1 activity
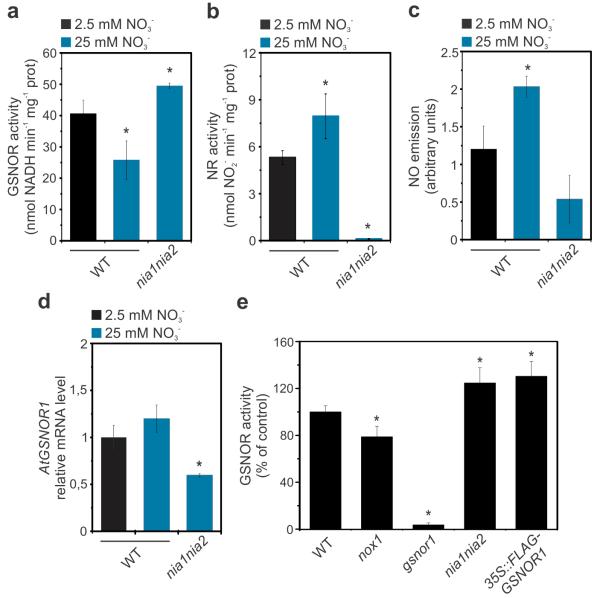
Given the impact of GSNOR1 on nitrate assimilation, we considered that GSNOR1 activity may be feedback regulated by nitrate. To examine this possibility, we grew WT plants under high nitrate availabilities that caused good growth vigour (25 and 40 mM), as well as lower nitrate availabilities (1 mM and 2.5 mM) that resulted in poor biomass accumulation (Supplementary Fig. 2). Because the irrigated nutrient solution was not compensated with any other N source, nitrate concentrations lower than 2.5 mM appeared undesirably detrimental for plant growth and development. Therefore we selected 2.5 mM and 25 mM nitrate concentrations for further experimentation. Additionally, as the nitrate concentration of nutrient solutions was composed of half KNO3 and half NH4NO3, we checked the possibility that the effects observed could partially be attributed to NH4+. However, when the concentration of NH4+ in the nutrient solution was reduced by ten times (from 12.5 mM to 1.25 mM), parameters of biomass growth of WT plants were comparable (Supplementary Fig. 4), indicating that the effect of ammonium in determining plant growth vigour was negligible under our conditions. Interestingly, increasing nitrate availability from 2.5 mM to 25 mM reduced mean GSNOR1 activity by ~35% ± 8% while significantly enhancing NR activity (Fig. 3a, 3b). Additional increase in nitrate availability to 40 mM did not suppress GSNOR activity any further, while intermediate nitrate levels (12.5 mM) reduced GSNOR activity by ~10% (Supplementary Fig. 5). These data suggest nitrate levels may regulate GSNOR1 activity. However, mutant nia1 nia2 plants that are void of NR activity (Figs. 1c, 3b) and accumulate elevated levels of endogenous nitrate15 (Fig. 1d), did not exhibit reduced but rather slightly elevated GSNOR1 activity (Fig. 3a). Thus, instead of nitrate, a metabolite downstream of NR-catalysed nitrate reduction may be responsible for inhibition of GSNOR1 activity.
Figure 3. Nitrate-derived NO suppresses activity of GSNOR1.
(a) GSNO Reductase (GSNOR) activity, (b) Nitrate Reductase (NR) activity, (c) NO emission and (d) GSNOR1 gene expression measured in leaf extracts of WT and nia1nia2 plants grown under low or high nitrate availability. Plants were grown in perlite:vermiculite (1:1) under a 12/12 hours light/dark period and irrigated three times a week with MS nutrient solution containing 2.5 mM or 25 mM nitrate (half KNO3 and half NH4NO3). All measurements were taken six hours post light exposure. Relative expression of GSNOR1 in (d) was determined by qRT-PCR and normalized to expression of ACT2. (e) GSNOR activity in genotypes with impaired and enhanced NO signalling. Plants were grown in soil with a photoperiod of 16/8 hours light/dark and irrigated with water as needed. Data points represent means ± SD of three independent experiments. Asterisks indicate significant differences from the WT (Student’s t test, P < 0.05).
Nitrite is the first reductive metabolite downstream of nitrate and in elevated concentrations can be converted into NO21,22,40. Therefore, we measured NO emission in plants grown under low and high nitrate availability. High concentrations of nitrate promoted NO emission in wild-type plants (Fig. 3c and Supplementary Fig. 6). Despite having elevated endogenous nitrate levels, nia1 nia2 plants did not show elevated NO emission when grown under higher nitrate, indicating that high nitrate availability leads to NR-mediated generation of NO.
Because nitrate-induced, NR-mediated NO production was associated with reduced enzymatic activity of GSNOR1 (Fig. 3a-c) but not gene expression (Fig. 3d), we considered a more direct role for NO in regulating the GSNOR1 enzyme. We examined this in genotypes with impaired and enhanced (S)NO signalling. Importantly, NO-overproducing nox1 mutants displayed significantly reduced GSNOR1 activity (Fig. 3e). Conversely, nia1 nia2 double mutants that cannot synthesize NO through the NR pathway, exhibited increased GSNOR1 activity to similar levels as 35S::FLAG-GSNOR1 plants. Collectively, these data suggest that the nitrogen assimilatory pathway inhibits GSNOR1 by a post-transcriptional, NO-dependent mechanism.
NO-induced S-nitrosylation inhibits GSNOR1
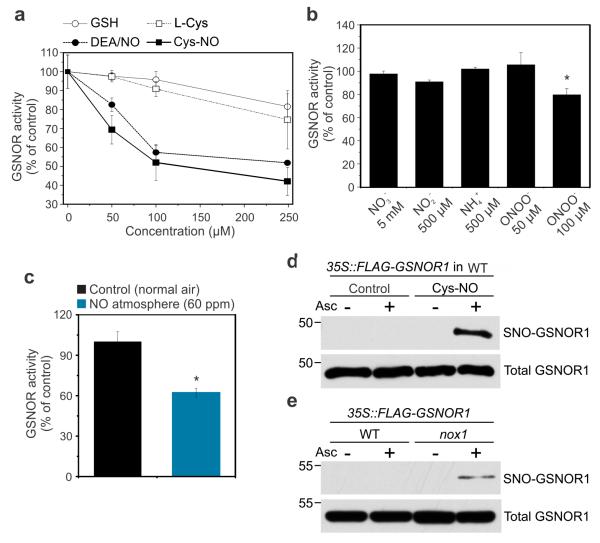
To further investigate if GSNOR1 is inhibited directly by NO or by other nitrogen assimilates, we measured its in vitro activity in pharmacological assays. Addition of the redox-active NO donors diethylamine NONOate (DEA/NO) and Cys-NO to WT leaf extracts resulted in dose-dependent inhibition of GSNOR1 with 15-30% decrease in activity already at only 50 μM of NO donors and over 60% at 250 μM (Fig. 4a). In contrast, the redox-active molecules glutathione (GSH) and L-Cysteine (L-Cys) that do not donate NO, had relatively little effect on GSNOR1 activity. Similarly, incubation with physiologically relevant concentrations of nitrogen assimilates (nitrate, nitrite, and ammonium) did not affect GSNOR1 activity with exception of high concentrations of the NO-related signal molecule peroxynitrite (ONOO−), which is formed by reaction of NO and O2− (Fig. 4b)41. Furthermore, when WT plants were fumigated for 12 hours with 60 ppm of NO gas, GSNOR activity in leaves was nearly 40% lower when compared to those exposed to normal air (44.6 ± 2.6 versus 71.7 ± 5.4 nmol NADH min−1 mg−1 prot) (Fig. 4c). Taken together, these data demonstrate a direct inhibitory effect of NO on GSNOR activity.
Figure 4. NO inhibits GSNOR1 by S-nitrosylation.
(a) Dose-response of GSNOR activity in WT leaf extracts supplemented with the S-nitrosylating agents DEA/NO and Cys-NO or the redox-active molecules GSH and L-Cys. (b) GSNOR activity in WT leaf extracts supplemented with indicated intermediates of the nitrogen assimilation pathway. (c) GSNOR activity in leaf extracts of WT plants fumigated with NO gas (60 ppm) or with normal air for 12h. (d) Cys-NO induced S-nitrosylation of GSNOR1 in vitro. Leaf extracts from 35S::FLAG-GSNOR1 plants in WT background were exposed to Cys-NO and subjected to the biotin switch technique. Total GSNOR1 protein ensures equal protein loading. The position of a 50 kDa marker is indicated. (e) In vivo S-nitrosylation of GSNOR1 using a 35S::FLAG-GSNOR1 construction in WT and nox1 background. SNO-GSNOR1 was analysed and detected as in (d) The position of a 55 kDa marker is indicated. Data points from (a) to (c) represent means ± SD of at least three independent experiments. Asterisks indicate significant differences from the controls (Student’s t test, P < 0.05).
NO has been well documented to regulate protein function by S-nitrosylation24,42. Therefore, we employed the biotin switch technique to examine if GNSOR1 is subjected to S-nitrosylation. This technique relies on specific reduction of SNO groups by ascorbate followed by their labelling with biotin43. Extracts of plants expressing 35S::FLAG-GSNOR1 (Supplementary Figs. 1 and 7) where treated with or without the NO donor Cys-NO and subjected to the biotin switch technique. Fig. 4d shows that Cys-NO induced strong S-nitrosylation of FLAG-GSNOR1 protein that was completely dependent on addition of ascorbate during biotin switching, indicating that GSNOR1 can be S-nitrosylated in vitro.
Next, we assessed if GSNOR1 is also S-nitrosylated in vivo by examining SNO modifications in NO overproducing nox1 plants that exhibit reduced GSNOR1 activity (Fig. 3e). To that end we crossed 35S::FLAG-GSNOR1 expressing plants with nox1 mutants and applied the biotin switch technique on the resulting homozygous progeny in which FLAG-GSNOR1 protein accumulated to comparable levels as the parent line (Supplementary Fig. 7). Whereas most FLAG-GSNOR1 protein was largely unmodified in wild-type plants, it was significantly S-nitrosylated in nox1 mutants (Fig. 4e). Taken together, these data indicate that nitrate-derived NO prevents scavenging of its major storage form by inhibitory S-nitrosylation of GSNOR1.
Discussion
As immobile organisms, plants have evolved to cope with environmental fluctuations by fine tuning metabolic pathways. Nitrogen metabolism is of particular importance as its intermediates influence plant development and responses to stress. Our study shows that NO, one of the end products of nitrogen metabolism, feedback regulates flux through nitrate assimilation pathways and controls its bioavailability by modulating its own consumption as depicted in Figure 5.
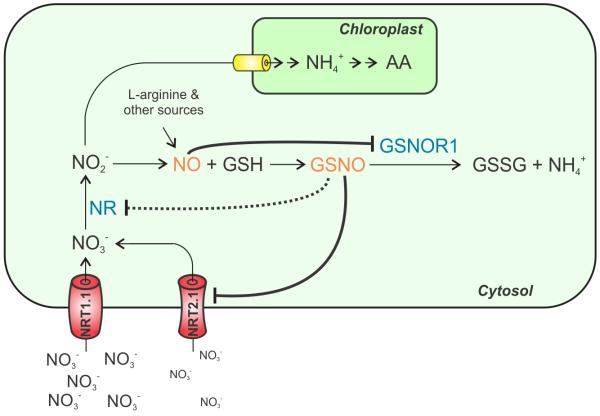
Figure 5. Schematic model for the control of nitrogen assimilation in plants through nitric oxide signalling.
Nitrate (NO3−) is taken up by nitrate transporters (NRT) in roots and reduced in leaves to nitrite (NO2−) by nitrate reductase (NR). Besides the transport to chloroplasts where it is reduced to ammonium (NH4+) and incorporated into amino acids (AA), nitrite can be reduced to nitric oxide (NO) by any favourable reducing power. NO is also thought to be generated from other sources, such as L-arginine. NO reacts with reduced glutathione (GSH) producing S-nitrosoglutathione (GSNO), the major cellular reservoir of NO. The levels of GSNO are controlled by the enzyme GSNO reductase (GSNOR), which catalyses the reduction of GSNO to oxidized glutathione (GSSG) and ammonium (NH3). GSNO inhibits nitrate uptake and reduction and NO S-nitrosylates and inhibits GSNOR preventing GSNO degradation. In this way NO, one of the end products of nitrogen metabolism, feedback regulates flux through nitrate assimilation pathways and controls its bioavailability by modulating its own consumption.
Previously it has been suggested that a metabolite resulting from nitrate reduction may feedback regulate nitrate uptake systems, but the identity of this metabolite remained unknown15,16,23. Genetic manipulation of NO signalling in our experiments illustrated that NO controls flux through nitrogen assimilatory pathway by modulating the expression of nitrate transporters and activity of NR (Fig. 1). Mutants that accumulate NO or GSNO displayed a classical switch in gene expression from high- to low-affinity transport, which is typically associated with decreased uptake of exogenous nitrate4. Moreover, genetically elevated levels of GSNO inhibited activity of NR, while reduced levels promoted its activity. We show that the cumulative effects of perturbed NO signalling on nitrate uptake and reduction determined leaf nitrate content (Fig. 1), homeostasis of primary transport amino acids (Fig. 2 e-g and Supplementary Fig. 3) and affected plant growth vigour (Fig. 2a-d). This NO-dependent mechanism may ensure the adjustment of plant growth according to nitrate availability.
In higher plants, NO is probably generated through a variety of mechanisms, including nitric oxide synthase-like activities, polyamine biosynthetic pathways, and mitochondrial or peroxisomal pathways20,23. Notably, however, significant amounts of NO are also thought to be generated through a NR-dependent process, which may be particularly important in root architecture44, floral transition45, responses to abiotic stresses46,47, and immune responses22,48. Thus, by suppressing nitrate uptake and reduction, NO may not only regulate nitrogen assimilation fluxes, it probably also feedback regulates its own generation.
Remarkably, NR activity was coupled to the level of functional GSNOR1 (Figs. 1c and 3e). As GSNO often regulates enzyme activity through S-nitrosylation, it is tempting to speculate NR is also subject to this post-translational modification. Indeed, NR is known to be regulated by other post-translational mechanisms, including phosphorylation and degradation. NIA2 was shown to interact with mitogen-activated protein kinase 6 (MPK6), resulting in site-specific phosphorylation that promoted NR activity44. Furthermore, phosphorylation of a distinct residue was shown to recruit inhibitory 14-3-3 proteins and may also promote NR proteolysis-49-51. While we observed impaired NR activity in gsnor1 plants that are deficient in functional GSNOR1, no effect was seen in NO overproducing nox1 plants (Fig. 1). Because NO donor stereochemistry and structure as well as allosteric effectors have a large influence on SNO reactivity52, these data imply that NO and GSNO do not always modify the same target proteins. Indeed, NO radicals are thought to S-nitrosylate proteins directly through a radical-mediated pathway or indirectly via higher oxides of NO, whereas GSNO trans-nitrosylates cysteine residues53.
Feeding experiments not only confirmed the previously described ability of nitrate to promote NR activity54, they also demonstrated that elevated nitrate levels suppress GSNOR1 activity (Fig. 3). GSNOR1 plays an important role in controlling the cellular levels of GSNO, which is thought to be the main NO reservoir in cells. Accordingly, mutation of GSNOR1 leads to elevated levels of protein-SNO31, indicating that GSNO functions as a potent cellular NO donor. As high NR activity promoted generation of NO (Fig. 3b, 3c), inhibition of GSNOR1 may be necessary to amplify SNO signals. Indeed, storing NO as GSNO dramatically prolongs its half-life38, perhaps enabling plants to utilize NO more efficiently while curbing loss due to emission. Taken together, our data illustrate that nitrate availability promotes formation of a more stable pool of NO, which in turn feedback regulates nitrate assimilation, allowing plants to finely tune nitrogen homeostasis. They also indicate that nitrogen-based nutrient availability may influence a variety of NO-mediated signalling events. This is supported by recent reports showing that the form of nitrogen assimilation determines NO-mediated immune responses55,56. Arabidopsis nia1 nia2 mutants are susceptible to the bacterial pathogen Pseudomonas syringae, even after amino acid recovery by feeding with Gln56. Furthermore, treatment of wild-type tobacco plants with ammonium bypassed NR-mediated generation of NO and consequently compromised immune responses55. By contrast, application of nitrate or nitrite promoted both NO formation and immune-induced hypersensitive cell death, a process that restricts pathogen growth and is known to be stimulated by SNO26,55,56.
It should be noted that although nitrate-induced NO emission was strongly reduced in absence of functional NR, residual NR-independent NO emission is still observed in nia1 nia2 (Fig. 3c, Supplementary Fig. 6). Taken together with the fact that NR-independent NO overproduction in nox1 mutants decreased nitrate content in part by suppressing nitrate transport (Fig. 1), these data indicate that NR-independent NO emission may also contribute to nitrate homeostasis (Fig. 5).
We showed that in vitro application of intermediates of nitrate assimilation did not affect GSNOR1 activity, whereas application of NO donors specifically blocked its activity even at low dosage (Fig. 4a,b). Accordingly, genetic manipulation of NO levels in nox1 and nia1 nia2 plants (Fig. 3e) and direct fumigation of NO gas on WT plants (Fig. 4c) also impacted GSNOR1 activity in planta. Remarkably, the inhibitory effect of NO was associated with S-nitrosylation of GSNOR1 both in vitro and in vivo (Fig. 4d, e), indicating that this NO scavenging enzyme is itself subject to direct regulation by NO. So how does S-nitrosylation inhibit the activity of GSNOR1? Elucidation of the crystal structure of tomato GSNOR1 indicated the presence of a number of important cysteine residues that might serve as sites for S-nitrosylation57. Two clusters of cysteine residues coordinate binding of two zinc atoms with catalytic and structural roles. The catalytic zinc atom may be necessary for coordination of the substrate and coenzyme NAD+. Thus, S-nitrosylation of any cysteine residue within the catalytic cluster would prevent coordination of zinc and disrupt the substrate or NAD+ binding pockets. Alternatively, S-nitrosylation of cysteines within the structural cluster may prevent GSNOR1 from folding appropriately. The exact site of S-nitrosylation of GSNOR1 and associated inhibitory mechanism remain to be determined. Regardless of these details, our data show NO directly regulates GSNOR1 through post-translational modification and suggest a novel mechanism by which NO controls its own bioavailability (Fig. 5).
Taken together with our biochemical and genetic evidence that nitrate assimilation is feedback repressed by NO, we conclude that NO is at the centre of fine-tuning nitrogen homeostasis in plants. These findings raise important considerations for the impacts of nitrogen-based fertilizers on redox-mediated traits in agricultural crops. Nitrogen is a major nutrient required for plant growth and development and for this reason insufficient N in soil severely restricts the use of potential agricultural lands. To circumvent this limitation, application of nitrate-based fertilizers has been the most widely used method to increase crop yields. However, the unutilized nitrate in agricultural fields is one of the main sources of environmental N pollution, as well economic losses58. Therefore, understanding the physiological basis involved in the adjustment of plant growth in response to nitrate availability is essential for the development of crop plants either adapted to N-limiting conditions or with high efficiency in nitrogen assimilation 59. The present identification of NO as a key element for adjustment in plant growth according to nitrate availability generates an important basis for future research programs to attain higher yields and promote a reduction in fertilizer-based environmental pollution.
Methods
Plant materials, growth conditions and treatments
Arabidopsis thaliana Columbia-0 wild-type (WT), and the mutants nia1 nia214, gsnor1 (par2-1)34 and nox1 (cue1-6)24, as well as the transgenic lines 35S::FLAG-GSNOR1 in WT and nox1 backgrounds were grown in soil in a controlled environmental chamber at 20-22 °C, 65% RH and a photoperiod of 16/8 hours light/dark. The form and content of N in the soil was not determined and the plants were irrigated with water as needed. Where indicated nitrate availability was controlled by growing plants in perlite:vermiculite (1:1) under a 12/12 hours light/dark period. Plants were irrigated with Murashige-Skoog (MS)60 nutrient solution three times a week. In these treatments the composition of inorganic N was altered from the original one in a way that nitrate supply was composed of half KNO3 and half NH4NO3. Four-week-old plants were used for the experiments.
For analysis of gene expression in roots, seeds were surface sterilized with 10% bleach for 5 min, washed 3 times with sterile water, and sown aseptically in petri dishes containing half-strength MS medium (in which the N source is composed of 9.4 mM KNO3 and 10.3 mM NH4NO3). Petri dishes were maintained vertically in a photoperiod of 16/8 hours light/dark at 20-22 °C. After 15 days seedlings were gently lifted from petri dishes using forceps and analysed immediately or incubated in 10 mL of sterile deionized water containing 1 mM KNO3 supplemented with or without GSNO or DEA/NO at room temperature for 3 hours. Roots were then separated from the shoot with the aid of a scalpel and RNA extraction was carried out as described below.
Construction of transgenic 35S::FLAG-GSNOR1 plants
The full-length GSNOR1 gene was multiplied from cDNA and TOPO cloned into the Gateway compatible pENTR/SD/D-TOPO vector according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The resulting pENTR/GSNOR1 clone was linearized with the restriction enzyme MluI to prevent subsequent transformation of the entry vector into E. coli. Using LR clonase (Invitrogen), the GSNOR1 sequence flanked by the entry vector’s attL recombination sites were recombined into the plant transformation vector pEarleyGate 20261, which contains an N-terminal FLAG epitope tag driven by a cauliflower mosaic virus 35S promoter. The resulting pEarleyGate 202/35S::FLAG-GSNOR1 vector was transformed into Agrobacterium tumefaciens strain GV3101(pMP90), which was subsequently used to transform WT plants by floral dipping62. Transgenic plants were selected on soil by repeated spraying with glufosinate ammonium. A homozygous transgenic line with appropriate transgene expression was isolated by immunoblotting for FLAG and crossed into nox1 mutants.
Plant NO fumigation
WT plants grown in perlite:vermiculite (1:1), irrigated with MS nutrient solution containing 12.5 mM nitrate and maintained in a growth chamber with a 12h photoperiod were fumigated with NO gas63. Briefly, plants were transferred to an acrylic fumigation chamber for 12 h. Exposure to NO (60 ppm) was performed by bubbling of NO gas with a continuous flow of 90 mL min−1 (200 ppm diluted in N2) plus 210 mL min−1 of commercial air. In the control assays, a total flow of 300 mL min−1 of air was applied. Subsequently, fumigated leaves were collected and prepared for measurement of GSNOR activity.
Measurement of GSNO Reductase activity
GSNO reductase activity in leaf extracts was measured spectrophotometrically as the rate of NADH oxidation in the presence of GSNO31. Briefly, total leaf protein was extracted in 20 mM HEPES buffer (pH 8.0), 0.5 mM EDTA and proteinase inhibitors (50 μg mL−1 TPCK; 50 μg mL−1 TLCK; 0.5 mM PMSF). Protein concentrations were measured with a Bio-Rad Protein Assay Kit II (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions and adjusted to either 62.5 μg (for nia1 nia2, which have very low protein content) or 125 μg (for all remaining genotypes). Protein extracts were incubated in 1 mL of reaction buffer containing 20 mM HEPES buffer (pH 8.0), 350 μM NADH and 350 μM GSNO. GSNO Reductase activity was determined by subtracting NADH oxidation in the absence of GSNO from that in the presence of GSNO. All samples were protected from light during the assay and tested for linearity. Where indicated protein extracts were pre-incubated for 20 min with intermediates of nitrogen metabolism or NO signalling molecules at the stated concentrations prior to addition of the reaction buffer.
Measurement of nitrate reductase activity
Nitrate reductase activity was measured as the rate of NO2− production64. Briefly, total leaf protein was extracted in 20 mM HEPES (pH 8.0), 0.5 mM EDTA, 10 μM FAD, 5 μM Na2MoO4, 6 mM MgCl2 and proteinase inhibitors (50 μg mL−1 TPCK; 50 μg mL−1 TLCK; 0.5 mM PMSF). A total of 50 μg protein was incubated in 300 μL of extraction buffer supplemented with 10 mM KNO3 and 1 mM NADH. Nitrite production was determined by adding equal volumes of 1% sulphanilamide and 0.02% N-(1-naphthyl) ethylenediamine dihydrochloride in 1.5 N HCl, and absorbance measured at 540 nm on a spectrophotometer. The obtained values were compared with those of a standard curve constructed using KNO2 and normalized by protein content. All samples were protected from light during the assay.
Determination of nitrate content
Nitrate content was determined by nitration of salicylic acid65. Briefly, leaves were ground in liquid nitrogen and resuspended in 20 mM HEPES (pH8.0). After centrifugation at 10,000 g for 10 min at 4 °C, aliquots of 5 μL of supernatant were mixed with 45 μL of 5% (v/v) salicylic acid in sulfuric acid for 20 min. The solution was neutralized by slowly adding 950 μL of NaOH (2 N). Absorbance was determined at 410 nm and the values obtained were compared with those of a standard curve constructed using KNO3 and normalized by protein content.
Measurement of NO emission
NO emission by leaves was determined by fluorometric analysis using 4,5-diamino-fluorescein-2 (DAF-2)66. Briefly, leaf samples of WT and nia1 nia2 plants were incubated in the dark with 10 μM DAF-2 dissolved in 10 mM phosphate buffer, pH 8.0. After 1h incubation fluorescence emission at 515 nm under an excitation at 495 nm was recorded using an F-4500 spectrofluorometer (Hitachi Ltd., Tokyo, Japan). As negative control, leaves were incubated in the presence of 200 μM of the NO scavenger 2-(4-carboxyphenyl)-4 4 5 5-tetramethyl imidazoline-1-oxyl-3-oxide (cPTIO) and the residual fluorescence subtracted.
Analysis of amino acid content
Leaf free amino acids were determined by reversed-phase HPLC56 after derivatization with o-phthaldialdehyde (OPA)67. Leaves were ground in liquid nitrogen and resuspended in methanol/chloroform/water (12:5:3, v/v). After incubation at room temperature for 24 h, the homogenate was centrifuged at 1,500 g for 30 min and the resulting supernatant mixed with chloroform/water (4:1:1.5, v/v/v). After decanting for 24 h the aqueous phase was separated and subjected to derivatization by mixing with 50 mM OPA, 1% 2-mercaptoethanol in 400 mM borate buffer pH 9.5 (1:3, v/v) for 2 min. The OPA derivatives content were determined by reverse-phase HPLC (Shimadzu Corporation, Kyoto, Japan) using a Waters Spherisorb ODS2 C-18 column (4.6 μm, 4.6 × 250 mm) eluted at 0.8 mL min−1 by a linear gradient formed by solutions A (65% methanol) and B (50 mM sodium acetate, 50 mM disodium phosphate, 1.5 mL acetic acid, 20 mL tetrahydrofuran, 20 mL methanol in 1 L water, pH 7.2). The gradient increased the proportion of solution A from 20 to 60% between 0 and 25 min, 60 to 75% from 25 to 30 min, and 75 to 100% from 30 to 50 min. The column effluent was monitored by a Shimadzu fluorescence detector (model RF-10AXL) operating at excitation of 250 nm and emission of 480 nm. Amino acids were identified by their respective retention times and values compared with those of an amino acid standard solution (AA-S-18, Sigma Aldrich, plus 250 mM asparagine, glutamine and gamma-aminobutyric acid) and normalized by fresh weight of leaf tissue.
Gene expression analysis
For real-time PCR analysis, total RNA was extracted with Trizol (Invitrogen) and ethanol precipitation, and subsequently treated with Amplification Grade DNAse I (Invitrogen, Carlsbad, CA). The cDNA was synthesized using Im-Prom II reverse transcriptase (Promega, Fitchburg, WI), as recommended by the manufacturer. Gene expression analysis was carried out using Platinum SYBR ® Green qPCR Supermix-UDG (Invitrogen, Carlsbad, CA) in a Real-Time PCR System 7500 (Applied Biosystem, Foster City, CA). Gene expression was calculated with the 2−ΔΔCt method68 with Actin2 as internal standard69. All gene-specific primers used in this study are shown in the Supplementary Table 1.
In vitro and in vivo S-nitrosylation assays
Leaf extracts from 35S::FLAG-GSNOR1 in WT or nox1 plants were mock-treated or S-nitrosylated in vitro with 500 μM of Cys-NO for 20 min in the dark. Excess Cys-NO was removed using Zeba desalting columns (Thermo Fisher Scientific) and proteins subjected to the biotin-switch technique as described previously43. Biotinylated proteins were pulled down with streptavidin agarose CL-6B (Thermo Fisher Scientific) and FLAG-GSNOR1 protein detected by western blotting with an anti-Flag M2 clone antibody (1:2,000 or 1:2,500, Sigma Aldrich, cat. no. F3165) (Supplementary Figure 8).
Supplementary Material
Acknowledgements
We thank Dr. Jianru Zuo for providing par2-1 seeds. This work was supported by a Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) PhD studentship to L.F., a Biotechnology and Biological Sciences Research Council PhD studentship to M.J.S., a Biotechnology and Biological Sciences Research Council grant to G.J.L. (no. BBH0009841), a Royal Society Research Fellowship and Research Grant to S.H.S (no. Uf090321 and Rg110495, respectively), and a Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Research grant to I.S. (no. 473090/2011-2).
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Crawford NM. Nitrate: nutrient and signal for plant growth. Plant Cell. 1995;7:859–868. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. Nitrate transporters and peptide transporters. FEBS Lett. 2007;581:2290–22300. doi: 10.1016/j.febslet.2007.04.047. [DOI] [PubMed] [Google Scholar]
- 3.Wang YY, Hsu PK, Tsay YF. Uptake, allocation and signalling of nitrate. Trends Plant Sci. 2012;17:458–467. doi: 10.1016/j.tplants.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Dechorgnat J, et al. From the soil to the seeds: the long journey of nitrate in plants. J. Exp. Bot. 2011;62:1349–1359. doi: 10.1093/jxb/erq409. [DOI] [PubMed] [Google Scholar]
- 5.Li W, et al. Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 2007;143:425–433. doi: 10.1104/pp.106.091223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu KH, Huang CY, Tsay YF. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell. 1999;11:865–874. doi: 10.1105/tpc.11.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, et al. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature. 2014;507:73–77. doi: 10.1038/nature13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker JL, Newstead S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature. 2014;507:68–72. doi: 10.1038/nature13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497:647–651. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lea PJ, Lea P, Leegood RC. Plant Biochem. Mol. Biol. John Wiley and Sons; 1993. pp. 155–180. [Google Scholar]
- 12.Martínez-Espinosa RM, Cole JA, Richardson DJ, Watmough NJ. Enzymology and ecology of the nitrogen cycle. Biochem. Soc. Trans. 2011;39:175–178. doi: 10.1042/BST0390175. [DOI] [PubMed] [Google Scholar]
- 13.Einsle O, Kroneck P. Structural basis of denitrification. Biol. Chem. 2004;385:875–883. doi: 10.1515/BC.2004.115. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson J, Crawford N. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol. Gen. Genet. 1993;239:289–297. doi: 10.1007/BF00281630. [DOI] [PubMed] [Google Scholar]
- 15.Lejay L, Tillard P, Lepetit M, Olive F. Molecular and functional regulation of two NO3-uptake systems by N-and C-status of Arabidopsis plants. Plant J. 1999;18:509–519. doi: 10.1046/j.1365-313x.1999.00480.x. [DOI] [PubMed] [Google Scholar]
- 16.Muños S, et al. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell. 2004;16:2433–2447. doi: 10.1105/tpc.104.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salgado I, Martínez MC, Oliveira HC, Frungillo L. Nitric oxide signaling and homeostasis in plants: a focus on nitrate reductase and S-nitrosoglutathione reductase in stress-related responses. Brazilian J. Bot. 2013;32:89–98. [Google Scholar]
- 18.Neill S, et al. Nitric oxide evolution and perception. J. Exp. Bot. 2008;59:25–35. doi: 10.1093/jxb/erm218. [DOI] [PubMed] [Google Scholar]
- 19.Yamasaki H, Sakihama Y, Takahashi S. An alternative pathway for nitric oxide production in plants: new features of an old enzyme. Trends Plant Sci. 1999;4:128–129. doi: 10.1016/s1360-1385(99)01393-x. [DOI] [PubMed] [Google Scholar]
- 20.Rockel P, Strube F, Rockel A. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002;53:103–110. [PubMed] [Google Scholar]
- 21.Modolo LV, Augusto O, Almeida IMG, Magalhaes JR, Salgado I. Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett. 2005;579:3814–3820. doi: 10.1016/j.febslet.2005.05.078. [DOI] [PubMed] [Google Scholar]
- 22.Modolo LV, et al. Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 2006;171:34–40. [Google Scholar]
- 23.Mur LAJ, et al. Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants. 2013;5:pls052. doi: 10.1093/aobpla/pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, et al. Nitric oxide represses the Arabidopsis floral transition. Science. 2004;305:1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- 25.Fernández-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18506–18511. doi: 10.1073/pnas.1108644108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun B, et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011;478:264–268. doi: 10.1038/nature10427. [DOI] [PubMed] [Google Scholar]
- 27.Spoel S, Loake G. Redox-based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol. 2011;14:358–364. doi: 10.1016/j.pbi.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Spadaro D, et al. The redox switch: dynamic regulation of protein function by cysteine modifications. Physiol. Plant. 2010;138:360–371. doi: 10.1111/j.1399-3054.2009.01307.x. [DOI] [PubMed] [Google Scholar]
- 29.Jensen DE, Belka GK, Du Bois GC. S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem. J. 1998;331:659–668. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 31.Feechan A, et al. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rustérucci C, Espunya MC, Díaz M, Chabannes M, Martínez MC. S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 2007;143:1282–1292. doi: 10.1104/pp.106.091686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell. 2008;20:786–802. doi: 10.1105/tpc.107.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R, et al. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19:1377–1387. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- 35.Frungillo L, et al. Modulation of mitochondrial activity by S-nitrosoglutathione reductase in Arabidopsis thaliana transgenic cell lines. Biochim. Biophys. Acta. 2013;1827:239–247. doi: 10.1016/j.bbabio.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Kwon E, et al. AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta. 2012;236:887–900. doi: 10.1007/s00425-012-1697-8. [DOI] [PubMed] [Google Scholar]
- 37.Mur LAJ, Hebelstrup KH, Gupta KJ. Striking a balance: does nitrate uptake and metabolism regulate both NO generation and scavenging? Front. Plant Sci. 2013;4:288. doi: 10.3389/fpls.2013.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamler JS, Singel DJ, Loscalzo J. Biochemistry of nitric oxide and its redox-activated forms. Science. 1992;258:1898–1902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- 39.Streatfield SJ, et al. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell. 1999;11:1609–1621. doi: 10.1105/tpc.11.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Planchet E, Jagadis Gupta K, Sonoda M, Kaiser WM. Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 2005;41:732–743. doi: 10.1111/j.1365-313X.2005.02335.x. [DOI] [PubMed] [Google Scholar]
- 41.Radi R, Cassina A, Hodara R. Nitric oxide and peroxynitrite interactions with mitochondria. Biol. Chem. 2002;383:401–409. doi: 10.1515/BC.2002.044. [DOI] [PubMed] [Google Scholar]
- 42.Skelly MJ, Loake GJ. Synthesis of redox-active molecules and their signaling functions during the expression of plant disease resistance. Antioxid. Redox Signal. 2013;19:990–997. doi: 10.1089/ars.2013.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic. Biol. Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang P, Du Y, Li Y, Ren D, Song CP. Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell. 2010;22:2981–2998. doi: 10.1105/tpc.109.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seligman K, Saviani EE, Oliveira HC, Pinto-Maglio CAF, Salgado I. Floral transition and nitric oxide emission during flower development in Arabidopsis thaliana is affected in nitrate reductase-deficient plants. Plant Cell Physiol. 2008;49:1112–1121. doi: 10.1093/pcp/pcn089. [DOI] [PubMed] [Google Scholar]
- 46.Desikan R, Griffiths R, Hancock J, Neill S. A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16314–16318. doi: 10.1073/pnas.252461999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao L, et al. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 2004;134:849–857. doi: 10.1104/pp.103.030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto-Katou A, Katou S, Yoshioka H, Doke N, Kawakita K. Nitrate reductase is responsible for elicitin-induced nitric oxide production in Nicotiana benthamiana. Plant Cell Physiol. 2006;47:726–735. doi: 10.1093/pcp/pcj044. [DOI] [PubMed] [Google Scholar]
- 49.Moorhead G, et al. Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr. Biol. 1996;6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- 50.Su W, Huber SC, Crawford NM. Identification in vitro of a post-translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell. 1996;8:519–527. doi: 10.1105/tpc.8.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaiser WM, Huber SC. Post-translational regulation of nitrate reductase: mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 2001;52:1981–1989. doi: 10.1093/jexbot/52.363.1981. [DOI] [PubMed] [Google Scholar]
- 52.Foster MW, Forrester MT, Stamler JS. A protein microarray-based analysis of S-nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kovacs I, Lindermayr C. Nitric oxide-based protein modification: formation and site specificity of protein S-nitrosylation. Front. Plant Sci. 2013;4:137. doi: 10.3389/fpls.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutiérrez RA, et al. Insights into the genomic nitrate response using genetics and the Sungear Software System. J. Exp. Bot. 2007;58:2359–2367. doi: 10.1093/jxb/erm079. [DOI] [PubMed] [Google Scholar]
- 55.Gupta KJ, et al. The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 2013;64:553–568. doi: 10.1093/jxb/ers348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliveira HC, Justino GC, Sodek L, Salgado I. Amino acid recovery does not prevent susceptibility to Pseudomonas syringae in nitrate reductase double-deficient Arabidopsis thaliana plants. Plant Sci. 2009;176:105–111. [Google Scholar]
- 57.Kubienová L, et al. Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum. Biochimie. 2013;95:889–902. doi: 10.1016/j.biochi.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 58.Hakeem KR, Ahmad A, Iqbal M, Gucel S, Ozturk M. Nitrogen-efficient rice cultivars can reduce nitrate pollution. Environ. Sci. Pollut. Res. Int. 2011;18:1184–1193. doi: 10.1007/s11356-010-0434-8. [DOI] [PubMed] [Google Scholar]
- 59.Guevara DR, El-Kereamy A, Yaish MW, Mei-Bi Y, Rothstein SJ. Functional characterization of rice UDP-glucose 4-epimerase 1, OsUGE1: A potential role in cell wall carbohydrate partitioning during limiting nitrogen conditions. Plos One. 2014;9:e96158. doi: 10.1371/journal.pone.0096158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murashige T, Skoog F. A Revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 61.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 62.Bent AF. Arabidopsis in planta transformation. Uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 2000;124:1540–1547. doi: 10.1104/pp.124.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vitor SC, et al. Nitrate reductase is required for the transcriptional modulation and bactericidal activity of nitric oxide during the defense response of Arabidopsis thaliana against Pseudomonas syringae. Planta. 2013;238:475–486. doi: 10.1007/s00425-013-1906-0. [DOI] [PubMed] [Google Scholar]
- 64.Lea US, Leydecker MT, Quilleré I, Meyer C, Lillo C. Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol. 2006;140:1085–1094. doi: 10.1104/pp.105.074633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cataldo DA, Haroon M, Schrader LE, Youngs VL. Rapid colorimetric determination of nitrate in plant tissues by nitration of salicylic acid. Commun. Soil Plant Anal. 1975;6:71–80. [Google Scholar]
- 66.Santos-Filho PR, et al. Nitrate reductase- and nitric oxide-dependent activation of sinapoylglucose:malate sinapoyltransferase in leaves of Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1607–1616. doi: 10.1093/pcp/pcs104. [DOI] [PubMed] [Google Scholar]
- 67.Puiatti M, Sodek L. Waterlogging affects nitrogen transport in the xylem of soybean. Plant Physiol. Biochem. 1999;37:767–773. [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.