Abstract
The insulin-like growth factor receptor-1 (IGF1R) plays an important role in cancer progression. Previous studies have been controversial with respect to the associations between IGF1R expression and non small cell lung cancer (NSCLC) prognosis. Thus, we performed a meta-analysis to investigate the prognostic value of IGF1R expression in NSCLC patients and the relationship between the expression of IGF1R and clinical characteristics. Two independent reviewers searched PubMed, Embase, Ovid Medline and CNKI to identify eligible studies. Overall survival (OS), disease free survival (DFS) and clinicopathological characteristics were collected from included studies. Pooled hazard ratios (HRs) or odds ratios (ORs) with 95% confidence interval (95% CI) were calculated to estimate the effect. 17 studies comprising 3,294 patients were included in this meta-analysis. The results showed IGF1R positive expression was associated with an unfavorable DFS in NSCLC patients on univariate analysis (HR = 1.26, 95% CI: 1.09-1.46, P = 0.002) and multivariate analysis (HR = 1.49, 95% CI: 1.01-2.20, p = 0.045), but the relationship between IGF1R expression and OS have no significant difference on univariate analysis (HR = 0.91, 95% CI: 0.82-1.01, P = 0.157) and multivariate analysis (HR = 0.79, 95% CI: 0.45-1.41, P = 0.427). Ever smoking and smaller tumor size (T1 or T2) were associated with IGF1R positive expression: pooled OR 1.45 (1.13-1.85) and pooled OR 0.61 (0.60-0.95). Our results suggested IGF1R positive expression as an unfavorable factor for DFS in NSCLC patients, and IGF1R expression was associated with smoking status and tumor size.
Keywords: Insulin-like growth factor receptor-1 (IGF1R), non-small cell lung cancer (NSCLC), prognosis, disease free survival (DFS), overall survival (OS), meta-analysis
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. About 1.5 million new cases of lung cancer are diagnosed annually [1] with 85% being non small-cell lung cancers (NSCLCs) [2]. Novel therapeutic developments in NSCLC have resulted in only minor improvement of patient outcomes, its 5-year overall survival (OS) is still only about 17% [3]. During the last few years, several new agents targeting critical and specific pathways for lung cancer have been evaluated in both preclinical and clinical models.
The insulin-like growth factor (IGF) pathway was regulated by a family of six IGF binding proteins (IGFBP), which were structurally related. The extracellular pathway components had two ligands (IGF1 and IGF2), two cell membrane receptors (IGF1R and IGF2R) and their binding proteins (IGFBP1-6) [4,5]. The insulin-like growth factor receptor-1 (IGF1R) is a transmembrane heterotetrameric protein encoded by a gene located on chromosome 15q25-26 [6]. It comprised two half-receptors, each consisting of one extracellular alpha-subunit and one transmembrane beta-subunit, which can possess tyrosine kinase activity [7]. By regulating its downstream signaling, IGF1R plays an important role in cancer cell growth, survival, metabolism and transformation [8-11]. Studies have demonstrated that IGF1R overexpression was associated with disease progression, poor prognosis and treatment resistance in breast cancer [12,13], esophagus adenocarcinoma [14], colorectal cancer [15], and the squamous cell carcinoma of head and neck [16]. In lung cancer, the plasma levels of IGF-1 have been associated with an increased risk of the disease and high plasma levels of IGFBP3 have been associated with reduced risk [17,18]. A meta-analysis by Huang indicated the genetic variations of IGF1R may be associated with increased risk of lung, especially among Asian populations [14]. Nevertheless, there was no meta-analysis of IGF1R protein expression in patients with non small cell lung cancer.
Given the impact of IGF-1R signaling on the development and progression of several types of cancer, researchers have long studied the prognostic significance of IGF1R protein expression in patients with NSCLC. However, the results of different studies are controversial and the prognostic role of IGF1R expression in NSCLC still remains unclear. Thus, the objective of our meta-analysis was to evaluate the potential relationship of the IGF1R expression with the clinical characteristics, disease free survival and overall survival in NSCLC patients.
Subjects and methods
Publication selection
Relevant studies were screened by an electronic search in PubMed, Embase, Ovid Medline and CNKI database from 1946 to March 2014, with a key word from amongst one of the following words: “non small cell lung cancer”, “lung cancer”, “lung carcinoma” or “lung neoplasm”. These were combined with “insulin-like growth factor-1 receptor,” “insulin like growth factor receptor-1” OR “IGF-1R”. Published studies were sought with no language restrictions or the minimum number of patients. Titles and abstracts were evaluated to identify related studies, and then full texts were read carefully. The eligible studies for inclusion in this meta-analysis had to meet the following criteria: 1. Expression of IGF1R was measured by immunohistochemistry (IHC), quantitative reverse transcription polymerase chain reaction (qRT-PCR) or fluorescence in situ hybridization (FISH). 2. Diagnosis of NSCLC was proven by histopathological methods and complied with the diagnosis criteria of the World Health Organization (WHO). Small-cell lung cancer was not included in our study, due to its highly malignant and undifferentiated cancer with a distinct pathology from NSCLC. 3. The patients did not undergo adjuvant therapy before surgery, and the tissue specimens were obtained prior to any treatment. The samples were surgically resected lung cancer tissues, rather than body fluids such as peritoneal fluid, serum and sputum. 4. The studies offered sufficient data for estimating hazard ratios (HRs) and their 95% confidence intervals (95% CIs).
Methodological assessment
To evaluate the methodological quality of each study, two independent investigators read and scored all the articles according to the assessment of European Lung Cancer Working Party quality scale for biological prognostic factors for lung cancer, which has been used in other similar meta-analyses widely [19]. The following four main dimensions were evaluated: scientific design, laboratory methodology, generalizability and results analysis. Each dimension had a maximal score of ten, with an overall maximum score of 40. Each item scored two points if it is clearly defined in the article, one point if its description is incomplete or unclear and zero point if it is not mentioned or inadequate. The each investigator’s scores were compared and disagreements were discussed. Finally, a consensus was reached. The final scores was calculated in the total of four main dimensions, which were expressed as percentages (0%-100%), and higher values indicated better methodological quality.
Data extraction
Two reviewers evaluated the articles, extracted data and checked all potentially relevant studies independently. All disagreements between the findings of the two reviewers in the data extraction were resolved by discussion and consensus, and if necessary, were adjudicated by a third reviewer. The following information from each article were extracted: first author, year of publication, country, number of patients, follow-up period, disease stage, cut-off score, detection method, IGF-1R positive ratio and HR estimation. From some published researches, HR and 95% CI could be directly obtained by using survival analysis. Otherwise, for articles which didn’t provide HR and 95% CI directly, two reviewers independently digitized and extracted the data through the Kaplan-Meier curves by using GetData Graph Digitizer 2.24 (http://getdata-graph-digitizer.com) and then extracted data were utilized to reconstruct the HR and its variance according to previously described methods [20,21].
Statistical methods
STATA 12.0 software (STATA Corp., College Station, TX) and Revman 5.2 software (Cochrane Collaboration, Copenhagen) were used to perform statistical analysis [22]. The associations between IGF1R expression and survival were described as HRs, and the strength of association between IGF1R and clinical characteristics were expressed as odds ratios (ORs). By convention, a pooled HR > 1.00 indicated an unfavourable survival for the group with IGF1R positive expression, and the effect of IGF1R expression on survival was considered to be statistically significant when the 95% CI for the overall HR did not overlap 1. The heterogeneity among studies was examined by the Cochrane’s Q test (Chi-squared test; Chi2) and inconsistency (I2) statistics, p < 0.05 was considered to be statistically significant [23]. When there was no significant heterogeneity among studies, the fixed effects model was employed to combine the individual HR estimates. Otherwise, the random effects model was used [24]. The distribution of score measurement according to the discrete variable was compared by non-parametric tests using SPSS 19.0. To evaluate the stability of the results, a sensitivity analysis was performed, in which one study was removed to know the influence of the individual study on the pooled HR [25]. Publication bias was investigated by Egger linear regression tests and funnel plots, P < 0.05 was considered to indicate statistically significant publication bias [26].
Results
Study selection and characteristics
715 studies were retrieved initially using the above search strategy. Titles and abstracts screened and the full-text articles reviewed, eventually, a total of 17 independent studies [27-43] were used in the present meta-analysis (15 in English and two in Chinese) (Figure 1).
Figure 1.
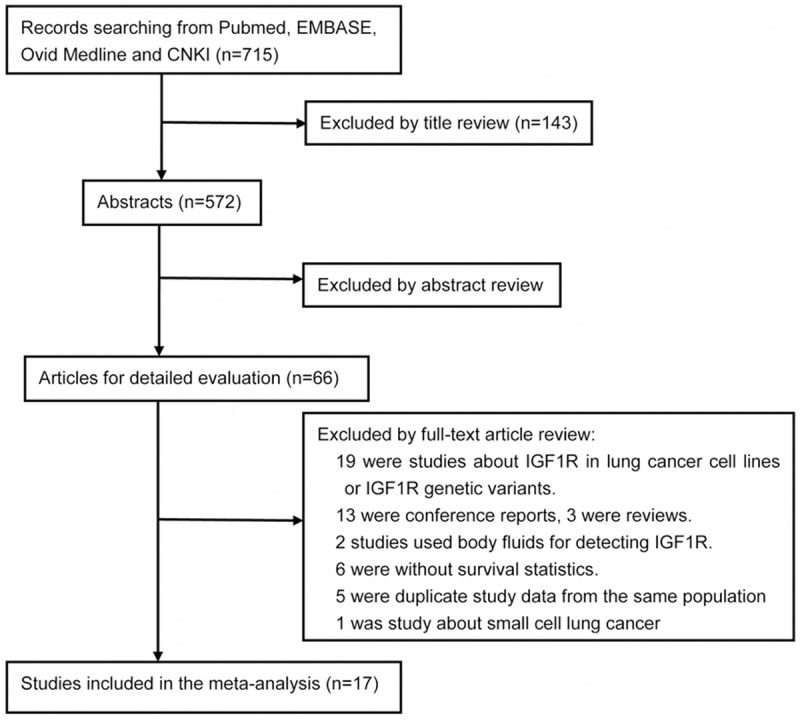
Flow chart of the literature search and selection of included studies.
The main characteristics of these studies are shown in Table 1. Of these studies, two were conducted in the United States, two in Europe and 13 in Asia. Overall, 3294 patients were included, with sample sizes ranging from 39 to 459 individuals. The mean follow-up period for the studies was 53.0 months (range 24 to 68.7 months). The proportion of cases with positive IGF1R expression ranged from 12.68% to 81.44%, with a median of 50.33%. Ten studies involved all disease stages, six studies included only early stage disease (I-III) and one studies included only late stage disease (III-IV). Among all the studies, four studies (23.5%) identified IGF1R positive expression as a significant poor prognosis factor, four studies (23.5%) reported that IGF1R positive expression as a good prognosis factor and nine studies (53.0%) reported that IGF1R expression as non-significant association with prognosis (P > 0.05).
Table 1.
Main characteristics of studies included in the meta-analysis
| Author | Year | Country | No. | Follow-up (median) | stage | Cut-off value | Detection method | IGF1R positive ratio (%) | Quality score (%) | Results |
|---|---|---|---|---|---|---|---|---|---|---|
| Gately K. | 2013 | Ireland | 184 | 65.09 | I-III | Score ≥ 200 | IHC | 53.80 | 70.78 | N* |
| Zhang XY | 2013 | China | 178 | 60 | I-III | Score ≥ 20 | IHC | 71.35 | 67.65 | N* |
| Xu C. | 2013 | China | 200 | 52.6 | I-IV | Score ≥ 4 | IHC | 78.00 | 55.59 | P |
| Ludovini V. | 2013 | Italy | 125 | 48.9 | I-III | > 10% | IHC, FISH | 36.80 | 61.67 | N* |
| Yamamoto T. | 2012 | Japan | 78 | 48.87 | I-III | Score ≥ 200 | IHC | 52.56 | 64.37 | P |
| Tsuta K | 2012 | Japan | 379 | 58.6 | I-IV | > 10% | IHC | 41.42 | 54.26 | N* |
| Kim YH | 2012 | Japan | 68 | 30 | III-IV | > 10% | IHC | 54.41 | 63.18 | N |
| Kim JS | 2012 | Houston | 459 | 49.2 | I-III | > 10% | IHC | 39.22 | 55.74 | P |
| Kikuchi R. | 2011 | Japan | 238 | 56.5 | I-IV | Score ≥ 2 | IHC | 55.04 | 60.21 | N |
| Nakagawa M. | 2011 | Japan | 182 | 68.7 | I-IV | > 10% | IHC | 23.62 | 74.64 | P |
| Dziadziuszko R. | 2010 | Poland | 189 | 63.6 | I-IV | Score ≥ 2 | IHC, qRT-PCR | 45.56 | 65.71 | N |
| Ning XH | 2010 | China | 39 | 35 | I-IV | Score ≥ 1 | IHC | 53.84 | 54.89 | N* |
| Cappuzzo F. | 2009 | Italy | 369 | 60 | I-III | score ≥ 100 | IHC | 76.4 | 58.01 | N* |
| Chang MH | 2009 | Korea | 194 | 60 | I-IV | Score ≥ 1 | IHC | 81.44 | 68.99 | N* |
| Gong YX | 2009 | Manhattan | 264 | 60 | I-IV | > 10% | IHC | 39.39 | 48.30 | N* |
| Lee CY | 2008 | Korea | 71 | 60 | I-IV | Score ≥ 1 | IHC | 12.68 | 57.82 | N* |
| Cappuzzo F. | 2006 | Italy | 77 | 24 | III-IV | Score ≥ 2 | IHC | 38.96 | 62.29 | N |
No.: Patients number; IHC: Immunohistochemistry; FISH: Fluorescence in situ hybridization; IGF1R: Insulin-like growth factor-1 receptor; qRT-PCR: Quantitative reverse transcription polymerase chain reaction. P: Studies identitying IGF1R positive expression as significant poor prognosis factor. N: Studies reporting IGF1R positive expression as good prognosis factor.
Studies reporting IGF1R positive expression as non-significant association with prognosis.
Quality assessment
The global quality score for all eligible studies ranged from 48.30% to 74.64% with a median of 61.42% (Table 1). There was no significant difference between studies with positive, negative and non-significant results (mean of 62.59% vs. 62.84% vs. 60.26%, P = 0.783). Similarly, no statistical difference was appeared in global score between studies involving Asian (N = 13) or non-Asian populations (N = 4), with scores of 61.81% and 60.13%, respectively (P = 0.681). Moreover, there was no statistically significant correlation between patient number and the global score (P = 0.762). There was also no significant association between publication year and the global score (P = 0.332). Thus, in all the studies, no significant methodological qualitative difference was observed between different subgroups. (Supporting Information Table S1).
IGF1R expression and clinical outcomes in patients with NSCLC
Among all the studies, 12 studies have analyzed the relationship between the IGF1R positive expression and overall survival (OS) in patients with NSCLC on univariate analysis, only four studies have sufficient data to estimate the HR and 95% CIs on multivariate analysis (Supporting Information Table S2). As for disease free survival (DFS) in NSCLC patients, we could have only gathered six studies to conduct the meta-analysis on univariate analysis, and two studies on multivariate analysis. As shown in Figure 2, IGF1R positive expression was significantly correlated with worse DFS according to univariate analysis, with a combined HR of 1.26 (95% CI: 1.09-1.46, P = 0.002). The fixed-effects model was used because of non-significant heterogeneity was observed among these researches (P = 0.080, I2 = 49.2%). Similarly, according to multivariate analysis of two studies, IGF1R positive expression was also significantly correlated with worse DFS (HR = 1.49, 95% CI: 1.01-2.20, P = 0.045). However, no statistically significant was observed between the positive expression of IGF1R and overall survival on univariate analysis (HR = 0.91, 95% CI: 0.82-1.01, P = 0.157, fixed-effect) or multivariate analysis (HR = 0.79, 95% CI: 0.45-1.41, P = 0.427, Random-effect) (Figure 3).
Figure 2.
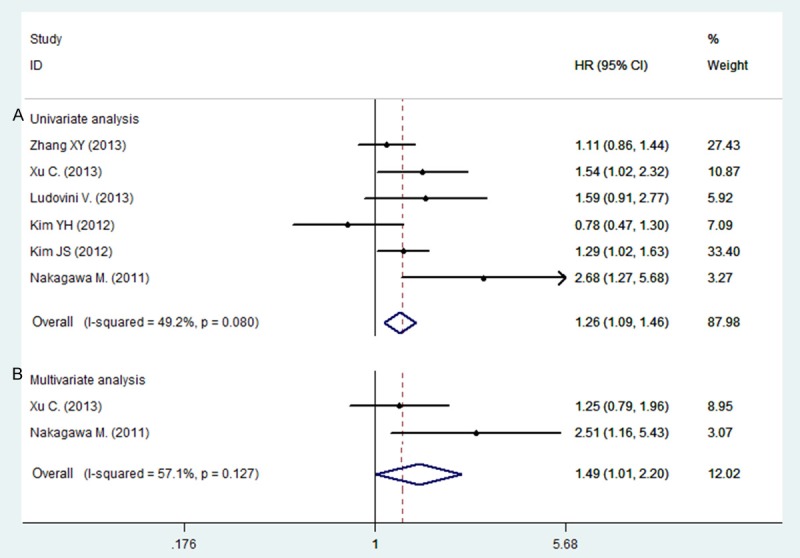
Forest plot showing the combined relative HR for disease free survival: A. univariate analysis; B. multivariate analysis.
Figure 3.
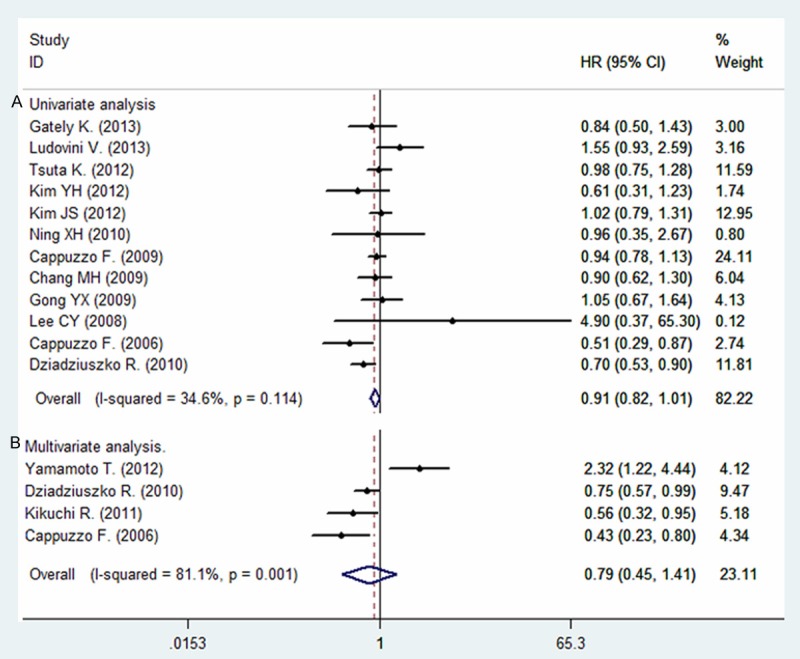
Forest plot showing the combined relative HR for overall survival: A. univariate analysis; B. multivariate analysis.
IGF1R expression and clinical characteristics
The associations between IGF1R positive expression and clinical characteristics were conducted among the available studies (Table 2). 13 studies assessed the relationship between IGF1R expression and smoking status, with a total number of 2169 patients. Four studies had sufficient data for assessing the relationship between IGF1R expression and tumor size, including 640 patients. The results suggested that the IGF1R positive expression was associated with smoking status (ever vs. none: pooled OR = 1.45, 95% CI = 1.13-1.85, P = 0.003) and tumor size (T1, 2 vs. T3, 4: pooled OR = 0.61, 95% CI: 0.60-0.95, P = 0.03). However, no significant correlations were found between IGF1R expression and gender (OR = 1.19, 95% CI = 0.98-1.43, P = p = 0.08), age (OR = 1.15, 95% CI = 0.47-2.81, P = 0.75), histological type (OR = 2.08, 95% CI = 0.88-4.93, P = 0.09), grade of tumor differentiation (OR = 1.04, 95% CI = 0.66-1.63, P = 0.87), TNM stage (OR = 0.93, 95% CI = 0.65-1.33, P = 0.71) or lymph node metastasis (OR = 1.31, 95% CI = 0.85-2.02, P = 0.22).
Table 2.
Meta-analysis assessing the association between IGF1R expression and clinical characteristics
| Clinical characteristics | No. of Studies | Cases | Pooled Data | Test for Heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| OR | 95% CI | P-value | Chi2 | P-value | I2 (%) | |||
| Gender (male/female) | 15 | 2554 | 1.19 | 0.98-1.43 | 0.08 | 16.49 | 0.28 | 15 |
| Smoking (ever/none) | 13 | 2169 | 1.45 | 1.13-1.85 | 0.003 | 22.39 | 0.03 | 46 |
| Age (< 60/≥ 60) | 3 | 397 | 1.15 | 0.47-2.81 | 0.75 | 5.43 | 0.07 | 63 |
| Tumor size (T1, 2/T3, 4) | 4 | 640 | 0.61 | 0.60-0.95 | 0.03 | 1.62 | 0.66 | 0 |
| Histological Type (SCC/ADC) | 10 | 1527 | 2.08 | 0.88-4.93 | 0.09 | 80.00 | < 0.00001 | 89 |
| Differentiation (poor/well-moderate) | 9 | 1585 | 1.04 | 0.66-1.63 | 0.87 | 23.09 | 0.003 | 65 |
| TNM Stage (INM Sta) | 7 | 1596 | 0.93 | 0.65-1.33 | 0.71 | 13.17 | 0.04 | 54 |
| Lymph Node Metastasis (NX/N0) | 6 | 1185 | 1.31 | 0.85-2.02 | 0.22 | 12.54 | 0.03 | 60 |
Abbreviation: No., number; SCC, Squamous cell carcinoma; ADC, Adenocarcinoma.
Subgroup analysis
We performed subgroup analysis in order to further explain the results of OS on univariate analysis. However, as for univariate analysis of DFS, and multivariate analysis of OS and DFS, we did not perform subgroup analysis due to their limited number of included literature. Ethnicity, patients number, follow-up period, tumor stage and quality score were included as factors in subgroup analysis (Table 3). After stratifying by patient number, the pooled HR in studies with smaller samples (N < 150) was 0.77 (95% CI: 0.63-0.94, P = 0.01), while there was no statistical significance in studies with large samples. In the subgroup analysis based on tumor stages, for late stage disease (III-IV), the association was statistically significant (HR = 0.55, 95% CI: 0.36-0.84, P = 0.006), but the pooled HR for all stages and early stages (I-III) was non-significant. Subgroup analysis stratified according to quality score, the pooled HRs of high quality studies (QS >60%) and low quality studies (QS < 60%) were 0.79 (95% CI: 0.67-0.94, P = 0.007) and 0.98 (95% CI: 0.87-1.11, P = 0.746), respectively. Similarly, when stratified by ethnicity and follow-up period, there was still no statistical significance.
Table 3.
A summary of HRs for the overall and subgroup analyses of IGF1R and overall survival of non-small cell lung cancer patients
| No. of studies | HR | 95% CI | Heterogeneity | |||
|---|---|---|---|---|---|---|
|
| ||||||
| I2 | P-value | |||||
| Overall | 12 | 0.91 | 0.82-1.01 | 34.6% | 0.114 | |
| Ethnicity | Asian | 8 | 0.93 | 0.82-1.06 | 39.8% | 0.113 |
| Non-Asian | 4 | 0.87 | 0.74-1.03 | 37.5% | 0.187 | |
| Patient numbers | > 150 | 6 | 0.96 | 0.86-1.08 | 0.0% | 0.976 |
| < 150 | 6 | 0.77 | 0.63-0.94 | 34.6% | 0.03 | |
| Follow-up period | > 60 month | 6 | 0.88 | 0.77-1.00 | 12.1% | 0.338 |
| < 60 month | 6 | 0.96 | 0.82-1.01 | 51.7% | 0.066 | |
| Tumor stage | I-III | 4 | 0.99 | 0.86-1.14 | 18.3% | 0.299 |
| I-IV | 6 | 0.87 | 0.75-1.02 | 14% | 0.325 | |
| III-IV | 2 | 0.55 | 0.36-0.84 | 0.0% | 0.690 | |
| Quality score | > 60 QS | 6 | 0.79 | 0.67-0.94 | 54.4% | 0.052 |
| < 60 QS | 6 | 0.98 | 0.87-1.11 | 0.0% | 0.867 | |
No.: Number; HR: Hazard ratio; CI: Confidence interval; IGF1R: Insulin-like growth factor-1 receptor.
Publication bias and sensitivity analysis
Publication bias was detected in this meta-analysis by using Egger linear regression tests. The association between IGF1R expression and the OS in NSCLC patients had no significant publication bias existed on univariate analysis (P = 0.268) and the funnel plot seemed symmetrical (Figure 4). However, on univariate analysis of DFS and multivariate analysis of OS and DFS, the number of studies included was no more than ten, so these tests have no power to detect publication bias. Thus, we did not detect the publication bias of them. In addition, the assessment of publication bias also showed that the Egger tests were not significant (P > 0.05) for studies included in analysis of clinicopathological characteristics and the funnel plots seemed symmetrical (figures were not shown). Sensitivity analysis showed that the pooled HRs were similar when one study was removed. Therefore, our results were statistically reliable.
Figure 4.
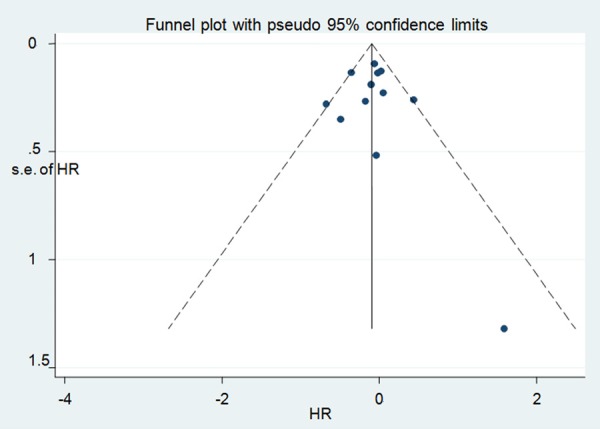
Funnel plot designed to visualize a potential publication bias on univariate analysis of overall survival.
Discussion
Our meta-analysis is based on published data and was performed using univariate analysis followed by further multivariate analysis. To the best of our knowledge, this study is the first meta-analysis focusing on IGF1R expression in resected NSCLC. Our results suggest that IGF1R positive expression is associated with an unfavourable DFS on both univariate and multivariate analysis, and IGF1R expression is also associated with smoking status and tumor size. However, IGF1R expression is not significantly associated with OS, but after subgroup analysis, IGF1R positive expression became associated with a favourable OS in studies with smaller samples, late stage disease (III-IV) and high quality.
IGF-1R is frequently disordered in human cancer and activation of IGF1R can activate the PI3K/AKT/mTOR and the Ras/Raf/MAPK pathways, which can promote proliferation, apoptosis, metastasis and resistance in cancer [44,45]. Moreover, IGF1R has become a target of anti-cancer therapy in solid tumors, including NSCLC. In a recent randomized phase II trial, 156 patients were randomized to paclitaxel-carboplatin with or without figitumumab, their results indicated a higher response rate and longer progression-free survival (PFS) in figitumumab-treated patients, especially in patients with squamous cell carcinoma [46]. Nevertheless, the phase III trials of figitumumab were terminated in 2010, because the HR was 1.1 towards the control arm. Furthermore, a randomized, phase II study comparing erlotinib plus R1507 (a monoclonal antibody against IGF1R) versus erlotinib plus placebo suggested that R1507 failed to show an improvement in PFS in unselected patients [47]. Thus, the reported clinical trials have given us serious concerns about the ability of IGF1R inhibition to serve as effective cancer treatments.
The prognostic significance of IGF1R expression has been examined in many cancers, including NSCLC. According to our study, IGF1R positive expression is associated with smoking status and tumor size, as well as an unfavourable DFS. Peled N. et al. considered that high IGF1R expression acted as an indicator for resistance to gefitinib in NSCLC cell lines and NSCLC patients, but did not seem to play a role in the intrinsic resistance to this drug [48]. Nevertheless, another study in 83 patients showed IGF1R expression measured by immunohistochemistry does not appear to be related to gefitinib resistance [49]. Gualberto A et al. found IGFIR was differentially expressed in histological subtypes (P = 0.04), with highest levels observed in squamous cell tumors [50]. However, our result suggested that IGF1R expression was irrelevant to histological subtypes. So far, the inconsistency of results for reported IGF-1R expression and outcomes may depend on the investigators and antibodies used for analysis, patient samples, disease stages, or the presence of other poorly understood pathways and regulators related to IGF-1R. Therefore, the results of our study provide useful information for clinicians assessing the prognosis of NSCLC patients and making personalized therapy decisions.
To be sure, there were some potential limitations in this study. Firstly, most included studies were retrospective studies, and no RCTs had been found. Owing to limitations in the original studies, we could not perform further subgroup analysis on univariate analysis of DFS, and on multivariate analysis of OS and DFS. Furthermore, we included studies with detection method by IHC, but the types or the dilutions of primary antibody were not the same in all the studies (Supporting Information Table S3). So, variability in protein expression assessment must be considered a potential source of bias. We also noticed that the cut-off values were arbitrarily selected and varied greatly between studies, which might produce the high heterogeneity. Nevertheless, due to the limited information of the original studies, we could not conduct further subgroup analysis by cut off values and histological type. Additionally, the HR was reconstructed from the survival curves when it was not reported directly in a study, this approach did not completely eliminate inaccuracy during extracting the survival rates despite being undertaken independently by two reviewers. The estimated HR might thus be less reliable than when obtained directly from published statistics. In our analysis, we don’t know whether previous adjuvant therapy has an impact on the prognostic significance of IGF1R. These issues had to be investigated by well designed prospective studies. Because none of the tests for funnel plot have power to detect publication bias when the number of studies included was no more than ten. Thus, we did not detect the publication bias on univariate analysis of DFS and multivariate analysis of OS and DFS, so publication bias might exist on them. Meanwhile, another potential source of bias cannot be ignored, we did not include unpublished papers, comment and abstracts, which may also lead to publication bias, since studies with positive results tend to be accepted by journals, whereas negative results often are rejected or even not be submitted.
In summary, positive expression of IGF1R was associated with unfavourable disease free survival in patients with NSCLC on univariate and multivariate analysis, but not associated with overall survival on univariate and multivariate analysis. With respect to clinical characteristics, IGF1R positive expression was related to smoking status and tumor size. However, since the limitations mentioned above, these findings need to be explained with cautions when applied to clinical practice. More prospective cohort studies with large samples are needed to further demonstrate the correlations between IGF1R expression and the survival in NSCLC patients.
Acknowledgements
This work was supported by grants from the Nature Science Foundation of China (81241068, 81372504).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32:669–92. doi: 10.1016/j.ccm.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 4.Camidge DR, Dziadziuszko R, Hirsch FR. The rationale and development of therapeutic insulin-like growth factor axis inhibition for lung and other cancers. Clin Lung Cancer. 2009;10:262–72. doi: 10.3816/CLC.2009.n.037. [DOI] [PubMed] [Google Scholar]
- 5.Badzio A, Wynes MW, Dziadziuszko R, Merrick DT, Pardo M, Rzyman W, Kowalczyk A, Singh S, Ranger-Moore J, Manriquez G, Gaire F, Jassem J, Hirsch FR. Increased insulin-like growth factor 1 receptor protein expression and gene copy number in small cell lung cancer. J Thorac Oncol. 2010;5:1905–11. doi: 10.1097/JTO.0b013e3181f38f57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dufourny B, Alblas J, van Teeffelen HA, van Schaik FM, van der Burg B, Steenbergh PH, et al. Mitogenic signalling of insulin-like growth factor I in MCF-7 human breast cancer cells requires phosphatidylinositol 3-kinase and is independent of mitogen-activated protein kinase. J Biol Chem. 1997;272:31163–71. doi: 10.1074/jbc.272.49.31163. [DOI] [PubMed] [Google Scholar]
- 7.Adams TE, Epa VC, Garrett TP, Ward CW. Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol Life Sci. 2000;57:1050–93. doi: 10.1007/PL00000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakesley VA, Stannard BS, Kalebic T, Helman LJ, LeRoith D. Role of the IGF-I receptor in mutagenesis and tumour promotion. J Endocrinol. 1997;152:339–44. doi: 10.1677/joe.0.1520339. [DOI] [PubMed] [Google Scholar]
- 9.Frasca F, Pandini G, Sciacca L, Pezzino V, Squatrito S, Belfiore A, Vigneri R. The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch Physiol Biochem. 2008;114:23–37. doi: 10.1080/13813450801969715. [DOI] [PubMed] [Google Scholar]
- 10.Khandwala HM, McCutcheon IE, Flyvbjerg A, Friend KE. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endocr Rev. 2000;21:215–44. doi: 10.1210/edrv.21.3.0399. [DOI] [PubMed] [Google Scholar]
- 11.LeRoith D, Roberts CT Jr. The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–37. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 12.Rocha RL, Hilsenbeck SG, Jackson JG, VanDenBerg CL, Weng Cn, Lee AV, Yee D. Insulin-like growth factor binding protein-3 and insulin receptor substrate-1 in breast cancer: correlation with clinical parameters and disease-free survival. Clin Cancer Res. 1997;3:103–9. [PubMed] [Google Scholar]
- 13.Gallardo A, Lerma E, Escuin D, Tibau A, Munoz J, Ojeda B, Barnadas A, Adrover E, Sánchez-Tejada L, Giner D, Ortiz-Martínez F, Peiró G. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3 K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br J Cancer. 2012;106:1367–73. doi: 10.1038/bjc.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang XP, Zhou WH, Zhang YF. Genetic variations in the IGF-IGFR-IGFBP axis confer susceptibility to lung and esophageal cancer. Genet Mol Res. 2014;13:2107–19. doi: 10.4238/2014.January.24.17. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura M, Miyamoto S, Maeda H, Zhang SC, Sangai T, Ishii G, Hasebe T, Endoh Y, Saito N, Asaka M, Ochiai A. Low levels of insulin-like growth factor type 1 receptor expression at cancer cell membrane predict liver metastasis in Dukes’ C human colorectal cancers. Clin Cancer Res. 2004;10:8434–41. doi: 10.1158/1078-0432.CCR-04-0430. [DOI] [PubMed] [Google Scholar]
- 16.Sun JM, Jun HJ, Ko YH, Park YH, Ahn YC, Son YI, Baek JH, Park K, Ahn MJ. Insulin-like growth factor binding protein-3, in association with IGF-1 receptor, can predict prognosis in squamous cell carcinoma of the head and neck. Oral Oncol. 2011;47:714–9. doi: 10.1016/j.oraloncology.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Spitz MR, Mistry J, Gu J, Hong WK, Wu X. Plasma levels of insulin like growth factor-I and lung cancer risk: a case-control analysis. J Natl Cancer Inst. 1999;91:151–6. doi: 10.1093/jnci/91.2.151. [DOI] [PubMed] [Google Scholar]
- 18.London SJ, Yuan JM, Travlos GS, Gao YT, Wilson RE, Ross RK, Yu MC. Insulin-like growth factor I, IGF-binding protein 3, and lung cancer risk in a prospective study of men in China. J Natl Cancer Inst. 2000;94:749–54. doi: 10.1093/jnci/94.10.749. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Luo J, Zhai X, Fu Z, Tang Z, Liu L, Chen M, Zhu Y. Prognostic value of phospho-Akt in patients with non-small cell lung carcinoma: A meta-analysis. Int J Cancer. 2014;135:1417–24. doi: 10.1002/ijc.28788. [DOI] [PubMed] [Google Scholar]
- 20.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–34. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions. Version 5.1.0. The Cochrane Collaboration; 2011. Available: www.cochrane-handbook.org. [updated March 2011] [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:11. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull. 1999;8:12. [Google Scholar]
- 26.Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Stat Med. 2011;20:641–54. doi: 10.1002/sim.698. [DOI] [PubMed] [Google Scholar]
- 27.Gately K, Forde L, Cuffe S, Cummins R, Kay EW, Feuerhake F, O’Byrne KJ. High coexpression of both EGFR and IGF1R correlates with poor patient prognosis in resected non-small-cell lung cancer. Clin Lung Cancer. 2014;15:58–66. doi: 10.1016/j.cllc.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Sun J, Wang H, Lou Y, Zhang Y, Sha H, Feng J, Han B. IGF-1R and Bmi-1 expressions in lung adenocarcinoma and their clinicopathologic and prognostic significance. Tumour Biol. 2014;35:739–45. doi: 10.1007/s13277-013-1100-9. [DOI] [PubMed] [Google Scholar]
- 29.Xu C, Xie D, Yu SC, Yang XJ, He LR, Yang J, Ping YF, Wang B, Yang L, Xu SL, Cui W, Wang QL, Fu WJ, Liu Q, Qian C, Cui YH, Rich JN, Kung HF, Zhang X, Bian XW. β-Catenin/POU5F1/SOX2 transcription factor complex mediates IGF-I receptor signaling and predicts poorprognosis in lung adenocarcinoma. Cancer Res. 2013;73:3181–9. doi: 10.1158/0008-5472.CAN-12-4403. [DOI] [PubMed] [Google Scholar]
- 30.Ludovini V, Flacco A, Bianconi F, Ragusa M, Vannucci J, Bellezza G, Chiari R, Minotti V, Pistola L, Tofanetti FR, Siggillino A, Baldelli E, Sidoni A, Daddi N, Puma F, Varella-Garcia M, Crinò L. Concomitant high gene copy number and protein overexpression of IGF1R and EGFR negatively affect disease-free survival of surgically resected non-small-cell-lung cancer patients. Cancer Chemother Pharmacol. 2013;71:671–80. doi: 10.1007/s00280-012-2056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto T, Oshima T, Yoshihara K, Nishi T, Arai H, Inui K, Kaneko T, Nozawa A, Adachi H, Rino Y, Masuda M, Imada T. Clinical significance of immunohistochemical expression of insulin-like growth factor-1 receptor and matrixmetalloproteinase-7 in resected non-small cell lung cancer. Exp Ther Med. 2012;3:797–802. doi: 10.3892/etm.2012.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuta K, Mimae T, Nitta H, Yoshida A, Maeshima AM, Asamura H, Grogan TM, Furuta K, Tsuda H. Insulin-like growth factor-1 receptor protein expression and gene copy number alterations in non-small cell lungcarcinomas. Hum Pathol. 2013;44:975–82. doi: 10.1016/j.humpath.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Kim YH, Sumiyoshi S, Hashimoto S, Masago K, Togashi Y, Sakamori Y, Okuda C, Mio T, Mishima M. Expressions of insulin-like growth factor receptor-1 and insulin-like growth factor binding protein 3 in advancednon-small-cell lung cancer. Clin Lung Cancer. 2012;13:385–90. doi: 10.1016/j.cllc.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Kim JS, Kim ES, Liu D, Lee JJ, Solis L, Behrens C, Lippman SM, Hong WK, Wistuba II, Lee HY. Prognostic impact of insulin receptor expression on survival of patients with non small cell lung cancer. Cancer. 2012;118:2454–65. doi: 10.1002/cncr.26492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikuchi R, Sonobe M, Kobayashi M, Ishikawa M, Kitamura J, Nakayama E, Menju T, Miyahara R, Huang CL, Date H. Expression of IGF1R is associated with tumor differentiation and survival in patients with lung adenocarcinoma. Ann Surg Oncol. 2012;19:S412–20. doi: 10.1245/s10434-011-1878-x. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa M, Uramoto H, Shimokawa H, Onitsuka T, Hanagiri T, Tanaka F. Insulin-like growth factor receptor-1 expression predicts postoperative recurrence in adenocarcinoma of the lung. Exp Ther Med. 2011;2:585–90. doi: 10.3892/etm.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dziadziuszko R, Merrick DT, Witta SE, Mendoza AD, Szostakiewicz B, Szymanowska A, Rzyman W, Dziadziuszko K, Jassem J, Bunn PA Jr, Varella-Garcia M, Hirsch FR. Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J. Clin. Oncol. 2010;28:2174–80. doi: 10.1200/JCO.2009.24.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ning XH, Wang YZ, Bai CM, Li J. Clinical significance of insulin-like growth factor-1 receptor in platinum-based chemotherapy for non-small celllung cancer. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2010;32:366–70. doi: 10.3881/j.issn.1000-503X.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Cappuzzo F, Tallini G, Finocchiaro G, Wilson RS, Ligorio C, Giordano L, Toschi L, Incarbone M, Cavina R, Terracciano L, Roncalli M, Alloisio M, Varella-Garcia M, Franklin WA, Santoro A. Insulin-like growth factor receptor 1 (IGF1R) expression and survival in surgically resected non-small-cell lung cancer (NSCLC) patients. Ann Oncol. 2010;21:562–7. doi: 10.1093/annonc/mdp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang MH, Lee J, Han J, Park YH, Ahn JS, Park K, Ahn MJ. Prognostic role of insulin-like growth factor receptor-1 expression in small cell lung cancer. APMIS. 2009;117:861–9. doi: 10.1111/j.1600-0463.2009.02545.x. [DOI] [PubMed] [Google Scholar]
- 41.Gong Y, Yao E, Shen R, Goel A, Arcila M, Teruya-Feldstein J, Zakowski MF, Frankel S, Peifer M, Thomas RK, Ladanyi M, Pao W. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507) PLoS One. 2009;4:e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee CY, Jeon JH, Kim HJ, Shin DH, Roh TW, Ahn CM, Chang YS. Clinical significance of insulin-like growth factor-1 receptor expression in stage I non-small-cell lung cancer: immunohistochemical analysis. Korean J Intern Med. 2008;23:116–20. doi: 10.3904/kjim.2008.23.3.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cappuzzo F, Toschi L, Tallini G, Ceresoli GL, Domenichini I, Bartolini S, Finocchiaro G, Magrini E, Metro G, Cancellieri A, Trisolini R, Crino L, Bunn PA Jr, Santoro A, Franklin WA, Varella-Garcia M, Hirsch FR. Insulin-like growth factor receptor 1 (IGFR-1) is significantly associated with longer survival in non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2006;17:1120–7. doi: 10.1093/annonc/mdl077. [DOI] [PubMed] [Google Scholar]
- 44.Gallardo A, Lerma E, Escuin D, Tibau A, Munoz J, Ojeda B, Barnadas A, Adrover E, Sánchez-Tejada L, Giner D, Ortiz-Martínez F, Peiró G. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3 K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br J Cancer. 2012;106:1367–73. doi: 10.1038/bjc.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ariga M, Medachi T, Akahori M, Sakamoto H, Ito Y, Hakuno F, Takahashi S. Signalling pathways of insulin-like growth factor-1 that are augmented by cAMP in FRTL-5 cells. Biochem J. 2000;348:409–16. [PMC free article] [PubMed] [Google Scholar]
- 46.Karp DD, Paz-Ares LG, Novello S, Haluska P, Garland L, Cardenal F, Blakely LJ, Eisenberg PD, Langer CJ, Blumenschein G Jr, Johnson FM, Green S, Gualberto A. Phase II study of the anti-insulin-like growth factor type 1 receptor antibody CP-751,871 in combination with paclitaxel and carboplatin in previously untreated, locally advanced, or metastatic non-small-cell lung cancer. J. Clin. Oncol. 2009;27:2516–22. doi: 10.1200/JCO.2008.19.9331. [DOI] [PubMed] [Google Scholar]
- 47.Ramalingam SS, Spigel DR, Chen D, Steins MB, Engelman JA, Schneider CP, Novello S, Eberhardt WE, Crino L, Habben K, Liu L, Jänne PA, Brownstein CM, Reck M. Randomized phase II study of erlotinib in combination with placebo or R1507, a monoclonal antibody to insulin-like growth factor-1 receptor, for advanced-stage non-small-cell lung cancer. J. Clin. Oncol. 2011;9:4574–80. doi: 10.1200/JCO.2011.36.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peled N, Wynes MW, Ikeda N, Ohira T, Yoshida K, Qian J, Ilouze M, Brenner R, Kato Y, Mascaux C, Hirsch FR. Insulin-like growth factor-1 receptor (IGF-1R) as a biomarker for resistance to the tyrosine kinase inhibitor gefitinibin non-small cell lung cancer. Cell Oncol. 2013;36:277–88. doi: 10.1007/s13402-013-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fidler MJ, Basu S, Buckingham L, Walters K, McCormack S, Batus M, Coon J 4th, Bonomi P. Utility of Insulin-like Growth Factor Receptor-1 Expression in Gefitinib-Treated Patients with Non-small Cell Lung Cancer. Anticancer. 2012;32:1705–10. [PubMed] [Google Scholar]
- 50.Gualberto A, Dolled-Filhart M, Gustavson M, Christiansen J, Wang YF, Hixon ML, Reynolds J, McDonald S, Ang A, Rimm DL, Langer CJ, Blakely J, Garland L, Paz-Ares LG, Karp DD, Lee AV. Molecular analysis of non-small cell lung cancer identifies subsets with different sensitivity to insulin-like growth factor I receptor inhibition. Clin Cancer Res. 2010;16:4654–65. doi: 10.1158/1078-0432.CCR-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


