Abstract
Resistance to chemotherapy is one of the key causal factors in cancer death and increasing evidence has revealed that microRNAs (miRNAs) are involved in chemoresistance in many kinds of human cancers. Paclitaxel has been used for treatment of advanced nasopharyngeal carcinoma (NPC); however, treatment failure often occurs due to development of acquired paclitaxel resistance. In this study, based on miRNA microarray screening and qRT-PCR validation, we found six differentially expressed miRNAs in our induced paclitaxel-resistant NPC CNE-1/Taxol cells. Furthermore, we clarified the role of miR-634, most significantly downregulated in the paclitaxel-resistant CNE-1/Taxol, in regulating the paclitaxel sensitivity in NPC cells. We restored miR-634 expression in the CNE-1/Taxol cells by lentivirus infection, and found restoration of miR-634 re-sensitized the CNE-1/Taxol cells to paclitaxel in vitro by MTT assay and colony formation assay. In xenograft mouse model, we found that miR-634 inhibited tumor growth and enhanced paclitaxel sensitivity. Thus, our findings provide important information for the development of targeted gene therapy for reversing paclitaxel resistance in NPC.
Keywords: Microarray, targeted therapy, paclitaxel resistance, nasopharyngeal carcinoma
Introduction
Epstein-Barr virus etiologically associated nasopharyngeal carcinoma (NPC) is endemic in Southern China and Southeast Asia with an annual incidence of up to 20 cases per 100,000 [1]. Thus, NPC is a highly prevalent malignant disease and a leading cause of death in these regions. Paclitaxel based chemotherapy is an extremely important therapeutic strategy in many types of human cancers [2,3], including advanced NPC [4-7]. However, in the clinical therapy, patients initially benefit from paclitaxel therapy, often develop acquired drug resistance leading to treatment failure. Thus, it is important to understand the underlying molecular mechanisms responsible for the development of paclitaxel resistance.
MicroRNAs (miRNAs) are a class of small non-coding regulatory RNAs that negatively regulate gene expression at the post-transcriptional level, and have been shown to play critical roles in a variety of pathological processes, including cancer pathogenesis [8,9]. More importantly, increasing evidence linked microRNAs to chemoresistance in human cancers. For example, upregulation of in miR-146a, miR-10a and miR-221/222 and decreased levels of the miR-200 family are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin [10]. The expression levels of miR-200 and let-7 were significantly downregulated in gemcitabine-resistant pancreatic cancer cells [11]. Considering that miRNAs regulating drug sensitivity are varied in different kinds of cancer and different anticancer agents, the identification of the role of specific miRNAs modulating a specific anticancer drug in a specific cancer has a clinical significance.
In the current study, we first performed miRNA microarray analysis to screen differentially expressed miRNAs in our induced paclitaxel-resistant NPC CNE-1/Taxol cells. Then, we clarified the role of miR-634, most significantly downregulated in the paclitaxel-resistant CNE-1/Taxol, in regulating the paclitaxel sensitivity in NPC cells.
Materials and methods
Cell line and culture
The human nasopharyngeal carcinoma CNE-1 cell line was obtained from Cancer Research Institute of Central South University. Cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin (Life Technologies, USA) in a humidified incubator at 37°C with 5% CO2.
MiRNA microarray analysis
Previously, we established paclitaxol-resistant CNE-1/Taxol cell sublines with a resistance index of 8.43 by treating the parental CNE-1 cells with increasing doses of paclitaxol for about five months. In this study, the parental CNE-1 cells (n = 3, Q1, Q3, Q5) and corresponding established paclitaxel-resistant CNE-1/Taxol cells (n = 3, N2, N4, N6) were sent to KangChen Bio-tech company (Shanghai, China) for miRNA isolation, quality control, chip hybridization, and microarray data analysis. In KangChen Bio-tech company, the samples were labeled using the miRCURY™ Hy3™/Hy5™ Power labeling kit and hybridized on the miRCURY™ LNA Array (version 18.0). Following the washing steps, the slides were scanned by the Agilent Scanner G2505C and scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs with more than 50 intensities in all samples were chosen for calculating normalization factor. Expressed data were normalized using the median normalization. After normalization, significant differentially expressed miRNAs were identified through Volcano Plot filtering.
Quantitative reverse transcribed PCR (qRT-PCR)
Total RNAs from cells were extracted using Trizol (Invitrogen, USA) according to the manufacturer’s instruction. About 500 ng of total RNAs was reversely transcribed into cDNA using miScript II RT Kit (Qiagen, USA) according to the manufacturer’s instructions. Real-time qRT-PCR was performed on ABI 7500 Sequence Detection System (Life Technologies, USA) using SYBR Green real-time PCR master mix (Toyobo Co., Japan) with a primer concentration of 200 nM under the conditions of 95°C for 1 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 15 sec, 72°C for 20 sec. The small nuclear U6 was used as internal control. The specific primers for miRNAs and U6 were purchased from Qiagen (Valencia, CA, USA). All experiments were performed in triplicate. Relative expression levels were calculated using the 2-∆∆Ct method.
Lentiviral stable infection
Lentiviruses containing miR-634 (Lv-miR-634) and scramble negative control (Lv-NC) were purchased from GeneChem Company (Shanghai, China). To get stably infected CNE-1/Taxol cells, the cells were cultured to about 70% of the plates, and then added by a concentration of 1.0 × 104 TU/well Lv-miR-634 or Lv-NC. Real-time qRT-PCR was performed to determinate expression levels of miR-634 after being infected for 6 days. The stably infected CNE-1/Taxol cells were expanded and harvested for further experiments.
MTT assay
Exponentially growing cells were seeded at 10,000 cells (100 μl culture medium) per well in 96-well plates and incubated for 12 h. The cells were then exposed to different concentrations of paclitaxel for 72 h, then 20 μl of MTT (Sigma Chemicals, St. Louis, MO, USA; 5 mg/ml in PBS) was added to each well, and the cells were cultured for an additional 4 h. Subsequently, 200 μl of DMSO was added to each well to dissolve the crystals. The values of the optical density at 570 nm were then measured using a microplate ELISA reader.
Colony formation assay
For the colony formation assay, Five hundred Lv-miR-634 or Lv-NC stably infected CNE-1/Taxol cells were placed in complete growth media in each 35 mm dish and allowed to grow for 6 h. A final concentration of 10 ng/ml paclitaxel was then added to each dish. After 24 h treatment, paclitaxel was removed by adding fresh complete growth media, and cells were allowed to grow until visible colonies formed (2 weeks). Cell colonies were fixed with 4% polysorbate, stained with Giemsa, washed, air dried, photographed and counted.
Animal treatments
Female nude mice of 3-5 weeks old, 17.9 ± 0.82 g in weight, were purchased from Shanghai Laboratory Animal Center (SLAC, Shanghai, China), and maintained under specific pathogen-free conditions. The parental CNE-1 cells and paclitaxel-resistant CNE-1/Taxol cells overexpressing miR-634 were harvested, resuspended in serum-free medium, and 1000,000 cells in 200 μl of cell suspension were injected into the proximal tibia of each anesthetized nude mice (n = 5 animals per group). Every 4 days post inoculation, individual orthotopic tumor from each mouse was measured with calipers, and the volume (mm3) of orthotopic tumor was calculated according to the formula: 1/2 × length × width2. 61 days after inoculation, the mice developed palpable tumors; the paclitaxel (10 mg kg-1) was intraperitoneally injected once a day for five days. Two days after complete paclitaxel treatments, all of the mice were euthanized and the tumors were excised and imaged under a light microscope.
Statistical analysis
The experiments were repeated at least three times, and the data are shown as the mean ± SD. Student’s t-test was used to analyze the differences in the experiments. Statistical analyses were performed using the SPSS 11.0 software, and P < 0.05 was considered to indicate a statistically significant difference.
Results
Screening and validation of miRNAs associated with paclitaxel resistance in CNE-1 cells
We performed miRNA microarray to screen differentially expressed miRNAs in our induced paclitaxel-resistant CNE-1/Taxol cell sublines. After normalizing the expression data with bioinformatical methods, we performed fold change filtering between the data for CNE-1/Taxol cells and the parental CNE-1 cells. The threshold for both the upregulated and downregulated miRNAs was at least 2-fold. Compared to the parental CNE-1 cells, there were 7 upregulated (miR-4788, miR-802, miR-3146, miR-4678, miR-377-3p, miR-208b, miR-22-5p) and 6 downregulated (miR-100-5p, miR-4478, miR-1204, miR-634, miR-4653-3p, miR-3198) miRNAs screened in CNE-1/Taxol cells (Figure 1A). To confirm the microarray findings, these differentially expressed miRNAs were validated by qRT-PCR in the parental CNE-1 cells and paclitaxol-resistant CNE-1/Taxol cells. Five significantly downregulated (miR-634, miR-4478, miR-3198, miR-100 and miR-4653) and one significantly upregulated (miR-802) miRNAs were consistent with the microarray results (Figure 1B).
Figure 1.
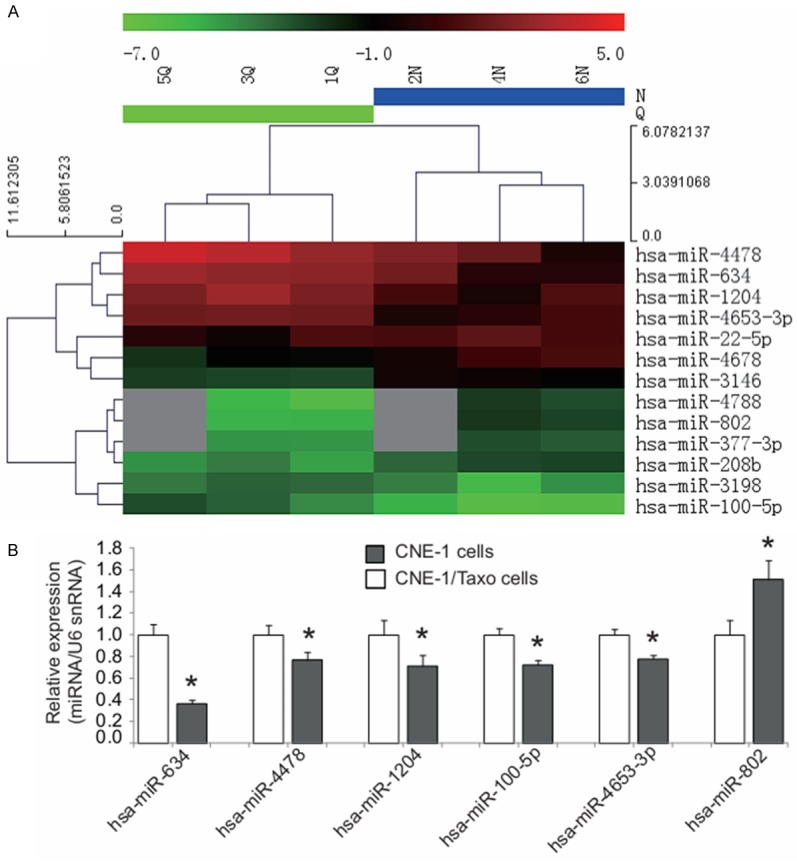
Screening and validation of miRNAs associated with paclitaxel resistance in CNE-1 cells. A. Hierarchical clustering analysis of 13 differentially expressed miRNAs detected by the miRNA microarray analysis in the paclitaxol-resistant CNE-1/Taxol cell sublines (n = 3, N2, N4, N6) compared to the parental CNE-1 cells (n = 3, Q1, Q3, Q5). The heat map diagram shows the results of the two-way hierarchical clustering analysis of miRNA expression levels and the cell lines. Each row represents a miRNA and each column represents a cell line. The miRNA-clustering tree is shown on the left and the cell line-clustering tree is at the top. The color scale shown at the top illustrates the relative expression level of a miRNA in a certain slide. A red color represents a high relative expression level and a green color represents a low relative expression level. B. Six differentially expressed miRNAs were validated by qRT-PCR in the paclitaxol-resistant CNE-1/Taxol cell sublines. There were five significantly downregulated (miR-634, miR-4478, miR-3198, miR-100 and miR-4653) and one significantly upregulated (miR-802) miRNAs consistent with the microarray results.
MiR-634 sensitizes nasopharyngeal carcinoma cells to paclitaxel in vitro
Among six validated miRNAs, we then focused on miR-634, considering it was the most downregulated in the paclitaxol-resistant CNE-1/Taxol cell sublines. To further confirm the involvement of miR-634 in regulating paclitaxel sensitivity in NPC cells, we exogenously upregulated miR-634 expression in CNE-1/Taxol cells via lentiviral infection and observed its impact on paclitaxel sensitivity using MTT assay and colony formation assay. As shown in Figure 2A, Lv-miR-634 stably infected CNE-1/Taxol cells were established, and had a significantly upregulated miR-634 expression level compared with the controls. Furthermore, the drug sensitivity was determined with MTT assay at 72 h with different paclitaxel doses (0, 2, 4, 6, 8 and 10 ng/ml). As shown in Figure 2B, paclitaxel sensitivities were significantly increased after forced overexpression of miR-634 in CNE-1/Taxol cells compared with negative controls. Colony formation assay revealed the similar result, in which paclitaxel (10 ng/ml) treatment for 24 h resulted in significant decreased colony formation percentages of CNE-1/Taxol cells infected with Lv-miR-634 lentivirus compared with the negative control (Figure 2C). Taken together, our findings suggest that miR-634 may modulate the sensitivity to paclitaxel in NPC cells.
Figure 2.
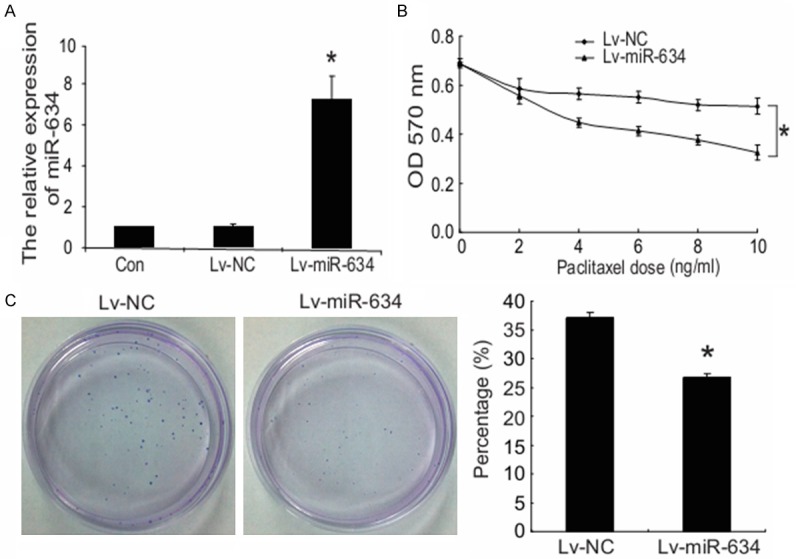
MiR-634 sensitizes nasopharyngeal carcinoma cells to paclitaxel in vitro. A. The CNE-1/Taxol cells were infected with Lv-miR-634 or Lv-NC, after being infected for 6 days, qRT-PCR was performed to determinate expression levels of miR-634 in stably infected CNE-1/Taxol cells, untreated groups were taken as the control. B. The impact of miR-634 on drug sensitivity at different paclitaxel doses (0, 2, 4, 6, 8 and 10 ng/ml) was determined by MTT assay. C. The CNE-1/Taxol cells were treated with a final concentration of 10 ng/ml paclitaxel for 24 h, and the impact of miR-634 on drug sensitivity was determined by colony formation assay. *P < 0.05.
MiR-634 inhibits tumor growth and sensitizes nasopharyngeal carcinoma cells to paclitaxel in vivo
To further investigate the role of miR-634 in regulating paclitaxel sensitivity in vivo, we injected subcutaneously Lv-miR-634 infected CNE-1/Taxol cells, as well as the negative control Lv-NC infected CNE-1/Taxol cells into 3- to 5-week female BALB/C nude mice (n = 5 per group), respectively. All mice developed palpable orthotopic tumors. 61 days after CNE-1/Taxol cells injection, the mice were treated with the paclitaxel (10 mg kg-1) once a day for five days, and euthanized two days later. All mice developed palpable orthotopic tumors, and the volume of tumor was calculated. As shown in Figure 3A, tumor growth was inhibited in miR-634 overexpression group compared to negative control group before paclitaxel administration (before 61 days post-injection). Upon 5-day paclitaxel treatment, tumor growth was more significantly inhibited in miR-634 overexpression group than that in negative control group (Figure 3A). By using qRT–PCR, we observed that miR-634 expression was significantly increased in tumor tissues grafted by stably overexpressed miR-634 cells compared with those negative controls (Figure 3C). Thus, our findings in vivo further support. Thus, our findings in vivo suggest that miR-634 may enhance paclitaxel sensitivity in nasopharyngeal cancer cells.
Figure 3.
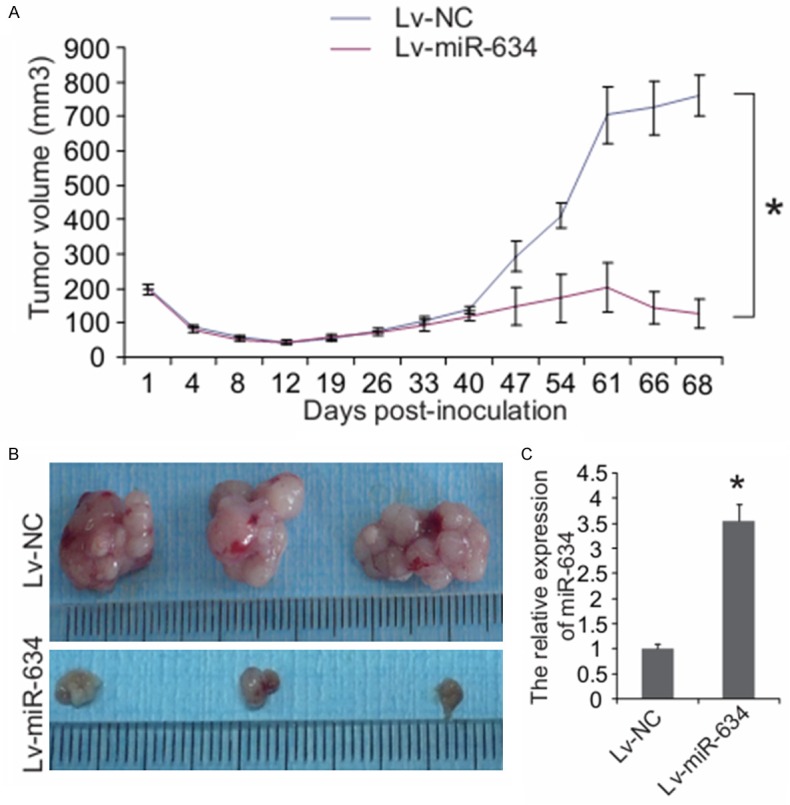
MiR-634 inhibits tumor growth and sensitizes nasopharyngeal carcinoma cells to paclitaxel in vivo. BALB/C nude mice were subcutaneously inoculated with CNE-1/Taxol cells with overexpressed miR-634 (Lv-miR-634, n = 5) and negative controls (Lv-miR-NC, n = 5), respectively. After 61 days, paclitaxel (10 mg kg-1) was then intravenously injected into mice once a day for five days. A. Tumor volume (mm3) was calculated every 4 days. B. Two days after complete paclitaxel treatments, all mice were euthanized and the tumors were excised and imaged under a light microscope. C. qRT-PCR analysis showed that miR-634 was upregulated in transplanted tumour tissues inoculated by miR-634-overexpressing CNE-1/Taxol cells. *P < 0.05.
Discussion
Paclitaxel has been widely used for the clinical treatment of many kinds of human cancers, including breast, ovarian, lung, bladder, esophagus and head and neck cancer [3,7,12]. Although paclitaxel-based therapy has a good initial response, the clinical effectiveness is limited by the emergence of paclitaxel-resistant cancer cells, which ultimately leads to tumor progression and relapse. Consequently, it is necessary to discover novel molecules regulating drug resistance for developing more effective targeted therapies. Abnormal expression of miRNAs can modulate drug resistance of cancers. Recent reports have documented that miR-200c, miR-125b, miR-21, miR-337-3p, miR-34a, miR-148a, miR-214 and miR-7 are involved in paclitaxel resistance or sensitivity in different cancers [13-20]. More importantly, manipulation of these miRNAs could alter drug sensitivity of cancer cells, which provides a promising tool to overcome cancer drug resistance. For instance, Ren et al have reported the miR-21 inhibitor could enhance the chemosensitivity of human glioblastoma cells to taxol [16].
In this study, miRNA microarray and qRT-PCR revealed that miR-634 was one of most down-regulated miRNAs in induced paclitaxel-resistant NPC CNE-1/Taxol cells, which suggests that loss of miR-634 is closely linked to the development of paclitaxel resistance in NPC. Then, we restored miR-634 in the CNE-1/Taxol cells by lentivirus infection to clarify the role of miR-634 in regulating the paclitaxel sensitivity in NPC cells. In vitro study revealed that restoration of miR-634 re-sensitized the CNE-1/Taxol cells to paclitaxel based on MTT assay and colony formation assay. In vivo study, miR-634 inhibited tumor growth and enhanced paclitaxel sensitivity of nasopharyngeal cancer upon paclitaxel treatment. Thus, our findings suggest that upregulating miR-634 might, at least partially, reverse paclitaxel resistance in NPC.
In recent studies, miR-634 has been reported to demonstrate anti-proliferative effects against prostate cancer cells [21], negatively affect cell proliferation, but not migration in glioblastoma LN229 cells [22], and function as an effective inhibitor of HER2 signaling and cell growth in breast cancer cells [23]. In addition, Yamamoto et al have reported that miR-634 negatively regulated the NRF2-mediated oncogenic pathway by directly targeting NRF2 and downregulation of miR-634 was associated with stabilized NRF2 and poor prognosis in esophageal squamous cell carcinoma [24]. In the present study, we found that restoration of miR-634 inhibited tumor growth and enhanced paclitaxel sensitivity of tumor cells in xenograft mouse model. Thereby, we hypothesized that miR-634 acts as a tumor suppressor, which could not only negatively regulate oncogenic pathway and growth of cancer cells but also reverse drug resistance to paclitaxel chemotherapy.
In summary, our present investigation suggests that loss of miR-634 is closely invovled in paclitaxel resistance development in NPC cells. Interestingly, we demonstrate that miR-634 sensitizes paclitaxel treatment and negatively affect tumor growth. Taken together, these findings suggest that miR-634 may be a useful biomarker for NPC paclitaxel resistance development and provide new information for using miR-634 based therapeutic strategies for NPC or other cancer treatments in the future.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 81372140, 81301688, 81272192, 81171882); Ph.D. Programs Foundation of Ministry of Education of China (No. 20130162110050 and 20130162120093); Post-doctoral Foundation of Central South University (No. 131425); China Postdoctoral Science Foundation (2014M552167); Natural Science Foundation of Hunan Province (Grant No. 12JJ4088); Technology Project of Hunan Province (2012SK3229); Research foundation of Health Department of Hunan Province (B2012-100); 125 Talent Project of the Third Xiangya Hospital of Central South University.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 3.Rowinsky EK. The development and clinical utility of the taxane class of antimicrotubule chemotherapy agents. Annu Rev Med. 1997;48:353–374. doi: 10.1146/annurev.med.48.1.353. [DOI] [PubMed] [Google Scholar]
- 4.He XY, Hu CS, Ying HM, Wu YR, Zhu GP, Liu TF. Paclitaxel with cisplatin in concurrent chemoradiotherapy for locally advanced nasopharyngeal carcinoma. Eur Arch Otorhinolaryngol. 2010;267:773–778. doi: 10.1007/s00405-009-1112-7. [DOI] [PubMed] [Google Scholar]
- 5.Leong SS, Wee J, Rajan S, Toh CK, Lim WT, Hee SW, Tay MH, Poon D, Tan EH. Triplet combination of gemcitabine, paclitaxel, and carboplatin followed by maintenance 5-fluorouracil and folinic acid in patients with metastatic nasopharyngeal carcinoma. Cancer. 2008;113:1332–1337. doi: 10.1002/cncr.23687. [DOI] [PubMed] [Google Scholar]
- 6.Leong SS, Wee J, Tay MH, Toh CK, Tan SB, Thng CH, Foo KF, Lim WT, Tan T, Tan EH. Paclitaxel, carboplatin, and gemcitabine in metastatic nasopharyngeal carcinoma: a Phase II trial using a triplet combination. Cancer. 2005;103:569–575. doi: 10.1002/cncr.20804. [DOI] [PubMed] [Google Scholar]
- 7.Tan EH, Khoo KS, Wee J, Fong KW, Lee KS, Lee KM, Chua ET, Tan T, Khoo-Tan HS, Yang TL, Au E, Tao M, Ong YK, Chua EJ. Phase II trial of a paclitaxel and carboplatin combination in Asian patients with metastatic nasopharyngeal carcinoma. Ann Oncol. 1999;10:235–237. doi: 10.1023/a:1008390929826. [DOI] [PubMed] [Google Scholar]
- 8.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, Liu M, Chen CT, Yu D, Hung MC. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127:1785–1794. doi: 10.1002/ijc.25191. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, VandenBoom TG 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain M, Gadgeel S, Kucuk O, Du W, Salwen W, Ensley J. Paclitaxel, cisplatin, and 5-fluorouracil for patients with advanced or recurrent squamous cell carcinoma of the head and neck. Cancer. 1999;86:2364–2369. [PubMed] [Google Scholar]
- 13.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule- targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, Schageman JJ, Greer R, Yang CR, Suraokar M, Wistuba II, Gazdar AF, Minna JD, Pertsemlidis A. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7:e39167. doi: 10.1371/journal.pone.0039167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita Y, Kojima K, Ohhashi R, Hamada N, Nozawa Y, Kitamoto A, Sato A, Kondo S, Kojima T, Deguchi T, Ito M. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem. 2010;285:19076–19084. doi: 10.1074/jbc.M109.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren Y, Zhou X, Mei M, Yuan XB, Han L, Wang GX, Jia ZF, Xu P, Pu PY, Kang CS. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. 2010;10:27. doi: 10.1186/1471-2407-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh S, Chitkara D, Mehrazin R, Behrman SW, Wake RW, Mahato RI. Chemoresistance in prostate cancer cells is regulated by miRNAs and Hedgehog pathway. PLoS One. 2012;7:e40021. doi: 10.1371/journal.pone.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, Xiong W, Li G, Lu J, Fodstad O, Riker AI, Tan M. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496–21507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu CZ, Shi RJ, Chen D, Sun YY, Wu QW, Wang T, Wang PH. Potential biomarkers for paclitaxel sensitivity in hypopharynx cancer cell. Int J Clin Exp Pathol. 2013;6:2745–2756. [PMC free article] [PubMed] [Google Scholar]
- 20.Xu CZ, Xie J, Jin B, Chen XW, Sun ZF, Wang BX, Dong P. Gene and microRNA expression reveals sensitivity to paclitaxel in laryngeal cancer cell line. Int J Clin Exp Pathol. 2013;6:1351–1361. [PMC free article] [PubMed] [Google Scholar]
- 21.Ostling P, Leivonen SK, Aakula A, Kohonen P, Makela R, Hagman Z, Edsjo A, Kangaspeska S, Edgren H, Nicorici D, Bjartell A, Ceder Y, Perala M, Kallioniemi O. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 22.Jeansonne D, Pacifici M, Lassak A, Reiss K, Russo G, Zabaleta J, Peruzzi F. Differential Effects of MicroRNAs on Glioblastoma Growth and Migration. Genes (Basel) 2013;4:46–64. doi: 10.3390/genes4010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leivonen SK, Sahlberg KK, Makela R, Due EU, Kallioniemi O, Borresen-Dale AL, Perala M. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol. 2014;8:93–104. doi: 10.1016/j.molonc.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto S, Inoue J, Kawano T, Kozaki K, Omura K, Inazawa J. The impact of miRNA-based molecular diagnostics and treatment of NRF2-stabilized tumors. Mol Cancer Res. 2014;12:58–68. doi: 10.1158/1541-7786.MCR-13-0246-T. [DOI] [PubMed] [Google Scholar]


