Abstract
Aim: To investigate the role of miR-101 in the regulation of tumor proliferation, invasion, apoptosis and to its target gene in human ESCC. Methods: The expression level of miR-101 in Eca109 cell line was determined by real-time polymerase chain reaction (PCR). After transfected with miR-101 mimics and inhibitor, proliferation, migration and apoptosis in ESCC cell line (Eca109) were detected by MTT, cell wound healing assay and flow cytometry, respectively. The expression of EZH2 in Eca109 cell was examined by immunohistochemical staining. Results: We found that miR-101 was significantly down-regulated in ESCC cell than in matched normal esophageal epithelium cell. The expression level of miR-101 was inversely correlated to EZH2 protein expression in ESCC cell. In Eca109 cells, over-expression of miR-101 significantly inhibited the migration and invasion of ESCC cells, and promotes cell apoptosis. Conclusions: These findings suggest that decreased expression of miR-101 might promote metastasis of human ESCC by inducing accumulation of EZH2 protein.
Keywords: MiR-101, ESCC, proliferation, invasion, EZH2
Introduction
Esophageal squamous cell carcinoma (ESCC) is the second most common cancer in China [1]. ESCC still remains the leading cancer-caused mortality in northern China, in particular in areas nearby Taihang Mountain [2]. In Xinjiang Uygur Autonomous Region which is a multi-ethnic area located in the Northwest of China, Kazakh ethnic group has a high incidence of ESCC being due to genetics and dietary. The recurrence rate of ESCC is extremely high after surgical treatment and the prognosis is usually poor (it is only 3-5% of diagnosed patients who can survive for 5 years) [3]. Ecidence supporting the critical role of proliferation, migration and apoptosis in the biological behavior of ESCC has led to a variety of studied on the basic mechanisms involed. Metastasis is a strong independent prognostic factor for ESCC [4]. Accordingly, the early recognition of the primary tumor and targeting agents to tumor are critical to successful treatment. Therefore, any insight into the mechanisms of ESCC metastasis may provide important clues for the development of clinical early diagnostic and effective therapeutics [5].
MicroRNAs (miRNAs) are a class of endogenous small, non-coding RNAs of 19-22 nucleotides that regulate a wide range of biological processes through changing the expression and translation of target mRNA genes [6]. Recently, miRNAs have been definitively linked to cancer development, including esophageal cancer. For example, it is reported that miR-142-3p is involved in the progression of ESCC and is a potential prognostic biomarker for ESCC [7]. miR-302b is a potential molecular marker of ESCC and functions as a tumor suppressor by post-transcriptionally regulating ErbB4 [8].
Considering that the expression and function of miRNAs may vary in different types of tumors, in this study we set out to investigate mechanism and function of miR-101 via altering its target gene expression in human ESCC. Here we report the EZH2 expression in ESCC is regulated by miR-101. We show that miR-101 is down-regulated in ESCC cells, resulting in increased EZH2 expression and enhance ESCC cell proliferation, migration and reduced apoptosis.
Materials and methods
Cell lines
The esophageal cancer cells (Eca109) were obtained from the Cell Bank of Shanghai. Cells were grown in DMEM medium and with 10% fetal bovine serum at 37°C in a humidified atmosphere of 95% air and 5% CO2. Cells were routinely passaged at 2-3 day intervals.
RNA isolation and real-time quantitative RT-PCR
Total RNA was extracted using the TRIzol kit (Invitrogen) according to the manufacturer’s instructions and To quantify mature miR-101 expression, the isolated total RNA was polyadenylated and reverse transcribed for use in a two-step quantitative RT-PCR. PCR conditions were 95°C for 3 minutes followed by 40 cycles of 95°C for 15 seconds, 56.7°C for 40 seconds. Real-time PCR analyses were performed with Real Master Mix using synthesized primers that were purchased from RiboBio. U6 small nuclear RNA was used as an internal control. The following primers were: microRNA-101, 5’-TACAGTACTGTGATAACTGAA-3’ (hsa-miR-101); 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTCAGT-3’ (hsa-miR-101-RT); 5’-GCGTGCTACAGTACTGTGATAACTG-3’ (hsa-miR-101-AS) and U6, 5’-CTCGCTTCGGCAGCACA-3’ (forward); 5’-AACGCTTCACGAATTTGCGT-3’ (reverse). The finally data were shown as fold change (2-ΔΔCt) and analyzed initially using GraphPad Prism 5.0 software. All samples were amplified in triplicate.
Scratch test
The Scratch test method was used to estimate cell’s Migration ability. Draw a trace in the bottom of the culture flasks, Take photos for three consecutive days and observe the cell around of the trace.
MTT proliferation assay
The capacity for cellular proliferation was measure with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. Eca109 cells were plated at 104 cells per well in 96-well plates with three replicate wells for each condition, transfected with oligonucleotides, and assayed 0 h, 24 h, 48 h, 72 h post-transfection. The cells were then incubated with 50 μl of MTT (5 mg/ml) for 4 h at 37°C and 150 μl of DMSO was added to solubilize the crystals for 20 min at room temperature. The optical density was determined with a spectrophotometer at a wavelength of 490 nm.
Apoptosis assays
The Annexin V-FITC Apoptosis Detection kit was used to detect and quantify apoptosis by flow cytometry. In brief, cells were harvested 24 h after transfection and collected by centrifugation for 5 min at 1000 × g. Cells were resuspended at a density of 1 × 106 cells/ml in 1X binding buffer, stained with FITC-labeled Annexin V for 5 min and immediately analyzed by FACScan Flow Cytometer (Becton Dickinson, San Jose, CA, USA). The data obtained were analyzed using CellQuest software.
Immunohistochemistry
Immunhistochemistry was performed according to the method described previously. Briefly, cells washed twice with 10 × PBS, 4% paraformaldehyde for 15 min, washed twice with 10 × PBS, 3% H2O2 for 15 minutes, washed twice with 10 × PBS. Serum closed, serum in blocking solution in the kit, 37°C, 20 minutes. Remove the serum closed, Add 1:150 primary antibody (EZH2 and COX-2), 4°C 12 h, Negative control antibody was replaced with PBS. Washed twice with 10 × PBS, Adding secondary antibody, 37°C, 20 minutes. DAB colorimetric method to color for 10 minutes. PBS to terminate and take photos. Experienced pathologists and graded as absent (0), weak (1+), moderate (2+) or strong (3+).
Statistical analysis
All mapping were using GraphPad Prism 5.0 software. All statistical analyses were carried out using SPSS version 17.0 statistical software (SPSS Inc, Chicago, IL, USA). P values < 0.05 were considered statistically significant and all of them are two-sided. Quantitative data were expressed as the mean ± SD. Student’s t test and ANOVA was used to determine statistical significance. Differences were considered significant at P < 0.01.
Results
miR-101 is downregulated in human Eca109 cell
Using a qRT-PCR method, miR-101 levels were detected in human Eca109 cell lines. miR-101 expression was reduced in human Eca109 cell lines compared with the normal esophagus cells. Approximately 20% lower than normal esophageal cells. (Figure 1A). The expression of miR-101 was significantly up-regulated in Eca109 cell which transfected into miR-101 mimic (P < 0.01). In addition, miR-101 mimics group level of expression was fifteen times rising than normal group (Figure 1B). In miR-101 inhibitor group, the level of miR-101’s expression was the sixth falling than normal group (Figure 1C).
Figure 1.
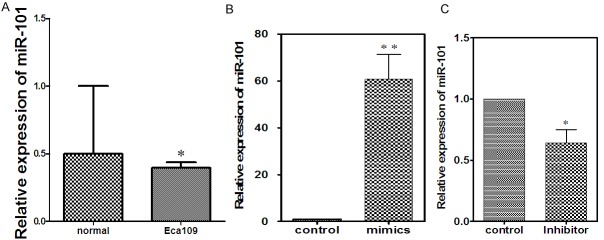
Expression of miR-101 is downregulated in human ESCC cell line. A: miR-101 expression was examined in Eca109 cell and normal esophageal cells by RT-PCR. B: Comparison the expression level of miR-101 before and after transfected with mimics. C: Comparison the expression level of miR-101 before and after transfected with inhibitor.
miRNA-101 inhibits the proliferation of Eca109 cells
The growth ability of Eca109 cell was determined by MTT assay in cells transfected with miR-101 inhibitor and mimic for 24, 48, and 72 h. There was no significant difference in the proliferation rate at the beginning of transfection. However, Eca109 cells treated with miR-101 mimic exhibited 30% decrease in growth rate compared with control group (P < 0.01) at 24 h, 24% decrease (P < 0.01) at 48 h and 13% decrease (P < 0.01) at 72 h (Figure 2A). Conversely, at 24, 48 and 72 h post transfection with the miR-101 inhibitor, cell proliferation rate was no significant (Figure 2B). Eca109 cells by MTT assay. At the time point of 0, 24, 48, and 72 h posttransfection of miR-101 mimics, the inhibition rates were 94%, 30%, 24% and 13%, respectively. The difference was statistically significant. At the time point of 0, 24, 48, and 72 h posttransfection of miR-101 inhibitor, the inhibition rates were 93%, 93%, 98% and 99% respectively. The difference was statistically significant.
Figure 2.
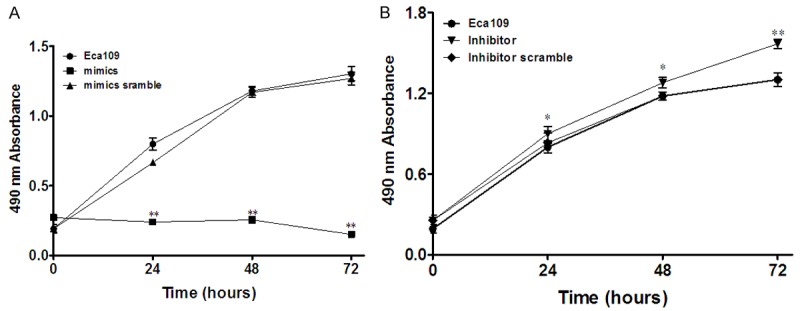
Ectopic expression of miR-101 inhibits tumor cell proliferation of Eca109 cells. A: Cell proliferation after transfection into miR-101mimics was measured by MTT assay at different time points. B: Cell proliferation after transfection into miR-101 inhibitor was measured by MTT assay at different time points.
miR-101 suppresses ESCC migration and invasion
We investigated the effect of miR-101 re-expression on the migration and invasion abilities of in ESCC cell line. The result of scratch detected were lower (55.14 μm ± 2 μm) than pre-transfection (137.71 μm ± 2 μm) (Figure 3E-G). The migration inhibition rates were 0%, 29%, 35% respectively. We found that the percentage of cells travelled through the micropore membrane was significantly decreased in cells transfected with miR-101 mimics as compared with control miRNA (Figure 3A).
Figure 3.
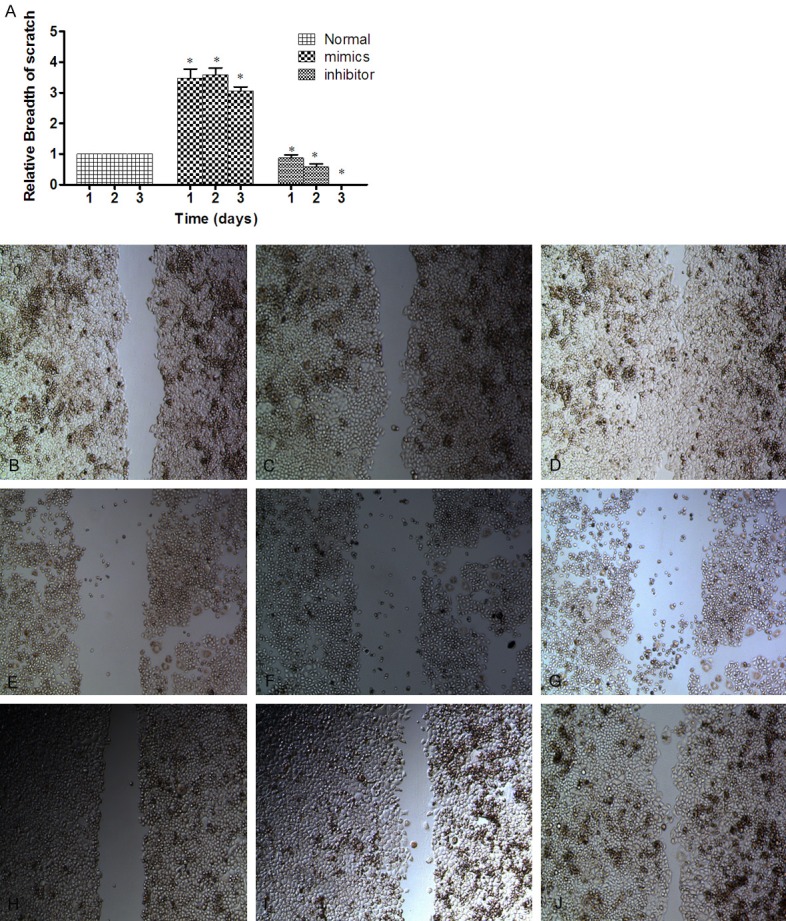
miR-101 inhibit the migration and invasion of ESCC cell line. In Eca109 cell, the expression of miR-101 was increases from scratch group (36.63 μm ± 2 μm) below the non-transfected cells in the experimental group (184.12 μm ± 2 μm). After 72 h, the width of scratches in the experimental group and the non-transfected cells experimental group were almost healed; whereas in the miR-101 up-regulated group, the width of scratches were basically no migration. Difference between the three groups was statistically significant after 72 h (P < 0.05). A. Each group of Eca109 cells by Cell wound healing assay. The migration inhibition rates were 0%, 29%, 35%, respectively. B-D. The picture of normal Eca109 cell three days scratches. E-G. The picture of after transfected the mimics three days scratches. H-J. The picture of after transfected the inhibitor three days scratches.
Overexpression of miR-101 promotes apoptosis of ESCC cell line
The Annexin V-positive early-phase apoptotic cells were significantly increased in cells transfected with miRNA-101 mimics when compared with miRNA-101 inhibitor cells. At 72 h after transfection, the early apoptosis rate in Eca109 cells transfected with miR-101 mimic (97.8%, Figure 4A) and inhibitor (6.3%, Figure 4B) After transfected mimics of miR-101, apoptosis rate was eight times than normal, After transfected inhibitor of miR-101, apoptosis rate was two times than normal, The difference was statistically significant. (P < 0.05, Figure 4D).
Figure 4.
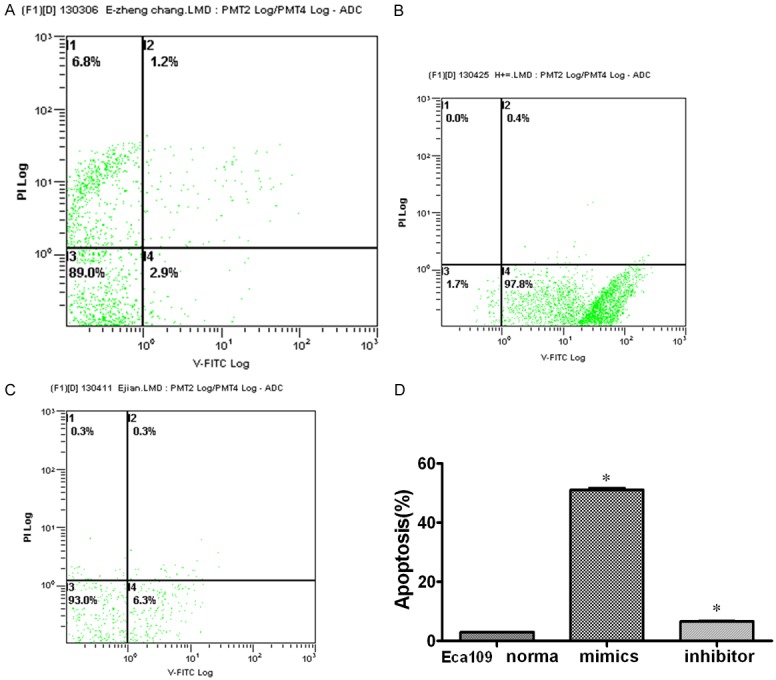
Overexpression of miR-101 promotes Cell Apoptosis. Eca109 cell group: the apoptotic rate of untransfected group was 2.9%; after transfection into miR-101mimics, the apoptotic rate was 97.8%. And after transfection into miR-101 inhibitor, the apoptotic rate decreased to 6.3%. GraphPad Prism 5.0 software was used to analyze the results. It was noted that up-regulated miR-101, the apoptotic rate increased about thirty-four times before transfection. The difference was statistically significant (P < 0.05). A. The apoptosis rate of Eca109 cell. B. The apoptotic rate when imported mimics of miR-101 into the cell. C. The apoptotic rate when imported inhibitor of miR-101 into the cell. D. Comparison of cell apoptotic rates (%). After transfected mimics of miR-101, apoptotic rate was thirty-four times higher than that before-trasfaction. After transfected inhibitor of miR-101, apoptotic rate was two times higher than that before-trasfaction. The difference was statistically significant.
MiR-101 posttranscriptionally down-regulates EZH2 expression in ESCC cell line
Since microRNA always regulates its target genes at the posttranscriptional level, we examined the level of EZH2 protein by immunohistochemistry in Eca109 cells. Observed under a microscope at 10 ×, Eca109 cells in the normal group, there are 860 cells is positive, 140 cells emerge negative (Provisions of its total number of observations is 1000 cells), while after transfected into miR-101 mimics, 626 cells were positive, 374 cells appear negative. And there were 721 cells were positive, 279 cells appear negative in miR-101 inhibitor group. We found that EZH2 protein expression in cells transfected with the miR-101 inhibitor was significantly higher than mimics groups (Figure 5B, 5C). Therefore, expression of EZH2 was elevated when miR-101 levels were decreased.
Figure 5.
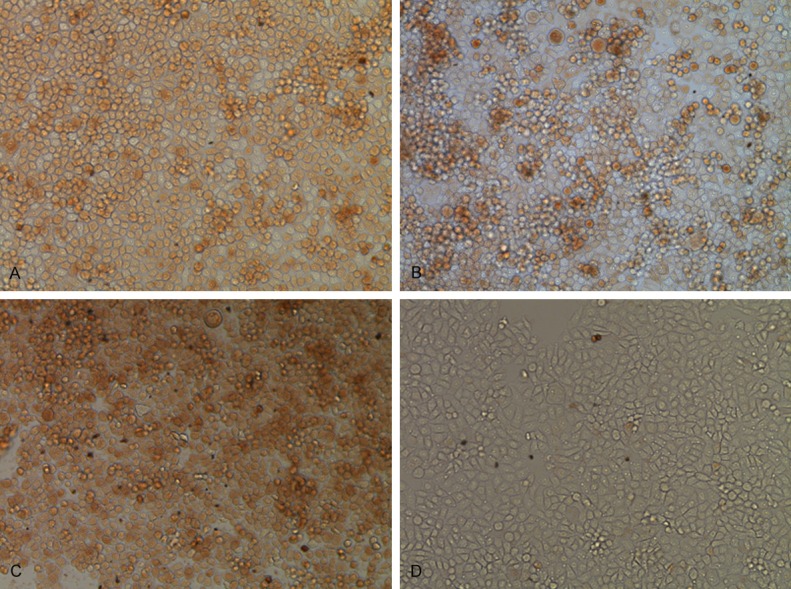
A. Immunohistochemistry of normal cells. B. Immunohistochemical of after transfected miR-101 mimics. C. Immunohistochemical of after transfected miR-101 inhibitor. D. Immunohistochemical of negative control.
Discussion
MiRNAs are evolutionarily conserved small noncoding RNAs (21-25 nucleotides) that regulate gene expression through modulation of translation efficiency or degradation of miRNAs. It was reported that miR-223, miR-25, miR-375 [9], miR-655 [10], miR-34b and miR-423 [11] were involved in regulation carcinogenesis in ESCC. MiR-101 was reported to be down-regulated in human colon cancer [12], prostate cancer [13], cervical cancer [14] and so on, and could repress the proliferation, invasion and metastasis of tumor cells. Here we show that miR-101 is down-regulated in ESCC cells Eca109.
MiR-101, belongs to a family of miRNAs that is involved in a series of cellular activities, e.g. cell proliferation, invasion, angiogenesis [15]. Previous studies have shown that the role of miR-101 in the progression of various types of tumors is controversial. The expression pattern and targets of miR-101 vary in different types of tumors. In recent years, relevant article been shown that miR-101 has to suppresses motility of bladder cancer cells [16], human hepatocellular carcinoma [17]. In our preliminary experiment, we found overexpression of miR-101 has obvious inhibitory effects on cell proliferation, migration and invasion in cervical cancer cells [18]. However, little information has been focused on the role of miR-101 in the regulation of its target gene expression in human ESCC. In our study, miR-101 expression in the Eca109 cells was down-regulated remarkably. After overexpression of miR-101 in Eca109 cells could significantly inhibit ESCC cell proliferation, migration and invasion and increas cellular apoptosis which are similar with human other cancers been reported. Such as, miR-101 may suppress HCC tumor progression by down-regulating SOX9 [19], miR-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2 [20]. These results suggested that miR-101 may be involved in the incidence of esophageal cancer as a role of tumor suppressor. That is to say, when we up-regulated miR-101, it can control the infiltration of cells into the surrounding tissue in order to inhibit a transfer of esophageal cancer.
To further probe the molecular mechanisms underlying miRNA-101 inhibition of ESCC cell growth and invasion, we searched for putative miRNA-101 targets through TargetScan, PicTar, and miRBase software. We identified EZH2 as a predictive target gene of miR-101. We further characterized EZH2 as the functional target of miR-101. In this study, we found that there was a significant correlation between expression levels of miR-101 and protein levels of EZH2 through Immunhistochemistry. In the EZH2 protein positive group, the expression levels of miR-101 was significantly lower than negative group. We found EZH2 protein can down-regulator expression by miR-101 in Eca109 cell. The enhancer of zestehomolog 2 (EZH2) is located at chromosome 7q35 and encodes a member of the Polycomb group proteins. We speculate that they may be involved in some of the tumor suppressor gene promoter methylation status, leading to tumor suppressor gene silencing, thereby contributing to the incidence and development of esophageal cancer. These findings suggest that miR-101 may play an important role in inhibiting the biological behavior of ESCCs by targeting EZH2. Although in Eca109 cells, over expression of miR-101 was found to suppress EZH2 expression, we did not detect significant correlation between the expression of miR-101 and EZH2 in clinical samples of ESSC tumor tissues, these researches will be need in further experiments.
In summary, we have demonstrated for the first time that miR-101 inhibits tumorigenesis in ESCC by attenuating cell proliferation and migration and promoting apoptosis. Our findings suggest that miR-101 and its target gene EZH2 may play important roles in the pathogenesis of ESCC, and miR-101 (and its targets) may be potential diagnostic or therapeutic targets for ESCC.
We propose that miR-101 is tumor suppressor genes in ESCC. It would be interesting to test whether these miRNAs act synergistically to regulate ESCC cell migration and invasion. Further experiments in animal models are warranted to establish the role of these miRNAs and EZH2 in regulating ESCC metastasis and EZH2-independent signal pathway, detect significant correlation between the expression of miR-101 and EZH2 in clinical samples of ESSC tumor tissues.
Acknowledgements
This research was supported by PHD Scientifc Research Foundation of Xinjiang Medical University (No. 2012-9).
Disclosure of conflict of interest
None.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–52. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Qi YJ, Chao WX, Chiu JF. An overview of esophageal squamous cell carcinoma proteomics. J Proteomics. 2012;75:3129–37. doi: 10.1016/j.jprot.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Gamliel Z, Krasna MJ. Multimodality treatment of esophageal cancer. Surg Clin North Am. 2005;85:621–30. doi: 10.1016/j.suc.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Xie F, Li K, Ouyang X. Twist, an independent prognostic marker for predicting distant metastasis and survival rates of esophageal squamous cell carcinoma patients. Clin Exp Metastasis. 2009;26:1025–32. doi: 10.1007/s10585-009-9292-5. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Tian X, Liu F, Zhao Y, Sun M, Chen D, Lu C, Wang Z, Shi X, Zhang Q, Zhang D, Shen Z, Li F, Harris CC, Cai H, Ke Y. Detection of HPV DNA in esophageal cancer specimens from different regions and ethnic groups: a descriptivestudy. BMC Cancer. 2010;10:19. doi: 10.1186/1471-2407-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Yang PY, Wang XF. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol. 2014;24:153–160. doi: 10.1016/j.tcb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, Xu LY. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–82. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 8.Zhang MX, Yang Q, Zhang LM, Zhou SN, Ye WG, Yao QL, Li ZF, Huang C, Wen QS, Wang JJ. miR-302b is a potential molecular marker of esophageal squamous cell carcinoma and functions as a tumor suppressor by targeting ErbB4. J Exp Clin Cancer Res. 2014;33:10. doi: 10.1186/1756-9966-33-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep. 2014;41:1257–66. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Zang W, Du Y, Ma Y, Li M, Li P, Chen X, Wang T, Dong Z, Zhao G. Mir-655 up-regulation suppresses cell invasion by targeting pituitary tumor-transforming gene-1 in esophageal squamous cell carcinoma. J Transl Med. 2013;11:301. doi: 10.1186/1479-5876-11-301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Yin J, Wang X, Zheng L, Shi Y, Wang L, Shao A, Tang W, Ding G, Liu C, Liu R, Chen S, Gu H. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk ofesophageal cancer in a Chinese population. PLoS One. 2013;8:e80570. doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L, Fiorentino M, Poggioli G, Rizzello F, Campieri M, Spisni E. Loss of miR-101 expression promotes Wnt/β-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229:379–389. doi: 10.1002/path.4097. [DOI] [PubMed] [Google Scholar]
- 13.Hao Y, Gu X, Zhao Y, Greene S, Sha W, Smoot DT, Califano J, Wu TC, Pang X. Enforced expression of miR-101 inhibits prostate cancer cell growth by modulating the COX-2 pathway in vivo. Cancer Prev Res (Phila) 2011;4:1073–83. doi: 10.1158/1940-6207.CAPR-10-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, Huang F, Zhang YJ, Tuokan T, Kuerban G. Roles of MiR-101 and its target gene Cox-2 in early diagnosis of cervical cancer of uygur women. Asian Pac J Cancer Prev. 2014;15:45–48. doi: 10.7314/apjcp.2014.15.1.45. [DOI] [PubMed] [Google Scholar]
- 15.Semaan A, Qazi AM, Seward S, Chamala S, Bryant CS, Kumar S, Morris R, Steffes CP, Bouwman DL, Munkarah AR, Weaver DW, Gruber SA, Batchu RB. MicroRNA-101 inhibits growth of epithelial ovarian cancer by relieving chromatin-mediated transcriptional Repression of p21(waf1/cip1) Pharm Res. 2013;28:3079–3090. doi: 10.1007/s11095-011-0547-x. [DOI] [PubMed] [Google Scholar]
- 16.Hu Z, Lin Y, Chen H, Mao Y, Wu J, Zhu Y, Xu X, Xu X, Li S, Zheng X, Xie L. MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem Biophys Res Commun. 2013;435:82–87. doi: 10.1016/j.bbrc.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, Zou L, Li Z, Zhao J, Lin N. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586:4362–4370. doi: 10.1016/j.febslet.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 18.Huang F, Lin C, Shi YH, Kuerban G. MicroRNA-101 inhibits cell proliferation, invasion, and promotes apoptosis by regulating cyclooxygenase-2 in hela cervical carcinoma cells. Asian Pac J Cancer Prev. 2013;14:5915–5920. doi: 10.7314/apjcp.2013.14.10.5915. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, Zou L, Li Z, Zhao J, Lin N. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586:4362–70. doi: 10.1016/j.febslet.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 20.Cho HM, Jeon HS, Lee SY, Jeong KJ, Park SY, Lee HY, Lee JU, Kim JH, Kwon SJ, Choi E, Na MJ, Kang J, Son JW. microRNA-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2. Exp Ther Med. 2011;2:963–967. doi: 10.3892/etm.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]


