Abstract
Introduction: Non-small cell lung cancer (NSCLC) is the major cause of cancer death worldwide. Increasing evidence shows that long non coding RNAs (lncRNAs) are widely involved in the development and progression of NSCLC. lncRNA PVT1 in several cancers has been studied, its role in lung cancer remains unknown. Our studies were designed to investigate the expression, biological role and clinical significance of PVT1 in lung cancer. Methods: lncRNA PVT1 expression in 82 NSCLC tissues and 3 lung cancer cell lines was measured by quantitative Real-time PCR (qRT-PCR). Its association with overall survival of patients was analyzed by statistical analysis. RNA interference (RNAi) approaches were used to investigate the biological functions of PVT1. The effect of PVT1 on proliferation was evaluated by MTT, cell migration and invasion ability was evaluated by cell migration and invasion assays. Results: lncRNA PVT1 expression was significantly upregulated in NSCLC tissues and lung cancer cells when compared with corresponding adjacent normal tissues and normal bronchial epithelial cells. Increased PVT1 expression was significantly correlated with histological grade and lymph node metastasis. In addition, NSCLC patients with PVT1 higher expression have shown significantly poorer overall survival than those with lower PVT1 expression. And PVT1 expression was an independent prognostic marker of overall survival in a multivariate analysis. In vitro assays our results indicated that knockdown of PVT1 inhibited cell proliferation, migration, and invasion. Conclusions: Our data indicated that lncRNA PVT1 is significantly upregulated in NSCLC tissues and may represent a new biomarker and a potential therapeutic target for NSCLC intervention.
Keywords: lncRNA PVT1, NSCLC, prognosis, proliferation, migration, invasion
Introduction
Lung cancer is the most common cause of cancer death, with more than 226,000 new cases in the United States in 2012, and the majority of lung cancer cases are non-small cell lung cancer (NSCLC), which accounts for approximately 80% of lung cancer [1,2]. Despite the enormous improvements made in chemotherapy and radiotherapy over the past few decades, the outlook for patients with NSCLC was dismal, with only slightly more than 15% of patients alive 5 years after diagnosis [3]. Thus, it is urgent to find new prognostic markers and therapeutic strategies to improve treatment of NSCLC.
Long non-coding RNAs (lncRNAs) are evolutionarily conserved non coding RNAs that are longer than 200 nucleotides in length with no protein-coding capacity [4]. Recent studies demonstrated that lncRNAs have important roles in diverse biological processes, such as cell growth and apoptosis, as well as in cancer progression and metastasis [5,6]. For example, Tu et al. demonstrated that the lncRNA GAS5 was down-regulated in hepatocellular carcinoma patients and GAS5 expression was an independent prognostic factor for patients with liver cancer, which might be a potential valuable biomarker for hepatocellular carcinoma [7]. Yan et al. showed lncRNA HOTAIR expression in stage Ta/T1 urothelial carcinoma of the bladder tissues was significantly upregulated compared with the matched nontumorous tissues, and the expression of HOTAIR is an independent prognostic factor of recurrence in stage Ta/T1 urothelial carcinoma of the bladder [8]. Ren et al. found that lncRNA MALAT-1 was increased in prostate cancer and higher MALAT-1 expression was correlated with Gleason score, prostate specific antigen, tumor stage and castration resistant prostate cancer, moreover down-regulation of MALAT-1 can inhibit prostate cancer cell growth, invasion and migration [9]. Unfortunately, the emerging functional role of lncRNAs in NSCLC remains largely unknown.
Mouse Pvt1 is an oncogene that was originally identified as a common retroviral integration site in murine leukemia virus (MLV)-induced T lymphomas [10]. The human PVT1 oncogene, which is located at 8q24.21, was found to be up regulated in a series of human tumors [11,12]. Wang et al. found that human lncRNA PVT1 was up-regulated in hepatocellular carcinoma tissues and that patients with higher lncRNA PVT1 expression had poor clinical prognosis. Moreover lncRNA PVT1 promotes cell proliferation, cell cycling and the acquisition of stem cell like properties in hepatocellular carcinoma cells by stabilizing NOP2 protein [13]. Takahashi et al. showed colorectal cancers patients with high PVT1 expression had a significantly poorer prognosis than those with low PVT1 expression, and down-regulated PVT1 promotes colorectal cancer cells apoptosis [14]. However, the lncRNA PVT1 expression in NSCLC and underlying mechanism is still unclear.
In the current study, we aim to explore the expression of lncRNA PVT1 in NSCLC and further investigate the clinical significance and biological functions of PVT1 in NSCLC.
Materials and methods
Patients and specimens
Eighty-two paired NSCLC tissues and matched adjacent normal tissues were obtained from the First Affiliated Hospital of Xinxiang Medical University (Weihui, China) between March 2007 and March 2009. All patients recruited in this study were not subjected to preoperative radiotherapy and/or chemotherapy and were diagnosed as infiltrating carcinoma by pathologists. Clinical and pathological variables analyzed are shown in Table 1. All patients were regularly followed up, with a mean observation period of 41 months. Tumor specimens and corresponding adjacent normal tissues were collected and stored in liquid nitrogen until use. The study was approved by the Medical ethics committee of the First Affiliated Hospital of Xinxiang Medical University.
Table 1.
Relationship between clinicopathological features and PVT1 in NSCLC
| Variable | Number | PVT1 expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age (years) | 0.564 | |||
| < 60 | 29 | 5 | 24 | |
| ≥ 60 | 53 | 12 | 41 | |
| Gender | 0.930 | |||
| Male | 49 | 10 | 39 | |
| Female | 33 | 7 | 26 | |
| Tumor size (cm) | 0.429 | |||
| < 3 | 20 | 6 | 14 | |
| ≥ 3 | 62 | 11 | 41 | |
| Histological grade | 0.002 | |||
| I | 40 | 14 | 26 | |
| II-III | 42 | 3 | 39 | |
| Lymph nodes metastasis | 0.001 | |||
| No | 43 | 15 | 28 | |
| Yes | 39 | 2 | 37 | |
For all patients, histological type and grade of cancer cell differentiation were reevaluated and determined by the classification system of the World Health Organization modified in 2004, and postsurgical pathological staging was determined based on the international staging system.
Cell culture
The human lung cell lines A549, H157, HEK-293T and normal bronchial epithelial cell line 16HBE were purchased from the American Type Culture Collection (ATCC, USA). All cell lines were routinely maintained in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 U/ml penicillin sodium, and 100 mg/ml streptomycin sulfate at 37°C in a humidified air atmosphere containing 5% CO2. Cells were used when they were in the logarithmic growth phase.
Cell transfection
The sequences of PVT1-specific siRNAs (si-PVT1) were 5’-GCUUGGAGGCUGA-GGAGUUTT-3’. Human NSCLC A549 cells were transfected with either 50 nmol si-PVT1 or si-NC using Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer’s instruction. After 48 hours, cells transfected with siRNA were harvested for qRT-PCR to determine the transfection efficiency.
Cell proliferation assays
To determine cell growth, 2 × 103 cells were seeded in 96-well plate and transfected with siRNA. And cell proliferation was determined using a MTT (3-(4,5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) according to the manufacturer’s protocol. The fluorescence intensity was measured using a fluorescence microplate reader and absorbance was measured at 570 nm (Molecular Devices). Three independent experiments (3 replicates in each) were performed.
In vitro migration and invasion assays
Cell migration and invasion assays were performed in a 24-well plate with 8 mm pore size chamber inserts (Corning). For migration assays, 5 × 104 cells were placed into the upper chamber per well with the non-coated membrane. For invasion assays, 1 × 105 cells were placed into the upper chamber per well with the Matrigel-coated membrane, which was diluted with serum-free culture medium. In both assays, cells were suspended in 200 ml of DMEM without fetal bovine serum when they were seeded into the upper chamber. In the lower chamber, 800 ml of DMEM supplemented with 10% fetal bovine serum was added. After incubation for some hours at 37°C and 5% CO2, the membrane inserts were removed from the plate, and non-invading cells were removed from the upper surface of the membrane. Cells that moved to the bottom surface of the chamber were fixed with 100% methanol for 20 min and stained with 0.1% crystal violet for 30 min. Then, the cells were imaged and counted in at least 5 random fields using a microscope (Olympus). The assays were conducted three independent times.
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA was isolated from tissues or cells using TRIZOL reagent according to the manufacturer’s protocol (Invitrogen). RNA was reverse transcribed using SuperScript First Strand cDNA System (Invitrogen) according to the manufacturer’s instructions. The PCR amplification were performed for 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, on a Applied Biosystems 7900HT (Applied Biosystems) with 1.0 μl of cDNA and SYBR Green Real-time PCR Master Mix (Takara). Data was collected and analyzed by SDS2.3 Software (Applied Biosystems). The expression level of each candidate gene was internally normalized against that of the RNU6B. The relative quantitative value was expressed by the 2-ΔΔCt method. Each experiment was performed in triplicates and repeated three times.
Statistical analysis
All data were presented as mean ± SD and analyzed by using SPSS 18.0. The analysis of data was conducted by using independent two-tailed t test. Categorical data were analyzed using the two-side chi-square test. Overall survival was estimated by using Kaplan-Meier method, and univariate analysis was conducted by log-rank test. The Cox proportional hazards model was used in the multivariate analysis. Values of P < 0.05 were considered statistically significant.
Results
LncRNA PVT1 is significantly upregulated in NSCLC
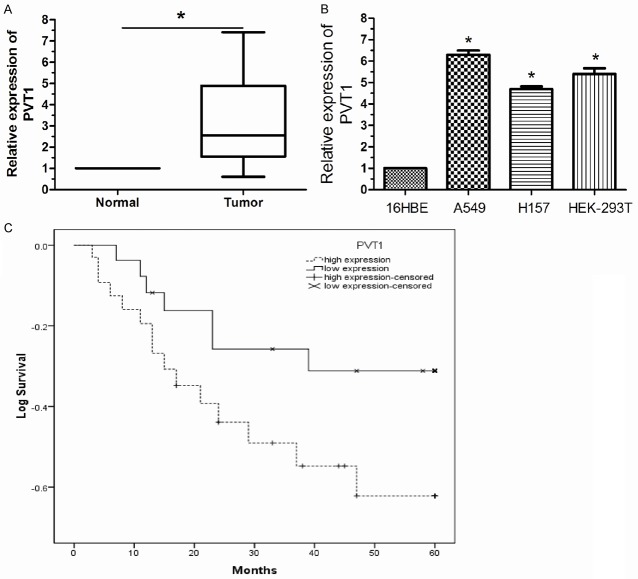
We firstly examined lncRNA PVT1 expression level in 82 paired human NSCLC and adjacent normal tissues by qRT-PCR. As shown in Figure 1A, after normalization to RNU6B expression levels, the expression level of PVT1 in NSCLC tissues was significantly higher than that in adjacent normal tissues (P < 0.05). Furthermore, expression was also examined by qRT-PCR in 3 lung cancer cell lines and human bronchial epithelial cell line 16HBE. This experiment showed that PVT1 expression was higher in lung cancer cell lines than in 16HBE (Figure 1B, P < 0.05). The data indicated that abnormal PVT1 expression may be related to NSCLC pathogenesis.
Figure 1.
Up-regulation of lncRNA PVT1 in NSCLC tissues is associated with poor prognosis in NSCLC. A. lncRNA PVT1 was significantly up-regulated in NSCLC tissues compared to adjacent normal tissues. B. Higher expression levels of lncRNA PVT1 were detected in lung cancer cell lines than normal bronchial epithelial cell line 16HBE. C. Kaplan-Meier curves revealed an association of higher lncRNA PVT1 levels with a short overall survival. The levels of lncRNA PVT1 were analyzed using qRT-PCR. Results are expressed as mean ± SD for three replicate determinations. *P < 0.05.
LncRNA PVT1 is a prognostic indicator for NSCLC patients
We divided the 82 patients with NSCLC into a high PVT1 expression group (n = 65) and a low PVT1 expression group (n = 17), classified as having expression levels higher or lower than the median expression level of PVT1 (2.87). Clinicopathological factors were then analyzed in the high and low PVT1 expression groups (Table 1). The high PVT1 expression group showed greater histological grade and lymph node metastasis compared with the low PVT1 expression group. With regard to overall survival, patients with high PVT1 expression had a significantly poorer prognosis than those with low PVT1 expression (Figure 1C, P < 0.05). Univariate and multivariate analysis showed that PVT1 expression level was an independent prognostic indicator of overall survival in patients with NSCLC (relative risk: 3.273, P < 0.05; Table 2).
Table 2.
Prognostic factors in Cox proportional hazards model
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Risk ratio | 95% CI | P | Risk ratio | 95% CI | P | |
| Age (years) | 0.719 | 0.417-1.614 | 0.283 | |||
| ≥ 60 vs. < 60 | ||||||
| Gender | 1.312 | 0.637-2.073 | 0.416 | |||
| Male vs. Female | ||||||
| Tumor size | 2.319 | 0.548-4.773 | 0.143 | |||
| ≥ 3 cm vs. < 3 cm | ||||||
| Histological grade | 2.929 | 1.837-7.146 | 0.005 | 2.417 | 1.611-6.271 | 0.009 |
| II, III vs. I | ||||||
| Lymph nodes metastasis | 4.479 | 2.817-8.692 | 0.004 | 3.641 | 2.382-6.817 | 0.007 |
| Yes vs. No | ||||||
| PVT1 | 3.581 | 2.141-7.793 | 0.011 | 3.273 | 2.184-6.937 | 0.016 |
| high vs. low | ||||||
Down-regulation of lncRNA PVT1 inhibited proliferation of NSCLC cells
We investigated the role of PVT1 in NSCLC cell proliferation by MTT assay. The efficiency of si-PVT1 was determined by qRT-PCR. As shown in Figure 2A, after transfection with si-PVT1 A549 cells showed a significant decreased mRNA expression of PVT1 compared to the si-NC group (P < 0.05). MTT assay showed that down-regulation of PVT1 remarkably inhibited proliferation of A549 cells (Figure 2B, P < 0.05). These data suggested that down-regulation of PVT1 inhibited proliferation of NSCLC cells.
Figure 2.
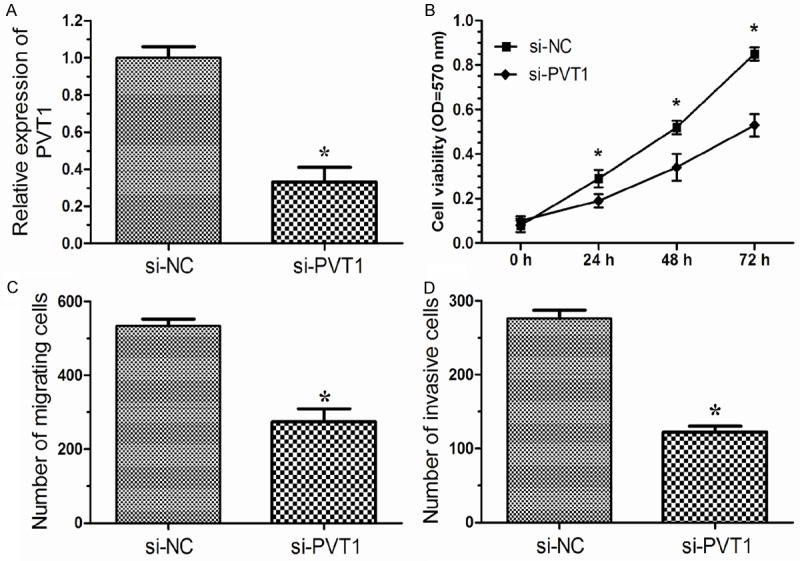
lncRNA PVT1 promotes cell growth, migration and invasion of NSCLC in vitro. A. A549 cells were transfected with si-PVT1 or si-NC for 48 hours, the expression of lncRNA PVT1 was measured by qRT-PCR. B. Cell proliferation of A549 cells was detected by MTT assay after transfected with si-PVT1 or si-NC. C. Migration assay showed inhibition of PVT1 in A549 cells produced a lower migration capacity than observed in controls tansfected with si-NC. D. Invasion assay showed A549 cells tansfected with si-PVT1 displayed a lower invasion capacity compared with those infected with si-NC. si-NC, cells transfected with nonspecific siRNA; si-PVT1, cells transfected with PVT1-specific siRNA. Results are expressed as means ± SD for three replicate determinations. *P < 0.05.
Down-regulation of lncRNA PVT1 inhibited migration and invasion of NSCLC cells
We then performed cell migration assays and invasion assays to investigate the role of PVT1 in the regulation of cell migration and invasion in human lung cancer cells. Cell migration assays showed that the migratory rate of cervical cancer cells transfected with si-PVT1 was significantly down-regulated compared with si-NC group (Figure 2C, P < 0.05). Cell invasion assays showed that the invasion of lung cancer cells transfected with si-PVT1 was notably down-regulated compared with si-NC group (Figure 2D, P < 0.05). These data indicated that PVT1 may promote the migration and invasion of lung cancer cells.
Discussion
Lung cancer is the primary cause of cancer-related deaths worldwide, so finding new mole-cular targets for its diagnosis, prognosis and treatment has the potential to improve the clinical strategies and outcomes of this disease [15]. In recent years, many studies have shown that the expression of lncRNAs is aberrant in human cancer [16]. Identification of tumor associated lncRNAs is critical for understanding the roles of lncRNAs in tumorigenesis and may be important for novel therapeutic targets [17]. In the present study, our attention focused on the lncRNA PVT1.
In this study, we characterized the function of lncRNA PVT1 in NSCLC development and progression. We found that lncRNA was up-regulated in NSCLC tissues and cell line to a greater extent than in corresponding normal tissues and human bronchial epithelial cell line 16HBE. Moreover, we also found that lncRNA PVT1 expression was positively correlated to the histological grade, and lymph node metastasis, although it showed no association with age, gender, and tumor size in patients with NSCLC. More importantly, PVT1 high expression was correlated with lower overall survival rates and could be an independent prognostic factor in patients with NSCLC. These results indicated that PVT1 expression was an independent prognostic factor for patients with NSCLC, and play an important role in tumorigenesis, and progression of NSCLC.
Takahashi et al. showed that PVT-1 expression levels in colorectal cancer tissues were significantly higher than those in non-cancerous tissues. Knockdown of PVT1 significantly inhibited colorectal cancer cells proliferation and reduced invasive abilities compared with negative control cells [14]. Guan et al. indicated PVT1 contribute independently to ovarian and breast pathogenesis when overexpressed because of genomic abnormalities. They also suggest that PVT1-mediated inhibition of apoptosis may explain why amplification of 8q24 is associated with reduced survival duration in patients treated with agents that act through apoptotic mechanisms. From our clinical pathological data, we found that high PVT1 expression is closely associated with histological grade and lymph node metastasis. Thus we suppose PVT1 may also regulate the growth and metastasis of NSCLC cells. So, it is necessary to identify the biological function of PVT1 in NSCLC cells.
PVT1-specific siRNA (si-PVT1) was used to investigate its biological functions in NSCLC cells. Our data showed that forced downregulation of PVT1 inhibited cellular proliferation, migration, and invasion in lung cancer cells, suggesting that PVT1 may play an oncogenic role in NSCLC. Further studies should focus on the underlying molecular mechanism of PVT1 in NSCLC.
In conclusion, the present study, for the first time, suggested that the up-regulation of lncRNA PVT1 expression was closely associated with the development, progression, and prognosis of NSCLC. Moreover, we preliminarily suggested a role lncRNA PVT1 plays in cell proliferation, migration, and invasion in lung cancer cells. As a result, these results suggested that PVT1 may become a novel promising candidate for the prognosis and therapy for NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 3.Pastorino U. Lung cancer screening. Br J Cancer. 2010;102:1681–1686. doi: 10.1038/sj.bjc.6605660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 8.Yan TH, Lu SW, Huang YQ, Que GB, Chen JH, Chen YP, Zhang HB, Liang XL, Jiang JH. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol. 2014 doi: 10.1007/s13277-014-2344-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 9.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190:2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Zeidler R, Joos S, Delecluse HJ, Klobeck G, Vuillaume M, Lenoir GM, Bornkamm GW, Lipp M. Breakpoints of Burkitt’s lymphoma t(8;22) translocations map within a distance of 300 kb downstream of MYC. Genes Chromosomes Cancer. 1994;9:282–287. doi: 10.1002/gcc.2870090408. [DOI] [PubMed] [Google Scholar]
- 11.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, Kalloger SE, Carlson JW, Ginzinger DG, Celniker SE, Mills GB, Huntsman DG, Gray JW. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 12.Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, Prives C. p53-Dependent induction of PVT1 and miR-1204. J Biol Chem. 2012;287:2509–2519. doi: 10.1074/jbc.M111.322875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, Yang N, Zhou WP, Li WL, Li W, Sun SH. Oncofetal Long Noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–90. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K. Amplification of PVT1is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2013;110:164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]