Abstract
Background and purpose: Previous studies observed the downregulation of microRNA (miR)-195 in esophageal squamous cell carcinoma (ESCC) tissues, confirmed cell division cycle 42 (Cdc42) as one target gene of miR-195, and demonstrated that miR-195 may act as a tumor suppressor in ESCC by regulating Cdc42 expression. This study aimed to explore the association of miR-195 and Cdc42 combined expression with clinicopathologic factors and prognosis. Methods: Expression of miR-195 and Cdc42 mRNA in 98 pairs of ESCC and paracancerous tissues were detected using real-time quantitative RT-PCR. Results: miR-195 downregulation and Cdc42 upregulation were both prevalent in ESCC tissues, and negatively correlated with each other. In addition, miR-195 expression negatively correlated with TNM stage (P=0.008) and lymphatic metastasis (P=0.022), while Cdc42 expression positively correlated with TNM stage (P=0.011) and tumor differentiation (P=0.024). Moreover, combined expression of miR-195 and Cdc42 (miR-195/Cdc42) was found to be prognostic indicators for progression-free survival and overall survival of ESCC patients both in univariate and multivariate analyses. Conclusion: The main findings of this study indicate the involvement of miR-195-Cdc42 axis in the progression of ESCC and suggest that the combined aberrant expression of miR-195 and Cdc42 mRNA can serve as a promising unfavorable prognostic biomarker in ESCC.
Keywords: microRNA-195, cell division cycle 42, esophageal squamous cell carcinoma, real-time quantitative PCR, prognosis
Introduction
Esophageal cancer ranks as the eighth most common malignancy worldwide and the fourth most cause of cancer-related death in China [1]. Its incidence varies greatly by geographic locations and ethnicity. Esophageal squamous cell carcinoma (ESCC) is the most common histological type of esophageal cancer, with a proportion of more than 90% cases [2]. ESCC development generally undergoes several phases, including atypical squamous cell hyperplasia, carcinoma in situ and infiltrating carcinoma [3]. Despite the tremendous progress in diagnosis and treatment, the prognosis of ESCC remains poor, with the average 5-year overall survival rate of approximately 40% [4]. In clinics, the main factors determining the clinical outcome of ESCC depend on clinicopathological characteristics, such as TNM stage and lymph nodal status, however, they are not very informative [5]. Accumulating studies have demonstrated that several dysregulated molecules may be involved in carcinogenesis of ESCC. Thus, it is imperative to identify the special biomarkers associated with different clinical phases of ESCC in order to predict the occurrence and survival rate of patients, to screen high-risk population and to evaluate the treatment efficiency.
MicroRNAs (miRNAs) represent a novel class of small, single-stranded, 19-25 nucleotide long non-coding RNA molecules [6]. miRNAs are cleaved from 60- to 110-nucleotide hairpin miRNA precursors in the cytoplasm by the RNase III enzyme Dicer, and post-transcriptionally regulate gene expression to suppress target messenger RNAs (mRNAs) by binding to their 3’- untranslated regions, leading to mRNA cleavage or translational repression [7]. Functionally, miRNAs play crucial roles in various cellular processes, including development, differentiation, proliferation, apoptosis and stress response [8]. Notably, accumulating studies have indicated that miRNA expression is closely related to carcinogenesis and have identified them as reliable diagnostic and prognostic biomarkers for various human cancer types. miRNAs participate in diverse biological pathways and may act as either tumor suppressor genes or oncogenes [9]. miR-195, a member of the miR-15/16/195 family, is localized at chromosome 17p13.1 [10]. There is growing evidence that miR-195 exhibits diverse expression pattern and function differently in different cancer including glioblastoma, chronic lymphocytic leukemia, breast cancer, gastric cancer, hepatocellular carcinoma, colorectal cancer, adrenocortical adenomas and bladder cancer [11-18]. More interestingly, Fu et al. [19] in 2013 observed that miR-195 was downregulated in patients with ESCC from Huaian, China. Similarly, another research group in the same year identified the downregulation of miR-195 in ESCC tissues compared with normal esophageal tissues [20]. They also confirmed cell division cycle 42 (Cdc42), which is a member of the Rho family and has been demonstrated to be overexpressed in a number of human cancers [21], as one target gene of miR-195. Although the previous studies have explored the involvement of miR-195-Cdc42 axis in carcinogenesis of ESCC in vitro system, its clinical significance in human ESCC remains unclear. Therefore, the current study aimed to investigate the association of miR-195 and Cdc42 combined expression with clinicopathologic factors and prognosis.
Materials and methods
Patients and tissue samples
The study was approved by the Ethics and Scientific Committees of Xinxiang Medical University. Informed consent was obtained from all patients before collection of the specimens.
A total of 98 patients with primary ESCC who had undergone radical resection of esophageal cancer were collected from October 2006 to February 2009 at the First Affiliated Hospital and the Third Affiliated Hospital of Xinxiang Medical University. Patients were enrolled in this study according to the following criteria: (1) histologically or cytologically confirmed ESCC; (2) resectability of the tumor in clinical evaluation; (3) medical ability of patient to tolerate major abdominal or thoracic surgical procedures or both; (4) no history of cancer or digestive diseases; (5) no chemotherapy or radiotherapy performed before the surgery. All patients’ tissue specimens were snap frozen in liquid nitrogen and stored at -80°C until the extraction of total RNA. All 98 ESCC patients consists 82 men and 16 women, whose median age was 65 years (range, 45-82 years). Their clinicopathologic stage was evaluated according to the TNM classification system of the American Joint Committee on Cancer-International Union Against Cancer staging manual. The clinicopathologic characteristics including patients’ gender, age, tumor size, tumor location, TNM stage, tumor differentiation, lymphatic metastasis, and venous invasion were summarized in Table 1.
Table 1.
Associations of microRNA (miR)-195 or Cdc42 mRNA expression with clinicopathological characteristics of 98 esophageal squamous cell carcinoma (ESCC) patients
| Clinical variables | No. of patients (n, %) | MiR-195-low (n, %) | P | Cdc42-high (n, %) | P |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 68 (69.39) | 36 (52.94) | NS | 36 (52.94) | NS |
| Female | 30 (30.61) | 16 (53.33) | 14 (46.67) | ||
| Age (year) | |||||
| > 60 | 56 (57.14) | 30 (53.57) | NS | 30 (53.57) | NS |
| ≤ 60 | 42 (42.86) | 22 (52.38) | 20 (47.62) | ||
| Tumor size (cm) | |||||
| > 5 | 58 (59.18) | 30 (51.72) | NS | 30 (51.72) | NS |
| ≤ 5 | 40 (40.82) | 22 (48.28) | 20 (50.00) | ||
| Tumor location | |||||
| Upper | 52 (46.67) | 32 (61.54) | NS | 30 (57.69) | NS |
| Middle/Lower | 46 (33.33) | 20 (43.48) | 20 (43.48) | ||
| TNM stage | |||||
| I | 15 (15.31) | 0 (0) | 0.008 | 2 (13.33) | 0.011 |
| II | 43 (43.88) | 17 (39.53) | 15 (34.88) | ||
| III | 25 (25.51) | 20 (80.00) | 18 (72.00) | ||
| IV | 15 (15.31) | 15 (100.0) | 15 (100.0) | ||
| Tumor differentiation | |||||
| Well~Moderately | 52 (53.06) | 26 (50.00) | NS | 22 (42.31) | 0.024 |
| Poorly | 46 (46.94) | 26 (56.52) | 28 (60.87) | ||
| Lymphatic metastasis | |||||
| Yes | 40 (16.67) | 30 (75.00) | 0.022 | 24 (60.00) | NS |
| No | 58 (37.50) | 22 (37.93) | 26 (44.83) | ||
| Venous invasion | |||||
| Yes | 42 (42.86) | 21 (50.00) | NS | 21 (50.00) | NS |
| No | 56 (57.14) | 31 (55.36) | 29 (51.79) |
All 98 ESCC patients received follow-up and the median follow-up period was 33.68 months (range, 0.82-62.96 months). The patients were detected every 3 months for the first year and then every 6 months for the next 2 years, and finally annually. The diagnostic examinations consisted of esophagography, computed tomography, chest x-ray, abdominal ultrasonography and bone scan when necessary to detect recurrence and/or metastasis. The overall survival (OS) was defined as the time period from the start date of follow-up to the date of death or the end date of follow-up. The distant progression-free survival (DPFS) was defined as the time from diagnosis to tumor distant progression.
Real-time quantitative RT-PCR for miR-195
Real-time quantitative RT-PCR was performed to detect expression levels of miR-195 in 98 self-paired specimens of ESCC and paracancerous tissues. Total RNA was isolated using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For miR-195 analysis, the stem-loop RT primer was 5’-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACG CCA AT-3’ and the amplifying primers were as follows: sense, 5’-CGT AGC AGC ACA GAA AT-3’ and antisense, 5’-GTG CAG GGT CCG AGG T-3’. RNU6B was used as an internal control. Primers for qRT-PCR of RNU6B were: sense, 5’-CTC GCT TCG GCA GCA CA-3’ and antisense, 5’-AAC GCT TCA CGA ATT TGC GT-3’. The reactions of quantitative PCR were incubated in 96-well optical plates at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 10 min and were performed in an ABI 7500 real-time PCR system. The delta-delta Ct method was employed to calculate the fold-change. For each sample, all experiments were done in triplicate.
Real-time quantitative RT-PCR for Cdc42 mRNA
Real-time quantitative RT-PCR was performed to detect expression levels of Cdc42 mRNA in 98 self-paired specimens of ESCC and paracancerous tissues. Total RNA was isolated using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Obtained cDNAs were amplified using specific primers, primers for human Cdc42: sense, 5’-GCC CGT GAC CTG AAG GCT GTC A-3’ and antisense, 5’-TGC TTT TAG TAT GAT GCC GAC ACC A-3’. GAPDH was used as an internal control. Primers for human GAPDH: sense, 5’-AAG GTG AAG GTC GGA GTC A-3’ and antisense, 5’-GGA AGA TGG TGA TGG GAT TT-3’. The reactions of quantitative PCR were incubated in 96-well optical plates at 95°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 1 min and 72°C for 1 min, and were performed in an ABI 7500 real-time PCR system. The delta-delta Ct method was employed to calculate the fold-change. For each sample, all experiments were done in triplicate.
Statistical analysis
Statistical analysis in the current study was performed using SPSS software (SPSS Standard version 13.0; SPSS Inc. Chicago, IL, USA). The differences in miR-195 or Cdc42 expression between ESCC and paracancerous tissues were analyzed using the Student’s t-test. The associations between miR-195, Cdc42 expression and various clinicopathological characteristics were assessed using the χ2 test. OS and DPFS were assessed using the Kaplan-Meier method and compared using the log-rank test. All parameters found to be significant in univariate analysis using the Cox proportional hazards model were entered into multivariate survival analysis. Differences were considered statistically significant when P was less than 0.05.
Results
miR-195 downregulation and Cdc42 upregulation in human ESCC tissues
The miR-195 and Cdc42 mRNA expression was tested in 98 pairs of primary ESCC and their corresponding paracancerous tissues by real-time quantitative RT-PCR. As shown in Figure 1, miR-195 expression was significantly downregulated in ESCC tissues compared with their normal counterparts (ESCC vs. paracancerous: 3.06±1.03 vs. 4.50±0.99, P < 0.001, Figure 1A), while the expression levels of Cdc42 mRNA in ESCC tissues were markedly higher than those in paracancerous tissues (ESCC vs. paracancerous: 4.54±0.93 vs. 2.59±0.90 P < 0.001, Figure 1B). More interestingly, the expression levels of miR-195 in ESCC tissues were negatively correlated with the expression levels of Cdc42 mRNA in ESCC tissues (Spearman’s correlation: r=-0.61, P < 0.001, Figure 1C).
Figure 1.
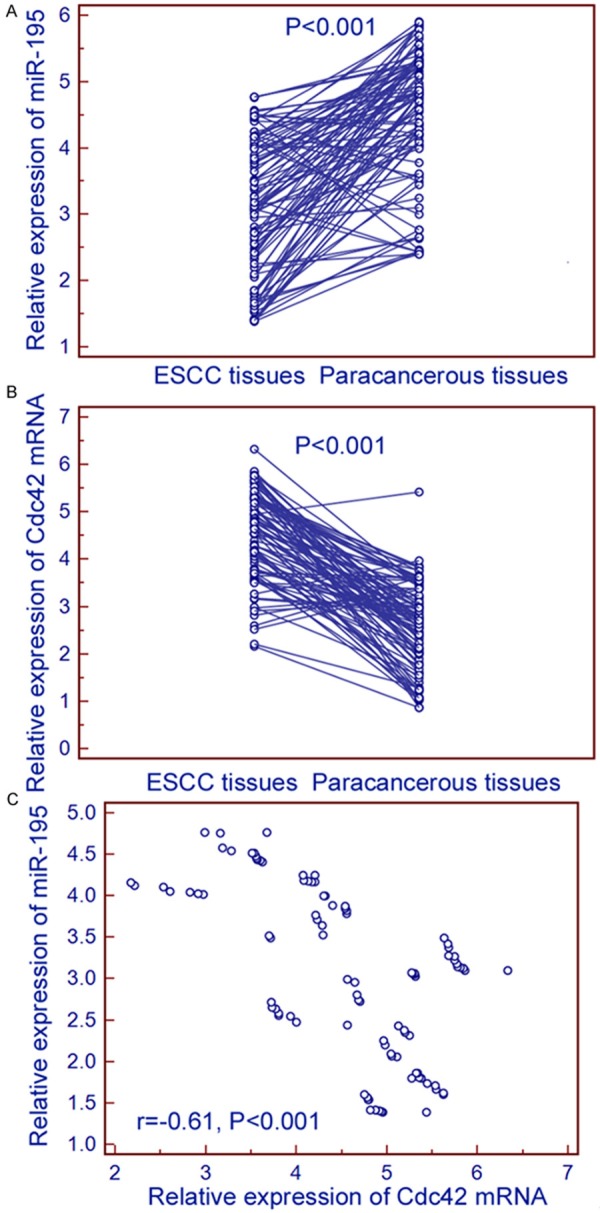
Relative expression of miR-195 and Cdc42 mRNA in 98 pairs of primary ESCC and their corresponding paracancerous tissues. miR-195 expression was significantly downregulated in ESCC tissues compared with their normal counterparts (ESCC vs. paracancerous: 3.06±1.03 vs. 4.50±0.99, P < 0.001, A), while the expression levels of Cdc42 mRNA in ESCC tissues were markedly higher than those in paracancerous tissues (ESCC vs. paracancerous: 4.54±0.93 vs. 2.59±0.90, P < 0.001, B). More interestingly, the expression levels of miR-195 in ESCC tissues were negatively correlated with the expression levels of Cdc42 mRNA in ESCC tissues (Spearman’s correlation: r=-0.61, P < 0.001, C).
miR-195 downregulation and Cdc42 upregulation significantly associate with aggressive clinicopathological characteristics of ESCC patients
In order to evaluate the associations of miR-195 downregulation and Cdc42 upregulation with the clinicopathological characteristics of ESCC patients, we divided all 98 ESCC patients into low miR-195, high miR-195, low Cdc42 and high Cdc42 expression groups using the median values of miR-195 and Cdc42 mRNA expression levels in ESCC tissues detected by real-time quantitative RT-PCR cutoff points, respectively. As a result, ESCC patients expressing miR-195 at levels less than the median value (3.10) were assigned to the low miR-195 expression group (mean expression value 2.67, n=52), and those samples with expression equal or above the median value were assigned to the high miR-195 expression group (mean expression value 3.58, n=46); Similarly, ESCC patients expressing Cdc42 mRNA at levels less than the median value (4.67) were assigned to the low Cdc42 expression group (mean expression value 3.26, n=48), and those samples with expression equal or above the median value were assigned to the high Cdc42 expression group (mean expression value 6.02, n=50). As shown in Table 1, miR-195 expression negatively correlated with TNM stage (P=0.008) and lymphatic metastasis (P=0.022), while Cdc42 expression positively correlated with TNM stage (P=0.011) and tumor differentiation (P=0.024). However, there were no significant associations of miR-195 or Cdc42 expression with patients’ age and gender, tumor size, tumor location, status of lymphatic metastasis and status of venous invasion (all P > 0.05).
Combined miR-195 downregulation and Cdc42 upregulation significantly associate with unfavorable prognosis of ESCC patients
Patients with low miR-195 expression or high Cdc42 mRNA expression showed a poorer prognosis than those with high miR-195 expression or low Cdc42 mRNA expression by the Kaplan-Meier analysis, respectively. The log-rank test revealed that the OS of ESCC patients with low miR-195 expression or high Cdc42 mRNA expression were markedly shorter than those with high miR-195 expression or low Cdc42 mRNA expression (both P < 0.001; Figure 2A and 2C). Similar results were also observed in the DPFS analysis (both P < 0.001; Figure 2B and 2D). In addition, the OS and DPFS of patients with combined low-miR195 and high-Cdc42 mRNA expression (miR-195-low/Cdc42-high) were the shortest (both P < 0.001, Figure 2E and 2F) when compared with patients in other three groups (miR-195-low/Cdc42-low, miR-195-high/Cdc42-high, miR195-high/Cdc42-low).
Figure 2.
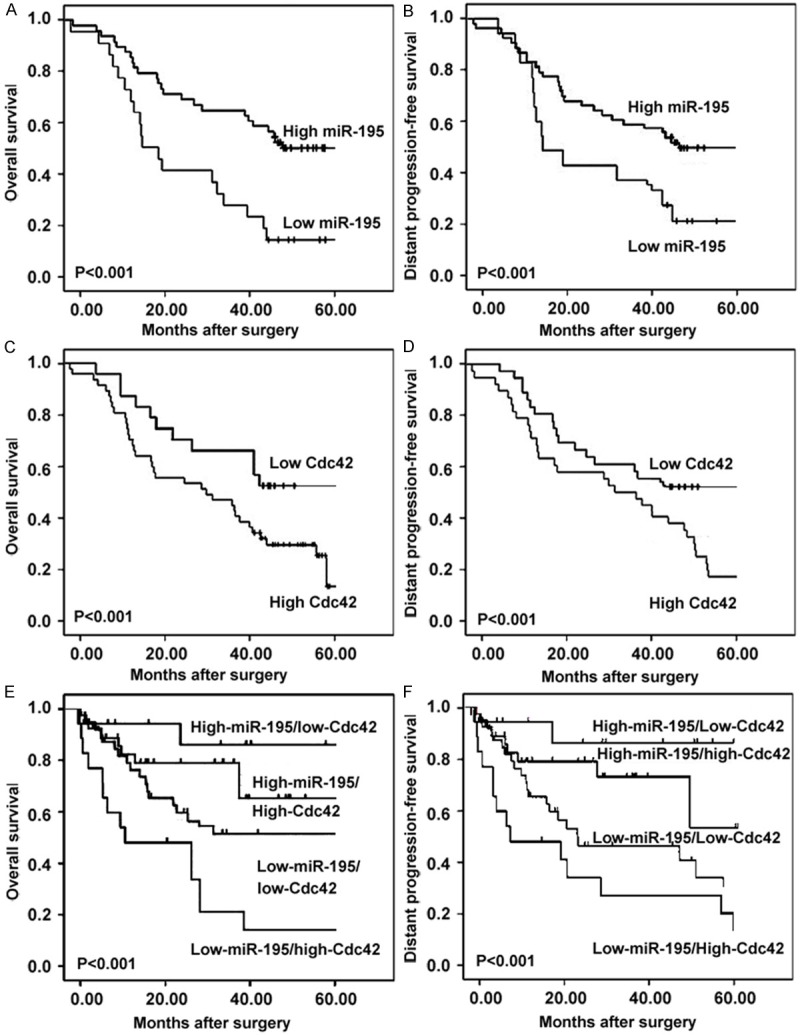
Survival curves for 98 esophageal squamous cell carcinoma (ESCC) patients according to expression patterns of miR-195 and Cdc42 mRNA in tumor tissues. Kaplan-Meier analysis showed that the OS of ESCC patients with low miR-195 expression or high Cdc42 mRNA expression were markedly shorter than that with high miR-195 expression or low Cdc42 mRNA expression (both P < 0.001; A and C). Similar results were also observed in the DPFS analysis (both P < 0.001; B and D). In addition, the OS and DPFS of patients with combined low-miR-195 and high-Cdc42 mRNA expression (miR-195-low/Cdc42-high) were the shortest (both P < 0.001, E and F) when compared with patients in other three groups (miR-195-low/Cdc42-low, miR-195-high/Cdc42-high, miR-195-high/Cdc42-low).
Cox proportional hazard model confirmed that miR-195 expression (for OS: RR 5.96, 95% CI, 1.26-11.93, P=0.01; for DPFS: RR 5.59, 95% CI, 1.13-11.16, P=0.01), Cdc42 expression (for OS: RR 5.33, 95% CI, 1.11-11.08, P=0.01; for DPFS: RR 5.18, 95% CI, 1.05-11.02, P=0.01) and miR-195/Cdc42 expression (for OS: RR 6.89, 95% CI, 1.66-14.19, P=0.001; for DFS: RR 6.69, 95% CI, 1.58-13.99, P=0.001) were all independent prognostic factors of unfavorable survival in human ESCC (Table 2).
Table 2.
Multivariate analysis of the impact of variables on overall survival (OS) and distant progression-free survival (DPFS) in esophageal squamous cell carcinoma (ESCC) patients
| Variable | OS | DPFS | ||
|---|---|---|---|---|
|
| ||||
| HR (95% CI) | P | HR (95% CI) | P | |
| TNM stage | 5.26 (1.02-10.63) | 0.01 | 5.12 (1.00-10.58) | 0.01 |
| Tumor differentiation | 3.92 (0.63-7.86) | 0.03 | 3.86 (0.60-7.69) | 0.03 |
| Lymphatic metastasis | 3.51 (0.58-7.26) | 0.03 | 3.48 (0.50-7.18) | 0.03 |
| miR-195 expression | 5.96 (1.26-11.93) | 0.01 | 5.59 (1.13-11.16) | 0.01 |
| Cdc42 expression | 5.33 (1.11-11.08) | 0.01 | 5.18 (1.05-11.02) | 0.01 |
| miR-195/Cdc42 expression | 6.89 (1.66-14.19) | 0.001 | 6.69 (1.58-13.99) | 0.01 |
Discussion
Treatment for ESCC remains one of the most challenging tasks for cancer clinicians. Thus, there is an urgent need for safer and more efficient therapies to improve the prognosis of ESCC patients. In the current study, the downregulation of miR-195 and the upregulation of Cdc42 mRNA were respectively detectable in 52 of 98 (53.06%) and 50 of 98 (51.02%) tumor tissues. In addition, we observed that ESCC patients with low miR-195 expression were more frequently TNM stage III-IV and positive lymphatic metastasis than those with high miR-195 expression, while ESCC patients with high Cdc42 expression were more frequently TNM stage III-IV and poor tumor differentiation than those with low Cdc42 expression. Further assessment demonstrated that OS and DPFS were better in patients with high miR-195 expression or low Cdc42 expression than those in patients with low miR-195 expression or high Cdc42 expression. Both Kaplan-Meier and multivariate analysis showed that miR-195 and Cdc42 expression, alone or combined, were all independent predictors of poor prognosis for both OS and DPFS. To the best of our knowledge, this is the first study to describe the prognostic relevance of miR-195 and its target gene Cdc42 expression in patients with ESCC.
miR-195 is located from 6881953 bp to 6862065 bp on the chromosome 17p13.1 [22]. Its sequence was predicted based on homology to a verified miRNA from mouse and was later verified in human [23]. It has been demonstrated that miR-195 expression is decreased, relative to nonmalignant tissue, in various solid tumors [11-18]. Functionally, an increasing number of evidence have shown that miR-195 can promote cell division and apoptosis while inhibit cell proliferation in diverse cancers. Its roles in cell cycle regulation and their aberrant expression in various cancers suggest that miR-195 may belong to a novel class of tumor suppressor gene depending on its regulatory targets. Jia et al. [24] found that the overexpression of miR-195 might inhibit cell cycle progression and promote apoptosis by reducing Cyclin D1 and Bcl-2 expression in two tongue squamous cell carcinoma cell lines; Liu et al. [12] provided evidence that miR-195 could promote apoptosis and suppress tumorigenicity of human colorectal cancer cells through targeting Bcl-2 expression; Xu et al. [13] showed that miR-195 may block the G1/S transition in hepatocellular carcinoma cells by repressing Rb-E2F signaling through targeting multiple molecules, including cyclin D1, CDK6, and E2F3; Similarly, Fu et al. [19] also indicated that miR-195 expression may inhibit tumor cell proliferation and invasion in ESCC cells by targeting of Cdc42. However, the previous studies did not clarify the relationship between miR-195 expression and tumor progression and patients’ prognosis in ESCC. In order to address this problem, the present study was performed using a large cohort of clinical samples with primary ESCC and verified the significant relationship between miR-195 downregulation and aggressive clinicopathological characteristics and unfavorable prognosis in patients with ESCC. These findings were in line with the reports of Wang et al. [15] on colorectal cancer, Jia et al. [24] on tongue squamous cell carcinoma, Chabre et al. [16] on adrenocortical cancer, but were opposite to the report of Heneghan et al. [25] on breast cancer, which suggest that miR-195 may display different roles under specific physiological conditions and in different types of cancers.
Cdc42, originally identified in Saccharomyces cerevisiae as a cell-cycle mutant involved in the regulation of budding and mating projection, maps to 1p36.1 and encodes a 25-kDa protein [26]. Functionally, it can regulate the dynamic organization of the cytoskeleton and membrane trafficking for physiologic processes such as cell proliferation, cell growth, motility, polarity and cytokinesis [27]. Considering its crucial roles in these diverse cellular functions, it is not surprising that the dysregulation of Cdc42 may be implicated in a number disorders and diseases, especially in human cancers. Aberrant expression of Cdc42 has been observed in a variety of malignancies, including breast cancer, lung cancer, gastric cancer, colon cancer and testicular cancer [28-30]. Growing evidence has suggested its key roles in the development and progression of cancer. For example, Gao et al. [29] reported that the active Rho GTPase Cdc42 could greatly enhance colorectal cancer cell SW480 to spread, migrate, and invade, which may contribute to colorectal cancer metastasis; Chen et al. [30] showed that the overexpression of Cdc42 may induce the invasion of lung cancer cells; Jung et al. [31] observed an increase of Cdc42 protein expression level in melanoma, and indicated that the surface antigen-induced Cdc42 activation could promote melanoma cell growth and cell invasion. It can interact with different effector molecules which, in turn, regulate actin cytoskeleton, microtubule networks, cell polarity, proliferation, apoptosis, endocytosis, and secretion. Especially in ESCC, Fu et al. [20] identified Cdc42 as one target of miR-195 and demonstrated that the ectopic expression of miR-195 in ESCC cells could downregulated Cdc42 by directly binding its 3’ untranslated regions, and induced G1 cell cycle arrest, leading to a significant decrease in cell growth, migration, and invasion in vitro. In agreement with these previous studies, our data, based on clinical samples, showed the upregulation of Cdc42 in ESCC tissues and the significant associations of Cdc42 overexpression with advanced tumor progression and poor prognosis in patients with ESCC.
In order to verify whether the miR-195/Cdc42 axis could be associated with patients’ prognosis of ESCC, we analyzed the relationship between miR-195 and Cdc42 expression in ESCC tissues by spearman’s rank correlation test and found there were a significant negative correlation between miR-195 and Cdc42. Then, we demonstrated that the prognosis, including OS and DPFS, in miR-195-low/Cdc42-high combined expression group was poorest. It is the most important finding of the current study that miR-195/Cdc42 combined expression was identified as an independent prognostic factor for patients with ESCC and its efficiency on prognosis was more significant than miR-195 and Cdc42 expression alone.
In conclusion, these findings indicate the involvement of miR-195-Cdc42 axis in the progression of ESCC and suggest that the combined aberrant expression of miR-195 and Cdc42 mRNA can serve as a promising unfavorable prognostic biomarker in ESCC.
Acknowledgements
The study was supported by National 863 Plan Project, No. 2012AA02A201-1; Sci-tech development Plan Funding Project of Sci-tech Agency of Henan Province, No. 112102310283 & 122102310198.
Disclosure of conflict of interest
None.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu M, Zaninotto G, Nagata K, Graham DY, Lauwers GY. Esophageal squamous cell carcinoma with special reference to its early stage. Best Pract Res Clin Gastroenterol. 2013;27:171–186. doi: 10.1016/j.bpg.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M, Kato H. Treatment options for esophageal squamous cell carcinoma. Expert Opin Pharmacother. 2013;14:1345–1354. doi: 10.1517/14656566.2013.801454. [DOI] [PubMed] [Google Scholar]
- 5.Mayne GC, Hussey DJ, Watson DI. MicroRNAs and esophageal cancer--implications for pathogenesis and therapy. Curr Pharm Des. 2013;19:1211–1226. doi: 10.2174/138161213804805702. [DOI] [PubMed] [Google Scholar]
- 6.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAswith a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Zang W, Wang Y, Du Y. Differential expression profiling of microRNAs and their potential involvement in esophageal squamous cell carcinoma. Tumor Biol. 2014;35:3295–3304. doi: 10.1007/s13277-013-1432-5. [DOI] [PubMed] [Google Scholar]
- 10.Yang G, Wu D, Zhu J. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep. 2013;30:877–889. doi: 10.3892/or.2013.2532. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Jiang H, Gu J. MicroRNA-195 targets ADP-ribosylation factor-like protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis. 2013;4:e695. doi: 10.1038/cddis.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M, Lang N, Qiu M. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer. 2011;128:1269–1279. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- 13.Xu T, Zhu Y, Xiong Y. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–121. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 14.Jiang HL, Yu H, Ma X. MicroRNA-195 regulates steroid receptor coactivator-3 protein expression in hepatocellular carcinoma cells. Tumour Biol. 2014;35:6955–60. doi: 10.1007/s13277-014-1933-x. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Qian L, Li X, Yan J. MicroRNA-195 inhibits colorectal cancer cell proliferation, colony formation and invasion through targeting CARMA3. Mol Med Rep. 2014;10:473–8. doi: 10.3892/mmr.2014.2178. [DOI] [PubMed] [Google Scholar]
- 16.Chabre O, Libé R, Assie G. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr Relat Cancer. 2013;20:579–594. doi: 10.1530/ERC-13-0051. [DOI] [PubMed] [Google Scholar]
- 17.Yongchun Z, Linwei T, Xicai W. MicroRNA-195 inhibits non-small cell lung cancer cell proliferation, migration and invasion by targeting MYB. Cancer Lett. 2014;347:65–74. doi: 10.1016/j.canlet.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Chen H, Fu Y. MiR-195 inhibits proliferation and growth and induces apoptosis of endometrial stromal cells by targeting FKN. Int J Clin Exp Pathol. 2013;6:2824–2834. [PMC free article] [PubMed] [Google Scholar]
- 19.Fu MG, Li S, Yu TT. Differential expression of miR-195 in esophageal squamous cell carcinoma and miR-195 expression inhibits tumor cell proliferation and invasion by targeting of Cdc42. FEBS Lett. 2013;587:3471–3479. doi: 10.1016/j.febslet.2013.08.036. [DOI] [PubMed] [Google Scholar]
- 20.Fu HL, Wu de P, Wang XF. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochem Biophys. 2013;67:657–668. doi: 10.1007/s12013-013-9554-3. [DOI] [PubMed] [Google Scholar]
- 21.Zins K, Lucas T, Reichl P, Abraham D, Aharinejad S. A Rac1/Cdc42 GTPase-specific small molecule inhibitor suppresses growth of primary human prostate cancer xenografts and prolongs survival in mice. PLoS One. 2013;8:e74924. doi: 10.1371/journal.pone.0074924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao JH, Zhou RP, Peng AF. microRNA-195 suppresses osteosarcoma cell invasion and migration in vitro by targeting FASN. Oncol Lett. 2012;4:1125–1129. doi: 10.3892/ol.2012.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Yu J, Yin J. MiR-195 regulates cell apoptosis of human hepatocellular carcinoma cells by targeting LATS2. Pharmazie. 2012;67:645–651. [PubMed] [Google Scholar]
- 24.Jia LF, Wei SB, Gong K, Gan YH, Yu GY. Prognostic implications of micoRNA miR-195 expression in human tongue squamous cell carcinoma. PLoS One. 2013;8:e56634. doi: 10.1371/journal.pone.0056634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heneghan HM, Miller N, Lowery AJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 26.Ma J, Xue Y, Liu W. Role of activated rac1/cdc42 in mediating endothelial cell proliferation and tumor angiogenesis in breast cancer. PLoS One. 2013;8:e66275. doi: 10.1371/journal.pone.0066275. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Chen Q, Chen X, Zhang M. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci. 2011;56:2009–2016. doi: 10.1007/s10620-010-1536-3. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Zhu X, Xu W, Wang D, Yan J. miR-330 regulates the proliferation of colorectal cancer cells by targeting Cdc42. Biochem Biophys Res Commun. 2013;431:560–565. doi: 10.1016/j.bbrc.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Gao L, Bai L, Nan Qz. Activation of Rho GTPase Cdc42 promotes adhesion and invasion in colorectal cancer cells. Med Sci Monit Basic Res. 2013;19:201–207. doi: 10.12659/MSMBR.883983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen QY, Jiao DM, Yao QH. Expression analysis of Cdc42 in lung cancer and modulation of its expression by curcumin in lung cancer cell lines. Int J Oncol. 2012;40:1561–1568. doi: 10.3892/ijo.2012.1336. [DOI] [PubMed] [Google Scholar]
- 31.Jung ID, Lee J, Yun SY. Cdc42 and Rac1 are necessary for autotaxin-induced tumor cell motility in A2058 melanoma cells. FEBS Lett. 2002;532:351–6. doi: 10.1016/s0014-5793(02)03698-0. [DOI] [PubMed] [Google Scholar]


