Abstract
Background: Several lines of evidence have reported that serum angiotensin-converting enzyme (CD143) levels are genetically regulated by insertion/deletion (ins/del) polymorphism in intron 16 of the CD143 gene. In addition, published data on the association of ins/del polymorphism and sepsis risk yielded contradictory conclusions. Therefore, we determined to perform a meta-analysis to validate the association of much debate. Methods and major findings: Relevant literature was identified through weekly searches in databases and references of systematic reviews and the single studies incorporated in this meta-analysis. We combined ORs and its 95% CIs for several genetic models to evaluate the risk of sepsis associated with ins/del polymorphism. A total of seven studies were considered eligible for this analysis. We found significantly increased risk of sepsis in relation to the homozygote ins/ins (OR: 1.32, 95% CI: 1.04-1.68, P: 0.4201, for ins/ins vs. del/del), heterozygote del/ins (OR: 1.33, 95% CI: 1.11-1.61, P: 0.7937, for del/ins vs. del/del) and the two genotypes combined (OR: 1.33, 95% CI: 1.11-1.59, P: 0.7018, for ins/ins + del/ins vs. del/del). Subgroup analysis by age group showed a significant association in pediatric sepsis, but not in adult sepsis. Conclusions: The statistical data suggest that the CD143 gene ins/del polymorphism may influence the risk of sepsis, especially pediatric sepsis.
Keywords: CD143-ins/del polymorphism-sepsis
Introduction
Sepsis is an infection-initiated systemic inflammatory response syndrome (SIRS) characterized by leukopenia or leukocytosis, hypothermia or fever, tachypnea or supranormal minute ventilation, and tachycardia [1]. There has been a continuous increase in the occurrence rate during the past decades and this makes sepsis and sepsis-induced organ failures enormous challenges for clinicians and scientists [2,3]. A number of groups have performed basic and clinic studies, attempting to identify the pathogens and to gain newest insights for the mechanism underlying sepsis [4-6]. These efforts, however, fail to unravel the nature of sepsis pathogenesis.
It is striking that Thomas nearly forty years ago first established a concept of host factors involved in susceptibility towards sepsis [7], and since then an increasing body of literature has emerged in an attempt to investigate the association of sepsis with sequence variations in the predisposition, low-penetrance genes [8].
Available data have shown that stimulation of inflammatory activities in conjunction with activation of coagulation leads to microvascular thrombosis, which ultimately causes multi-organ dysfunction and exacerbates the severity of sepsis [9,10]. The renin-angiotensin system is of fundamental importance for the regulation of inflammatory reactions in addition to systemic blood pressure and intravascular volume. Angiotensin-converting enzyme (CD143) converts angiotensin I to angiotensin II, a major biological effector synthesized in various organs and functioning as a potent inducer of apoptosis in human endothelial cells [11-13].
Polymorphisms in the CD143 gene regulate the plasma CD143 levels, including the insertion/deletion (ins/del) polymorphism (dbSNPs: rs4340) at intron 16 [14]. Individuals with ins/ins or ins/del genotype have markedly lower CD143 levels compared to the individuals with del/del genotype [15]. Earlier research provided ample evidence supporting an association of higher CD143 levels with del/del genotype [16,17]. However, serum CD143 activity varies considerably between individuals. For example, significantly higher levels are detected in type II diabetic patients with macroalbuminuria relative to the patients with normoalbuminuria [18]. So it is interesting to investigate the effects of this CD143 polymorphism on sepsis. Nevertheless, there are controversial results with respect to the contribution of the CD143 polymorphism to sepsis [19-21].
In this meta-analysis, we incorporated all literature on the subject of interest to determine the association between CD143 ins/del polymorphism and sepsis risk.
Methods
This meta-analysis was conducted in agreement with the guidelines of preferred reporting items for systematic reviews and meta-analyses [22].
Search strategy and study eligibility
We performed weekly searches from Dec. 2013 to Feb. 2014 in multiple databases (PubMed, Embase, Web of Knowledge, Science Direct, Cochrane Library) to identify the literature addressing the association of CD143 ins/del polymorphism with sepsis risk. The keywords embraced polymorphism, polymorphisms, angiotensin converting enzyme, CD143, and sepsis. The online searches were supplemented by hand searching the references of two types of study: 1) systematic reviews; 2) the single studies incorporated in this meta-analysis. All publications, no matter what language was used, were included if: 1) the study addressed the association being investigated; 2) sepsis patients were analyzed; 3) reference group consisted of healthy controls; 4) genotype distribution between cases and controls were detailed; 5) subjects investigated must be unique. In cases of duplicates, we selected the largest study.
According to the inclusion criteria, two authors evaluated the titles, abstracts and full-texts whenever necessary to pick out the studies that provided usable data for this metaanalysis.
Data abstraction
A consensus on all items was reached prior to data abstraction. For each of the studies included, the pre-mentioned authors gathered first author’s surname, year of publication, country where the study was completed, ethnicity of study population (Asian or Caucasian), genotyping assay, genotype count, study design (retrospective or prospective), age group (pediatric or adult), and Hardy-Weinberg equilibrium (HWE) wherever available.
Statistical analysis
Deviations from HWE were determined using a goodness-of-ft X2 -test in controls and P < 0.05 was considered statistically significant. The crude OR and its 95% CI were calculated for several genetic models to evaluate the association between CD143 ins/del polymorphism and sepsis risk. A Z test was used to assess the significance of pooled ORs, with P < 0.05 being considered significant. Between-study heterogeneity was examined by the chi-square-based Q-test, and the significance level was fixed at P < 0.05. We also quantified the variance across studies using the I2 statistic and the I2 values ≥ 50% represented large heterogeneity [23]. When lack of significant heterogeneity was indicated, we used the Mantel-Haenszel method (the fixed effect model) to combine effect estimates. Otherwise, the DerSimonian Laird method (the random-effects model) was performed.
In addition to global analysis, we also performed subgroup analyses according to age group. The funnel plots, described by Begg, were applied to detect the publication bias in this analysis [24]. The Egger’s test was used to examine the symmetry of each funnel plot (P < 0.05 indicated asymmetric plots) [25]. Sensitivity analysis was performed to assess the stability of the combined results. Statistical analyses were done by R software (version 2.15.0) and Stata software (version 12.0).
Results
Characteristics of included studies
Database and hand searches yielded 63 publications. After excluding the evidently irrelevant study, we were left with 11 studies. We then read through the full-texts, and found three case-only studies [26-28] and one narrative review [29]. Our final dataset therefore combined seven research articles [19-21,30-33]. A flow chat showing the selection process is displayed in Figure 1.
Figure 1.
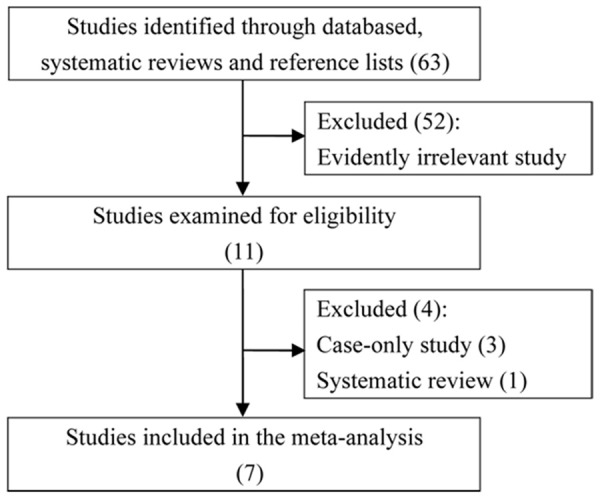
A flow diagram of study identification, exclusion and inclusion.
As described in Table 1, there were seven Caucasian studies, of which four studies investigated sepsis in children and three in adults. Five retrospective studies were conducted in four different countries: Netherlands, Turkey, Greece, and Spain. The two prospective studies were from the United States and Germany, respectively. Significant deviation from HWE was revealed in a Turkish study [31]. Genotyping methods, including multiplex polymerase chain reaction (multiplex PCR), PCR-restriction fragment length polymorphism (PCR-RFLP), reverse-hybridization technique and Taqman were performed in identifying the genotypes of CD143 polymorphism.
Table 1.
Summary information for the studies analyzed in the meta-analysis
| Authors | Cases-controls | Age group | Study design | Country of origin | Ethnicity | Genotyping method | Deviation from HWE |
|---|---|---|---|---|---|---|---|
| Bunker-Wiersma et al. | 53-135 | Pediatric | Retrospective | Netherlands | Caucasian | Taqman | No |
| Celik et al. | 98-100 | Pediatric | Retrospective | Turkey | Caucasian | NA※ | Yes |
| Cogulu et al. | 98-287 | Pediatric | Retrospective | Turkey | Caucasian | multiplex PCR | No |
| Davis et al. | 28-53 | Adult | Prospective | United States | Caucasian | Taqman | No |
| Spiegler et al. | 246-963 | Pediatric | Prospective | Germany | Caucasian | PCR-RFLP | No |
| Tsantes et al. | 186-180 | Adult | Retrospective | Greece | Caucasian | PCR, reverse-hybridisation technique | No |
| Villar et al. | 212-364 | Adult | Retrospective | Spain | Caucasian | PCR | No |
| Total | 921-2082 |
NA: not available.
Association between CD143 polymorphism and sepsis
The association of CD143 polymorphism with sepsis risk was assessed in a meta-analysis combining 921 patients and 2 082 controls. All genetic models, with the exception of ins/ins vs. del/ins + del/del, provided statistical evidence for a significant association between the ins/del polymorphism and sepsis risk (OR: 1.32, 95% CI: 1.04-1.68, P: 0.4201, for ins/ins vs. del/del; OR: 1.33, 95% CI: 1.11-1.59, P: 0.7018, for ins/ins + del/ins vs. del/del; OR: 1.33, 95% CI: 1.11-1.61, P: 0.7937, for del/ins vs. del/del) (Figures 2, 3 and 4; Table 2).
Figure 2.
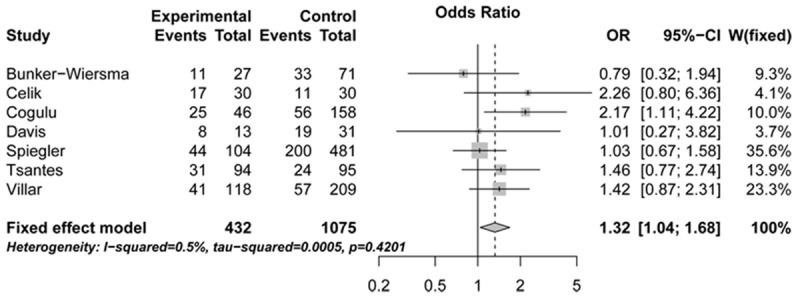
Forest plots illustrating the association between sepsis risk and CD143 gene ins/del polymorphism under ins/ins vs. del/del genetic model (fixed-effects model). OR, indicates odds ratio; CI, confidence interval.
Figure 3.
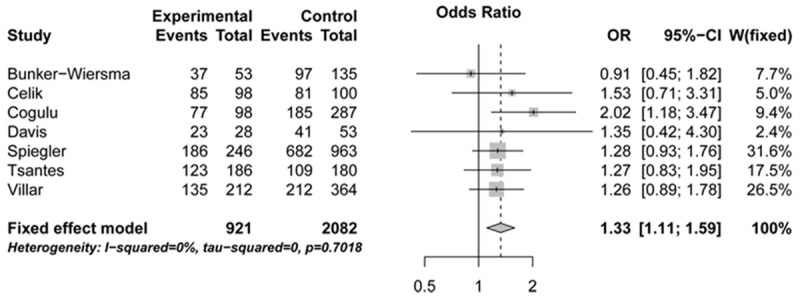
Forest plots illustrating the association between sepsis risk and CD143 gene ins/del polymorphism under ins/ins + del/ins vs. del/del genetic model (fixed-effects model). OR, indicates odds ratio; CI, confidence interval.
Figure 4.
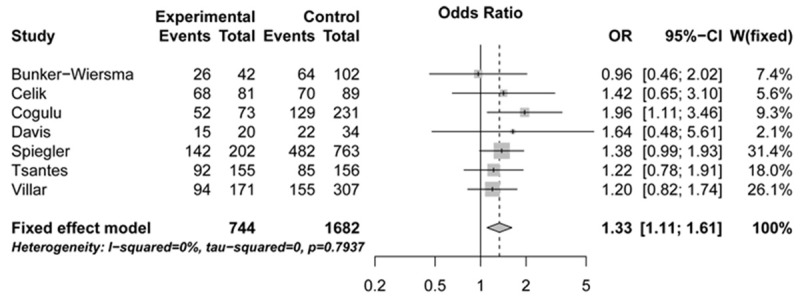
Forest plots illustrating the association between sepsis risk and CD143 gene ins/del polymorphism under del/ins vs. del/del genetic model (fixed-effects model). OR, indicates odds ratio; CI, confidence interval.
Table 2.
Meta-analysis of the association between CD143 I/D polymorphism and sepsis risk
| Comparisons | Subgroups | No. of cases and controls | Effect size | Heterogeneity test | Model※※ | |
|---|---|---|---|---|---|---|
|
|
|
|||||
| OR (95% CI) | PHet | I2 (%) | ||||
| (ins/ins vs. del/del) | Total | 921-2082 | 1.32 (1.04, 1.68) | 0.4201 | 0.5 | FEM |
| Pediatric | 495-1485 | 1.27 (0.93, 1.74) | 0.1302 | 46.9 | FEM | |
| Adult | 426-597 | 1.39 (0.96, 2.02) | 0.8833 | 0.0 | FEM | |
| (ins/ins + del/ins vs. del/del) | Total | 921-2082 | 1.33 (1.11, 1.59) | 0.7018 | 0.0 | FEM |
| Pediatric | 495-1485 | 1.38 (1.08, 1.76) | 0.3056 | 17.1 | FEM | |
| Adult | 426-597 | 1.27 (0.97, 1.65) | 0.9936 | 0.0 | FEM | |
| (ins/ins vs. del/ins + del/del) | Total | 921-2082 | 1.08 (0.88, 1.33) | 0.3832 | 5.8 | FEM |
| Pediatric | 495-1485 | 1.01 (0.78, 1.31) | 0.2143 | 33.0 | FEM | |
| Adult | 426-597 | 1.21 (0.87, 1.68) | 0.5448 | 0.0 | FEM | |
| (del/ins vs. del/del) | Total | 921-2082 | 1.33 (1.11, 1.61) | 0.7937 | 0.0 | FEM |
| Pediatric | 495-1485 | 1.43 (1.11, 1.84) | 0.5133 | 0.0 | FEM | |
| Adult | 426-597 | 1.23 (0.93, 1.62) | 0.8928 | 0.0 | FEM | |
Model: fixed-effects model.
We subsequently performed subgroup analysis according to age group. The statistical data revealed significantly increased risk of sepsis in relation to the combined ins/ins and del/ins genotypes (OR: 1.38, 95% CI: 1.08-1.76, P: 0.3056) or del/ins genotype alone (OR: 1.43, 95% CI: 1.11-1.84, P: 0.5133) (Table 2).
Sensitivity analysis
To examine if the combined results were affected by some single study, we performed sensitivity analyses through removing one study each time. The removals did not cause quantitative differences compared to the original results. This process confirmed the stability of our findings.
Publication bias
We evaluated the publication bias by use of the funnel plots along with the Egger’s test. The studies were symmetrically distributed within the funnel plots (z: 0.30, P: 0.764 for ins/ins vs. del/del) (Figure 5). Evidence of asymmetry was not revealed by performing the Egger’s test (z: -0.24, P: 0.822 for ins/ins vs. del/del).
Figure 5.
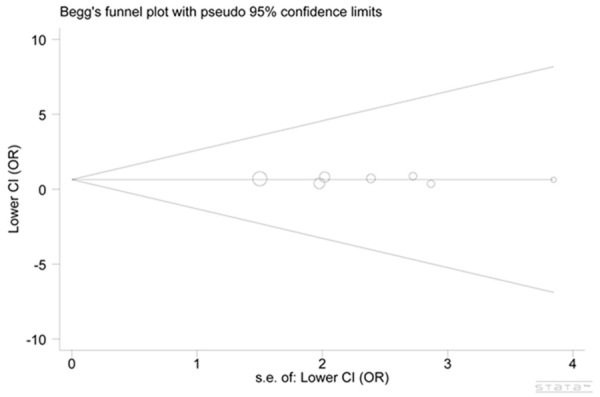
Egger’s test and Begg’s test of publication bias.
Discussion
Single nucleotide polymorphisms in pro-inflammatory genes that catalyze the stimuli of bacterial pathogens affect the biological function of the protein and thus confer individual susceptibility to the development of various human diseases, such as sepsis [34]. The human CD143 at chromosome 17q23 is an exopeptidase with a pivotal role in regulating inflammatory responses. Several studies have investigated the genetic alternations in CD143 gene and demonstrate that these variations impair the innate immune system by upregulating CD143 activity and consequently promote inflammation process [35,36]. All genotypes of the ins/del polymorphism, including ins/ins, ins/del, and del/del are associated with plasma CD143 activities, with the heterozygote ins/del accounting for almost half (47%) of the total phenotypic variance, indicating most circulating CD143 concentration is determined by its genetic variations [16].
Recently, the effects of a functional polymorphism, CD143 ins/del, have been extensively studied in sepsis community because of the regulatory role in plasma CD143 levels [19-21,33]. The previous studies, however, may have reported false-negative or false-positive results most likely due to poor study design and methodological limitations [37]. In an effort to clarify whether or not CD143 ins/del polymorphism affects susceptibility to sepsis, we determined to carry out a meta-analysis.
Meta-analysis as a known analytical method could supply strong evidence of genotype-phenotype relationships for human ma-lignancies [38]. Herein, we analyzed 3 003 subjects and found a significant association between the ins/del polymorphism and overall sepsis risk. To be more specific, the carriage of ins/ins or ins/del alone, or ins/ins and ins/del combined had approxi-mately 33% greater risk of sepsis compared to the carriage of del/del. Strikingly, the observed association only persisted in pe-diatric subjects and was lost in adult subjects when data were stratified by age group. As reported by Felehgari et al., CD143 levels are significantly lower in patients harboring normoalbuminuria in comparison to patients with macroalbuminuria [18], showing plasma CD143 activities vary substantially even in patients with the same type of disease. Therefore, we hypothesize that the ins/del polymorphism modify susceptibility towards common diseases possibly by interacting with various confounders, such as age, gender, environmental pathogens and other predisposition genes. In addition, several lines of evidence have shown that there is no clear connection between the ins/del polymorphism and circulating CD143 levels. Thus we cannot rule out the possibility that the ins/del polymorphism exerts no influence on CD143 function, and does not modulate sepsis susceptibility [39-41]. Whether the ins/del polymorphism plays a major role in sepsis remains to be elucidated.
Previous reports have demonstrated wide differences in terms of the impact of ins/del polymorphism on CD143 activities as a result of disparate ethnicities. Tiret et al. found increased CD143 levels in Caucasian populations [16,17]. A study by Bloem et al. revealed a clear association between serum CD143 activity and CD143 gene ins/del polymorphism in whites, but not in blacks, suggesting ethnic variation plays an important role in genetic regulation of serum CD143 levels and the relation of ins/del polymorphism with human diseases [42]. In this meta-analysis, only Caucasian studies were identified and the effects on other ethnic populations remain unknown. We expect that studies of various ethnicities will be conducted to determine the role of the CD143 gene ins/del polymorphism in sepsis.
Several limitations should be noted when interpreting the results of this meta-analysis. First, the small number of included studies, with each of them conducted with an inadequate sample, may likely make our analysis less powerful. Second, a positive association which requires further studies to validate was observed among Caucasian subjects in this work. It is still unclear whether the individuals of different ethnicities have higher risk of sepsis. Third, while no asymmetry was revealed in the funnel plots and no statistical evidence of significant publication bias was shown by performing the Egger’s test, the bias may be introduced as we failed to include the unpublished studies and the studies written in other languages.
However, we incorporated all genetic association studies reporting on CD143 ins/del polymorphism and sepsis risk and the results were more powerful than any of the previously published studies concerning the same topic. We noted that although the ins/del polymorphism showed a significant association with sepsis risk in general populations, it seemed to not associate with the adult sepsis.
To sum up, we found some evidence supporting that the ins/del polymorphism in intron 16 of the CD143 gene was associated with risk of sepsis, pediatric sepsis in particular. The inadequate sample size of this investigation emphasizes the necessity for future studies to be larger and better designed to evaluate the effects the ins/del polymorphism has on sepsis.
Disclosure of conflict of interest
None.
References
- 1.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 2009;101:1644–55. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 2.Dombrovskiy VY, Martin AA, Sunderram J, Paz HL. Rapid increase in hospitalization and mortality rates for severe sepsis in the United States: a trend analysis from 1993 to 2003. Crit Care Med. 2007;35:1244–50. doi: 10.1097/01.CCM.0000261890.41311.E9. [DOI] [PubMed] [Google Scholar]
- 3.Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8:776–87. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remick DG. Cytokine therapeutics for the treatment of sepsis: why has nothing worked? Curr Pharm Des. 2003;9:75–82. doi: 10.2174/1381612033392567. [DOI] [PubMed] [Google Scholar]
- 5.Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, Abraham E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279:7370–7. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 6.Hotchkiss RS, Nicholson DW. Apoptosis and caspases regulate death and inflammation in sepsis. Nat Rev Immunol. 2006;6:813–22. doi: 10.1038/nri1943. [DOI] [PubMed] [Google Scholar]
- 7.Thomas L. Germs. N Engl J Med. 1972;287:553–5. doi: 10.1056/NEJM197209142871109. [DOI] [PubMed] [Google Scholar]
- 8.Namath A, Patterson AJ. Genetic polymorphisms in sepsis. Crit Care Nurs Clin North Am. 2011;23:181–202. doi: 10.1016/j.ccell.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Schouten M, Wiersinga WJ, Levi M, van der Poll T. Inflammation, endothelium, and coagulation in sepsis. J Leukoc Biol. 2008;83:536–45. doi: 10.1189/jlb.0607373. [DOI] [PubMed] [Google Scholar]
- 10.van der Poll T, Levi M. Crosstalk between inflammation and coagulation: the lessons of sepsis. Curr Vasc Pharmacol. 2012;10:632–8. doi: 10.2174/157016112801784549. [DOI] [PubMed] [Google Scholar]
- 11.Rossig L, Hermann C, Haendeler J, Assmus B, Zeiher AM, Dimmeler S. Angiotensin II-induced upregulation of MAP kinase phosphatase-3 mRNA levels mediates endothelial cell apoptosis. Basic Res Cardiol. 2002;97:1–8. doi: 10.1007/s395-002-8381-2. [DOI] [PubMed] [Google Scholar]
- 12.Kamiyama M, Farragut KM, Garner MK, Navar LG, Kobori H. Divergent localization of angiotensinogen mRNA and protein in proximal tubule segments of normal rat kidney. J Hypertens. 2012;30:2365–72. doi: 10.1097/HJH.0b013e3283598eed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramkumar N, Stuart D, Ying J, Kohan DE. A possible interaction between systemic and renal angiotensinogen in the control of blood pressure. Am J Hypertens. 2013;26:473–80. doi: 10.1093/ajh/hps078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villard E, Tiret L, Visvikis S, Rakotovao R, Cambien F, Soubrier F. Identification of new polymorphisms of the angiotensin I-converting enzyme (ACE) gene, and study of their relationship to plasma ACE levels by two-QTL segregation-linkage analysis. Am J Hum Genet. 1996;58:1268–78. [PMC free article] [PubMed] [Google Scholar]
- 15.Abbas D, Ezzat Y, Hamdy E, Gamil M. Angiotensin-converting enzyme (ACE) serum levels and gene polymorphism in Egyptian patients with systemic lupus erythematosus. Lupus. 2012;21:103–10. doi: 10.1177/0961203311418268. [DOI] [PubMed] [Google Scholar]
- 16.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiret L, Rigat B, Visvikis S, Breda C, Corvol P, Cambien F, Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet. 1992;51:197–205. [PMC free article] [PubMed] [Google Scholar]
- 18.Felehgari V, Rahimi Z, Mozafari H, Vaisi-Raygani A. ACE gene polymorphism and serum ACE activity in Iranians type II diabetic patients with macroalbuminuria. Mol Cell Biochem. 2011;346:23–30. doi: 10.1007/s11010-010-0587-2. [DOI] [PubMed] [Google Scholar]
- 19.Cogulu O, Onay H, Uzunkaya D, Gunduz C, Pehlivan S, Vardar F, Atlihan F, Ozkinay C, Ozkinay F. Role of angiotensin-converting enzyme gene polymorphisms in children with sepsis and septic shock. Pediatr Int. 2008;50:477–80. doi: 10.1111/j.1442-200X.2008.02583.x. [DOI] [PubMed] [Google Scholar]
- 20.Spiegler J, Gilhaus A, Konig IR, Kattner E, Vochem M, Kuster H, Moller J, Muller D, Kribs A, Segerer H, Wieg C, Nikischin W, von der Wense A, Gebauer C, Herting E, Gopel W. Polymorphisms in the Renin-Angiotensin system and outcome of very-low-birthweight infants. Neonatology. 2010;97:10–4. doi: 10.1159/000226602. [DOI] [PubMed] [Google Scholar]
- 21.Tsantes A, Tsangaris I, Kopterides P, Nikolopoulos G, Kalamara E, Antonakos G, Kapsimali V, Gialeraki A, Dimopoulou I, Orfanos S, Dima K, Travlou A, Armaganidis A. Angiotensin converting enzyme (ACE) insertion/deletion (I/D) polymorphism and circulating ACE levels are not associated with outcome in critically ill septic patients. Clin Chem Lab Med. 2012;50:293–9. doi: 10.1515/CCLM.2011.752. [DOI] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62:1006–12. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. [PubMed] [Google Scholar]
- 25.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardinal-Fernandez P, Ferruelo A, El-Assar M, Santiago C, Gómez-Gallego F, Martín-Pellicer A, Frutos-Vivar F, Peñuelas O, Nin N, Esteban A, Lorente JA. Genetic predisposition to acute respiratory distress syndrome in patients with severe sepsis. Shock. 2013;39:255–60. doi: 10.1097/SHK.0b013e3182866ff9. [DOI] [PubMed] [Google Scholar]
- 27.Cardinal-Fernandez P, Ferruelo A, El-Assar M, Santiago C, Gómez-Gallego F, Martín-Pellicer A, Frutos-Vivar F, Peñuelas O, Nin N, Esteban A, Lorente JA. Genetic predisposition to acute kidney injury induced by severe sepsis. J Crit Care. 2013;28:365–70. doi: 10.1016/j.jcrc.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 28.John Baier R, Loggins J, Yanamandra K. Angiotensin converting enzyme insertion/deletion polymorphism does not alter sepsis outcome in ventilated very low birth weight infants. J Perinatol. 2005;25:205–9. doi: 10.1038/sj.jp.7211231. [DOI] [PubMed] [Google Scholar]
- 29.Dahmer MK, Randolph A, Vitali S, Quasney MW. Genetic polymorphisms in sepsis. Pediatr Crit Care Med. 2005;6:S61–73. doi: 10.1097/01.PCC.0000161970.44470.C7. [DOI] [PubMed] [Google Scholar]
- 30.Bunker-Wiersma HE, Koopmans RP, Kuipers TW, Knoester H, Bos AP. Single nucleotide polymorphisms in genes of circulatory homeostasis in surviving pediatric intensive care patients with meningococcal infection. Pediatr Crit Care Med. 2008;9:517–23. doi: 10.1097/PCC.0b013e318184985b. [DOI] [PubMed] [Google Scholar]
- 31.Celik U YD, Celik T, et al. Relationship between angiotensin-converting enzyme gene polymorphism (insertion/deletion) and the clinical condition of sepsis in Turkish children. Turkiye Klinikleri J Med Sci. 2010;30:591–597. [Google Scholar]
- 32.Davis SM, Clark EA, Nelson LT, Silver RM. The association of innate immune response gene polymorphisms and puerperal group A streptococcal sepsis. Am J Obstet Gynecol. 2010;202:308.e1–8. doi: 10.1016/j.ajog.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Villar J, Flores C, Pérez-Méndez L, Maca-Meyer N, Espinosa E, Blanco J, Sangüesa R, Muriel A, Tejera P, Muros M, Slutsky AS GRECIA group; GEN-SEP group. Angiotensin-converting enzyme insertion/deletion polymorphism is not associated with susceptibility and outcome in sepsis and acute respiratory distress syndrome. Intensive Care Med. 2008;34:488–95. doi: 10.1007/s00134-007-0937-z. [DOI] [PubMed] [Google Scholar]
- 34.Lin MT, Albertson TE. Genomic polymorphisms in sepsis. Crit Care Med. 2004;32:569–79. doi: 10.1097/01.CCM.0000110878.49476.42. [DOI] [PubMed] [Google Scholar]
- 35.Rice CL, Kohler JP, Casey L, Szidon JP, Daise M, Moss GS. Angiotensin-converting enzyme (ACE) in sepsis. Circ Shock. 1983;11:59–63. [PubMed] [Google Scholar]
- 36.Phillips MI, Kagiyama S. Angiotensin II as a pro-inflammatory mediator. Curr Opin Investig Drugs. 2002;3:569–77. [PubMed] [Google Scholar]
- 37.Clark MF, Baudouin SV. A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med. 2006;32:1706–12. doi: 10.1007/s00134-006-0327-y. [DOI] [PubMed] [Google Scholar]
- 38.Salanti G, Sanderson S, Higgins JP. Obstacles and opportunities in meta-analysis of genetic association studies. Genet Med. 2005;7:13–20. doi: 10.1097/01.gim.0000151839.12032.1a. [DOI] [PubMed] [Google Scholar]
- 39.Zhu X, McKenzie CA, Forrester T, Nickerson DA, Broeckel U, Schunkert H, Doering A, Jacob HJ, Cooper RS, Rieder MJ. Localization of a small genomic region associated with elevated ACE. Am J Hum Genet. 2000;67:1144–53. doi: 10.1016/s0002-9297(07)62945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox R, Bouzekri N, Martin S, Southam L, Hugill A, Golamaully M, Cooper R, Adeyemo A, Soubrier F, Ward R, Lathrop GM, Matsuda F, Farrall M. Angiotensin-1-converting enzyme (ACE) plasma concentration is influenced by multiple ACE-linked quantitative trait nucleotides. Hum Mol Genet. 2002;11:2969–77. doi: 10.1093/hmg/11.23.2969. [DOI] [PubMed] [Google Scholar]
- 41.Sayed-Tabatabaei FA, Oostra BA, Isaacs A, van Duijn CM, Witteman JC. ACE polymorphisms. Circ Res. 2006;98:1123–33. doi: 10.1161/01.RES.0000223145.74217.e7. [DOI] [PubMed] [Google Scholar]
- 42.Bloem LJ, Manatunga AK, Pratt JH. Racial difference in the relationship of an angiotensin I-converting enzyme gene polymorphism to serum angiotensin I-converting enzyme activity. Hypertension. 1996;27:62–6. doi: 10.1161/01.hyp.27.1.62. [DOI] [PubMed] [Google Scholar]


