Abstract
Cutaneous squamous cell carcinoma (cSCC), the second most common form of human cancer, is an epithelial skin tumor, which can result in metastasis with lethal consequences accounting for about 20% of all skin cancer-related deaths. The metastasis is the main reason for cSCC-related deaths with an overall 5-year survival rate < 30% in cases that spread systemically. The role of miRNAs has been involved in SCC of different origins. Recent data have revealed that the expression of miRNA-199a was changed in many human cancers. In this study, we found that miR-199a was significantly decreased in cSCC tissues, which had an inverse relationship with CD44. MiR-199a specifically regulated the expression of CD44 at mRNA and protein levels, and the interaction between CD44 and Ezrin in cSCC cells. Moreover, the suppressive role of miR-199a in cell migration in cSCC cells was also associated with the activity of MMP2 and MMP9. Taken together, our data indicated that increased expression of endogenous mature miR-199a might prevent the growth and migration of human cSCC via decreasing the expression of CD44 and regulating the interaction between CD44 and Ezrin, which may provide a potentially important therapeutic target for human cSCC.
Keywords: Cutaneous squamous cell carcinoma, miR-199a, CD44, Ezrin, MMP2, MMP9
Introduction
Cutaneous squamous cell carcinoma (cSCC), the second most common form of human cancer, is an epithelial skin tumor [1], which can result in metastasis with lethal consequences accounting for about 20% of all skin cancer-related deaths. Some locally invasive cSCCs show an aggressive progress [2]. The metastasis is the main reason for cSCC-related deaths with an overall 5-year survival rate < 30% in cases that spread systemically.
MicroRNAs (miRNAs), short non-coding RNA molecules containing about 20 nucleotides (nt), can regulate gene expression at a post-transcriptional level. They play an important role in a variety of physiologic cellular functions and diseases, including cutaneous squamous cell carcinoma. miRNA regulation, as tumor suppressors or oncogenes, affected approximately 30%-60% of all human genes with a gradually increasing. The role of miRNAs has been involved in SCC of different origins, such as cervical, oral and tongue tissue [3,4]. The involvement of miRNA dysregulation in cSCC has just recently begun to be researched. It has reported an ectopic expression of miRNAs, including increased expression of hsamiR-21 and hsa-miR-184, and the decreased expression of hsa-miR-203, in cSCC [5]. Yamane et al. showed that miR-124 and miR-214 were shown to be dramatically down-regulated in SCC, which miR-214 is the regulator of extracellular-signal- regulated kinase 1 (ERK1) and miR-124 and miR-214 are both regulators of ERK2 [6].
Recent data have revealed that the expression of miRNA-199a was changed in many human cancers [7,8]. For example, the increased expression of miRNA-199a was observed in ovarian cancer cells, pancreatic adenocarcinomas, and cervical carcinomas [7,9]. Meanwhile, the downregulated expression of miRNA-199a was observed in liver, breast, and bladder cancer [10,11]. Furthermore, miRNA-199a could decrease cell resistance to cytotoxic drugs [9]. Moreover, the miRNA-199a level was significantly downregulated in human osteosarcoma cell lines and transfection of miRNA-199a mimic into those cells repressed cell proliferation and invasion [12]. MiRNA-199a was also involved in the progression of gastric cancer [13]. According these previous findings, we propose that miRNA-199a functions as a tumor suppressor gene and plays an important role in the clinical pathology of cSCC carcinogenesis. In the present study, therefore, we investigated the expression and mechanisms of miRNA-199a in cSCC. We found that miRNA-199a expression was decreased in human cSCC cancer tissues, compared with normal matched tissues, and miRNA-199a inhibited the proliferation and migration of cSCC cells (A-431). Furthermore, we identified CD 44 as a direct target gene of miRNA-199a by luciferase reporter assay. Moreover, although miR-199a cannot directly regulate the expression of Ezrin by targeting 3’UTR, we found that miR-199a regulated the interaction between CD44 and Ezrin. Thus, our findings suggested that miRNA-199a plays a critically suppressive role in cSCC by targeting CD44 and regulating the interaction between CD44 and Ezrin and provided a candidate strategy for treatment of cutaneous squamous cell carcinoma.
Materials and methods
Tissue samples
All patients signed an informed consent, approved by the Independent Ethical Committee of Central South University. The skin scale cancer and the matched adjacent normal tissues used in the study were collected from 10 patients between 2011 and 2014 in our hospital. All samples were stored at -80°C until tissue analysis.
Cells culture and treatment
SCC cell line, A-431 cells, was obtained from American type culture collection (ATCC) and cultivated in RPMI-1640 medium with a final concentration of 10% fetal bovine serum. All cells cultured in the conditions: 95% air and 5% carbon dioxide (CO2) at 37°C.
Ectopic expression of miR-199a in cells was achieved by transfection with miR-199a mimics or inhibitors using Lipofectamine 2000 (Invitrogen, USA). Cells were plated in 24-well plates and transfected for 24 hours or 48 hours. Total RNA or protein was extracted from the indicated cells for analysis.
Reagents
Rabbit polyclonal antibody of β-actin was purchased from Assay Biotech (Assay Biotech, USA). Rabbit monoclonal antibody of CD44 was purchased from Biorbyt (Biorbyt, USA). Mouse monoclonal antibody of Ezrin was purchased from Santa Cruze.
Quantitative PCR (qPCR) assay
Total RNA was extracted from the indicated cells, according to the manufacturer’s instructions using Qiagen RNeasy Kit. Expression of CD44 and Ezrin mRNA was detected by SYBR green qPCR assay (BioRad, USA). Expression of β-actin was used as an endogenous control. The specific primers are as follow: CD44, F: agcaaccaagaggcaagaaa, R: gtgtggttgaaatggtgctg; Ezrin, F: ggctgcaggactatgaggag, R: tggcagtgtattctgcaagc; β-actin, F: cattaaggagaagctgtgct, R: gttgaaggtagtttcgtgga. MiScript Reverse Transcription kit was used to reverse transcribe RNA into cDNA and MiScript SYBR-Green PCR Kit was used for real time PCR to detect the expression of miR-199a. The specific primers sets for miRNA-199a and U6 were purchased from GeneCopoeia. Expression of U6 was used as an endogenous control. Data were analyzed through 2-ΔΔCT method.
Western blotting
Total protein (60μg) extracted from indicated cells was separated on SDS-polyacrylamide gels for CD44, Ezrin and β-actin detection. β-actin was used as loading control. The protein in gels was transferred to nitrocellulose membranes, and then incubated with the indicated antibodies in recommended dilution for overnight at 4°C. Then membranes were washed with 0.1 M PBST and incubated with HRP-conjugated secondary antibody. Signals were visualized using ECL Substrates and quantified using Optiquant software.
Dual luciferase reporter assay
The indicated cells were seeded in 96-well plates the day before transfection. Then, cells were cotransfected with 25 ng pmiR-report 3’UTR CD44 target gene and 10 ng pRL-TK (Renilla plasmid), and 15 nM miR-199a mimics or miR-199a inhibitors using Lipofectamine 2000. Cells were transfected for 48 hours with Dual-Luciferase-Reporter Assay System and luminescence was detected using a luminometer (Promega). The luciferase activity was normalized to the Renilla activity.
CCK-8 cell proliferation assay
Cell Counting Kit-8 (CCK-8) (Sigma, USA) was used to measure cell proliferation rates. 0.5 × 104 cells transfected with the indicated miRNA were seeded in each 96-well plate for 24 h, and further incubated for oh, 24 h, 48 h, 72 h, respectively. 1 h before ending of incubation, 10 μl CCK-8 reagents was added to each well. OD 490 nm value in each well was determined by an enzyme immunoassay analyzer.
Transwell assay
A-431 cells were transfected with miR-199a mimics, miR-199a inhibitors or scramble and starved in serum-free medium for 24 h, then resuspended in serum-free medium. The cells were added to the upper chamber, while the lower chamber was filled with completed medium containing 10% FBS. Incubation for 24 hours later, cells attached to the bottom were fixed with 95% alcohol for 15 min and stained with hematoxylin for 20 min. The redundant hematoxylin was washed by water, and dried in air. Then, the stained cells were photographed and counted in the inverted microscope.
Immunoprecipitation
The indicated cells were added 300 μl cell lysate and shaking for 1 h, and centrifuged at 10000 g for 15 min. The concentration of protein extracts was measured by the BCA protein Assay Kit (Merck Millipore). Protein extracts were incubated with 50 μl of normal rabbit IgG for 1 h, and precleared with proteinA-agarose beads (Bio-Rad, Germany) at 4°C for 30 min, then centrifuged at 12000 g for 10 min at 4°C. The total 60 μl of the supernatant were incubated with 2 μg of CD44 antibody at 4°C overnight, washed in lysis buffer and centrifugated at 2500 g for 5 min at 4°C. The precipitates were then detected by Western blot with the Ezrin antibody.
Immunofluorescence staining
The cell slices were washed with PBS for three times and fixed with 4% paraformaldehyde for 20 min. The slices were then washed with PBST, and blocked with 5% normal goat serum for 60 min. The first primary antibody was rabbit against CD44 antibody (1:200). After overnight incubation at 4°C, the slices were washed with PBST and incubated with Alexa Fluor 488-conjugated goat anti-rabbit antibody for 40 min at 37°C. Then the slices were washed with PBST and incubated with another primary antibody mouse against Ezrin antibody (1:200) for 1 h at 37°C. After incubation, the slices were washed with PBST and incubated with Alexa Fluor 555-conjugated goat anti-mouse antibody for 40 min at 37°C. The slices were washed and counterstained with DAPI (1:1000) for 30 min. The samples were visualized under a microscope (Nikon) through the program DP2-BSW.
Gelatin zymography
MMP zymography assay kit (for MMP-2 and MMP-9) (Applygen Technologie, China) was used to detect the activity of MMP2 and MMP9. Protein extracts and positive mixture were mixed with an equal volume of 2 × SDS-PAGE non-reducing buffer, and electrophoresed on 8% SDS polyacrylamide gels containing 2 mg/ml of gelatin. Gels were then washed twice for 30 min in buffer A at room temperature, and incubated for 4 hours at room temperature in incubation buffer B. Gels were then stained for 2 hour with 0.25% Coomassie brilliant blue and then destained in destaining buffer (10% acetic acid and 20% methanol) for 60 min.
Statistical analysis
Data are expressed as mean ± SD and analyzed by Student’s t-test. Compared with respective controls, P values of < 0.05 were considered statistically significant.
Results
Expression of miR-199a, CD44 and Ezrin in cSCC tissues
Western blot was used to analyze the expression of CD44 and Ezrin in SCC tissues. As the results shown in Figure 1A and 1B, we found that CD44 was significantly increased in SCC tissues when compared with that in the normal tissues (P = 0.0181). However, we found that Ezrin was slightly, but not significantly, increased in SCC tissues when compared with that in the normal tissues (P = 0.49) (Figure 1C and 1D). In addition, we detected the expression of miR-199a using qPCR. It was showed a notable down-regulation of miR-199a (P < 0.0001) (Figure 1E).
Figure 1.
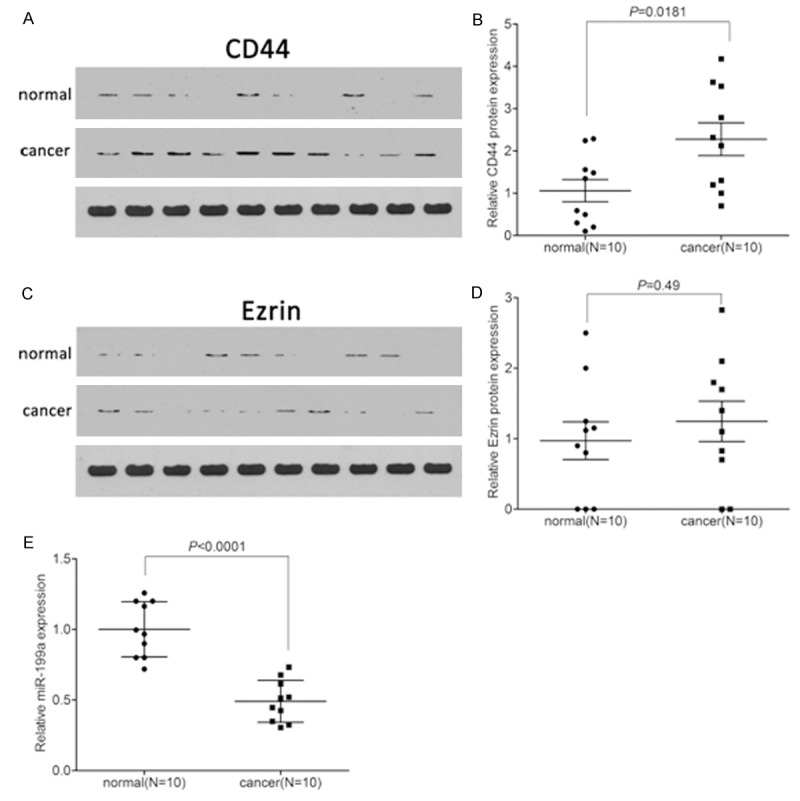
Expression of CD44, Ezrin and miR-199a in cutaneous squamous cell carcinoma tissues. A. The CD44 protein levels were detected by western blot. B. Quantification of CD44 protein in cSCC tissues. C. The Ezrin protein levels were detected by western blot. D. Quantification of Ezrin protein in cSCC tissues. E. The expression of miR-199a was detected by qPCR.
MiR-199a directly targeted CD44
QPCR was used to detect the expression of miR-199a after miR-199a mimics or inhibitor treatment. The results showed that miR-199a mimics induced efficiently the expression of miR199a, and miR-199a inhibitor downregulated dramatically the expression of miR199a (Figure 2A). Furthermore, miR-199a mimics reduced efficiently the expression of CD44 mRNA, and miR-199a inhibitor upregulated dramatically the expression of CD44 mRNA (Figure 2B). However, there were no significant changes of Ezrin mRNA after miR-199a mimics or inhibitor treatment (Figure 2C). To investigate if miR-199a directly targeted to 3’UTR of CD44, we cloned the 3’UTR of CD44 downstream to a luciferase reporter gene (CD44). CD44 vectors were co-transfected with miR-199a mimics or inhibitor into A-431 cells. The luciferase activity of miR-199a mimics transfected cells was significantly decreased compared with inhibitor and scramble control cells (Figure 2D). These results suggested miR-199a could inhibit CD44 expression at transcriptional level. Moreover, we further analyzed the protein expression of CD44 and Ezrin in cells transfected with miR-199a mimics or inhibitor. Similarly, the expression of CD44 was decreased or induced by miR-199a mimics or inhibitor, respectively (Figure 2E, 2G). There were also no significant changes of Ezrin protein after miR-199a mimics or inhibitor treatment (Figure 2F, 2H).
Figure 2.
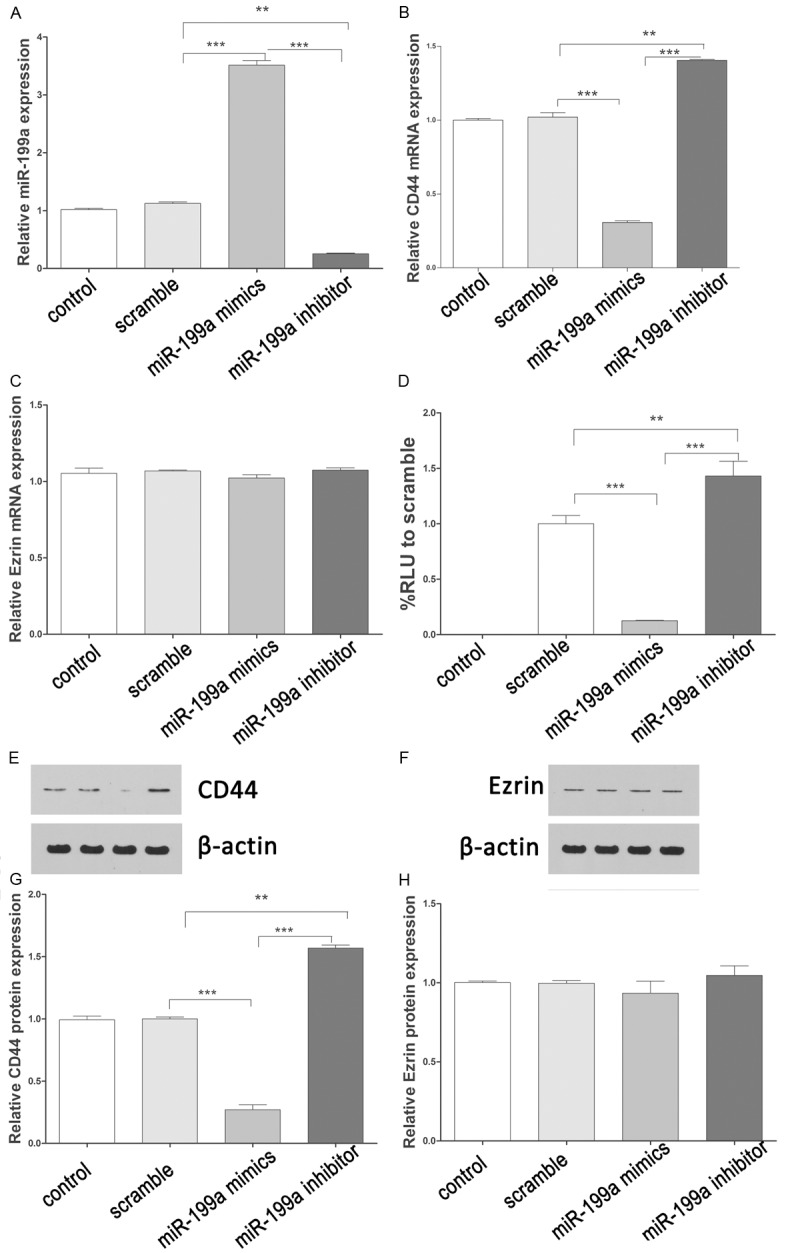
miR-199a directly targets CD44. A. The expression of miR-199a after transfecting with miR-199a mimics or inhibitors. B. The expression of CD44 mRNA after transfecting with miR-199a mimics or inhibitors. C. The expression of Ezrin mRNA after transfecting with miR-199a mimics or inhibitors. D. The luciferase activity was repressed by miR-199a mimics, and induced by miR-199a inhibitors. E, G. The CD44 protein levels were detected by western blot. Quantification of CD44 protein after transfection. F, H. The Ezrin protein levels were detected by western blot. Quantification of Ezrin protein after transfection. Data present as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 VS scramble.
MiR-199a regulated the interaction of CD44 with Ezrin
According above results, we found that the CD44 was significantly higher after miR-199a mimics treatment, which was abolished by miR-199a inhibitor treatment, whereas the expression of Ezrin showed no significant change with treatment of miR-199a mimics or inhibitor. Thus, we investigated the interaction between CD44 and Ezrin using immunoprecipitation and immunofluorescence. As shown in Figure 3C, interaction of CD44 and Ezrin was decreased in the group receiving miR-199a mimics treatment as compared to the scramble and control group, whereas miR-199a inhibitor treatment induced the interaction of CD44 and Ezrin. Immunoprecipitation analysis result was confirmed by the Immunofluorescence result. The results showed that the CD44 co-immunoprecipitated with Ezrin and the binding affinity was decreased significantly by miR-199a mimic treatment, which was increased by miR-199a inhibitor transfection (Figure 3A, 3B).
Figure 3.
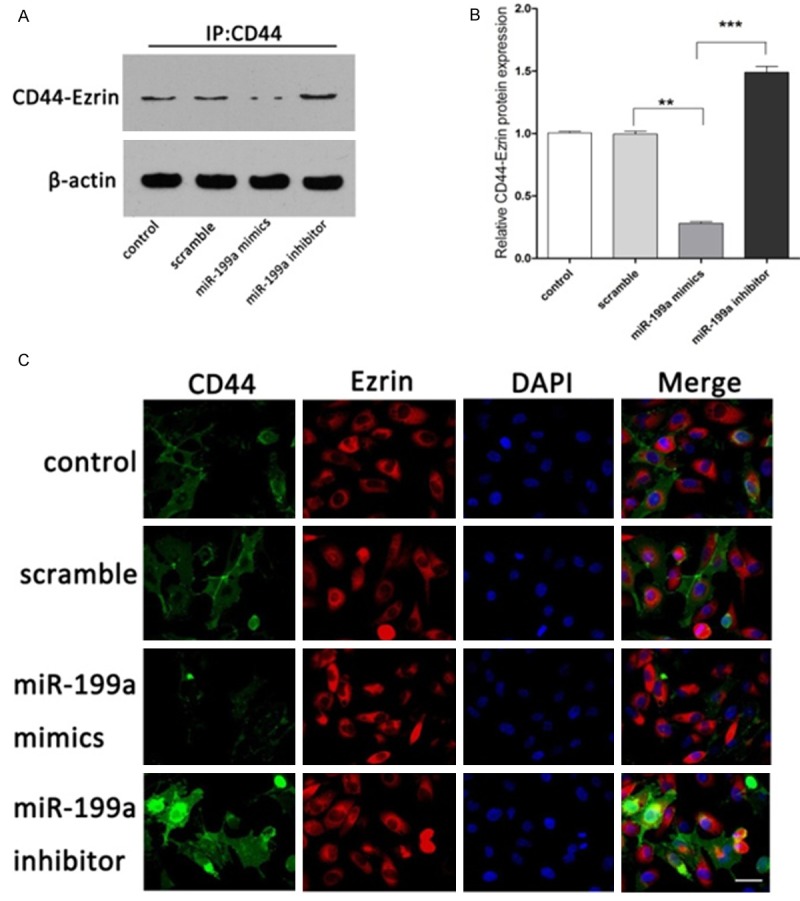
MiR-199a regulates the interaction of CD44 and Ezrin. A. Interaction of CD44 with Ezrin by IP assays. B. Quantification of CD44-Ezrin protein. C. Interaction of CD44 with Ezrin by immunofluorescence. Magnification, 400 ×. Data present as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 VS scramble.
Overexpression of miR-199a repressed proliferation and migration in A-431 cells
To further explore the function of miR-199a in SCC cells, we evaluated the cell proliferation and migration by CCK-8 and transwell after miR-199a mimics or inhibitor transfection. As the data shown, we observed that overexpression of miR-199a significantly decreased cell proliferation, and inhibition of miR-199a significantly increased cell growth (Figure 4A). Inhibition of cell migration ability in A-431 cells was observed in the cells treated with miR-199a mimics, whereas miR-199a inhibitor increased the cell migration ability compared to scramble control cells (Figure 4C, 4D). Besides, we observed that the activity of MMP2 and MMP9 was induced significantly by downregulation of miR-199a, and reduced by upregulation of miR-199a compared to scramble control (Figure 4B). It suggested that up-regulation of miR-199a inhibited the proliferation and invasion of SCC cells.
Figure 4.
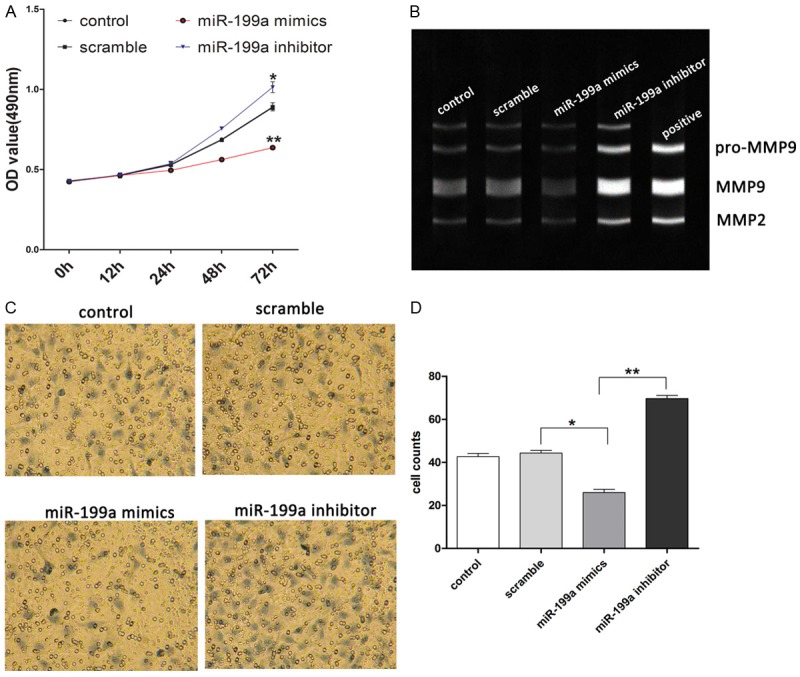
Effects of miR-199a on cell proliferation and migration. (A) Cell proliferation was measured by MTT after transfection. (B) Activation of MMP2/9 was evaluated by gelatin zymography. (C) Cell migration was measured by transwell after transfection. (D) Cell counts of (C). Magnification, 400 ×. Data present as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 VS scramble.
Discussion
Overall, the morbility of cSCC have been increasing in the world over the past 10 years. 5-year overall survival (OS) for patients with metastatic cSCC was less approximately 50% than patients with regional or localized disease respectively [14]. A better understanding of the gene or molecular alterations involved in cSCC progression, particularly from localized tumors to metastasis, will be helpful in early detection and future targeted treatment strategies. Recently, multiple studies have revealed that microRNA expression have been shown to be important regulators of cSCC progression [15-16]. In this study, we found that miR-199a was significantly downregulated in cSCC tissues compared with the matched normal tissues. Moreover, upregulated CD44 expression was observed in cSCC tissues. There was an inverse relationship between miR-199a and CD44. Using luciferase reporter assay, we found that miR-199a regulated the expression of CD44 by targeting its 3’URT. Interesting, the expression of Ezrin had no significant alterations in cSCC tissues, and could not be regulated by miR-199a, but there was an interaction between CD44 and Ezrin, regulating by miR-199a. It was confirmed by immunoprecipitation and immunofluorescence.
According the previous study, the downregulated expression of miRNA-199a was observed in liver, breast, and bladder cancer [10]. It was reported that induced the expression of miRNA-199a was capability of decreasing the proliferation of gastric cancer cells by targeting the mTOR signaling pathway [17]. The abundant expression of miR-199a was observed in liver tissue and in epithelial as well as non epithelial tissues [18]. In HCC cell lines, a recent study showed that miR-199a targeted the hyaluronic acid (HA) receptor CD44, and overexpression of the miR-199a reduced the proliferation of HCC cells [19]. Similarly, another recent investigation found that miR-199a specifically regulated CD44, and that upregulation of miR-199a repressed the proliferation of ovarian cancer-initiating cells in vitro and in vivo [20]. Consistence with the previous data, we found that miR-199a specifically regulated the expression of CD44 and the interaction between CD44 and Ezrin in cSCC cells, indicating that increased expression of endogenous mature miR-199a might prevent the growth of human cSCC via increasing the expression of CD44, which may provide a potentially important therapeutic target for human cSCC.
CD44, a transmembrane glycoprotein widely expressed in cell surface, was highly conserved in mammalian species. CD44 was involved in cell-matrix and cell-cell interactions through interactions with its major ligand and other extracellular factors, such as Ezrin [21]. Moreover, CD44 was very important on cellular adhesion, migration, tumorigenesis, and metastasis [22]. CD44, as a single marker or combined with other markers, has been widely used to investigate many cancers including skin cancer [23]. In animal model, it has demonstrated that blocking CD44 with antibodies or antisense oligonucleotides reduced the malignant activities of the tumor [24]. Ezrin belongs to the ezrin-moesin-radixin (ERM) family. The ERM proteins formed a bridge between CD44 and the actin cytoskeleton, mediating cell morphology changes that were important for cell migration. Ezrin interacted with CD44 through its conserved N-terminal band four-point-one, ezrin, radixin, moesin domain and C-terminal ERM Association Domain domain [25]. Ezrin was involved in diverse cellular processes including cell adhesion, polarization and morphogenesis [26]. And more importantly, it has been identified as a potent regulator of tumor cell invasion and metastasis [27]. A positive correlation between Ezrin expression and cervical lymph node metastasis and clinical staging has been found in various tumors [28].
Henry and his colleagues have showed that there was an inverse correlation between the expression of miR-199a and that of CD44 protein, not only in hepatocellular carcinoma specimens, but also in hepatocellular carcinoma cell lines. Moreover, miR-199a was capability of killing the CD44+ hepatocellular carcinoma cells specially by regulating the expression of CD44 [29]. Recently, another study also demonstrated that miR-199a targets the 3’-UTR of CD44, and confirmed that induced the expression of miR-199a by mimic transfection leads to reduced CD44 expression at mRNA and protein levels in cancer-initiating cells. Besides, MTT assays and the transwell migration invasion assay showed that overexpression of miR-199a inhibited the proliferation, migration and invasion of ovarian cancer-initiating cells, which leads to reduced expression of CD44 [30].
In accordance with the previous study, in our study we found that miR-199a targeted CD44 to repress the proliferation, migration and invasion of cSCC cells confirmed by CKK-8 and transwell migration invasion assay. It has demonstrated that knocking down Ezrin expression suppresses cell growth, invasion, tumor progression and metastasis by RNA interference in cancer cell lines [31]. In a 726 breast cancer cases study, it was demonstrated that the Ezrin protein was expressed at a higher level in CD44 (+) breast cancer cells compared with that of in the control cells. Ezrin and CD44 co-expression was observed in 235 (32.37%) of the 726 cases examined, associating with a poorer prognosis may be as an independent prognostic factor of breast cancer [32]. Moreover, a recent study showed that overexpression of ezrin in the CD44 (+) subpopulation of SCC cells contributed to the metastatic behavior of malignant oral squamous cell carcinoma [33]. MiR-199a could not directly regulate the expression of Ezrin at mRNA and protein levels; however, we found that miR-199a regulated the interaction CD44 with Ezrin evaluated by immunoprecipitation and immunofluorescence. It seems that miR-199a regulated the functions of Ezrin through regulating the expression of CD44.
MMP (metalloproteinase) is believed to play an important role in metastasis of tumor cells by decomposing extracellular matrix and destroying the basement membrane of blood vessels. The dissolution of basement membrane caused by MMP in tumors was an important process in the movement of tumor cells. It was reported that the activation of MMP was in a significant correlation with the migration and invasion of tumor cells [34,35]. It was found that the expression of MMP2 and MMP9 were higher in patients with lymph node metastases comparing patients without lymph node involvement in oropharyngeal squamous cell carcinoma [36]. By Gelatin zymography, we found that transfection of miR-199a mimic reduced the activity of MMP2 and MMP9. Inversely, the activity of MMP2 and MMP9 were activated by transfection of miR-199a inhibitor. Thus, the suppressive role of miR-199a in cell migration in cSCC cells might be associated with the activity of MMP2 and MMP9.
In conclusion, we found that miR-199a specifically regulated the expression of CD44 at mRNA and protein levels, and the interaction between CD44 and Ezrin in cSCC cells. Moreover, the suppressive role of miR-199a in cell migration in cSCC cells was also associated with the activity of MMP2 and MMP9. Taken together, our data indicated that increased expression of endogenous mature miR-199a might prevent the growth and migration of human cSCC via decreasing the expression of CD44 and regulating the interaction between CD44 and Ezrin, which may provide a potentially important therapeutic target for human cSCC.
Disclosure of conflict of interest
None.
References
- 1.Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122:464–472. doi: 10.1172/JCI57415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassarino DS, Derienzo DP, Barr RJ. Cutaneous squamous cell carcinoma: a comprehensive clinicopathologic classification. Part one. J Cutan Pathol. 2006;33:191–206. doi: 10.1111/j.0303-6987.2006.00516_1.x. [DOI] [PubMed] [Google Scholar]
- 3.Muralidhar B, Winder D, Murray M, Palmer R, Barbosa-Morais N, Saini H, Roberts I, Pett M, Coleman N. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol. 2011;224:496–507. doi: 10.1002/path.2898. [DOI] [PubMed] [Google Scholar]
- 4.Lajer CB, Nielsen FC, Friis-Hansen L, Norrild B, Borup R, Garnaes E, Rossing M, Specht L, Therkildsen MH, Nauntofte B, Dabelsteen S, von Buchwald C. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: a prospective translational study. Br J Cancer. 2011;104:830–840. doi: 10.1038/bjc.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dziunycz P, Iotzova-Weiss G, Eloranta JJ, Lauchli S, Hafner J, French LE, Hofbauer GF. Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J Invest Dermatol. 2010;130:2686–2689. doi: 10.1038/jid.2010.169. [DOI] [PubMed] [Google Scholar]
- 6.Yamane K, Jinnin M, Etoh T, Kobayashi Y, Shimozono N, Fukushima S, Masuguchi S, Maruo K, Inoue Y, Ishihara T, Aoi J, Oike Y, Ihn H. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J Mol Med (Berl) 2013;91:69–81. doi: 10.1007/s00109-012-0935-7. [DOI] [PubMed] [Google Scholar]
- 7.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, Bae DS. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 10.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 12.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, Eggermont AM, Flaherty KT, Gimotty PA, Kirkwood JM, McMasters KM, Mihm MJ, Morton DL, Ross MI, Sober AJ, Sondak VK. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanemura A, Terando AM, Sim MS, van Hoesel AQ, de Maat MF, Morton DL, Hoon DS. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res. 2009;15:1801–1807. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 17.Peng W, Chen ZY, Wang L, Wang Z, Li J. MicroRNA-199a-3p is downregulated in gastric carcinomas and modulates cell proliferation. Genet Mol Res. 2013;12:3038–3047. doi: 10.4238/2013.August.20.5. [DOI] [PubMed] [Google Scholar]
- 18.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry JC, Park JK, Jiang J, Kim JH, Nagorney DM, Roberts LR, Banerjee S, Schmittgen TD. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2010;403:120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279:2047–2059. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 21.Blacking TM, Waterfall M, Argyle DJ. CD44 is associated with proliferation, rather than a specific cancer stem cell population, in cultured canine cancer cells. Vet Immunol Immunopathol. 2011;141:46–57. doi: 10.1016/j.vetimm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Hou NY, Yang K, Chen T, Chen XZ, Zhang B, Mo XM, Hu JK. CD133+ CD44+ subgroups may be human small intestinal stem cells. Mol Biol Rep. 2011;38:997–1004. doi: 10.1007/s11033-010-0195-y. [DOI] [PubMed] [Google Scholar]
- 23.Hartmann-Petersen S, Tammi RH, Tammi MI, Kosma VM. Depletion of cell surface CD44 in nonmelanoma skin tumours is associated with increased expression of matrix metalloproteinase 7. Br J Dermatol. 2009;160:1251–1257. doi: 10.1111/j.1365-2133.2009.09031.x. [DOI] [PubMed] [Google Scholar]
- 24.Zheng S, Franzmann EJ. Comments on ‘CD44-negative cells in head and neck squamous carcinoma also have stem-cell like traits’, Se-Yeong Oh et al,European Journal of Cancer, published online 6 July 2012. Eur J Cancer. 2013;49:3380–3381. doi: 10.1016/j.ejca.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 25.Donatello S, Babina IS, Hazelwood LD, Hill AD, Nabi IR, Hopkins AM. Lipid raft association restricts CD44-ezrin interaction and promotion of breast cancer cell migration. Am J Pathol. 2012;181:2172–2187. doi: 10.1016/j.ajpath.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, Crothers JJ, Zhou R, Forte JG. A possible mechanism for ezrin to establish epithelial cell polarity. Am J Physiol Cell Physiol. 2010;299:C431–C443. doi: 10.1152/ajpcell.00090.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legg JW, Lewis CA, Parsons M, Ng T, Isacke CM. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat Cell Biol. 2002;4:399–407. doi: 10.1038/ncb797. [DOI] [PubMed] [Google Scholar]
- 28.Meng Y, Lu Z, Yu S, Zhang Q, Ma Y, Chen J. Ezrin promotes invasion and metastasis of pancreatic cancer cells. J Transl Med. 2010;8:61. doi: 10.1186/1479-5876-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry JC, Park JK, Jiang J, Kim JH, Nagorney DM, Roberts LR, Banerjee S, Schmittgen TD. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2010;403:120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alaniz L, Cabrera PV, Blanco G, Ernst G, Rimoldi G, Alvarez E, Hajos SE. Interaction of CD44 with different forms of hyaluronic acid. Its role in adhesion and migration of tumor cells. Cell Commun Adhes. 2002;9:117–130. doi: 10.1080/15419060214522. [DOI] [PubMed] [Google Scholar]
- 31.Xie JJ, Xu LY, Xie YM, Zhang HH, Cai WJ, Zhou F, Shen ZY, Li EM. Roles of ezrin in the growth and invasiveness of esophageal squamous carcinoma cells. Int J Cancer. 2009;124:2549–2558. doi: 10.1002/ijc.24216. [DOI] [PubMed] [Google Scholar]
- 32.Ma L, Jiang T. Clinical implications of Ezrin and CD44 coexpression in breast cancer. Oncol Rep. 2013;30:1899–1905. doi: 10.3892/or.2013.2641. [DOI] [PubMed] [Google Scholar]
- 33.Wu G, Sun X, Yuan H, Hu M. Ezrin gene expression and protein production in the CD44(+) subpopulation of SCC-9 cells in a malignant oral cancer cell line in vitro. J Oral Maxillofac Surg. 2013;71:e151–e157. doi: 10.1016/j.joms.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Ryzhakova OS, Solov’Eva NI. [Matrix metalloproteinases (MMP)--MMP-1,-2,-9 and its endogenous activity regulators in transformed by E7 oncogene HPV16 and HPV18 cervical carcinoma cell lines] . Biomed Khim. 2013;59:530–540. doi: 10.18097/pbmc20135905530. [DOI] [PubMed] [Google Scholar]
- 35.Mohtasham N, Babakoohi S, Shiva A, Shadman A, Kamyab-Hesari K, Shakeri MT, Sharifi-Sistani N. Immunohistochemical study of p53, Ki-67, MMP-2 and MMP-9 expression at invasive front of squamous cell and verrucous carcinoma in oral cavity. Pathol Res Pract. 2013;209:110–114. doi: 10.1016/j.prp.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Burduk PK, Bodnar M, Sawicki P, Szylberg L, Wisniewska E, Kazmierczak W, Martynska M, Marszalek A. Expression of metalloproteinases 2 and 9 and tissue inhibitors 1 and 2 as predictors of lymph node metastases in oropharyngeal squamous cell carcinoma. Head Neck. 2014 doi: 10.1002/hed.23618. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]


