Abstract
Background: Left ventricular dysfunction (LVD) occurs with myocardial ischemia and coronary artery disease (CAD). The natriuretic peptide system has compensatory vasodilatory, natriuretic and paracrine effects on LVD and subsequent heart failure. The aim of this study was to investigate the relationship between natriuretic peptide polymorphisms and risk of LVD in CAD patients. Methods: We recruited 747 consecutive Southern Han Chinese patients with angiographically confirmed CAD, 201 had a reduced left ventricle ejection fraction (LVEF ≤45%, LVD group) and 546 had a preserved left ventricle ejection fraction (LVEF >45%). The reduced and preserved LVEF groups were matched by gender and age. Taqman assays were performed to identify five polymorphisms in the NPPA-NPPB locus (rs5065, rs5063, rs632793, rs198388 and rs198389). Results: Single-locus analyses found no significant difference in the allele and genotype frequencies of the reduced and preserved LVEF group, even after adjusting for confounding factors. Subgroup analyses performed by hyperlipidemia (HLP) demonstrated 3 polymorphisms, rs632793 (OR = 0.31, 95% CI 0.1-0.93, P = 0.04), rs198388 (OR = 0.26, 95% CI 0.09-0.79, P = 0.02) and rs198389 (OR = 0.26, 95% CI 0.09-0.80, P = 0.02) were associated with the reduced risk of LVD. No CAD-susceptible haplotypes were identified. Multifactor dimensionality reduction analysis did not detect any gene-to-gene interactions among the five loci. Three loci (rs5063, rs632793 and rs198388) formed the best model with the maximum testing accuracy (39.89%) and cross-validation consistency (10/10). Conclusion: Three NPPA-NPPB polymorphisms (rs632793, rs198388 and rs198389) were associated with reduced risk of LVD in CAD patients with HLP.
Keywords: Left ventricular dysfunction, coronary artery disease, natriuretic peptide, polymorphism, heart failure
Introduction
Left ventricular dysfunction (LVD) is an early stage of heart failure (HF) characterized by a reduced ejection fraction and a depressed level of left ventricular wall motility [1]. Coronary artery disease (CAD) and myocardial infarction (MI), the cardiac polygenic disorders [2-4], are contributing factors to the development of LVD. Other diseases, such as cardiomyopathy, hypertension (HTN) and valvular disease, may also lead to LVD. LVD with subsequent HF is characterized by a continuous interaction between the underlying myocardial dysfunction and a series of compensatory mechanisms. A host of hemodynamic and neurohormonal factors, such as the adrenergic and renin-angiotensin-aldosterone systems (RAAS) are triggered to modulate left ventricle remodeling of the vascular tree once LVD occurs. One of the key neuroendocrine axes is the natriuretic peptide system. This system consists of five peptides, atrial peptide (ANP), urodilatin (an isoform of ANP), B-type natriuretic peptide (BNP), C-type natriuretic peptide (CNP), and dendroaspis natriuretic peptide (DNP) [5]. These peptides share a similar molecular structure and biological functions (natriuresis, diuresis and vasodilation) [6]. ANP and BNP are mainly synthesized and secreted by cardiomyocytes in response to increased cardiac wall tension and volume loading [7]. The secretion of natriuretic peptides from cardiac ventricles is increased with LVD. The extent of ANP and BNP release is in line with the degree of cardiac dysfunction and left ventricular failure, making them useful markers in evaluating the severity of these phenomena [8]. Circulating natriuretic peptide levels are helpful to assess the prognosis in patients with CAD and MI, with or without LVD [9,10]. The persistence of elevated levels of natriuretic peptides for several months after MI suggests a continued risk of pathologic remodeling and the development of LVD and HF [11].
The NPPA ( natriuretic peptide precursor A) and NPPB ( natriuretic peptide precursor B) genes lie in tandem 9.7kb apart on chromosome 1. Mice with homozygous mutations of the NPPA gene have right and left ventricular hypertrophy. This hypertrophy is increased disproportionately (relative to controls) in response to transverse aortic constriction, suggesting that ANP negatively regulates matrix remodeling in the myocardium [12]. In contrast, NPPB gene knockout mice exhibit multifocal fibrotic lesions in the cardiac ventricles. These increase in size and number in response to ventricular overload, indicating that BNP acts as an anti-fibrotic factor [13]. Several polymorphisms within the NPPA-NPPB locus have been reported to be correlated with inter-individual variation in circulating natriuretic peptide concentrations [14,15], contributing to ambulatory cardiovascular disease states such as stroke [16], HF [17], CAD [18] and HTN [15,19]. The nonsynonymous coding polymorphism rs5065 is significantly associated with increased circulating BNP and amino-terminal BNP (NT-proBNP) levels in severe HF patients [20]. The rs5065 and rs5063 polymorphisms correlate with increased left ventricular mass index and left ventricular septal thickness in HTN patients [21]. The minor NPPA-NPPB allele, rs632793, rs198388 and rs198389 are associated with increased circulating ANP and BNP concentrations and a reduced rate of cardiovascular readmission in CAD patients [15,22]. Surprisingly, these polymorphisms have not been evaluated as markers for LVD. Therefore, we investigated the relationship between common NPPA-NPPB polymorphisms and the presence of LVD in CAD patients.
Materials and methods
Ethics statement
The ethics committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine approved this study. All the authors followed the guidelines of the World’s Association Declaration of Helsinki. Written informed consent for the study was obtained from each patient.
Study cohort
A total of 747 consecutive patients undergoing coronary angiography for the diagnosis and intervention of CAD were admitted to Ruijin Hospital from January 2008 to December 2012. Coronary angiography was performed via the femoral or radial artery approach, according to the clinical standards of the American College of Cardiology/American Heart Association guidelines for coronary angiography [23]. Standard Judkins techniques were used for angiography [24]. Physicians performing the studies were blinded to the study protocol. CAD was defined as ≥50% luminal obstruction at least one or more major coronary epicardial coronary vessel. The patients were grouped according to the number of significantly stenotic vessels, single-, double- or triple-vessel disease. Any patient with cardiomyopathy, congenital heart disease, pulmonary heart disease, valvular heart disease, stroke, infection, tumor or immune system disorders was excluded. Left ventricular (LV) function was calculated by the echocardiography staff just before angiography. Imaging (e.g., M-mode, 2D) and color-flow Doppler echocardiography were performed. LV ejection fraction (EF), volumes and internal dimensions were measured according to the American Society of Echocardiography recommendations [25]. LVD was defined as (modified Simpson’s rule) LVEF≤ 45% [26]. All patients were interviewed to ascertain their sociodemographic, economic and health status characteristics as well as lifestyle (e.g., habits, tobacco usage). Ultrasound staff was blinded to polymorphism analysis results. Subjects were residents of Shanghai, a population that is primarily of Southern Han Chinese ethnicity.
Data collection
Complete clinical history and information on conventional cardiovascular risk factors were obtained by reviewing the patients’ medical records. HTN was defined as a systolic blood pressure >140 mmHg, a diastolic blood pressure >90 mmHg or the use of anti-hypertensive drugs. Smoking habit was classified as smokers (ex-smoker and current smokers) or non-smokers. Diabetes mellitus (DM) was defined as a fasting serum glucose level above 7.0 mmol/L, a two-hour postprandial glucose greater than 11.1 mmol/L, or the use of diabetic medications. The diagnosis and management of Hyperlipidemia (HLP) was performed according to the National Cholesterol Education Program Adult Treatment Panel III Guidelines (NCEP ATIII) [27]. Stroke was defined and classified based according to Trial of Org 10172 in Acute Stroke Treatment (TOAST) criteria [28].
DNA isolation and genotyping
Blood was drawn using EDTA-collection tubes. Genomic DNA was extracted from peripheral blood leukocytes according to standard phenol-chloroform methods, and stored at -20°C until batch genotyping. Taqman assay was used to genotype SNPs (Single nucleotide polymorphisms). SNP Taqman probes and primers were designed using the Applied Biosystems Assay-by-Design Service for SNP genotyping. The sample DNA was amplified by PCR following the recommendations of the manufacturer. Thermal cycling was done on a Gene Amp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA). Allele detection and genotype determination were performed using the fluorescence mode of an ABI PRISM 7900HT Sequence Detector (Applied Biosystems, Foster City, CA, USA). Genotyping was performed without knowledge of the case-control status of the subjects. Ten percent of samples from patients and controls were sequenced to estimate the quality of genotyping. No discrepancy was found. Five SNPs located in the NPPA-NPPB locus (rs5065, rs5063, rs632793, rs198388 and rs198389) were investigated.
Statistical analysis
Continuous variables were reported as the mean ± standard deviation (SD) or median with 5th and 95th percentiles. Categorical measures were reported as percentages. The chi-square test or Fisher’s exact test were used to examine the goodness of fit between the observed allelic frequencies and the allelic frequencies expected by Hardy Weinberg Equilibrium (HWE). HWE was determined using the web program- http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl. Differences in the allelic and genotypic frequencies of cases and controls were evaluated using the chi-square test. An unpaired t-test was used to test group differences of continuous variables. Genotype frequency of the subjects specified by different genetic models (allelic, additive, dominant, recessive and homozygote comparison) was analyzed by multivariate logistic regression adjusted for confounding factors. A two-tailed P<0.05 was regarded as statistically significant. Data management and statistical analyses were performed using SPSS software version 20.0 (SPSS, Chicago, IL, USA).
Linkage disequilibrium (LD) and haplotype blocks within 5 SNPs of the NPPA-NPPB locus were identified using the online computer platform SHEsis- http://analysis.bio-x.cn/myAnalysis.php [29]. LD coefficients were calculated using the formula D’=D/Dmax or D/Dmin. Haplotypes with a frequency higher than 1% were examined and significance was estimated by reference to the most frequent haplotype. Haplotype analysis was performed using the Haplo.stats program developed by R language version 3.0.2 (http://www.r-project.org). The main functions in Haplo.stats were implemented: Haplo.em was used to calculate maximum likelihood estimates of haplotype probability using a “progressive insertion” algorithm that progressively inserted batches of loci into haplotypes of growing lengths. Haplo.cc and Haplo.glm were used to calculate crude and adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for each haplotype, respectively. These two approaches computed the regression of a trait on haplotypes and other covariates based on a generalized linear model [30,31]. Haplo.score was used to calculate score statistics and test the difference in haplotype frequencies between cases and controls on the basis of simulated P (Psim) values that were obtained from 1000 replicates [32].
Gene-to-gene interactions in the occurrence of LVD were evaluated using the open-source multifactor dimensionality reduction (MDR) software version 3.0 (www.epistasis.org) [33,34]. MDR collapsed high-dimensional multilocus-genotype variables into a single dimensional multilocus-genotype variable by sorting the genotypes into two levels, high- or low- risk. All possible combinations of these 5 polymorphisms were tested. A probabilistic naïve Bayes classifier with 10-fold cross-validation was used to estimate the prediction accuracy and the empirical P-value. The data are divided into 10 divisions equally. 9/10 of the data is regarded as training set and then the remaining 1/10 is tested. A single best model with maximal testing accuracy and cross-validation consistency was determined by measuring the number of times of 10 divisions of the data. The P-value <0.05 was considered statistically significant using a 1000-fold permutation test.
Results
Patient characteristics
Clinical characteristics of CAD patients are shown in Table 1. Of the total 747 CAD patients, 201 had reduced (≤45%) LVEF and 546 had preserved LVEF (>45%). The reduced LVEF group was matched with the preserved LVEF group for age (P = 0.97) and gender (P = 0.62). There was no significant difference in the distribution of other confounding factors including DM (P = 0.91), smoking habit (P = 0.15) and stroke history (P = 0.49). The reduced LVEF group had a significantly lower proportion of HTN (P = 0.01) and HLP (P = 0.02) than the preserved LVEF group. The angiographic profile of the reduced LVEF group was significantly different from that of the preserved LVEF group (P = 0.01) (Table 1).
Table 1.
The baseline characteristics of study population
| Characteristics | Reduced LVEF (n = 201) | Preserved LVEF (n = 546) | P valueb |
|---|---|---|---|
| Age, yeara | 64.26±11.76 | 64.29±8.98 | 0.97 |
| Male sex, n (%) | 157 (78.1) | 417 (76.4) | 0.62 |
| Risk factors | |||
| Hypertension, n(%) | 135 (67.2) | 418 (76.6) | 0.01 |
| Diabetes, n (%) | 58 (28.9) | 160 (29.3) | 0.91 |
| Hyperlipidaemia, n (%) | 37 (18.4) | 145 (26.6) | 0.02 |
| Stroke history, n (%) | 21 (10.4) | 67 (12.3) | 0.49 |
| Smoking habit (%) | 89 (44.3) | 209 (38.4) | 0.15 |
| Angiographic profile | |||
| Single vessel disease (SVD), n (%) | 46 (22.9) | 184 (33.7) | 0.01 |
| Double vessel disease (DVD), n (%) | 68 (33.8) | 186 (34.1) | |
| Triple vessel disease (TVD), n (%) | 87 (43.3) | 176 (32.2) |
Data are expressed as mean ± SD.
The unpaired t-test is used for age and the x2 test is used for other categorical characteristics.
Single-locus analysis
Ninety-nine percent of the samples were successfully identified. The genotype and allele frequencies of these 5 polymorphisms, as well as their risk prediction under different genetic models, are listed in Table 2. No deviation from HWE was observed in both the reduced LVEF group and the preserved LVEF group (P>0.05). There was no difference in the allele or genotype frequencies of the two groups. The risk estimates for LVD with these 5 polymorphisms did not change after adjusting for age, gender, HTN, DM, HLP, stroke history, smoking habit and angiographic profile.
Table 2.
Genotype distributions and allele frequencies of the 5 examined polymorphisms between the reduced and preserved LVEF groups in CAD patients and the risk prediction under various genetic models
| SNP ID (rs number) | Genotype and allele | Cases | Control | P Χ2 a | Genetic models | Unadjusted | Adjustedb | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| OR | 95% CI | P value | OR | 95% CI | P value | ||||||
| rs5065 | GG | 0 | 0 | 0.75 | Allelic comparison | 0.81 | 0.22-2.97 | 0.75 | 0.84 | 0.23-3.11 | 0.80 |
| AG | 3 | 10 | Dominant model | 0.81 | 0.22-2.98 | 0.75 | 0.84 | 0.23-3.12 | 0.80 | ||
| AA | 198 | 535 | Recessive model | –c | – | – | – | – | – | ||
| G (%) | 0.7 | 0.9 | 0.75 | Homozygote comparison | – | – | – | – | – | – | |
| A (%) | 99.3 | 99.1 | Additive model | 0.81 | 0.22-2.98 | 0.75 | 0.84 | 0.23-3.12 | 0.80 | ||
| rs5063 | TT | 4 | 7 | 0.78 | Allelic comparison | 1.09 | 0.74-1.61 | 0.65 | 1.06 | 0.72-1.58 | 0.76 |
| CT | 32 | 86 | Dominant model | 1.06 | 0.69-1.62 | 0.79 | 1.04 | 0.67-1.60 | 0.87 | ||
| CC | 165 | 452 | Recessive model | 1.56 | 0.45-5.39 | 0.48 | 1.43 | 0.40-5.13 | 0.58 | ||
| T (%) | 10.0 | 9.2 | 0.65 | Homozygote comparison | 1.57 | 0.45-5.42 | 0.48 | 1.46 | 0.41-5.20 | 0.56 | |
| C (%) | 90.0 | 90.8 | Additive model | 1.09 | 0.75-1.58 | 0.66 | 1.06 | 0.72-1.55 | 0.77 | ||
| rs632793 | GG | 6 | 9 | 0.50 | Allelic comparison | 1.07 | 0.76-1.50 | 0.69 | 1.06 | 0.75-1.50 | 0.75 |
| AG | 42 | 120 | Dominant model | 1.01 | 0.69-1.48 | 0.95 | 1.02 | 0.69-1.50 | 0.93 | ||
| AA | 153 | 416 | Recessive model | 1.83 | 0.64-5.22 | 0.26 | 1.56 | 0.53-4.56 | 0.42 | ||
| G (%) | 13.4 | 12.7 | 0.69 | Homozygote comparison | 1.81 | 0.64-5.18 | 0.27 | 1.54 | 0.52-4.52 | 0.43 | |
| A (%) | 86.6 | 87.3 | Additive model | 1.07 | 0.77-1.49 | 0.70 | 1.06 | 0.75-1.48 | 0.75 | ||
| rs198388 | TT | 5 | 14 | 0.50 | Allelic comparison | 0.84 | 0.61-1.17 | 0.30 | 0.81 | 0.58-1.13 | 0.22 |
| CT | 45 | 145 | Dominant model | 0.8 | 0.56-1.16 | 0.25 | 0.78 | 0.53-1.14 | 0.20 | ||
| CC | 151 | 386 | Recessive model | 0.97 | 0.34-2.72 | 0.95 | 0.85 | 0.30-2.44 | 0.76 | ||
| T (%) | 13.7 | 15.9 | 0.30 | Homozygote comparison | 0.91 | 0.32-2.58 | 0.86 | 0.82 | 0.28-2.35 | 0.71 | |
| C (%) | 86.3 | 84.1 | Additive model | 0.84 | 0.61-1.17 | 0.30 | 0.81 | 0.59-1.13 | 0.22 | ||
| rs198389 | GG | 6 | 10 | 0.43 | Allelic comparison | 0.96 | 0.68-1.33 | 0.79 | 0.95 | 0.68-1.34 | 0.77 |
| AG | 42 | 132 | Dominant model | 0.89 | 0.61-1.29 | 0.54 | 0.89 | 0.61-1.31 | 0.56 | ||
| AA | 153 | 402 | Recessive model | 1.64 | 0.59-4.58 | 0.34 | 1.49 | 0.52-4.27 | 0.45 | ||
| G (%) | 13.4 | 14.0 | 0.79 | Homozygote comparison | 1.58 | 0.56-4.41 | 0.39 | 1.44 | 0.51-4.13 | 0.49 | |
| A (%) | 86.6 | 86.0 | Additive model | 0.96 | 0.69-1.33 | 0.79 | 0.95 | 0.68-1.33 | 0.77 | ||
P values were calculated by x2 test for differences in genotypes and alleles between the two groups.
ORs adjusted for age, gender, HTN, DM, HLP, Stroke history, smoking habit and angiographic profile.
data not available.
Stratification analyses were performed to investigate the interactive effect of NPPA-NPPB polymorphisms and confounding factors on LVD. When data were classified by HLP (Table 3) and adjusted for other confounding factors, three polymorphisms of the NPPA-NPPB locus were significantly associated with LVD (rs632793: P = 0.04, OR=0.31, 95% CI 0.10-0.93; rs198388: P = 0.02, OR = 0.26, 95% CI 0.09-0.79; rs198389: P = 0.02, OR=0.26, 95% CI 0.09-0.80). No such difference was found in the non-HLP population.
Table 3.
Stratified analyses of the 5 NPPA-NPPB polymorphisms with the risk of LVD in CAD patients with HLP under the additive genetic model
| Study ID (rs number) | Group | Unadjusted | Adjusteda | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| OR | 95% CI | P value | OR | 95% CI | P value | ||
| rs5065 | HLP | –b | – | – | – | – | – |
| Non-HLP | 0.91 | 0.24-3.49 | 0.89 | 0.97 | 0.25-3.77 | 0.96 | |
| rs5063 | HLP | 0.69 | 0.28-1.71 | 0.42 | 0.69 | 0.26-1.81 | 0.45 |
| Non-HLP | 1.24 | 0.82-1.88 | 0.31 | 1.19 | 0.78-1.82 | 0.41 | |
| rs632793 | HLP | 0.35 | 0.12-1.00 | 0.05 | 0.31 | 0.10-0.93 | 0.04 |
| Non-HLP | 1.31 | 0.91-1.89 | 0.15 | 1.30 | 0.90-1.89 | 0.16 | |
| rs198388 | HLP | 0.30 | 0.10-0.86 | 0.03 | 0.26 | 0.09-0.79 | 0.02 |
| Non-HLP | 0.99 | 0.69-1.40 | 0.93 | 0.97 | 0.68-1.39 | 0.86 | |
| rs198389 | HLP | 0.31 | 0.11-0.88 | 0.03 | 0.26 | 0.09-0.80 | 0.02 |
| Non-HLP | 1.17 | 0.82-1.68 | 0.39 | 1.17 | 0.81-1.69 | 0.40 | |
ORs adjusted for age, gender, HTN, DM, Stroke history, smoking habit and angiographic profile.
data not available.
Haplotype analysis
Considering these 5 polymorphisms were located on the same chromosome, we performed a linkage analysis (Figure 1). Strong linkage patterns were observed between rs632793, rs198388 and rs198389 (D’≥0.91), suggesting that these 3 loci in the NPPA-NPPB locus were in the same block (block 1, Figure 1). Haplotype analyses were performed to investigate combinational effects of these 5 polymorphisms on LVD risk (Table 4). Four haplotypes (alleles in order of rs5065, rs5063, rs632793, rs198388 and rs198389) had frequencies ≥1% and were included in the haplotype analyses. The A-C-A-C-A haplotype was the most frequent (79.13%in the reduced LVEF group and 78.86% in the preserved LVEF group). Using the A-C-A-C-A haplotype as a reference, no significant difference was found between the other three allele combinations and the reference haplotype in prediction of LVD. Similarly, no difference was found for all the haplotypes in block1 (alleles in order of rs632793, rs198388 and rs198389), even after adjusting for confounding factors.
Figure 1.
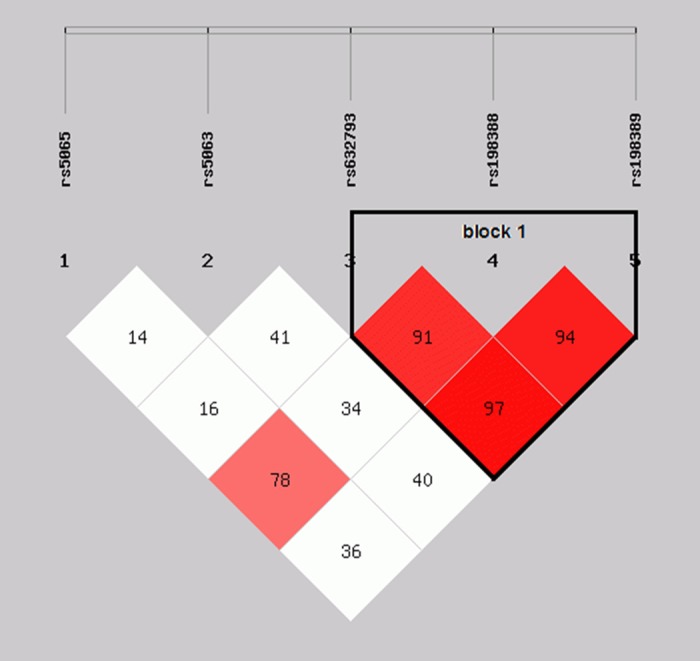
Block structure of linkage disequilibrium (LD) for five genotyped single nucleotide polymorphisms (SNPs) in the NPPA-NPPB locus. Stronger correlations between these SNPs are noted by red color in the intersecting squares linking each pair of SNPs.
Table 4.
Haplotype frequencies of the 5 NPPA-NPPB polymorphisms and their risk prediction of LVD in CAD patients
| Haplotype | Cases (%) | Controls (%) | P sim | Unadjusted | Adjusteda | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| OR | 95% CI | P value | OR | 95% CI | P value | ||||
| Totalb | |||||||||
| A-C-G-T-G | 7.09 | 7.76 | 0.95 | 1.02 | 0.66-1.58 | 0.96 | 1.05 | 0.67-1.63 | 0.84 |
| A-T-A-C-A | 5.44 | 4.44 | 0.47 | 1.19 | 0.70-2.02 | 0.47 | 1.24 | 0.72-2.12 | 0.44 |
| A-T-G-T-G | 4.03 | 3.93 | 0.90 | 1.01 | 0.54-1.87 | 0.90 | 0.92 | 0.49-1.74 | 0.80 |
| A-C-A-C-A | 79.13 | 78.86 | 0.86 | Reference | Reference | ||||
| Block1c | |||||||||
| A-T-A | 1.26 | 2.40 | 0.17 | 0.51 | 0.19-1.34 | 0.16 | 0.41 | 0.15-1.11 | 0.08 |
| G-T-G | 11.93 | 12.03 | 0.99 | 0.97 | 0.68-1.38 | 0.96 | 0.97 | 0.68-1.38 | 0.86 |
| A-C-A | 84.81 | 83.44 | 0.51 | Reference | Reference | ||||
Psim: simulated P value.
ORs adjusted for age, gender, HTN, DM, HLP, Stroke history, smoking habit and angiographic profile.
Alleles in total haplotype were arrayed in order of rs5065, rs5063, rs632793, rs198388 and rs198389.
Alleles in block 1 haplotype were arrayed in order of rs632793, rs198388 and rs198389.
Gene-to-gene Interactions
An exhaustive MDR analysis that estimated possible interactions of different gene polymorphisms with LVD was performed (Table 5). Each best model was accompanied with its testing accuracy, cross-validation consistency and significant level determined by permutation testing. The single-locus model including rs198388 generated a testing accuracy of 39.63% and a cross-validation consistency of 9/10. Three polymorphisms, rs5063, rs632793 and rs198388 constituted the best overall MDR model with the highest testing accuracy of 39.89% and a maximal cross-validation consistency of 10. None of these models was significant in predicting LVD risk in CAD patients (P>0.05).
Table 5.
Summary of the multifactor dimensionality reduction analysis
| Best combination of each model | Testing accuracy | Cross-validation consistency | P valuea |
|---|---|---|---|
| rs198388 | 0.3963 | 9 | 0.8834 |
| rs632793, rs198388 | 0.3788 | 5 | 0.9281 |
| rs5063, rs632793, rs198388 | 0.3989 | 10 | 0.7708 |
P value based on 1000 permutations.
Discussion
We evaluated the relationship of 5 common NPPA-NPPB locus polymorphisms with LVD in a large Chinese population. To the best of our knowledge, this is the first such study performed in CAD patients. The principal finding of the study indicated none of the NPPA-NPPB polymorphisms was associated with LVD in overall analysis. Haplotype analysis confirmed the lack of association of these 5 examined polymorphisms with LVD. A subgroup analysis of HLP patients identified a significant association between 3 polymorphisms (rs632793, rs198388 and rs198389) and a reduced risk of LVD after adjusting for environmental covariates. Our results confirmed a previous study of 1,164 Europeans undergoing primary coronary artery bypass graft (CABG) surgery with cardiopulmonary bypass (CPB) [35]. No significant associations were reported between the 2 NPPA-NPPB polymorphisms (rs5065 and rs5063) and ventricular dysfunction after CABG with CPB. Three polymorphisms (rs632793, rs198388 and rs198389) were protective of the ventricular dysfunction after CABG with CPB. These 3 polymorphisms are located in the NPPA-NPPB promoter region that has a tandem array of possible cis regulatory elements. These are known to be gene regulators and targets for gene up regulation via different signaling pathways [36]. Various physiologic stimuli including mechanical stretch, ischemic injury and hypoxia, as well as inflammatory mediators activate regulation of the NPPA-NPPB promoter, resulting in increased secretion of natriuretic peptide [37]. These SNPs are arranged in tandem on chromosome 1 and may coordinately regulate gene expression, through shared enhancer elements [15]. Further work will be required to determine the mechanisms by which these polymorphisms alter transcript stability or biological activity of the peptides.
A relationship was identified between the NPPA-NPPB polymorphisms in CAD patients with HLP and LVD. Natriuretic peptides are believed to be important in lipid metabolism, probably promoting adipose tissue lipolysis through increased cyclic guanosine monophosphate (cGMP) production [38]. The migration of human mesangial cells, may play a role in the pathogenesis of atherosclerosis. Oxidized LDL and lysophosphatidylcholine stimulate the migration of human mesangial cells. This migration is inhibited by ANP and BNP, possibly via a cGMP-dependent process [39]. BNP broadly inhibits Ang II-stimulated steroidogenesis by a number of mechanisms including the modulation of cholesterol biosynthesis, inhibition of Ang II- induced expression of scavenger receptor class B type I (SR-BI) , LDL receptors (LDLR), uptake of cholesterol from HDL and LDL into adrenocortical cells, inhibition of cholesterol transfer through the mitochondrial inner membrane, and reduction of steroid synthesis in primary human adrenocortical cells [40].
MDR is a novel method of analyzing genotype-genotype and genotype-phenotype associations. The advantage of MDR is that high-order gene-gene interactions can be detected in a relatively small sample population without the influence of dimensionality and genetic models. We did not detect any gene-gene interactions affecting LVD risk in CAD patients examined with MDR. This may be explained by the possibilities that the effect of the NPPA-NPPB promoter on LVD is predominant, and the interactive effects among genes may be overwhelmed by this main effect. Racial genetic diversity of the 5 examined polymorphisms could also explain this finding. There is a relatively lower genotype frequency of these 5 NPPA-NPPB polymorphisms in Chinese than in Caucasians [14,15,22]. The NPPA-NPPB polymorphisms probably had more complex genetic effects on the Chinese population than the Caucasian population. Additional studies of the pleiotropic effect of NPPA-NPPB polymorphisms are warranted.
This study had several limitations. First, the study was case-control in design, which precludes comments on causality. Second, only 5 NPPA-NPPB loci were evaluated. We could not exclude the possibility that other common SNPs or rare variations would affect disease progression. Third, we only evaluated genes related to the natriuretic peptide systems. LVD is a complex process that involves interactions of the adrenergic nervous system, RAAS and natriuretic peptides [41-43]. The interaction of natriuretic peptide genes with other neurohumoral factors, such as angiotensin and adrenergic genes, merits further study.
In conclusion, rs632798, rs198388 and rs198389 polymorphisms of the NPPA-NPPB locus might be additive genetic factors influencing disease progression of LVD in CAD patients with HLP. Rs5065 and rs5063 polymorphisms were not independent determinants of LVD in CAD patients. Our study reinforces the belief that natriuretic peptides may be protective of LVD, especially those with atherogenic dyslipidemia.
Acknowledgements
This work was supported by the Youth Science and Technology Talents “Sail” Program of Shanghai Municipal Science and Technology Commission (14YF1402700), the New Hundred Talents Program of the Shanghai Municipal Health Bureau (XBR2013100) and the National Natural Science Foundation of China (81070177 & 81370397).
Disclosure of conflict of interest
None.
References
- 1.Armstrong PW. Left ventricular dysfunction: causes, natural history, and hopes for reversal. Heart. 2000;84(Suppl 1):i15–17. doi: 10.1136/heart.84.suppl_1.i15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z, Lou Y, Jin W, Liu Y, Lu L, Lu G. The C161T polymorphism in the peroxisome proliferator-activated receptor gamma gene (PPARgamma) is associated with risk of coronary artery disease: a meta-analysis. Mol Biol Rep. 2013;40:3101–3112. doi: 10.1007/s11033-012-2384-3. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, Lou Y, Lu L, Liu Y, Chen Q, Chen X, Jin W. Heterogeneous effect of two selectin gene polymorphisms on coronary artery disease risk: a meta-analysis. PLoS One. 2014;9:e88152. doi: 10.1371/journal.pone.0088152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z, Lou Y, Jin W, Liu Y, Lu L, Chen Q, Zhang R. The Connexin37 gene C1019T polymorphism and risk of coronary artery disease: a meta-analysis. Arch Med Res. 2014;45:21–30. doi: 10.1016/j.arcmed.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Cea LB. Natriuretic peptide family: new aspects. Curr Med Chem Cardiovasc Hematol Agents. 2005;3:87–98. doi: 10.2174/1568016053544309. [DOI] [PubMed] [Google Scholar]
- 6.Rubattu S, Sciarretta S, Valenti V, Stanzione R, Volpe M. Natriuretic peptides: an update on bioactivity, potential therapeutic use, and implication in cardiovascular diseases. Am J Hypertens. 2008;21:733–741. doi: 10.1038/ajh.2008.174. [DOI] [PubMed] [Google Scholar]
- 7.Lang RE, Tholken H, Ganten D, Luft FC, Ruskoaho H, Unger T. Atrial natriuretic factor--a circulating hormone stimulated by volume loading. Nature. 1985;314:264–266. doi: 10.1038/314264a0. [DOI] [PubMed] [Google Scholar]
- 8.Mohammed AA, Januzzi JL Jr. Natriuretic peptides in the diagnosis and management of acute heart failure. Heart Fail Clin. 2009;5:489–500. doi: 10.1016/j.hfc.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Epshteyn V, Morrison K, Krishnaswamy P, Kazanegra R, Clopton P, Mudaliar S, Edelman S, Henry R, Maisel A. Utility of B-type natriuretic peptide (BNP) as a screen for left ventricular dysfunction in patients with diabetes. Diabetes Care. 2003;26:2081–2087. doi: 10.2337/diacare.26.7.2081. [DOI] [PubMed] [Google Scholar]
- 10.Morrow DA, de Lemos JA, Blazing MA, Sabatine MS, Murphy SA, Jarolim P, White HD, Fox KA, Califf RM, Braunwald E. Prognostic value of serial B-type natriuretic peptide testing during follow-up of patients with unstable coronary artery disease. Jama. 2005;294:2866–2871. doi: 10.1001/jama.294.22.2866. [DOI] [PubMed] [Google Scholar]
- 11.Clerico A, Emdin M. Diagnostic accuracy and prognostic relevance of the measurement of cardiac natriuretic peptides: a review. Clin Chem. 2004;50:33–50. doi: 10.1373/clinchem.2003.024760. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Oparil S, Feng JA, Li P, Perry G, Chen LB, Dai M, John SW, Chen YF. Effects of pressure overload on extracellular matrix expression in the heart of the atrial natriuretic peptide-null mouse. Hypertension. 2003;42:88–95. doi: 10.1161/01.HYP.0000074905.22908.A6. [DOI] [PubMed] [Google Scholar]
- 13.Tamura N, Ogawa Y, Chusho H, Nakamura K, Nakao K, Suda M, Kasahara M, Hashimoto R, Katsuura G, Mukoyama M, Itoh H, Saito Y, Tanaka I, Otani H, Katsuki M. Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc Natl Acad Sci U S A. 2000;97:4239–4244. doi: 10.1073/pnas.070371497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Greco MF, Pattaro C, Luchner A, Pichler I, Winkler T, Hicks AA, Fuchsberger C, Franke A, Melville SA, Peters A, Wichmann HE, Schreiber S, Heid IM, Krawczak M, Minelli C, Wiedermann CJ, Pramstaller PP. Genome-wide association analysis and fine mapping of NT-proBNP level provide novel insight into the role of the MTHFR-CLCN6-NPPA-NPPB gene cluster. Hum Mol Genet. 2011;20:1660–1671. doi: 10.1093/hmg/ddr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, Morgenthaler NG, Bergmann A, Blankenberg S, Kee F, Nilsson P, Yin X, Peltonen L, Vartiainen E, Salomaa V, Hirschhorn JN, Melander O, Wang TJ. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41:348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubattu S, Stanzione R, Di Angelantonio E, Zanda B, Evangelista A, Tarasi D, Gigante B, Pirisi A, Brunetti E, Volpe M. Atrial natriuretic peptide gene polymorphisms and risk of ischemic stroke in humans. Stroke. 2004;35:814–818. doi: 10.1161/01.STR.0000119381.52589.AB. [DOI] [PubMed] [Google Scholar]
- 17.Liguori A, Di Gregorio F, Napoli C, D’Armiento FP, Posca T, Di Benedetto A, Di Ieso N, Di Paolo E, Ferrara A. Atrial natriuretic factor and sympathetic activation in human heart failure. Riv Eur Sci Med Farmacol. 1994;16:61–67. [PubMed] [Google Scholar]
- 18.Larifla L, Maimaitiming S, Velayoudom-Cephise FL, Ferdinand S, Blanchet-Deverly A, Benabdallah S, Donnet JP, Atallah A, Roussel R, Foucan L. Association of 2238T >C polymorphism of the atrial natriuretic peptide gene with coronary artery disease in Afro-Caribbeans with type 2 diabetes. Am J Hypertens. 2012;25:524–527. doi: 10.1038/ajh.2011.233. [DOI] [PubMed] [Google Scholar]
- 19.Lynch AI, Boerwinkle E, Davis BR, Ford CE, Eckfeldt JH, Leiendecker-Foster C, Arnett DK. Pharmacogenetic association of the NPPA T2238C genetic variant with cardiovascular disease outcomes in patients with hypertension. Jama. 2008;299:296–307. doi: 10.1001/jama.299.3.296. [DOI] [PubMed] [Google Scholar]
- 20.Vassalle C, Andreassi MG, Prontera C, Fontana M, Zyw L, Passino C, Emdin M. Influence of ScaI and natriuretic peptide (NP) clearance receptor polymorphisms of the NP System on NP concentration in chronic heart failure. Clin Chem. 2007;53:1886–1890. doi: 10.1373/clinchem.2007.088302. [DOI] [PubMed] [Google Scholar]
- 21.Rubattu S, Bigatti G, Evangelista A, Lanzani C, Stanzione R, Zagato L, Manunta P, Marchitti S, Venturelli V, Bianchi G, Volpe M, Stella P. Association of atrial natriuretic peptide and type a natriuretic peptide receptor gene polymorphisms with left ventricular mass in human essential hypertension. J Am Coll Cardiol. 2006;48:499–505. doi: 10.1016/j.jacc.2005.12.081. [DOI] [PubMed] [Google Scholar]
- 22.Ellis KL, Newton-Cheh C, Wang TJ, Frampton CM, Doughty RN, Whalley GA, Ellis CJ, Skelton L, Davis N, Yandle TG, Troughton RW, Richards AM, Cameron VA. Association of genetic variation in the natriuretic peptide system with cardiovascular outcomes. J Mol Cell Cardiol. 2011;50:695–701. doi: 10.1016/j.yjmcc.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Scanlon PJ, Faxon DP, Audet AM, Carabello B, Dehmer GJ, Eagle KA, Legako RD, Leon DF, Murray JA, Nissen SE, Pepine CJ, Watson RM, Ritchie JL, Gibbons RJ, Cheitlin MD, Gardner TJ, Garson A Jr, Russell RO Jr, Ryan TJ, Smith SC Jr. ACC/AHA guidelines for coronary angiography: executive summary and recommendations. A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Committee on Coronary Angiography) developed in collaboration with the Society for Cardiac Angiography and Interventions. Circulation. 1999;99:2345–2357. doi: 10.1161/01.cir.99.17.2345. [DOI] [PubMed] [Google Scholar]
- 24.Kushner FG, Hand M, Smith SC Jr, King SB 3rd, Anderson JL, Antman EM, Bailey SR, Bates ER, Blankenship JC, Casey DE Jr, Green LA, Hochman JS, Jacobs AK, Krumholz HM, Morrison DA, Ornato JP, Pearle DL, Peterson ED, Sloan MA, Whitlow PL, Williams DO American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. 2009 Focused Updates: ACC/AHA Guidelines for the Management of Patients With ST-Elevation Myocardial Infarction (updating the 2004 Guideline and 2007 Focused Update) and ACC/AHA/SCAI Guidelines on Percutaneous Coronary Intervention (updating the 2005 Guideline and 2007 Focused Update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2009;120:2271–2306. doi: 10.1161/CIRCULATIONAHA.109.192663. [DOI] [PubMed] [Google Scholar]
- 25.Gottdiener JS, Bednarz J, Devereux R, Gardin J, Klein A, Manning WJ, Morehead A, Kitzman D, Oh J, Quinones M, Schiller NB, Stein JH, Weissman NJ American Society of Echocardiography. American Society of Echocardiography recommendations for use of echocardiography in clinical trials. J Am Soc Echocardiogr. 2004;17:1086–1119. doi: 10.1016/j.echo.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Nadir MA, Dow E, Davidson J, Kennedy N, Lang CC, Struthers AD. Myocardial ischaemia is associated with an elevated brain natriuretic pepide level even in the presence of left ventricular systolic dysfunction. Eur J Heart Fail. 2014;16:56–67. doi: 10.1093/eurjhf/hft130. [DOI] [PubMed] [Google Scholar]
- 27.Grundy SM, Cleeman JI, Merz CN, Brewer HB Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC Jr, Stone NJ. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 28.Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, Marsh EE 3rd. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 29.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 30.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 31.Stram DO, Leigh Pearce C, Bretsky P, Freedman M, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Thomas DC. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum Hered. 2003;55:179–190. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 32.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattin KA, White BC, Barney N, Gui J, Nelson HH, Kelsey KT, Andrew AS, Karagas MR, Moore JH. A computationally efficient hypothesis testing method for epistasis analysis using multifactor dimensionality reduction. Genetic epidemiology. 2009;33:87–94. doi: 10.1002/gepi.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 35.Fox AA, Collard CD, Shernan SK, Seidman CE, Seidman JG, Liu KY, Muehlschlegel JD, Perry TE, Aranki SF, Lange C, Herman DS, Meitinger T, Lichtner P, Body SC. Natriuretic peptide system gene variants are associated with ventricular dysfunction after coronary artery bypass grafting. Anesthesiology. 2009;110:738–747. doi: 10.1097/aln.0b013e31819c7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Rumayor A, Richards AM, Burnett JC, Januzzi JL Jr. Biology of the natriuretic peptides. Am J Cardiol. 2008;101:3–8. doi: 10.1016/j.amjcard.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Onuoha GN, Nicholls DP, Patterson A, Beringer T. Neuropeptide secretion in exercise. Neuropeptides. 1998;32:319–325. doi: 10.1016/s0143-4179(98)90054-3. [DOI] [PubMed] [Google Scholar]
- 38.Dessi-Fulgheri P, Sarzani R, Rappelli A. Role of the natriuretic peptide system in lipogenesis/lipolysis. Nutr, Metab Cardiovasc Dis. 2003;13:244–249. doi: 10.1016/s0939-4753(03)80018-2. [DOI] [PubMed] [Google Scholar]
- 39.Kohno M, Yasunari K, Maeda K, Kano H, Minami M, Hanehira T, Yoshikawa J. Effects of cardiac natriuretic peptides on oxidized low-density lipoprotein- and lysophosphatidylcholine-induced human mesangial cell migration. Hypertension. 2000;35:971–977. doi: 10.1161/01.hyp.35.4.971. [DOI] [PubMed] [Google Scholar]
- 40.Liang F, Kapoun AM, Lam A, Damm DL, Quan D, O’Connell M, Protter AA. B-Type natriuretic peptide inhibited angiotensin II-stimulated cholesterol biosynthesis, cholesterol transfer, and steroidogenesis in primary human adrenocortical cells. Endocrinology. 2007;148:3722–3729. doi: 10.1210/en.2006-1599. [DOI] [PubMed] [Google Scholar]
- 41.Weber KT. Aldosterone in congestive heart failure. N Engl J Med. 2001;345:1689–1697. doi: 10.1056/NEJMra000050. [DOI] [PubMed] [Google Scholar]
- 42.Sisson DD. Neuroendocrine evaluation of cardiac disease. Vet Clin North Am Small Anim Pract. 2004;34:1105–1126. doi: 10.1016/j.cvsm.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Schrier RW, Abraham WT. Hormones and hemodynamics in heart failure. N Engl J Med. 1999;341:577–585. doi: 10.1056/NEJM199908193410806. [DOI] [PubMed] [Google Scholar]


