Abstract
Background: Small cell lung cancer (SCLC) is one of highly aggressive cancers with poor prognosis. Unfortunately, there are as yet no molecular targets that can be exploited to prolong survival in patients with SCLC. This study aimed to investigate possible molecular markers associated with prognosis in limited-stage small cell lung cancer (LS-SCLC). Methods: The demographic and clinical data for LS-SCLC patients treated in a tertiary care hospital between January 2008 and December 2012 were retrospectively reviewed. NQO1 polymorphism and the expression of p53, SOD2, PARP1 were examined in biopsy specimens, and the factors affecting prognosis were identified. Results: 79 patients with LS-SCLC having available pathologic tissues were analyzed. 84.8% of them received both chemotherapy and radiotherapy. NQO1 polymorphism was detected in 60.0% (45/79; heterozygous in 26 patients, homozygous in 19 patients). Over-expression of p53, SOD2, PARP1 was seen in 45.6% (36/79), 38.0% (30/79) and 41.8% (33/79) of the patients, respectively. The univariate Cox proportional hazards model revealed that serum lactate dehydrogenase (LDH) levels and PARP1 expression were associated with disease progression. In the multivariate analysis, only PARP1 expression was a significant independent prognostic factor for progression-free survival (hazard ratio: 0.494; 95% CI, 0.267-0.913, P = 0.025). Conclusions: PARP1 expression is correlated with longer progression-free survival in LS-SCLC requiring further studies to clarify the precise role of PARP1 and the relevance of PARP1-targeted therapy.
Keywords: Small cell lung cancer, NQO1 polymorphism, PARP1, prognosis
Introduction
Small cell lung cancers (SCLCs) represent approximately 15% of all lung cancers [1]. Although initial treatment response is favorable, overall prognosis is very poor, with median survival of less than 2 years [2,3]. A simple staging system that classifies patients into a limited or an extensive stage of SCLC is generally accepted in clinical practice and affects treatment decisions [4]. In limited-stage small cell lung cancer (LS-SCLC), combined radiotherapy and chemotherapy improve clinical outcomes significantly more than chemotherapy alone [5,6]. Although there have been many studies of the optimal timing of radiation and the total radiation dose [7-9], 5-year survival of LS-SCLC patients remains below 25% at best [10]. Therefore, many workers have sought to identify molecular targets for treating SCLC [11,12]. However, currently there are no targeted therapies with beneficial effects on SCLC patients.
NAD(P)H: quinoneoxidoreductase 1 (NQO1) is a cytosolic flavoenzyme that reduces quinones to less toxic hydroquinones in a single two-electron step [13]. It protects cells against endogenous and exogenous quinines, and there is some evidence that NQO1 activity can induce apoptotic cell death in cancer tissues by regulating the tumor suppressor gene p53 [14,15]. Recent studies have revealed an NQO1 polymorphism that affects NQO1 activity: the NQO1*2 allele is a C to T alteration in codon 609 of NQO1 DNA that leads to weak NQO1 activity [16,17], and there is evidence that the C609T polymorphism can increase cancer risk and reduce treatment responses [18-20].
Superoxide dismutase 2 (SOD2) is an antioxidant enzyme that transforms toxic superoxide into hydrogen peroxide and diatomic oxygen [21], and studies suggest that it is associated with the development and prognosis of cancers [22,23]. There have also been many studies on the effect of p53 expression on cancer susceptibility, with conflicting results [24,25].
Poly (ADP-Ribose) polymerase 1 (PARP1) is a DNA binding enzyme involved in DNA repair via the base excision repair (BER) pathway [26]. It is overexpressed in many cancers and its level has been associated with prognosis [27,28]. There are indications that PARP1 inhibitors may be novel therapeutic tools in many cancers [29], and one study showed that the antitumor agent β-lapachone induced PARP1-mediated cell death in an NQO1-dependent manner [30].
In this study we aimed to investigate the frequency of the NQO1 C609T polymorphism and expression of p53, SOD2, PARP1 in LS-SCLC, and to examine their effects on prognosis.
Methods
Study population
The study population consisted of 79 patients with limited stage small cell lung cancer with available pathologic tissue. These patients were treated at the Asan Medical Center, a 2,700-bed tertiary referral hospital in Seoul, Republic of Korea, between January 2008 and December 2012. The data were retrospectively collected from medical records and a special data system in the Medical Center designated ABLE (Asan Biomedical Research Environment). Patients were followed-up until May 2014. As this was a retrospective study written informed consent was waived. The study protocol was approved by the Institutional Review Board of Asan Medical Center.
Evaluation of NQO1 polymorphism
DNA purification and detection of NQO1 gene polymorphism were performed as previously reported [31]. The following forward and reverse primers were used to amplify a 230 bp NQO1 gene fragment: 5’-TCCTCAGAGTGGCATTCTGC-3’ and 5’-TCTCCTCATCCTGTACCTCT-3’. Amplification was carried with an AccuPower TagPCR PreMix (Bioneer Corp., Daejeon, Korea). PCR mixtures contained primers (0.5 pmoL, each) and 50 ng of genomic DNA in a final volume of 20 μL. PCR conditions consisted of initial denaturing at 94°C for 5 min, 35 amplification cycles (95°C for 30 s, 58°C for 30 s, and 72°C for 30 s), and a final extension at 72°C for 5 min. To examine restriction fragment length polymorphism (RFLP), the amplified fragments were digested with Hinf1 (Thermo Scientific, Rockford, IL) and analyzed by agarose gel electrophoresis. Wild type (Pro187Ser) NQO1 formed a 191 bp band while the homozygous (Ser/Ser) and heterozygous (Pro/Ser) NQO1*2 variants produced a single 151 bp band and a191 bp and 151 bp band, respectively (Figure 1A).
Figure 1.
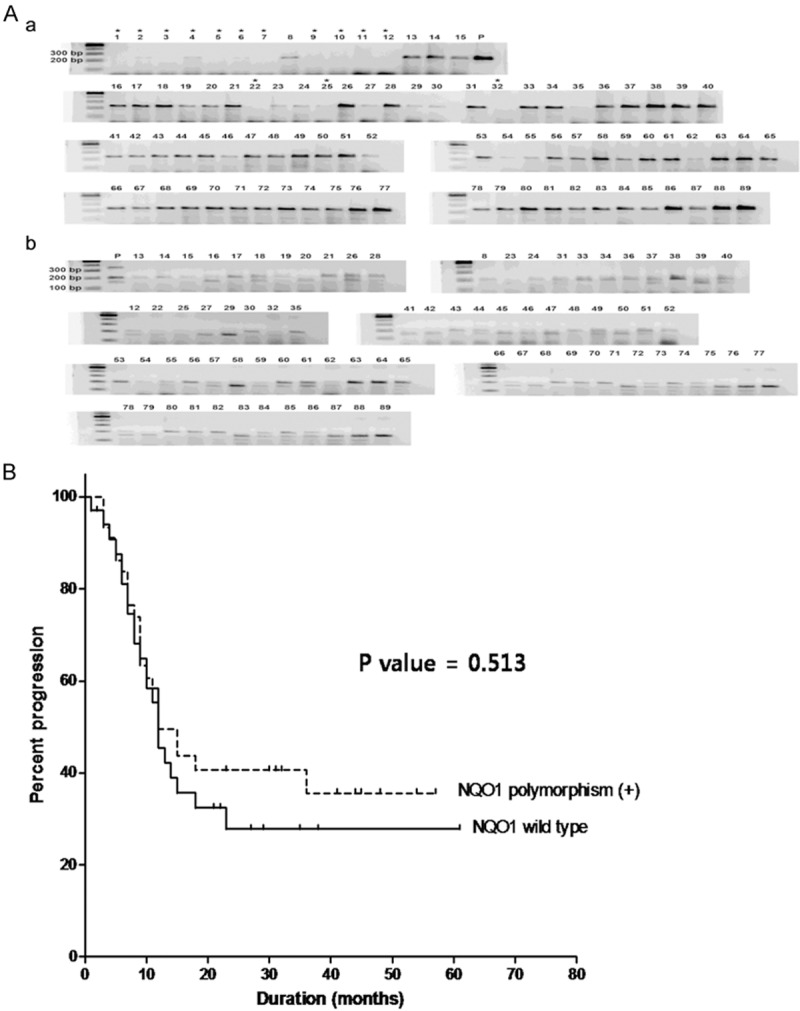
NQO1 polymorphism (A) and progression-free survival in 79 patients with LS-SCLC according to NQO1 polymorphism (B). NQO1 gene fragments were amplified (A) and digested with HInf1 endonuclease (B). *insufficient DNA available for analysis; P: positive control (heterozygous NQO1).
Immunohistochemistry of NQO1, p53, SOD2, and PARP1
Immunohistochemistry was performed with anautomated IHC staining device (Benchmark XT; Ventana Medical Systems, Tucson, AZ). Briefly, 4-μm thick sections of formalin-fixed paraffin-embedded (FFPE) tissue were made and subjected to antigen retrieval by microwave boiling in 0.01 M sodium citrate buffer (pH 6.0) for 30 min. Endogenous peroxidase activity was eliminated by exposure to 3% hydrogen peroxide for 30 min. The slides were then incubated sequentially with antibodies toNQO1 (sc-32793, 1:400, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), p53 (M7001; 1:1,500; Dako, Glostrup, Denmark), SOD2 (sc-133134, 1:5,000, Santa Cruz Biotechnology Inc.) and PARP1 (IHC-00279, 1:200, Bethly Laboratories Inc., Montgomery, TX, USA) followed byperoxidase-labeled secondary antibody. Peroxidase activity was visualized using an aminoethylcarbazole substrate kit (Zymed Laboratories, Inc., San Francisco, California), and the slides were counterstained with hematoxylin.
Antibody staining was analyzed semiquantitatively by classifying the area of tissue stained into four levels (0, < 10% of tumor area stained; 1, 10%-30% of the area stained; 2, 30-60% of the area stained; and 3, > 60% of the area stained). P53 and PARP1 expression was evaluated at the nuclear level and NQO1 and SOD2 expressions at the cytoplasmic level (Figure 2).
Figure 2.
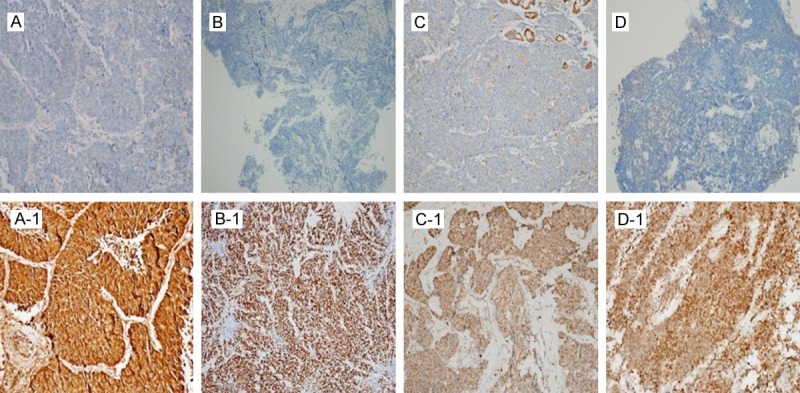
Immunohistochemical staining of SCLC specimens. TOP, (A-D) Negative results for NQO1, p53, SOD2 and PARP1. Bottom, (A1-1-D1-1) Positive results for NQO1, p53, SOD2 and PARP1.
Statistical analysis
For statistical analysis antibody staining was divided into two levels (negative including the above scores 0, 1 and 2 vs. positive including score 3). Comparisons of baseline characteristics and treatments were made using Student’s t-test or the Mann-Whitney U test for continuous variables, and the chi-square or Fisher’s exact test for categorical data. All P-values were two-tailed, with statistical significance set at P < 0.05. Kaplan-Meier analysis was used to evaluate differences in progression-free survival, and risk factors for progression were analyzed with Cox proportional hazards models. Statistical analyses were performed with SPSS version 18.0 for Windows (SPSS, Inc., Chicago, IL).
Results
Patient characteristics
The mean age of the 79 LS-SCLC patients was 62.6 ± 8.0 years, and 89.9% (71/79) were male. Eastern cooperative oncology group (ECOG) performance was 1 (78.5%) or 2 (21.5%), and diabetes mellitus was the most common comorbidity (17.7%). Other characteristics are presented in Table 1.
Table 1.
Baseline characteristics of small cell lung cancer limited stage (79 patients)
| Characteristics | |
|---|---|
| Age (mean), years ± SD | 62.6 ± 8.0 |
| Gender, male | 71 (89.9) |
| Body mass index, kg/m2 (mean ± SD) | 23.8 ± 4.0 |
| Smoking (Ever smoker) | 73 (92.4) |
| Pack years | 42.1 ± 22.1 |
| ECOG performance | |
| 1 | 62 (78.5) |
| 2 | 17 (21.5) |
| 3 | 0 |
| 4 | 0 |
| Underlying disease | |
| Diabetes mellitus | 14 (17.7) |
| Heart failure | 1 (1.3) |
| Chronic kidney disease | 0 |
| Liver cirrhosis | 1 (1.3) |
| Cerebrovascular accident | 4 (5.1) |
| Previous tuberculosis history | 6 (7.6) |
| Paraneoplastic syndrome | 4 (5.1) |
| LDH levels* (median, interquartile range) | 222 (191-253) |
| TNM stage | |
| 1 | 5 (6.3) |
| 2 | 9 (11.4) |
| 3 | 65 (82.3) |
| 4 | 0 |
Values are reported as mean ± standard deviation or as frequency (%), ECOG = Eastern cooperative oncology group; LDH = Lactate dehydrogenase.
LDH levels were checked in 44 patients.
Treatments and clinical course of the patients
Table 2 gives the treatment and clinical courses of the LS-SCLC patients. All 79 LS-SCLC patients received chemotherapy. The first chemotherapy regimen was chosen by the attending physician (Etoposide + Cisplatin in 17.7% of the patients, Etoposide + Carboplatin in 82.3%). 84.8% of the patients (67/79) received thoracic radiotherapy, and 31.6% (25/79) underwent prophylactic cranial irradiation treatment. 15.2% of patients (12/79) did not receive thoracic radiotherapy because of refusal (5 patients), old age (3 patients), or poor general condition (4 patients). The median follow-up duration was 535 days (interquartile range 301-874 days). All-cause mortality was 50.6% (40/79).
Table 2.
Treatment and clinical course of small cell lung cancer limited stage (79 patients)
| Characteristics | |
|---|---|
| Initial chemotherapy regimen | |
| Etoposide + Cisplatin | 14 (17.7) |
| Etoposide + Carboplatin | 65 (82.3) |
| Thoracic radiotherapy | 67 (84.8) |
| Operation | 6 (7.6) |
| Sequential chemotherapy only | 4 |
| Sequential chemotherapy and radiation therapy | 2 |
| Concurrent chemoradiation therapy | 46 (58.2) |
| Chemotherapy + sequential radiation therapy | 21 (26.6) |
| Prophylactic cranial irradiation | 25 (31.6) |
| Median follow up days (range, interquartile range) | 535 (301-874) |
| Overall death | 40 (50.6) |
Values are reported as frequency (%).
NQO1 polymorphism, expression of p53, SOD2, PARP1
NQO1 polymorphism was detected in 57.0% of the patients (45/79, heterozygous in 26 patients, homozygous in 19 patients). NQO1 IHC staining was positive in 29.4% of the wild type tissues (10/34), 11.5% of the heterozygous tissues (3/26) and 5.3% of the homozygous tissues (1/19). P53, SOD2, and PARP1 IHC staining was positive in 45.6% (35/79), 38.0% (30/79) and 41.8% (33/79) of the total study population, respectively.
Factors associated with progression-free survival
The univariate Cox proportional hazards model revealed that only serum lactate dehydrogenase (LDH) levels and PARP1 expression were associated with progression. In the multivariate analysis, PARP1 expression was a significant independent prognostic factor for disease progression (hazard ratio: 0.494; 95% CI, 0.267-0.913, P = 0.025). Covariates in the full model were age, ECOG performance, prophylactic cranial irradiation and PARP1 expression. LDH was not included because values were frequently missing (44.3%), and TNM stage was excluded because of its strong correlation with ECOG performance (Table 3). Also, PARP1 expression was related to progression-free survival of the LS-SCLC patients (Figure 3).
Table 3.
Prediction factors for progression in patients with small cell lung cancer limited stage assessed by Cox proportional hazard model
| Parameters | Hazard ratio | 95% CI | P value |
|---|---|---|---|
| Univariate analysis | |||
| Age | 1.004 | 0.966-1.044 | 0.825 |
| Gender, male | 1.101 | 0.466-2.603 | 0.826 |
| Smoking (Ever smoker) | 0.710 | 0.279-1.806 | 0.472 |
| ECOG performance | 1.327 | 0.658-1.327 | 0.430 |
| Paraneoplastic syndrome | 0.044 | 0.000-6.186 | 0.215 |
| LDH value | 1.003 | 1.000-1.005 | 0.020 |
| TNM stage | 1.615 | 0.388-6.725 | 0.804 |
| Thoracic radiotherapy | 1.491 | 0.534-4.164 | 0.446 |
| Prophylactic cranial irradiation | 0.603 | 0.321-1.133 | 0.116 |
| Concurrent chemoradiation therapy | 0.897 | 0.466-1.727 | 0.745 |
| NQO1 polymorphism | 0.670 | 0.299-1.502 | 0.331 |
| Positive p53 | 0.675 | 0.370-1.231 | 0.200 |
| Positive SOD2 | 0.866 | 0.475-1.578 | 0.638 |
| Positive PARP1 | 0.500 | 0.271-0.925 | 0.027 |
| Multivariate analysis | |||
| Age | 1.001 | 0.964-1.039 | 0.964 |
| ECOG performance | 1.163 | 0.566-2.391 | 0.681 |
| Prophylactic cranial irradiation | 0.611 | 0.317-1.178 | 0.141 |
| Positive PARP1 | 0.494 | 0.267-0.913 | 0.025 |
Figure 3.
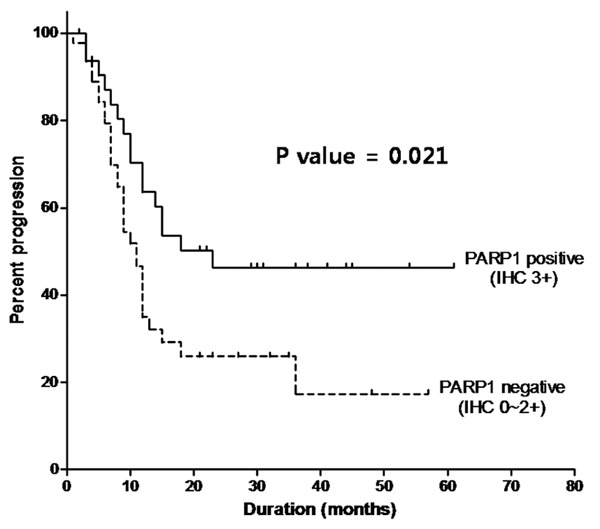
Progression-free survival in 79 patients with LD-SCLC according to PARP1.
Relationship between NQO1 polymorphism, expression of other genes and progression- free survival
NQO1 genotype was not associated with p53, SOD2, PARP1 expression or TNM stage (see Table 4). Also, NQO1 polymorphism was not related to progression-free survival of the LS-SCLC patients (Figure 1B).
Table 4.
Association with NQO1 polymorphism and other gene expression, TNM stage
| Homozygote (N = 19) | Heterozygote (N = 26) | Wild type (N = 34) | P value | |
|---|---|---|---|---|
| P53 expression | 9 (47.4) | 10 (38.5) | 17 (50.0) | 0.663 |
| SOD2 expression | 8 (42.1) | 10 (38.5) | 12 (35.3) | 0.886 |
| PARP1 expression | 5 (26.3) | 10 (38.5) | 18 (52.9) | 0.155 |
| TNM stage | 0.700 | |||
| 1 | 2 (10.5) | 2 (7.7) | 1 (2.9) | |
| 2 | 1 (5.3) | 3 (11.5) | 5 (14.7) | |
| 3 | 16 (84.2) | 21 (80.8) | 28 (82.4) | |
| 4 | 0 | 0 | 0 |
Values are reported as frequency (%).
Discussion
Our current study showed that while NQO1 polymorphism did not affect prognosis and expression of the other genes examined, PARP1 expression was related to longer progression-free survival. We focused on the limited stage of small cell lung cancer, and found that age, sex, ECOG performance, TNM stage, and prophylactic cranial irradiation were unrelated to cancer progression. This underscores the importance of identifying new genetic markers in small cell lung cancer.
Although chemotherapy and radiation therapy have developed remarkably, the treatment of SCLC remains a challenge. It is difficult to develop novel therapeutic strategies because SCLC can arise by numerous distinct routes [11]. Also, there is still a lack of data concerning prognosis and survival in SCLC patients; our study was aimed at establishing the clinical significance of several genetic markers.
We found that NQO1 polymorphism was not associated with p53, SOD2, or PARP1 expression or with TNM stage. Also, we did not detect any association between NQO1 polymorphism and prognosis. There is still controversy concerning the relation between NQO1 polymorphism and non-small cell lung cancer (NSCLC). Studies have suggested that the presence of the NQO1*2 allele is associated with NSCLC susceptibility [19], locoregional recurrence [20] or even poor overall survival [32] whereas others have reported no relationship with NSCLC susceptibility [33] or that it has a protective effect on NSCLC [34]. In the case of SCLC, there is a lack of evidence in relation to NQO1 polymorphism, but one study has claimed that it is associated with the development of SCLC [35]. Thus, further study of NQO1 polymorphism in SCLC patients is needed.
There have been many studies of PARP1 expression in human cancers [36-38]. In breast cancer, there is growing evidence that PARP1 expression is related to a worse prognosis [27] and PARP1 inhibitors are potential treatments especially in patients harboring BRCA mutations [39]. There are very little data on PARP1 expression in lung cancer. One study has suggested that PARP1 expression is associated with a poor prognosis in NSCLC [28], but another found PARP1 and other marker expression related with prolonged survival within untreated patients [40]. In the case of SCLC, there is limited evidence that PARP1 expression is higher in SCLC than NSCLC, and PARP1 inhibitors could provide potential treatment strategies via several pathways [41,42]. PARP1 inhibition causes down-regulation of essential DNA repair genes including RAD51, and PARP1 inhibitor alone or in combination with chemotherapy seem to be effective in cell lines. However, the study referred to only compared protein expression in cell lines and there were no clinical data. To the best of our knowledge, our study is the first to evaluate the association between PARP1 expression and prognosis in SCLC.
There are limitations to this study. First, since it was retrospective in design and non-interventional, there could be selection bias. Second, as it was performed at a single center in Korea and involved a relatively small number of patients, it would be unwise to generalize our results. Further studies with larger numbers are needed.
In summary, NQO1 polymorphism and p53 and SOD2 expression were not associated with prognosis in LS-SCLC. However, PARP1 expression was independently related to longer progression-free survival.
Acknowledgements
This work was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare (HI12C1146000013).
Disclosure of conflict of interest
None.
References
- 1.Stupp R, Monnerat C, Turrisi AT 3rd, Perry MC, Leyvraz S. Small cell lung cancer: state of the art and future perspectives. Lung Cancer. 2004;45:105–117. doi: 10.1016/j.lungcan.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 3.Chua YJ, Steer C, Yip D. Recent advances in management of small-cell lung cancer. Cancer Treat Rev. 2004;30:521–543. doi: 10.1016/j.ctrv.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Micke P, Faldum A, Metz T, Beeh KM, Bittinger F, Hengstler JG, Buhl R. Staging small cell lung cancer: Veterans Administration Lung Study Group versus International Association for the Study of Lung Cancer-what limits limited disease? Lung Cancer. 2002;37:271–276. doi: 10.1016/s0169-5002(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 5.Turrisi AT 3rd, Kim K, Blum R, Sause WT, Livingston RB, Komaki R, Wagner H, Aisner S, Johnson DH. Twice-daily compared with once-daily thoracic radiotherapy in limited small-cell lung cancer treated concurrently with cisplatin and etoposide. N Engl J Med. 1999;340:265–271. doi: 10.1056/NEJM199901283400403. [DOI] [PubMed] [Google Scholar]
- 6.Takada M, Fukuoka M, Kawahara M, Sugiura T, Yokoyama A, Yokota S, Nishiwaki Y, Watanabe K, Noda K, Tamura T, Fukuda H, Saijo N. Phase III study of concurrent versus sequential thoracic radiotherapy in combination with cisplatin and etoposide for limited-stage small-cell lung cancer: results of the Japan Clinical Oncology Group Study 9104. J. Clin. Oncol. 2002;20:3054–3060. doi: 10.1200/JCO.2002.12.071. [DOI] [PubMed] [Google Scholar]
- 7.Fried DB, Morris DE, Poole C, Rosenman JG, Halle JS, Detterbeck FC, Hensing TA, Socinski MA. Systematic review evaluating the timing of thoracic radiation therapy in combined modality therapy for limited-stage small-cell lung cancer. J. Clin. Oncol. 2004;22:4837–4845. doi: 10.1200/JCO.2004.01.178. [DOI] [PubMed] [Google Scholar]
- 8.Huncharek M, McGarry R. A meta-analysis of the timing of chest irradiation in the combined modality treatment of limited-stage small cell lung cancer. Oncologist. 2004;9:665–672. doi: 10.1634/theoncologist.9-6-665. [DOI] [PubMed] [Google Scholar]
- 9.Watkins JM, Fortney JA, Wahlquist AE, Shirai K, Garrett-Mayer E, Aguero EG, Sherman CA, Turrisi AT 3rd, Sharma AK. Once-daily radiotherapy to > or =59.4 Gy versus twice-daily radiotherapy to > or =45.0 Gy with concurrent chemotherapy for limited-stage small-cell lung cancer: a comparative analysis of toxicities and outcomes. Jpn J Radiol. 2010;28:340–348. doi: 10.1007/s11604-010-0429-x. [DOI] [PubMed] [Google Scholar]
- 10.Sculier JP, Lafitte JJ, Efremidis A, Florin MC, Lecomte J, Berchier MC, Richez M, Berghmans T, Scherpereel A, Meert AP, Koumakis G, Leclercq N, Paesmans M, Van Houtte P. A phase III randomised study of concomitant induction radiochemotherapy testing two modalities of radiosensitisation by cisplatin (standard versus daily) for limited small-cell lung cancer. Ann Oncol. 2008;19:1691–1697. doi: 10.1093/annonc/mdn354. [DOI] [PubMed] [Google Scholar]
- 11.D’Angelo SP, Pietanza MC. The molecular pathogenesis of small cell lung cancer. Cancer Biol Ther. 2010;10:1–10. doi: 10.4161/cbt.10.1.12045. [DOI] [PubMed] [Google Scholar]
- 12.Sattler M, Salgia R. Molecular and cellular biology of small cell lung cancer. Semin Oncol. 2003;30:57–71. doi: 10.1053/sonc.2003.50019. [DOI] [PubMed] [Google Scholar]
- 13.Ross D, Kepa JK, Winski SL, Beall HD, Anwar A, Siegel D. NAD(P)H:quinone oxidoreductase 1 (NQO1): chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem Biol Interact. 2000;129:77–97. doi: 10.1016/s0009-2797(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 14.Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A. 2002;99:3099–3104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci U S A. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anwar A, Siegel D, Kepa JK, Ross D. Interaction of the molecular chaperone Hsp70 with human NAD(P)H:quinone oxidoreductase 1. J Biol Chem. 2002;277:14060–14067. doi: 10.1074/jbc.M111576200. [DOI] [PubMed] [Google Scholar]
- 17.Ross D, Traver RD, Siegel D, Kuehl BL, Misra V, Rauth AM. A polymorphism in NAD(P)H:quinone oxidoreductase (NQO1): relationship of a homozygous mutation at position 609 of the NQO1 cDNA to NQO1 activity. Br J Cancer. 1996;74:995–996. doi: 10.1038/bjc.1996.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Zhang D. The NQO1 C609T polymorphism and risk of lung cancer: a meta-analysis. Asian Pac J Cancer Prev. 2011;12:3091–3095. [PubMed] [Google Scholar]
- 19.Tian G, Wang M, Xu X. The role of NQO1 polymorphisms in the susceptibility and chemotherapy response of Chinese NSCLC patients. Cell Biochem Biophys. 2014;69:475–479. doi: 10.1007/s12013-014-9820-z. [DOI] [PubMed] [Google Scholar]
- 20.Song SY, Jeong SY, Park HJ, Park SI, Kim DK, Kim YH, Shin SS, Lee SW, Ahn SD, Kim JH, Lee JS, Choi EK. Clinical significance of NQO1 C609T polymorphisms after postoperative radiation therapy in completely resected non-small cell lung cancer. Lung Cancer. 2010;68:278–282. doi: 10.1016/j.lungcan.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Mates JM, Segura JA, Alonso FJ, Marquez J. Oxidative stress in apoptosis and cancer: an update. Arch Toxicol. 2012;86:1649–1665. doi: 10.1007/s00204-012-0906-3. [DOI] [PubMed] [Google Scholar]
- 22.Li N, Huang HQ, Zhang GS. Association between SOD2 C47T polymorphism and lung cancer susceptibility: a meta-analysis. Tumour Biol. 2014;35:955–959. doi: 10.1007/s13277-013-1127-y. [DOI] [PubMed] [Google Scholar]
- 23.Chen PM, Wu TC, Wang YC, Cheng YW, Sheu GT, Chen CY, Lee H. Activation of NF-kappaB by SOD2 promotes the aggressiveness of lung adenocarcinoma by modulating NKX2-1-mediated IKKbeta expression. Carcinogenesis. 2013;34:2655–2663. doi: 10.1093/carcin/bgt220. [DOI] [PubMed] [Google Scholar]
- 24.Paik KH, Park YH, Ryoo BY, Yang SH, Lee JC, Kim CH, Ki SS, Kim JM, Park MJ, Ahn HJ, Choi W, Chung JH. Prognostic value of immunohistochemical staining of p53, bcl-2, and Ki-67 in small cell lung cancer. J Korean Med Sci. 2006;21:35–39. doi: 10.3346/jkms.2006.21.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JS, Yoon A, Kalapurakal SK, Ro JY, Lee JJ, Tu N, Hittelman WN, Hong WK. Expression of p53 oncoprotein in non-small-cell lung cancer: a favorable prognostic factor. J. Clin. Oncol. 1995;13:1893–1903. doi: 10.1200/JCO.1995.13.8.1893. [DOI] [PubMed] [Google Scholar]
- 26.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojo F, Garcia-Parra J, Zazo S, Tusquets I, Ferrer-Lozano J, Menendez S, Eroles P, Chamizo C, Servitja S, Ramirez-Merino N, Lobo F, Bellosillo B, Corominas JM, Yelamos J, Serrano S, Lluch A, Rovira A, Albanell J. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Ann Oncol. 2012;23:1156–1164. doi: 10.1093/annonc/mdr361. [DOI] [PubMed] [Google Scholar]
- 28.Xie KJ, He HE, Sun AJ, Liu XB, Sun LP, Dong XJ. Expression of ERCC1, MSH2 and PARP1 in non-small cell lung cancer and prognostic value in patients treated with platinum-based chemotherapy. Asian Pac J Cancer Prev. 2014;15:2591–2596. doi: 10.7314/apjcp.2014.15.6.2591. [DOI] [PubMed] [Google Scholar]
- 29.Kummar S, Chen A, Parchment RE, Kinders RJ, Ji J, Tomaszewski JE, Doroshow JH. Advances in using PARP inhibitors to treat cancer. BMC Med. 2012;10:25. doi: 10.1186/1741-7015-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bey EA, Bentle MS, Reinicke KE, Dong Y, Yang CR, Girard L, Minna JD, Bornmann WG, Gao J, Boothman DA. An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc Natl Acad Sci U S A. 2007;104:11832–11837. doi: 10.1073/pnas.0702176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sameer AS, Shah ZA, Syeed N, Rasool R, Afroze D, Siddiqi MA. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and colorectal cancer predisposition in the ethnic Kashmiri population. Asian Pac J Cancer Prev. 2010;11:209–213. [PubMed] [Google Scholar]
- 32.Kolesar JM, Dahlberg SE, Marsh S, McLeod HL, Johnson DH, Keller SM, Schiller JH. The NQO1*2/*2 polymorphism is associated with poor overall survival in patients following resection of stages II and IIIa non-small cell lung cancer. Oncol Rep. 2011;25:1765–1772. doi: 10.3892/or.2011.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo S, Gao M, Li X, Li Y, Chu S, Zhu D, Niu W. Lack of association between NADPH quinone oxidoreductase 1 (NQO1) gene C609T polymorphism and lung cancer: a case-control study and a meta-analysis. PLoS One. 2012;7:e47939. doi: 10.1371/journal.pone.0047939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock CH, Wenzlaff AS, Cote ML, Land SJ, Schwartz AG. NQO1 T allele associated with decreased risk of later age at diagnosis lung cancer among never smokers: results from a population-based study. Carcinogenesis. 2005;26:381–386. doi: 10.1093/carcin/bgh314. [DOI] [PubMed] [Google Scholar]
- 35.Lewis SJ, Cherry NM, Niven RM, Barber PV, Povey AC. Polymorphisms in the NAD(P)H: quinone oxidoreductase gene and small cell lung cancer risk in a UK population. Lung Cancer. 2001;34:177–183. doi: 10.1016/s0169-5002(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 36.Csete B, Lengyel Z, Kadar Z, Battyani Z. Poly (adenosine diphosphate-ribose) polymerase-1 expression in cutaneous malignant melanomas as a new molecular marker of aggressive tumor. Pathol Oncol Res. 2009;15:47–53. doi: 10.1007/s12253-008-9086-0. [DOI] [PubMed] [Google Scholar]
- 37.Nosho K, Yamamoto H, Mikami M, Taniguchi H, Takahashi T, Adachi Y, Imamura A, Imai K, Shinomura Y. Overexpression of poly (ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur J Cancer. 2006;42:2374–2381. doi: 10.1016/j.ejca.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 38.Brustmann H. Poly (adenosine diphosphate-ribose) polymerase expression in serous ovarian carcinoma: correlation with p53, MIB-1, and outcome. Int J Gynecol Pathol. 2007;26:147–153. doi: 10.1097/01.pgp.0000235064.93182.ec. [DOI] [PubMed] [Google Scholar]
- 39.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 40.Olaussen KA, Adam J, Vanhecke E, Vielh P, Pirker R, Friboulet L, Popper H, Robin A, Commo F, Thomale J, Kayitalire L, Filipits M, Le Chevalier T, Andre F, Brambilla E, Soria JC. PARP1 impact on DNA repair of platinum adducts: preclinical and clinical read-outs. Lung Cancer. 2013;80:216–222. doi: 10.1016/j.lungcan.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, Giri U, Peyton M, Fan YH, Diao L, Masrorpour F, Shen L, Liu W, Duchemann B, Tumula P, Bhardwaj V, Welsh J, Weber S, Glisson BS, Kalhor N, Wistuba II, Girard L, Lippman SM, Mills GB, Coombes KR, Weinstein JN, Minna JD, Heymach JV. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosell R, Wannesson L. A genetic snapshot of small cell lung cancer. Cancer Discov. 2012;2:769–771. doi: 10.1158/2159-8290.CD-12-0346. [DOI] [PubMed] [Google Scholar]


