Abstract
EIF3e is a component of the eukaryotic translation initiation factor 3 (eIF-3) complexes, which is an essential factor for initiation of protein synthesis in mammalian cells. Translational control plays key roles in the complex mechanism of cancer development and progression. However, the clinical significance of eIF3e in colon cancer remains to be elucidated. We analyzed the eIF3e expression in a tissue microarray (TMA), which contained 173 colon cancer tissues paired with adjacent normal mucosa and lymph node metastasis. The expression of eIF3e was significantly elevated in colon cancer tissues in comparison with those in adjacent normal mucosa (P < 0.001) and lymph node metastasis (P < 0.001). The high expression of eIF3e in colon cancer was significantly correlated with tumor size (P < 0.001), lymph node involvement (P < 0.001), distant metastasis (P < 0.001), clinical stage (P < 0.001), histopathologic classification (P < 0.001), and vessel invasion (P = 0.036). Univariate and multivariate analysis revealed that eIF3e is an independent prognosis factor for overall survival and disease-free survival in colon cancer. Down-regulation of eIF3e in vitro inhibited colon cancer cell proliferation, clonality and promoted cell apoptosis. Taken together, high eIF3e expression may contribute to tumor progression and predict poor prognosis in colon cancer.
Keywords: Colon cancer, clinicopathological features, prognosis, eIF3e, proliferation, apoptosis
Introduction
Colon cancer claims a third rank in the list of most common cancers diagnosed worldwide [1]. More developed countries suffer higher morbidity of colon cancer due to the unhealthy lifestyle [2]. But in recent years, the risk of colon cancer significantly increased in several less developed and economically transitioning countries, such as China, due to the adoption of unhealthy western lifestyles such as smoking and physical inactivity and consumption of calorie-dense food [3,4]. Colon cancer arises through a multistep process in which genetic and epigenetic aberrations accumulate in a sequential order to drive malignant transformation of normal colon cells [5]. Some of these aberrations, for instance, microsatellite instability, BRAF mutation, 18q loss of heterozygosity, KRAS, p53, guanylyl cyclase 2, ERCC-1, had been validated as biomarkers for predicting chemoresistance and prognostic in colon cancer [6,7].
Screening of differentially expressed genes (DEGs) and pathways in the pathogenesis through constructing and analyzing high-throughput genomic data can contribute to the understanding on molecular mechanisms of colon cancer. These DEGs could act as new biomarkers for diagnosis and therapy of colon cancer [8]. In our previous studies, a pure population of colon cancer and normal colon cells was obtained and the global gene-expression differences were compared in the 2 cell types using combined cDNA microarray and antisense RNA-long serial analysis of gene expression (aRNA-LongSAGE) approaches. Several genes (IFITM3, FERMT1, MT1F, IMP3, PER 3, etc) that were differentially expressed in those 2 types of cells were identified as potential colon cancer related genes in further studies [9-13]. Eukaryotic translation initiation factor 3 subunit E (eIF3e) was overexpressed in colon cancer cells compared with normal colonic cells in our microarray data (signal: 394.8 vs. 124.9) and LongSAGE analysis (tag count: 49 vs. 14).
EIF3e is a component of the eukaryotic translation initiation factor 3 (eIF-3) complexes, which is required for the regulation of translation initiation of protein synthesis. Alterations in the protein synthesis machinery and translational control could lead to selective translation of specific mRNAs that promote tumor cell survival, angiogenesis, transformation, invasion and metastasis. Translation factors also functionally interact with oncogenes and are often primary targets of signal transduction pathways that underlie most human cancers. Therefore, translational control plays key roles in the complex mechanism of cancer development and progression [14]. Human eIF3 is an 800-kDa molecular mass assembly of 13 subunits that are designated eIF3a-eIF3m. The functional core of human eIF3 is composed of three conserved subunits (eIF3a, eIF3b, and eIF3c), and three non-conserved subunits (eIF3e, eIF3f, and eIF3h). The eIF3-abc subcomplex performed most eIF3 functions. The presence of the eIF3e was required for formation of a stable eIF3 complex [15]. The role of eIF3e in carcinogenesis is still controversial in the literature and could depend on the tumor stage and type. EIF3e was generally considered as tumor suppressor in breast and lung carcinomas [14,16]. However, eIF3e silencing inhibits human glioma cell proliferation via induction of cell cycle arrest and apoptosis [17]. The clinical significance and function of eIF3e in colon cancer remain largely unknown.
In this study, we evaluated the expression pattern of eIF3e, and the relationship between eIF3e expression and clinicopathological characteristics in colon cancer. We investigated whether eIF3e could act as an independent prognosis predictor in colon cancer patients. Furthermore, we revealed that the mechanism of eIF3e promoting tumor progression and predicting poor prognosis was partially dependent on the ability to promote cancer cell proliferation, enhance colony formation, and inhibits cell apoptosis.
Materials and methods
Patients and specimens
Tissue specimens were obtained from colon cancer patients who underwent colectomy between January 2001 and December 2003 at Shanghai Jiaotong University Affiliated First People’s Hospital. None of these patients received adjuvant therapy prior to the surgery. 173 formalin-fixed, paraffin-embedded tissues were applied for histological and immunohistochemical analysis. All specimens were analyzed and diagnosed by two pathologists who were blinded to clinical information. Tumor staging were carried out according to the American Joint Committee on Cancer (AJCC) classification system based on the tumor size (T), lymph node involvement (N), and distant metastasis (M). The disease-free survival (DFS) was defined as the interval from the surgery to clinically or radiologically proven recurrence or metastasis while the overall survival (OS) was defined as the interval from initial surgery to death. The follow-up was carried out according to the National Comprehensive Cancer Network Practice guidelines and the end date was June 29, 2008.
Bioinformatics analysis
The transcriptome database for colon cancer based on LCM-LongSAGE was constructed as described previously [13], and upregulated genes were identified according the following conditions: P < 0.05, fold change > 3, and number of expression tags > 0.
RNA extraction and quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted according to the manufacturer’s protocol (RNAEasy kit; Qiagen; Hilden, Germany). One microgram of total RNA was subjected to first-strand cDNA synthesis according to the manufacturer’s instruction (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems; Carlsbad, CA, USA). Quantitative PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems) and Mastercycler ep Realplex (Eppendorf; Hamburg, Germany). EIF3e was amplified using the sense primer 5’-TTCTTCAATCACCCCAAAGG-3’ and antisense primer 5’-TAGAACCTGCCGACGTTTTC-3’. Glyceraldehydes-3-phosphate dehydrogenase (GAPDH) was amplified as endogenous control using the sense primer 5’-AGCAAGAGCACAAGAGGAAG-3’ and the antisense primer 5’-AACTGGTTGAGCACAGGGTA-3’. These reactions were repeated three times. The results were calculated using the formulas below: eIF3eΔCt = (mean eIF3e_Ct-mean GAPDH_Ct), eIF3eΔΔCt = (eIF3eΔCt_tumor-eIF3eΔCt_normal).
In these formulas, Ct represents the thresholds for eIF3e or GAPDH in the tumor or normal group. The fold-change was represented by the logarithmic scale of 2-ΔΔCt.
Tissue microarray (TMA) construction and immunohistochemistry
The TMAs were constructed in cooperation with Outdo Biotech Co. (Shanghai, China). Cores were archived from optimal tumor specimens with the help of a 2.0 mm diameter punch instrument. The samples archived from the same patient were placed next to each other on the TMA for assuring the uniform treatment. Immunohistochemistry was performed as previously reported to investigate eIF3e expression [12]. Briefly, immunostaining was performed using Envision kit (Dako; Glostrup, Denmark). After antigen retrieval, sections were incubated overnight at 4°C with the primary antibody against eIF3e (Anti-eIF3e antibody ab36766, Abcam; Cambridge, MA, USA; diluted 1:200). The slides were then incubated with secondary antibody for 30 min at room temperature. The immunohistochemical staining was evaluated on the basis of the staining intensity and extent by two pathologists without knowledge of the patient clinical information. Positive staining was classified into three groups: negative, weakly positive and strongly positive, representing negative, low and high eIF3e expression, respectively.
Lentiviral-mediated RNA interference (RNAi)
The lentiviral-mediated RNA interfering delivery system was constructed with the assistance of GenePharma Biotech Co. (Shanghai, China). The human colon cancer cell lines, HCT116 were cultured in DMEM (Gibco; Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Gibco), at 37°C with 5% CO2. Exponentially growing cells were divided into two groups: negative control (Scr-shRNA-HCT116) and knockdown (eIF3e-shRNA-HCT116), in which the normal target cells were infected with negative control virus and target RNA interfering virus, respectively. After incubation for three days, the expression of GFP was observed under the fluorescence microscope to evaluate the infection efficiency.
Cell proliferation assay
Cells were trypsinized at the logarithmic phase and resuspended in complete medium. After adjusting density to 2 × 104/ml, cells were seeded into plates (2000 cells/well). Each group contained three compound perforations. The cells were incubated at 37°C with 5% CO2 and the plates were read with Cellomics ArrayScan system for five days. With the appliance of Cellomics ArrayScan (Thermo Fisher Scientific Inc, USA), the quantity of cells with green fluorescence were accurately calculated. The data were collected and analyzed to establish a proliferation curve for the five days.
Plate colony formation assay
Exponentially growing cells were collected and seeded in 6-well plates with 800 cells per well. The medium was changed every 3 days, for 14 days incubation. After fixed by paraformaldehyde, the colonies were stained with Giemsa for 20 min. The plates were photographed, and the colonies were counted.
Cell apoptosis assay
Cell apoptosis assay was conducted using Annexin V Apoptosis Detection Kit APC (ebioscience, San Diego, USA) according to the manufacturer’s protocol. Briefly, cells were collected at logarithmic phase with three compound perforations in each group. After the cells were washed by binding buffer, staining buffer was added to resuspend the precipitate. Five microlitre Annexin V-APC was added to 100 μl cell suspension for dyeing. After incubation for 15 min away from light, the mixture was analyzed using FACS Calibur (BD, USA).
Statistical analysis
The χ2 test or Fisher’s exact test was used to estimate the statistical significance of difference between eIF3e expression and clinicopathological features. Kaplan-Meier method was used for analyzing patients’ cumulative survival rate. Furthermore, univariate and multivariate Cox proportional hazard model was applied to evaluate the hazard ratios for the variables. Differences between experimental groups were analyzed by t-test or one-way analysis of variance (ANOVA) followed by Bonferroni test. We used the statistical software SPSS19.0 (Chicago, IL, USA) to carry out all statistical analyses. A P-value of less than 0.05 was regarded to be statistically significant.
Results
Bioinformatics results
Six eIF subunits were significantly overexpressed through bioinformatics analysis of 2 transcriptome databases (Table 1). Both of these genes were upregulated, including eIF3e (P < 0.001). Among these subunits, eIF3B, eIF3I, eIF4E and eIF5A had been validated to be associated with tumor progression. The role of eIF3e in colon cancer progression was urgent to be elucidated.
Table 1.
Overexpressed eIF subunits in colon cancer specimens identified by bioinformatics analysis of LCM LongSAGE libraries and LCM microarray
| Ca-Count | N-Count | Total | AVG_P_Chance | Unigene | Description | |
|---|---|---|---|---|---|---|
| 53 | 14 | 67 | < 0.001 | 371001 | Eukaryotic translation initiation factor 3, subunit 9 eta, 116 kDa | EIF3B |
| 49 | 14 | 63 | < 0.001 | 405590 | EIF3S6 Eukaryotic translation initiation factor 3, subunit 6 48 kDa | EIF3E |
| 16 | 2 | 18 | 0.000713 | 539684 | EIF2S3 Eukaryotic translation initiation factor 2, subunit 3 gamma, 52 kDa | EIF2G |
| 8 | 1 | 9 | 0.019967 | 530096 | EIF3S2 Eukaryotic translation initiation factor 3, subunit 2 beta, 36 kDa | EIF3I |
| 5 | 0 | 5 | 0.031014 | 558324 | EIF4E Eukaryotic translation initiation factor 4E | EIF4E |
| 10 | 3 | 13 | 0.044844 | 534314 | EIF5A Eukaryotic translation initiation factor 5A | EIF5A |
LCM, laser capture microdissection; Long SAGE, long serial analysis of gene expression.
Aberrant overexpression of eIF3e in colon cancer tissues
Quantitative real-time polymerase chain reaction (RT-PCR) revealed that the eIF3e was overexpressed at mRNA level in tumor tissues. Thirty five of forty tumor tissues showed higher eIF3e mRNA expression compared with paired normal tissues. The ration of eIF3e mRNA expression was 3.04 ± 2.65 and the difference of eIF3e mRNA expression between tumor tissues and paired normal tissues was significant (P < 0.01, Figure 1). TMA containing cancerous tissues paired with normal mucosa and lymph node metastasis was constructed and immunohistochemstry was performed to investigate the eIF3e protein expression in colon cancer. EIF3e was mainly observed brown staining in the cytoplasm of colon cancer cells, colon epithelial and mesenchymal (Figure 2). Significant differences were found between normal mucosa and cancerous tissue (P < 0.001, Table 2), normal mucosa and LNM metastasis (P < 0.001, Table 2). Only 6 of 173 (3.5%) normal mucosa showed strong eIF3e expression with an obvious contrast to 42 of 173 (24.3%) in cancerous tissue and 31 of 54 (57.4%) in LNM tissue.
Figure 1.
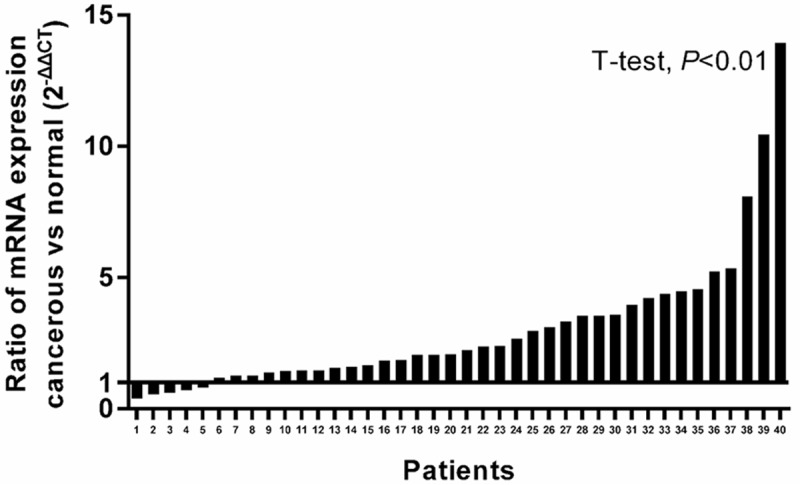
Analysis of eIF3e mRNA expression in normal mucosa and tumor tissue. Quantitative real-time PCR assessed eIF3e mRNA expression in 40 paired fresh specimens. GAPDH was amplified as endogenous control, 2-ΔΔCt was calculated to represent the fold-change. Tumor tissues had a significantly higher eIF3e mRNA expression than that in normal mucosa (P < 0.01).
Figure 2.
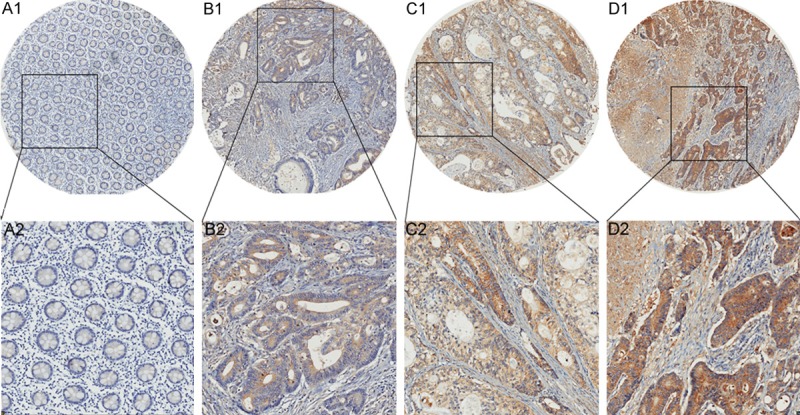
Analysis of eIF3e protein expression in normal mucosa and tumor tissue. A1, A2. Negative eIF3e expression in normal mucosa. B1, B2. Weakly positive staining of eIF3e in colon cancer tissue. C1, C2. Moderately positive staining of eIF3e in colon cancer tissue. D1, D2. Strongly positive staining of eIF3e in colon cancer tissue.
Table 2.
Expression of eIF3e in normal mucosa, cancerous tissues and lymph node metastasis tissues
| Tissue sample | n | eIF3e expression | P value | ||
|---|---|---|---|---|---|
|
| |||||
| Negative (n, %) | Weak (n, %) | Strong (n, %) | |||
| Normal mucosa | 173 | 117 (67.6) | 50 (28.9) | 6 (3.5) | |
| Cancerous tissue | 173 | 32 (18.5) | 99 (57.2) | 42 (24.3) | < 0.001a |
| LNM tissue | 54 | 5 (9.3) | 18 (33.3) | 31 (57.4) | < 0.001b |
eIF3e, eukaryotic translation initiation factor 3 subunit E; LNM, lymph node metastasis.
Significantly different eIF3e expression between cancerous tissues and normal mucosa;
significantly different eIF3e expression between LNM tissues and normal mucosa.
Relationship of eIF3e expression with clinicopathological features in colon cancer
The relationship between eIF3e expression and clinicopathological features is displayed in (Table 3). As shown, we found no significant differences between eIF3e expression and age, gender or tumor location (P > 0.05). On the contrary, eIF3e expression was significantly correlated with T classification (P < 0.001), lymph node involvement (P < 0.001), distant metastasis (P < 0.001), clinical stage (P < 0.001), differentiation (P < 0.001), and vessel invasion (P = 0.036). These data strongly indicated that eIF3e expression play an important role in the progression of colon cancer.
Table 3.
Associations of eIF3e expression with clinicopathological features in colon cancer (n = 173)
| Variables | n | eIF3e expression | P value | ||
|---|---|---|---|---|---|
|
| |||||
| Negative (n = 32) | Weak (n = 99) | Strong (n = 42) | |||
| Age | |||||
| < 65 y | 71 | 10 | 44 | 17 | 0.417 |
| ≥ 65 y | 102 | 22 | 55 | 25 | |
| Gender | |||||
| Male | 73 | 14 | 42 | 17 | 0.959 |
| Female | 100 | 18 | 57 | 25 | |
| Location | |||||
| Right | 73 | 10 | 46 | 17 | 0.209 |
| Transverse | 13 | 2 | 10 | 1 | |
| Left | 87 | 20 | 43 | 24 | |
| T stage | |||||
| T1 | 7 | 3 | 3 | 1 | < 0.001 |
| T2 | 21 | 8 | 12 | 1 | |
| T3 | 64 | 11 | 45 | 8 | |
| T4 | 81 | 10 | 39 | 32 | |
| N stage | |||||
| N0 | 90 | 24 | 55 | 11 | < 0.001 |
| N1 | 55 | 7 | 34 | 14 | |
| N2 | 28 | 1 | 10 | 17 | |
| M stage | |||||
| M0 | 157 | 31 | 96 | 30 | < 0.001 |
| M1 | 16 | 1 | 3 | 12 | |
| AJCC Stage | |||||
| I | 21 | 9 | 10 | 2 | < 0.001 |
| II | 67 | 15 | 44 | 8 | |
| III | 69 | 7 | 42 | 20 | |
| IV | 16 | 1 | 3 | 12 | |
| Differentiation | |||||
| Well | 82 | 24 | 50 | 8 | < 0.001 |
| Moderate | 63 | 6 | 38 | 19 | |
| Poorly | 28 | 2 | 11 | 15 | |
| Vessel invasion | |||||
| No | 162 | 30 | 96 | 36 | 0.036 |
| Yes | 11 | 2 | 3 | 6 | |
eIF3e, Eukaryotic translation initiation factor 3 subunit E; AJCC, American Joint Committee on Cancer. P values are based on chi-square test or Fisher’s Exact test if necessary.
Prognostic value of eIF3e expression in colon cancer patients
Kaplan-Meier curve analysis indicated that patients with high eIF3e expression were associated with poor prognosis (Figure 3). The estimated mean OS was 86.5 months, 72.67 months and 37.1 months in patients with negative, weak and strong eIF3e expression, respectively. Similar results were observed while analyzing DFS in patients with different eIF3e expression (84.33 months, 66.69 months and 29.14 months).
Figure 3.
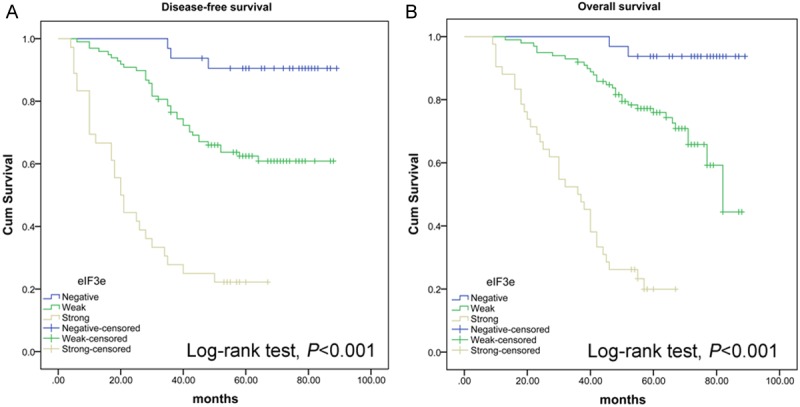
High eIF3e expression was associated with poor prognosis of colon cancer patients. A Kaplan-Meier analysis of: (A) disease-free survival (n = 173) and (B) overall survival (n = 173) in colon cancer patients according to the expression level of eIF3e. Patients with higher eIF3e expression had a significantly poorer prognosis.
To further evaluate the prognostic value of eIF3e expression in colon cancer patients, we performed univariate analysis for clinicopathological features and eIF3e expression level. In univariate analysis, lymph node involvement, distant metastasis, clinical stage, histopathological grade, vessel invasion and eIF3e expression were associated with both DFS and OS (Tables 4 and 5). Multivariate analysis was carried out using the Cox proportional hazards model for significant variables in univariate analysis. N stage and M stage were excluded from the multivariate analysis for their collinearity with AJCC stage. As shown in Tables 4 and 5, high eIF3e expression remained significantly prognostic for decreased survival and increased disease recurrence. Strong eIF3e expression was associated with approximately 10.3-fold risk of disease recurrence than negative eIF3e expression. Meanwhile, patients with strong eIF3e suffer nearly 17-fold risk of decreased survival than those with negative eIF3e expression.
Table 4.
Cox regression model analysis of disease-free survival (DFS) in 173 colon cancer patients
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | ||||
| < 65 y | 1 | |||
| ≥ 65 y | 1.133 (0.695, 1.848) | 0.616 | ||
| Gender | ||||
| Male | 1 | |||
| Female | 1.198 (0.737, 1.948) | 0.466 | ||
| Location | ||||
| Right | 1 | |||
| Transverse | 1.082 (0.417, 2.810) | 0.872 | ||
| Left | 1.055 (0.640, 1.738) | 0.834 | ||
| T stage | ||||
| T1 | 1 | |||
| T2 | 0.463 (0.077, 2.772) | 0.399 | ||
| T3 | 1.066 (0.248, 4.576) | 0.932 | ||
| T4 | 2.784 (0.674, 11.490) | 0.157 | ||
| N stage | ||||
| N0 | 1 | NR | ||
| N1 | 2.911 (1.622, 5.223) | < 0.001 | ||
| N2 | 10.685 (5.699, 20.034) | < 0.001 | ||
| M stage | ||||
| M0 | 1 | NR | ||
| M1 | 9.400 (4.489, 19.683) | < 0.001 | ||
| AJCC stage | ||||
| I | 1 | 1 | ||
| II | 1.726 (0.503, 5.924) | 0.386 | 1.435 (0.416, 4.944) | 0.567 |
| III | 6.017 (1.857, 19.495) | 0.003 | 4.092 (1.250, 13.395) | 0.020 |
| IV | 31.329 (8.245, 119.048) | < 0.001 | 11.759 (2.535, 54.556) | 0.002 |
| Differentiation | ||||
| Well | 1 | 1 | ||
| Moderate | 2.133 (1.221, 3.728) | 0.008 | 1.347 (0.749, 2.423) | 0.320 |
| Poorly | 4.967 (2.604, 9.477) | < 0.001 | 2.259 (1.021, 4.999) | 0.044 |
| Vessel invasion | ||||
| No | 1 | 1 | ||
| Yes | 3.173 (1.508, 6.673) | 0.002 | 0.851 (0.343, 2.108) | 0.727 |
| eIF3e | ||||
| Negative | 1 | 1 | ||
| Weak | 4.817 (1.484, 15.631) | 0.009 | 3.581 (1.079, 11.884) | 0.037 |
| Strong | 19.343 (5.842, 64.047) | < 0.001 | 10.281 (2.959, 35.728) | < 0.001 |
HR, Hazard ratio; CI, confidence interval; NR, variable was not include in the resultant model; P < 0.05 indicated that the 95% CI of HR was not including 1.
Table 5.
Cox regression model analysis of overall survival (OS) in 173 colon cancer patients
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | ||||
| < 65 y | 1 | |||
| ≥ 65 y | 1.212 (0.732, 2.006) | 0.455 | ||
| Gender | ||||
| Male | 1 | |||
| Female | 1.316 (0.794, 2.179) | 0.287 | ||
| Location | ||||
| Right | 1 | |||
| Transverse | 1.076 (0.413, 2.805) | 0.881 | ||
| Left | 1.011 (0.606, 1.687) | 0.967 | ||
| T stage | ||||
| T1 | 1 | |||
| T2 | 0.262 (0.037, 1.868) | 0.181 | ||
| T3 | 0.761 (0.174, 3.334) | 0.717 | ||
| T4 | 2.512 (0.609, 10.362) | 0.203 | ||
| N stage | ||||
| N0 | 1 | NR | ||
| N1 | 4.447 (2.278, 8.684) | < 0.001 | ||
| N2 | 16.131 (8.069, 32.245) | < 0.001 | ||
| M stage | ||||
| M0 | 1 | NR | ||
| M1 | 13.222 (7.068, 24.734) | < 0.001 | ||
| AJCC stage | ||||
| I | 1 | 1 | ||
| II | 1.548 (0.334, 7.176) | 0.576 | 1.133 (0.243, 5.276) | 0.874 |
| III | 8.786 (2.113, 36.530) | 0.003 | 5.183 (1.232, 21.803) | 0.025 |
| IV | 57.871 (12.843, 260.767) | < 0.001 | 15.843 (3.033, 82.751) | 0.001 |
| Differentiation | ||||
| Well | 1 | 1 | ||
| Moderate | 2.140 (1.153-3.975) | 0.016 | 1.174 (0.613, 2.247) | 0.628 |
| Poorly | 7.853 (4.142-14.889) | < 0.001 | 2.928 (1.331, 6.441) | 0.008 |
| Vessel invasion | ||||
| No | 1 | 1 | ||
| Yes | 3.926 (1.989, 7.752) | < 0.001 | 0.801 (0.359, 1.784) | 0.587 |
| eIF3e | ||||
| Negative | 1 | 1 | ||
| Weak | 6.979 (1.647, 29.573) | 0.008 | 4.940 (1.141, 21.379) | 0.033 |
| Strong | 40.933 (9.437, 177.537) | < 0.001 | 17.027 (3.744, 77.424) | < 0.001 |
HR, Hazard ratio; CI, confidence interval; NR, variable was not include in the resultant model; P < 0.05 indicated that the 95% CI of HR was not including 1.
Knockdown of eIF3e expression leads to the inhibition of colon cancer cell proliferation
As high eIF3e expression is associated with poor prognosis of colon cancer patients, we knocked down the eIF3e expression in human colon cancer cell line to investigate the biological behavior variation. After lentiviral-mediated RNAi in HCT116, which is a human colon cell line, cell proliferation variation was evaluated by cellomics (Figure 4). As shown in Figure 4, the cell count in the knockdown group was significantly reduced in comparison with the negative control. The results indicated that downregulation of eIF3e gene inhibited the proliferation of HCT116 cells.
Figure 4.
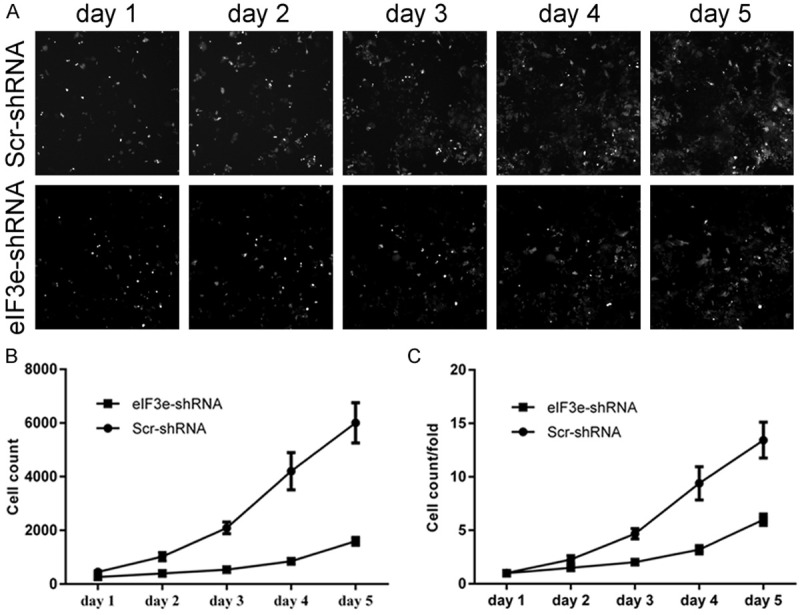
Downregulation of eIF3e inhibits the proliferation of HCT116 cells. A. Cellomics demonstrates the cell growth in Scr-shRNA and eIF3e-shRNA cells. B. Proliferation volume of HCT116 cells in eIF3e-shRNA cells is reduced with the Scr-shRNA cells. C. Proliferation fold change of HCT116 cells in eIF3e-shRNA group is decreased with the Scr-shRNA group.
Knockdown of eIF3e expression leads to the inhibition of colon cancer cell clonality
Since eIF3e gene silencing was associated with a decrease in the cell count of HCT116 cell, we performed plate colony formation assay to investigate the variation of clonality. The results showed that the clone number decreased, which was in obvious contrast to the negative control group (Figure 5, P < 0.01). These findings suggest that downregualtion of eIF3e expression signally inhibited the clonality of HCT116.
Figure 5.
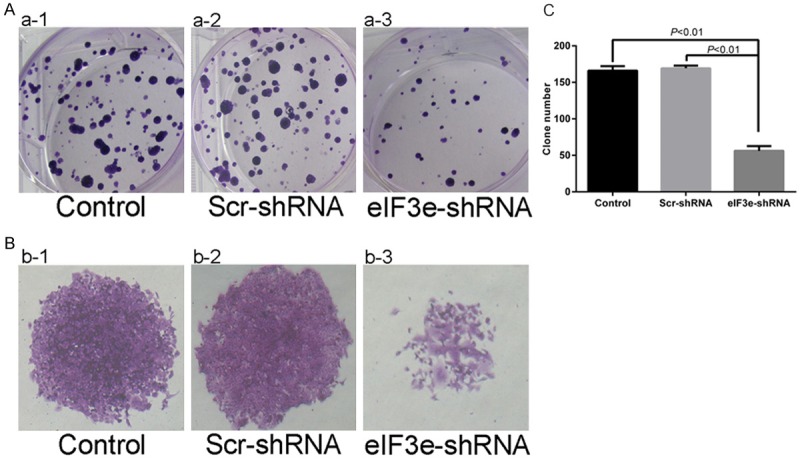
Downregulation of eIF3e inhibits clonality of HCT116. A and B. Plate colony formation assay reveals the weakness of clonality in eIF3e-shRNA HCT116 cells. C. Significant differences of clone number between control and eIF3e-shRNA cells (P < 0.01), Scr-shRNA and eIF3e-shRNA cells (P < 0.01).
Knockdown of eIF3e expression reduce the apoptosis of colon cancer cell
FACS analysis was performed to further explore the effect of eIF3e expression on cell apoptosis. The apoptosis percentage of HCT116 cells was significantly increased in eIF3e-shRNA cells compared to Scr-shRNA (Figure 6, P < 0.01). It indicates that knockdown of eIF3e gene promotes the apoptosis of HCT116 cells.
Figure 6.
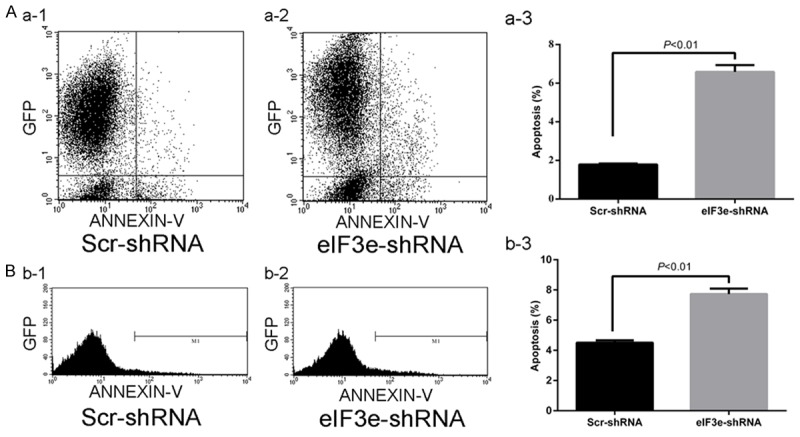
Downregulation of eIF3e gene promotes apoptosis of HCT116. A and B. Scatter diagram and Peak diagram shows the significant differences of apoptosis percentage between Scr-shRNA and eIF3e-shRNA cells.
Discussion
According to our digital gene expression profiling data by LCM-LongSAGE, 6 eIF subunits (eIF2G, eIF3B, eIF3E, eIF3I, eIF4E and eIF5A) were significantly overexpressed in colon cancer cells (Table 1). The contribution of eIF4E to cellular transformation, tumorigenesis and metastasis has been well documented over the past few years. EIF-4E was overexpressed in a variety of human tumors, and has been considered as a therapeutic target and as a marker for human cancer progression [18]. Decreased expression of EIF3B gene resulted in the inhibition of human colon cancer cell proliferation [19]. Overexpression of eIF3I drives colon oncogenesis by directly upregulating the synthesis of cyclooxygenase-2 (COX-2) protein and activating the β-catenin signaling pathway [20]. The EIF5A mRNA expression level was a significant prognostic factor in CRC patients [21]. The function and clinical significance of eIF2G in cancer has not been reported. We can infer that the abnormal expression of eIF subunits was an important molecular event in colon cancer progression.
Regulation of the activity of eIF3 by its subunits resulted in hypo- and hyperactivation of the rate of the protein synthesis. A suitable level of translation initiation is necessary to regulate proliferation and cell cycle and contributes importantly to oncogenic transformation [22]. Tumorigenesis in diverse tissue types is often associated with alterations in the expression of various eIF3 subunits [23]. The expression and function of eIF3e, an up-regulated gene in our expression profiling database, is not yet characterized in colon carcinoma [13]. In the present study, the elevated expression of elF3e at protein level in colon adenocarcinoma tissues was validated by immunohistochemistry staining. The overexpression of eIF3e protein was significantly associated with worse overall survival time and disease free survival time of colon cancer patients. Knockdown of eIF3e inhibits proliferation and promotes apoptosis in colon cancer cells. To our knowledge, this is the first study showed that eIF3e might be a colon cancer-related gene and its value as a potential novel prognostic marker of the disease.
As a component of the eIF3 translation initiation factor, the pattern of protein synthesis changes mediated by eIF3e abnormal expression should be clarified in carcinogenesis. Recently, a microarray analysis revealed that mRNAs regulated either positively or negatively by eIF3e in human breast cancer cells were markedly enriched in genes associated with cell proliferation, invasion and apoptosis. In addition, RT–PCR analysis of RNA immunoprecipitated using an eIF3b-specific anti-body showed that apoptotic regulator BCLXL mRNA was decreased and the mitotic checkpoint component MAD2L1 mRNA enriched following eIF3e knockdown in MDA-MB-231 cells. Finally, eIF3e-depleted breast carcinoma cells showed reduced in vitro invasion and proliferation. These data suggest that eIF3e has an oncogenic role in breast cancer progression [24]. The target genes regulated by eIF3e in colon cancer cells should be screened and validated in further study.
The potential mechanisms of eIF3e in tumorigenesis were explored in some studies. EIF3e was identified as a tissue-specific modulator of MEK-ERK signaling through genetic and chemical-genetic approaches [25]. The MEK protein levels reduced in the eIF3e-siRNA transfected cells. MEK, a dual specificity kinase, is a key player in this pathway; it is downstream of both Ras and Raf and activates ERK1/2 through phosphorylation of key tyrosine and threonine residues [26]. Aberrant regulation of the MEK-ERK pathway can contribute to uncontrolled cell growth and lead to malignant transformation [27]. The small-molecule targeting the MEK-ERK pathway was potential method for the treatment of cancer [28]. A study showed that eIF3e suppression affects cell proliferation, cell cycle and apoptosis of various Glioblastomas (GBM) cells. These phenotypes are independent of global cell translation inhibition and are accompanied by decreased HIF expression when eIF3e is silenced [17]. Further study should been made to clarify the precise role of eIF3e in colon cancer progression.
In conclusion, this study reveals the clinical significance of eIF3e expression in colon cancer patients for the first time. Our data suggests that eIF3e is an independent prognostic factor in colon cancer patients influencing DFS and OS. Experiments in vitro indicate that eIF3e promotes colon cancer cell proliferation, colony formation, and inhibits apoptosis, which can partially explain the mechanism of eIF3e to promote tumor progression and predict poor prognosis. These results may provide important diagnostic and therapeutic value of eIF3e in colon cancer patients in the future.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Number 30901423 and 81272744), Medical engineering crossing project grant funded by Shanghai Jiaotong University (YG2011MS59).
Disclosure of conflict of interest
None.
References
- 1.Neri E, Faggioni L, Cini L, Bartolozzi C. Colonic polyps: inheritance, susceptibility, risk evaluation, and diagnostic management. Cancer Manag Res. 2010;3:17–24. doi: 10.2147/CMR.S15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanavos P. The rising burden of cancer in the developing world. Ann Oncol. 2006;17(Suppl 8):viii15–viii23. doi: 10.1093/annonc/mdl983. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 4.Jiang T, Tang HM, Lu S, Yan DW, Yang YX, Peng ZH. Up-regulation of tripartite motif-containing 29 promotes cancer cell proliferation and predicts poor survival in colorectal cancer. Med Oncol. 2013;30:715. doi: 10.1007/s12032-013-0715-4. [DOI] [PubMed] [Google Scholar]
- 5.Mathonnet M, Perraud A, Christou N, Akil H, Melin C, Battu S, Jauberteau MO, Denizot Y. Hallmarks in colorectal cancer: angiogenesis and cancer stem-like cells. World J Gastroenterol. 2014;20:4189–4196. doi: 10.3748/wjg.v20.i15.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangadhar T, Schilsky RL. Molecular markers to individualize adjuvant therapy for colon cancer. Nat Rev Clin Oncol. 2010;7:318–325. doi: 10.1038/nrclinonc.2010.62. [DOI] [PubMed] [Google Scholar]
- 7.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, Morikawa T, Shen J, Meyerhardt JA, Fuchs CS, Ogino S. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zheng T. Screening of hub genes and pathways in colorectal cancer with microarray technology. Pathol Oncol Res. 2014;20:611–618. doi: 10.1007/s12253-013-9739-5. [DOI] [PubMed] [Google Scholar]
- 9.Li D, Yan D, Tang H, Zhou C, Fan J, Li S, Wang X, Xia J, Huang F, Qiu G, Peng Z. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16:3499–3506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Yan D, Teng M, Fan J, Zhou C, Li D, Qiu G, Sun X, Li T, Xing T, Tang H, Peng X, Peng Z. Reduced expression of PER3 is associated with incidence and development of colon cancer. Ann Surg Oncol. 2012;19:3081–3088. doi: 10.1245/s10434-012-2279-5. [DOI] [PubMed] [Google Scholar]
- 11.Yan DW, Fan JW, Yu ZH, Li MX, Wen YG, Li DW, Zhou CZ, Wang XL, Wang Q, Tang HM, Peng ZH. Downregulation of metallothionein 1F, a putative oncosuppressor, by loss of heterozygosity in colon cancer tissue. Biochim Biophys Acta. 2012;1822:918–926. doi: 10.1016/j.bbadis.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 12.Fan J, Peng Z, Zhou C, Qiu G, Tang H, Sun Y, Wang X, Li Q, Le X, Xie K. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses: identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer. 2008;113:266–275. doi: 10.1002/cncr.23551. [DOI] [PubMed] [Google Scholar]
- 13.Fan J, Yan D, Teng M, Tang H, Zhou C, Wang X, Li D, Qiu G, Peng Z. Digital transcript profile analysis with aRNA-LongSAGE validates FERMT1 as a potential novel prognostic marker for colon cancer. Clin Cancer Res. 2011;17:2908–2918. doi: 10.1158/1078-0432.CCR-10-2552. [DOI] [PubMed] [Google Scholar]
- 14.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 15.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershey JW. Regulation of protein synthesis and the role of eIF3 in cancer. Braz J Med Biol Res. 2010;43:920–930. doi: 10.1590/s0100-879x2010007500098. [DOI] [PubMed] [Google Scholar]
- 17.Sesen J, Cammas A, Scotland SJ, Elefterion B, Lemarie A, Millevoi S, Mathew LK, Seva C, Toulas C, Moyal EC, Skuli N. Int6/eIF3e is essential for proliferation and survival of human glioblastoma cells. Int J Mol Sci. 2014;15:2172–2190. doi: 10.3390/ijms15022172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Chen J, Sun J, Cui Z, Wu H. RNA interference-mediated silencing of eukaryotic translation initiation factor 3, subunit B (EIF3B) gene expression inhibits proliferation of colon cancer cells. World J Surg Oncol. 2012;10:119. doi: 10.1186/1477-7819-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi J, Dong Z, Liu J, Zhang JT. EIF3i promotes colon oncogenesis by regulating COX-2 protein synthesis and beta-catenin activation. Oncogene. 2014;33:4156–63. doi: 10.1038/onc.2013.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tunca B, Tezcan G, Cecener G, Egeli U, Zorluoglu A, Yilmazlar T, Ak S, Yerci O, Ozturk E, Umut G, Evrensel T. Overexpression of CK20, MAP3K8 and EIF5A correlates with poor prognosis in early-onset colorectal cancer patients. J Cancer Res Clin Oncol. 2013;139:691–702. doi: 10.1007/s00432-013-1372-x. [DOI] [PubMed] [Google Scholar]
- 22.Goh SH, Hong SH, Lee BC, Ju MH, Jeong JS, Cho YR, Kim IH, Lee YS. eIF3m expression influences the regulation of tumorigenesis-related genes in human colon cancer. Oncogene. 2011;30:398–409. doi: 10.1038/onc.2010.422. [DOI] [PubMed] [Google Scholar]
- 23.Joseph P, O’Kernick CM, Othumpangat S, Lei YX, Yuan BZ, Ong TM. Expression profile of eukaryotic translation factors in human cancer tissues and cell lines. Mol Carcinog. 2004;40:171–179. doi: 10.1002/mc.20033. [DOI] [PubMed] [Google Scholar]
- 24.Grzmil M, Rzymski T, Milani M, Harris AL, Capper RG, Saunders NJ, Salhan A, Ragoussis J, Norbury CJ. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene. 2010;29:4080–4089. doi: 10.1038/onc.2010.152. [DOI] [PubMed] [Google Scholar]
- 25.Grzmil M, Whiting D, Maule J, Anastasaki C, Amatruda JF, Kelsh RN, Norbury CJ, Patton EE. The INT6 cancer gene and MEK signaling pathways converge during zebrafish development. PLoS One. 2007;2:e959. doi: 10.1371/journal.pone.0000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace EM, Lyssikatos JP, Yeh T, Winkler JD, Koch K. Progress towards therapeutic small molecule MEK inhibitors for use in cancer therapy. Curr Top Med Chem. 2005;5:215–229. doi: 10.2174/1568026053507723. [DOI] [PubMed] [Google Scholar]
- 27.McCubrey JA, Milella M, Tafuri A, Martelli AM, Lunghi P, Bonati A, Cervello M, Lee JT, Steelman LS. Targeting the Raf/MEK/ERK pathway with small-molecule inhibitors. Curr Opin Investig Drugs. 2008;9:614–630. [PubMed] [Google Scholar]
- 28.Friday BB, Adjei AA. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin Cancer Res. 2008;14:342–346. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]


