Abstract
Background
Family history of prostate cancer is a well-recognized risk factor. Previous linkage studies have reported a putative prostate cancer susceptibility locus at chromosome 17q21-22. SPOP (Speckle-type POZ protein)maps to the 17q21-22 candidate linkage region and is one of the most frequently mutated genes in sporadic prostate cancers.
Methods
We performed targeted next generation sequencing to analyze 2009 exons from 202 genes in a candidate linkage region on chromosome 17q21-22 using 94 unrelated familial prostate cancer cases from the University of Michigan Prostate Cancer Genetics Project (n= 54) and Johns Hopkins University (n=40) including the exons and UTRs of SPOP.
Results
We identified anovel SPOP missense mutation (N296I)in a man with prostate cancer diagnosed at age 43. This mutation completely segregates with prostate cancer affection status among the men in this family. The N296I mutation resides within the evolutionarily conserved Bric-a-brac, Tram track, Broad-complex (BTB) domain, involved in recruiting targets to Cul3 for degradation. Analysis of the prostate tumor from this individual verified the presence of heterozygous N296I as well as an ERG fusion.
Conclusions
We have discovered a novel mutation in SPOP that tracks with prostate cancer within a family and is predicted to be deleterious. Taken together, our results implicate SPOP as a candidate gene for hereditary prostate cancer.
Keywords: familial, gene, candidate linkage region
Introduction
Prostate cancer is the most common non-cutaneous cancer diagnosed among American men and the second leading cause of cancer death with an estimated 233,000 new cases and 29,480 deaths expected in the United States in 2014. Known risk factors for prostate cancer are increasing age, African American race, and positive family history of the disease. Studies performed with the objective of elucidating a heritable component have succeeded in identifying a region of genetic susceptibility on chromosome 17q first reported by Lange et al.[1] on the basis of linkage analysis of 175 pedigrees of families with hereditary prostate cancer from the University of Michigan Prostate Cancer Genetics Project (UM-PCGP). Fine mapping of chromosome 17q, using 453 pedigrees from the UM-PCGP and Johns Hopkins University (JHU) refined this region of interest to chromosome 17q21-22[2]. Further analysis of a subset of 147 families with four or more affected men and an average age of prostate cancer diagnosis ≤ 65 years narrowed the candidate interval (1-LOD support interval) for a putative susceptibility gene to a 10cM region of 17q that contains over 75 known genes. We have previously reported the identification of a rare but recurrent G84E (rs138213197) missense mutation in HOXB13 from the chromosome 17q-linked families described herein [3]. Carriers of G84E were reported to have a 10-20 fold increased risk of developing prostate cancer with the highest frequency in men with early-onset and familial prostate cancer. Subsequent studies have confirmed this report providing further evidence that rare genetic variants play a role in prostate cancer susceptibility [4–13]. Genome wide association studies of prostate cancer in populations of men of African and European descent have also identified risk variantswithin17q21. Specifically, rs7210100[14] andrs1165049[15] located in intron 1 and downstream of ZNF652 respectively. ZNF652 is located within a gene-dense region of 17q21 that harbors prostate cancer susceptibility genes HOXB13PRAC and SPOP. To date, chromosome 17q21-22 remains one of the most reproducible linkage regions for prostate cancer susceptibility loci.
The Speckle-type POZ protein or SPOP gene, which maps to the 17q21-22 candidate linkage region, is one of the most frequent somatically mutated genes in prostate cancer. Initially described by Kan et al. [16], SPOP mutations have been observed in (2/58) [16]and 14/111[17] prostate cancers and(2/7) tumors from men with high risk disease [18]. SPOP encodes the substrate-binding subunit of a Cullin-based E3 ubiquitin ligase. Studies using the MCF-7 breast cancer cell line have shown SPOP directly impacts cancer cell growth and invasion, suggesting that SPOP may function as a tumor suppressor gene (TSG) in breast and possibly other cancers[19]. Tissue microarray screening for SPOP expression in 18 cancer types from different organs revealed high expression of SPOP in kidney, endometrial, and germ cell cancers when compared to normal tissues [20]. Additionally, recent evidence has shown that the previously reported prostate-cancer-associated mutants of SPOP cannot promote androgen receptor (AR) ubiquitination [21], suggesting that SPOP is part of the degradation system for AR, an important driver of prostate carcinogenesis.
In the present study, we set out to analyze SPOP germline sequence data from 94 familial prostate cancer cases with evidence of linkage to chromosome 17 compiled at the University of Michigan and the Johns Hopkins University.
Methods
Patient Selection
University of Michigan Prostate Cancer Genetics Project (UM-PCGP)
UM-PCGP prostate cancer cases were restricted to (1) men diagnosed with prostate cancer with at least one living first- or second-degree relative also diagnosed with prostate cancer or (2) men diagnosed with prostate cancer at <56 years of age. We confirmed the diagnosis of prostate cancer by medical record review whenever possible. All subjects provided written informed consent to participate in the study. The protocol and consent documents were approved by the University of Michigan Medical School Institutional Review Board Health Insurance Portability and Accountability Act (HIPAA) regulations and all subjects gave written informed consent.
Johns Hopkins University (JHU)
Hereditary prostate cancer (HPC) families each had at least three first-degree relatives affected with prostate cancer. We verified diagnosis of prostate cancer by medical records.) The protocol and consent documents were approved by the Johns Hopkins School of Medicine Institutional Review Board and all subjects gave written informed consent.
Discordant Sibling Pairs
The details of the discordant sibling pair (DSP) project have been described elsewhere [22]. For the present study, 569 families were identified in which DNA was available from at least one pair of brothers discordant for prostate cancer.
Targeted sequencing of SPOP
We selected the youngest prostate cancer case with available DNA from 94 prostate cancer families (40 families from JHU and 54 from the UM-PCGP) as described previously [23]. Seven of the families were of African descent, 2 were of Asian descent, and the remaining 85 described themselves as being of European descent. A primer library was designed for amplification of 7053 base pairs (bp) of SPOP including all exons, intron/exon boundaries, and the 5’ and 3’ untranslated regions. We then used the Rain Dance RDT 1000 system (Rain Dance Technologies, Inc., Lexington, MA) to amplify 3 µg of sheared genomic DNA from each sample using our primer library. Purified amplicons were used as template for sequencing using the Life Technologies SOLiD™ system version 4.0 fragment library methodology (Life Technologies Corporation, Carlsbad, CA). Sequence data processing was performed using Bioscope to align the sequences to the genomic reference (Build 36, hg18). Variant detection was performed using Sam Tools 1.3[24] and Sol SNP 1.1. We confirmed and tested all variant sequences in family members using standard Sanger sequencing, capillary electrophoresis technology and BigDye® Terminator chemistry (Life Technologies, Carlsbad, CA).
Tumor Sequencing
Prostate tumor DNA from the index case was extracted from using the QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA). Sanger sequencing of N296I was performed using custom SPOP primers as described above.
Immunohistochemistry
Hematoxylin and Eosin (H&E) slides from the prostatectomy of the index case were reviewed and re-graded according to current ISUP guidelines. ERG immunostaining was performed essentially as described [25, 26] using pre-diluted ERG antibody (provided by Ventana Medical Systems) using the automated Discovery XT staining platform (Ventana Medical Systems).
Genotyping Assays
We genotyped N296I in the DSP cohort using a custom TaqMan SNP assay (Life Technologies). Allelic discrimination was performed on an ABI Prism 7900HT Sequence Detection System and SDS version 2.1 software (Applied Biosystems). Genotyping call rate was 99%.
Results
Molecular study result of the index case
Targeted next-generation sequencing revealed a novel heterozygous 1358 A>T mutation (Figure 1) in an individual of European descent diagnosed with Gleason 3+4=7 prostate cancer at age 43 (NM_001007226). The missense mutation is a single A to T trans version in exon 11resulting in an amino acid substitution of asparagine to isoleucine at codon 296 (N296I). This variant was confirmed by Sanger sequencing. No additional SPOP missense variants were observed in the remaining 93 HPC probands.
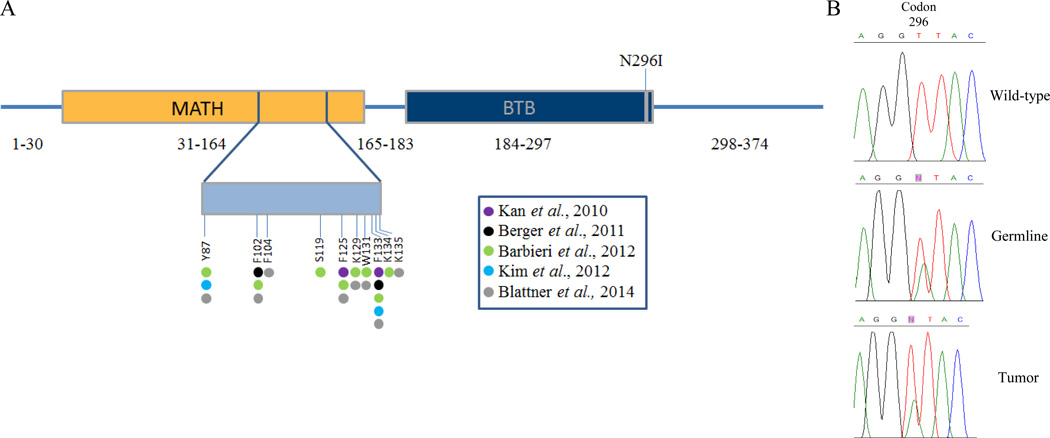
Figure 1. Location of the SPOP N296I mutation.
A. The 374 amino acid SPOP protein consists of an N-terminal meprin and TRAF-homology (MATH) domain, a BTB substrate binding, otherwise known as a pox virus and zinc finger (POZ), domain, and a C-terminal nuclear localization sequence. The SPOP N296I mutation resides in the Bric-a-brac, Tramtrack, Broad-complex (BTB) binding domain. The somatic mutations identified in prostate tumors are located in the MATH domain. B) Chromatograms showing the presence of the N296I missense mutation in both germline and tumor DNA from our index case.
Molecular study result of other family members
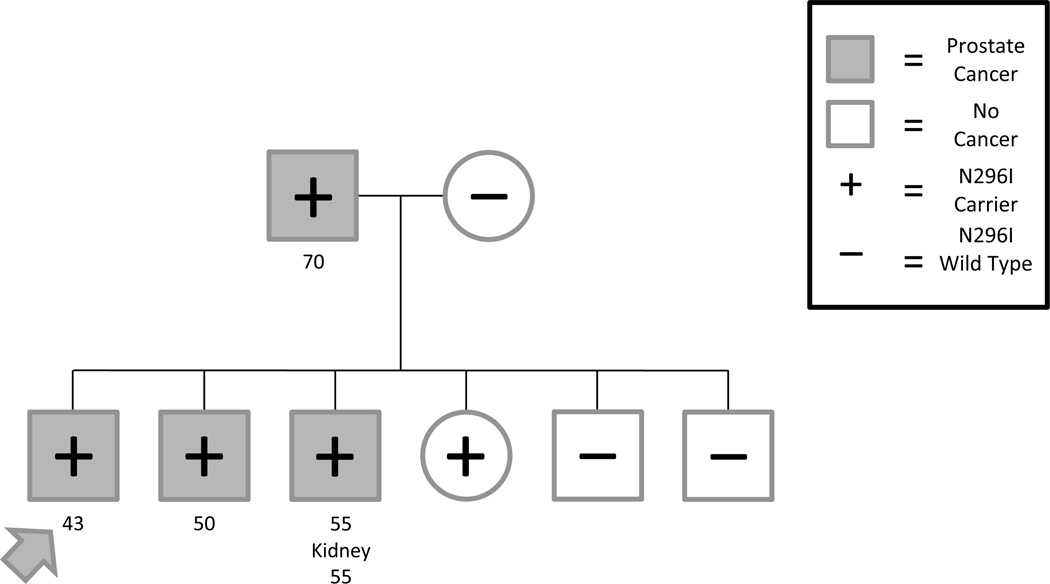
Sanger sequencing of additional family members revealed complete segregation of N296I and prostate cancer affection status amongst the men in this family (Figure 2). Sequence data from five additional male relatives revealed that the father and a brother with prostate cancer carry the mutation, as does another brother affected with both prostate and kidney cancer. Two unaffected brothers, ages 46 and 56, do not carry the N296I allele. Female relatives with DNA were also genotyped, the mother of the pro band did not carry the N296I, yet the sister of the pro band was positive for the variant. To our knowledge, neither of the female relatives has been diagnosed with cancer.
Figure 2.
Pedigree of the family harboring the SPOP N296I mutation. The pro band initially selected for sequencing is indicated by the arrow. With the exception of the pro band, the ages of diagnosis have been rounded to the nearest 5-year interval and are shown under the subject.
Molecular and histopathologic characterization of the prostate tumor
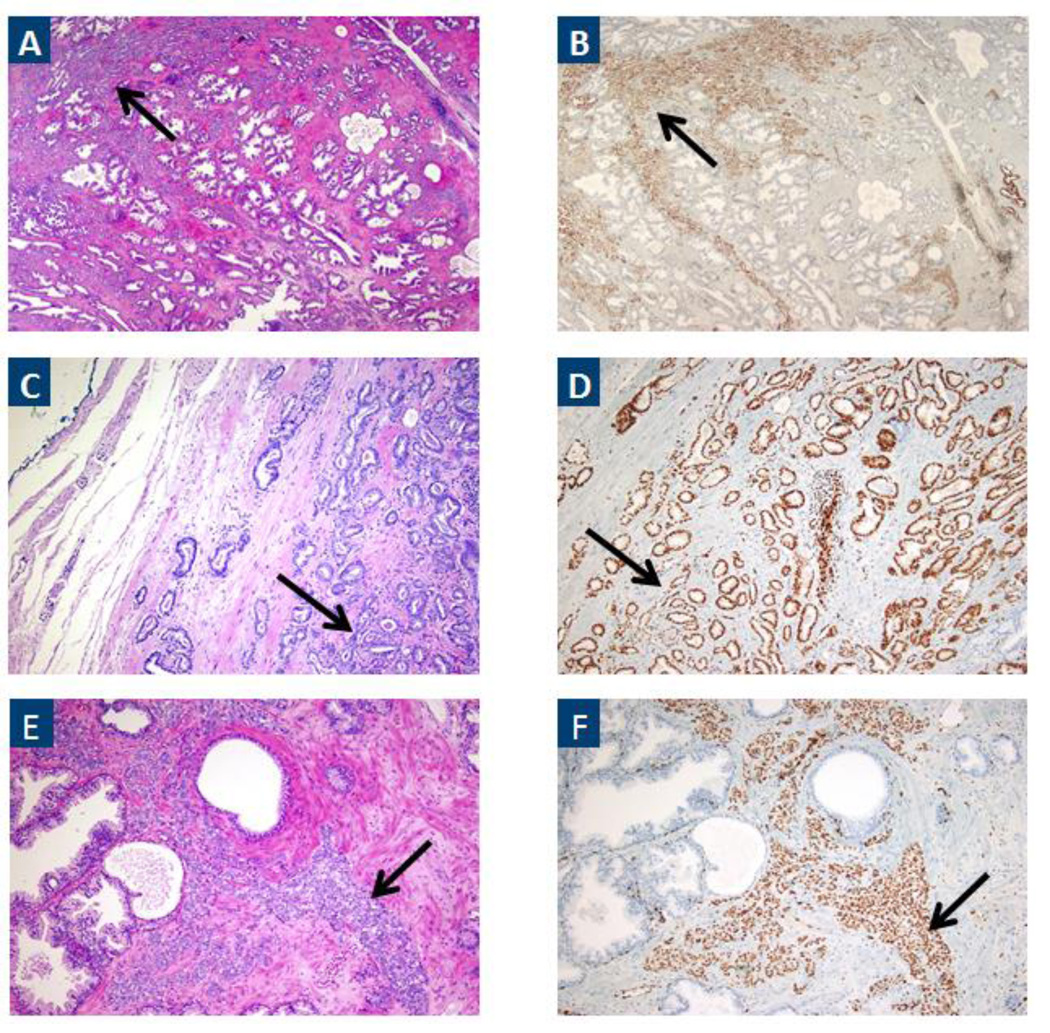
Sanger sequencing of DNA extracted from archival tissue confirmed the presence of the heterozygous N296I variant in the prostate tumor from the pro band. Additionally, ERG expression was observed in a Gleason 3+4=7 focus of prostate cancer from the index case consistent with the presence of an ERG rearrangement[27] (Figure 3). SPOP expression was also observed in the prostate tissue (data not shown).
Figure 3.
A) H&E staining of the index focus (Gleason score 3+4=7) on prostatectomy. Prostatic adenocarcinoma is indicated by the black arrow. B) Consecutive sections were stained using a monoclonal antibody against ERG (EPR3864). Specific, diffuse moderate to strong nuclear staining was observed in cancer (black arrow). Original magnification 4X. C&D) As in A&B except higher power (10X) of an area of Gleason pattern 3 carcinoma. E&F) As in A & B, except higher power (10X) of an area of Gleason pattern 4 carcinoma.
Molecular study result of DSPs
No additional N296I carriers were detected amongst 786 affected and 654 unaffected men from 569 families.
Discussion
SPOP encodes a 374 amino acid protein that contains three domains: an N-terminal meprin and TRAF-homology (MATH) domain (amino acids 31-164), a Bric-a-brac, Tramtrack, Broad-complex (BTB) substrate binding, otherwise known as a pox virus and zinc finger (POZ), domain (amino acids 184-297) and a C-terminal nuclear localization sequence (amino acids 365-374)[28, 29]. SPOP missense mutations have been described in up to 15% of prostate tumors; classifying SPOP as one of the genes most commonly affected by somatic missense mutations in prostate cancer [16, 17, 30, 31]. Additionally, targeted sequencing of exons 6 (amino acids 80-106) and 7 (amino acids 120-140) of SPOP in prostate tumors from a cohort of demographically diverse patients showed an overall mutation rate of 8.1% [32]. In the present study, we describe the identification of a novel germline SPOP mutation (N296I) in a family with early-onset and hereditary prostate cancer that showed evidence of linkage to chromosome 17. The N296I mutation was found to completely co-segregate with prostate cancer disease status amongst the men in this family. Furthermore, a family member with both prostate and kidney cancer was also shown to carry N296I. Although tumor tissue from the kidney cancer was unavailable for analysis, it should be noted that SPOP has been previously implicated in renal cell carcinoma (RCC). Specifically, SPOP was shown to be over expressed in 77% (199/258) of RCCs, where as all normal kidney samples were negative [20]. In addition, staining for SPOP also indicated its utility as a highly specific and sensitive biomarker capable of distinguishing histological subtypes of RCCs.
Molecular characterization of prostate tumors has generated evidence of distinct molecular subtypes defined by the presence or absence of TMPRSS2-ERG fusions, SPINK1, and somatic SPOP non-synonymous point mutations [25, 33–39]. Previous studies have indicated that TMPRSS2-ERG fusions are mutually exclusive to SPINK1 over expression and somatic SPOP mutations. Immunohistochemistry for ERG expression revealed diffuse moderate to strong nuclear staining associated with ERG+ carcinoma in the tumor of our index case, which also harbored an N296I mutation. Although N296I is exceedingly rare, this report expands the knowledge of variation in SPOP and provides evidence in opposition of a mutually exclusive relationship between SPOP point mutations, specifically those in the BTB domain, and TMPRSS2-ERG fusions.
Recent studies of localized and advanced prostate tumors have identified a cluster of SPOP mutations exclusive to the MATH domain (Figure 1). Studies conducted to gain insight on the effect of these mutations have concluded SPOP mutants lack the ability to interact with or degrade SRC-3, thus attenuating the tumor suppressor gene function of wild-type SPOP [31]. SRC-3 is known to bind directly to the SPOP MATH domain prior to ubiquitination and degradation via the Cul3 based ubiquitin ligase complex [19]. Over-expression of SRC-3 has been noted in several human cancers including prostate, breast, and ovarian [40–43] and is often associated with poor prognosis [44, 45]. In prostate cancer, SRC-3 over expression is thought to promote tumorigenesis and more rapid progression to castrate resistant prostate cancer.
Recently, similar to SRC-3, SPOP has been shown to interact with AR in prostate cancer cells and target AR for degradation [21]. Interestingly, mutated alleles of SPOP found in prostate cancers were unable to bind or target AR for degradation, suggesting that the accumulation of AR due to this mechanism may play an important role in prostate carcinogenesis and perhaps disease progression. It will be of interest to determine if the N296I mutant observed in this study is similarly deficient in targeting AR, SRC-3, or other protein substrates for degradation.
The N296I mutation is located in a highly conserved region of the SPOP BTB domain which binds directly to Cul3, a scaffold protein required for substrate ubiquitination. Overall, this mutation appears to be rare, as it was not observed in our familial and early-onset prostate cancer DSP population nor was it seen in large public databases including the 1000 Genomes [46] project and the Exome Sequencing Project (ESP) which includes 4300 individuals of European ancestry and 2203 individuals of African American ancestry [47]. Despite its low frequency and its position outside of the structurally defined substrate-binding MATH domain, data exists in support of the notion that N296I may be involved in carcinogenesis. Geng et al. have recently demonstrated that expression vectors lacking the BTB domain cannot bind Cul3 and subsequently cannot promote the degradation of SRC-3 in prostate cancer PC-3 cells [31].
In summary, we have identified a novel germline SPOP mutation in a hereditary prostate cancer family which exhibits complete co-segregation with disease status. The early-on set nature of prostate cancer diagnoses and the co-occurrence of kidney cancer in one of the mutation carriers suggest that germline mutations in SPOP may be increasing cancer risk in this family. As additional hereditary prostate cancer families are studied, SPOP should be considered a candidate prostate cancer susceptibility gene worthy of additional focus.
References
- 1.Lange EM, Gillanders EM, Davis CC, Brown WM, Campbell JK, Jones MP, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Giri V, Beebe-Dimmer J, Montie JE, Trent JM, Cooney KA. Genome-wide scan for prostate cancer susceptibility genes using families from the University of Michigan Prostate Cancer Genetics Project finds evidence for linkage on chromosome 17 near BRCA1. The Prostate. 2003;57(4):326–334. doi: 10.1002/pros.10307. [DOI] [PubMed] [Google Scholar]
- 2.Lange EM, Robbins CM, Gillanders EM, Zheng SL, Xu J, Wang Y, White KA, Chang BL, Ho LA, Trent JM, Carpten JD, Isaacs WB, Cooney KA. Fine-mapping the putative chromosome 17q21-22 prostate cancer susceptibility gene to a 10 cM region based on linkage analysis. Hum Genet. 2007;121(1):49–55. doi: 10.1007/s00439-006-0274-2. [DOI] [PubMed] [Google Scholar]
- 3.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, Gielzak M, Partin AW, Shanmugam V, Izatt T, Sinari S, Craig DW, Zheng SL, Walsh PC, Montie JE, Xu J, Carpten JD, Isaacs WB, Cooney KA. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366(2):141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbari MR, Trachtenberg J, Lee J, Tam S, Bristow R, Loblaw A, Narod SA, Nam RK. Association Between Germline HOXB13 G84E Mutation and Risk of Prostate Cancer. Journal of the National Cancer Institute. 2012 doi: 10.1093/jnci/djs288. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson R, Aly M, Clements M, Zheng L, Adolfsson J, Xu J, Gronberg H, Wiklund F. A Population-based Assessment of Germline HOXB13 G84E Mutation and Prostate Cancer Risk. European Urology. 2012 doi: 10.1016/j.eururo.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 6.Breyer JP, Avritt TG, McReynolds KM, Dupont WD, Smith JR. Confirmation of the HOXB13 G84E germline mutation in familial prostate cancer. Cancer Epidemiol Biomarkers Prev. 2012;21(8):1348–1353. doi: 10.1158/1055-9965.EPI-12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Greenwood C, Isaacs WB, Foulkes WD, Sun J, Zheng SL, Condreay LD, Xu J. The G84E mutation of HOXB13 is associated with increased risk for prostate cancer: results from the REDUCE trial. Carcinogenesis. 2013;34(6):1260–1264. doi: 10.1093/carcin/bgt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laitinen VH, Wahlfors T, Saaristo L, Rantapero T, Pelttari LM, Kilpivaara O, Laasanen SL, Kallioniemi A, Nevanlinna H, Aaltonen L, Vessella RL, Auvinen A, Visakorpi T, Tammela TL, Schleutker J. HOXB13 G84E mutation in Finland: population-based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22(3):452–460. doi: 10.1158/1055-9965.EPI-12-1000-T. [DOI] [PubMed] [Google Scholar]
- 9.Stott-Miller M, Karyadi DM, Smith T, Kwon EM, Kolb S, Stanford JL, Ostrander EA. HOXB13 mutations in a population-based, case-control study of prostate cancer. Prostate. 2013;73(6):634–641. doi: 10.1002/pros.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Lange EM, Lu L, Zheng SL, Wang Z, Thibodeau SN, Cannon-Albright LA, Teerlink CC, Camp NJ, Johnson AM, Zuhlke KA, Stanford JL, Ostrander EA, Wiley KE, Isaacs SD, Walsh PC, Maier C, Luedeke M, Vogel W, Schleutker J, Wahlfors T, Tammela T, Schaid D, McDonnell SK, DeRycke MS, Cancel-Tassin G, Cussenot O, Wiklund F, Gronberg H, Eeles R, Easton D, Kote-Jarai Z, Whittemore AS, Hsieh CL, Giles GG, Hopper JL, Severi G, Catalona WJ, Mandal D, Ledet E, Foulkes WD, Hamel N, Mahle L, Moller P, Powell I, Bailey-Wilson JE, Carpten JD, Seminara D, Cooney KA, Isaacs WB. HOXB13 is a susceptibility gene for prostate cancer: results from the International Consortium for Prostate Cancer Genetics (ICPCG) Hum Genet. 2013;132(1):5–14. doi: 10.1007/s00439-012-1229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin X, Qu L, Chen Z, Xu C, Ye D, Shao Q, Wang X, Qi J, Zhou F, Wang M, Wang Z, He D, Wu D, Gao X, Yuan J, Wang G, Xu Y, Dong P, Jiao Y, Yang J, Ou-Yang J, Jiang H, Zhu Y, Ren S, Zhang Z, Yin C, Wu Q, Zheng Y, Turner AR, Tao S, Na R, Ding Q, Lu D, Shi R, Sun J, Liu F, Zheng SL, Mo Z, Sun Y, Xu J. A novel germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate. 2013;73(2):169–175. doi: 10.1002/pros.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kluzniak W, Wokolorczyk D, Kashyap A, Jakubowska A, Gronwald J, Huzarski T, Byrski T, Debniak T, Golab A, Gliniewicz B, Sikorski A, Switala J, Borkowski T, Borkowski A, Antczak A, Wojnar L, Przybyla J, Sosnowski M, Malkiewicz B, Zdrojowy R, Sikorska-Radek P, Matych J, Wilkosz J, Rozanski W, Kis J, Bar K, Bryniarski P, Paradysz A, Jersak K, Niemirowicz J, Slupski P, Jarzemski P, Skrzypczyk M, Dobruch J, Domagala P, Akbari MR, Lubinski J, Narod SA, Cybulski C. The G84E mutation in the HOXB13 gene is associated with an increased risk of prostate cancer in Poland. Prostate. 2013;73(5):542–548. doi: 10.1002/pros.22594. [DOI] [PubMed] [Google Scholar]
- 13.Beebe-Dimmer J, Isaacs WB, Zuhlke KA, Yee C, Walsh PC, Isaacs SD, Johnson AM, Ewing CE, Humphreys EB, Chowdhury WH, Montie JE, Cooney KA. The Prevalence of the HOXB13 G84E Prostate Cancer Risk Allele in Men Treated with Radical Prostatectomy. BJU Int. 2013 doi: 10.1111/bju.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, Diver WR, Witte JS, Hsing AW, Nemesure B, Rebbeck TR, Cooney KA, Xu J, Kibel AS, Hu JJ, John EM, Gueye SM, Watya S, Signorello LB, Hayes RB, Wang Z, Yeboah E, Tettey Y, Cai Q, Kolb S, Ostrander EA, Zeigler-Johnson C, Yamamura Y, Neslund-Dudas C, Haslag-Minoff J, Wu W, Thomas V, Allen GO, Murphy A, Chang BL, Zheng SL, Leske MC, Wu SY, Ray AM, Hennis AJ, Thun MJ, Carpten J, Casey G, Carter EN, Duarte ER, Xia LY, Sheng X, Wan P, Pooler LC, Cheng I, Monroe KR, Schumacher F, Le Marchand L, Kolonel LN, Chanock SJ, Van Den Berg D, Stram DO, Henderson BE. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43(6):570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PD, Pashayan N, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TL, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Roder MA, Nielsen SF, Bojesen SE, Siddiq A, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin HY, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC, Kote-Jarai Z, Easton DF Initiative CO-CRUG-E, Australian Prostate Cancer B, Oncology UKGPCSCBAoUSSo, Collaborators UKPS, Consortium P. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45(4):385–391. doi: 10.1038/ng.2560. 391e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 17.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature Genetics. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, O'Malley BW. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30(42):4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Ghanim M, Xue L, Brown CD, Iossifov I, Angeletti C, Hua S, Negre N, Ludwig M, Stricker T, Al-Ahmadie HA, Tretiakova M, Camp RL, Perera-Alberto M, Rimm DL, Xu T, Rzhetsky A, White KP. Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science. 2009;323(5918):1218–1222. doi: 10.1126/science.1157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An J, Wang C, Deng Y, Yu L, Huang H. Destruction of Full-Length Androgen Receptor by Wild-Type SPOP, but Not Prostate-Cancer-Associated Mutants. Cell reports. 2014;6(4):657–669. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas JA, Zuhlke KA, Beebe-Dimmer J, Levin AM, Gruber SB, Wood DP, Cooney KA. Identifying susceptibility genes for prostate cancer--a family-based association study of polymorphisms in CYP17, CYP19, CYP11A1, and LH-beta. Cancer Epidemiol. Biomarkers Prev. 2005;14(8):2035–2039. doi: 10.1158/1055-9965.EPI-05-0170. [DOI] [PubMed] [Google Scholar]
- 23.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, Gielzak M, Partin AW, Shanmugam V, Izatt T, Sinari S, Craig DW, Zheng SL, Walsh PC, Montie JE, Xu J, Carpten JD, Isaacs WB, Cooney KA. Germline mutations in HOXB13 and prostate-cancer risk. The New England Journal of Medicine. 2012;366(2):141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. American Journal of Human Genetics. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomlins SA, Palanisamy N, Siddiqui J, Chinnaiyan AM, Kunju LP. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch Pathol Lab Med. 2012;136(8):935–946. doi: 10.5858/arpa.2011-0424-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young A, Palanisamy N, Siddiqui J, Wood DP, Wei JT, Chinnaiyan AM, Kunju LP, Tomlins SA. Correlation of urine TMPRSS2:ERG and PCA3 to ERG+ and total prostate cancer burden. Am J Clin Pathol. 2012;138(5):685–696. doi: 10.1309/AJCPU7PPWUPYG8OH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Srivastava A, Tewari AK, Sathyanarayana U, Nagy D, Pestano G, Kunju LP, Demichelis F, Chinnaiyan AM, Rubin MA. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12(7):590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bunce MW, Boronenkov IV, Anderson RA. Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem. 2008;283(13):8678–8686. doi: 10.1074/jbc.M710222200. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP, Schulman BA. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36(1):39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 31.Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, Zimmermann M, Bond R, Shou J, Li C, Blattner M, Lonard DM, Demichelis F, Coarfa C, Rubin MA, Zhou P, O'Malley BW, Mitsiades N. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci U S A. 2013;110(17):6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blattner M, Lee DJ, O'Reilly C, Park K, Macdonald TY, Khani F, Turner KR, Chiu YL, Wild PJ, Dolgalev I, Heguy A, Sboner A, Ramazangolu S, Hieronymus H, Sawyers C, Tewari AK, Moch H, Yoon GS, Known YC, Andren O, Fall K, Demichelis F, Mosquera JM, Robinson BD, Barbieri CE, Rubin MA. SPOP Mutations in Prostate Cancer across Demographically Diverse Patient Cohorts. Neoplasia. 2014;16(1):14–20. doi: 10.1593/neo.131704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams M, Cookson VJ, Higgins J, Martin HL, Tomlinson DC, Bond J, Morrison EE, Bell SM. A high-throughput assay to identify modifiers of premature chromosome condensation. Journal of biomolecular screening. 2014;19(1):176–183. doi: 10.1177/1087057113495443. [DOI] [PubMed] [Google Scholar]
- 34.Barbieri CE, Tomlins SA. The prostate cancer genome: Perspectives and potential. Urol Oncol. 2014;32(1):53. doi: 10.1016/j.urolonc.2013.08.025. e15-22. [DOI] [PubMed] [Google Scholar]
- 35.Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, Schalken JA. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. European Urology. 2009;56(2):275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 36.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10(2):177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66(7):3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 38.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 39.Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, Demichelis F, Helgeson BE, Laxman B, Morris DS, Cao Q, Cao X, Andren O, Fall K, Johnson L, Wei JT, Shah RB, Al-Ahmadie H, Eastham JA, Eggener SE, Fine SW, Hotakainen K, Stenman UH, Tsodikov A, Gerald WL, Lilja H, Reuter VE, Kantoff PW, Scardino PT, Rubin MA, Bjartell AS, Chinnaiyan AM. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13(6):519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277(5328):965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 41.Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, Kimura A, Nakamura Y, Inazawa J, Abe T, Yamagishi H. Amplification and over-expression of the AIB1 nuclear receptor co-activator gene in primary gastric cancers. Int J Cancer. 2000;89(3):217–223. [PubMed] [Google Scholar]
- 42.Wang Y, Wu MC, Sham JS, Zhang W, Wu WQ, Guan XY. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer. 2002;95(11):2346–2352. doi: 10.1002/cncr.10963. [DOI] [PubMed] [Google Scholar]
- 43.Gnanapragasam VJ, Leung HY, Pulimood AS, Neal DE, Robson CN. Expression of RAC 3, a steroid hormone receptor co-activator in prostate cancer. Br J Cancer. 2001;85(12):1928–1936. doi: 10.1054/bjoc.2001.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agoulnik IU, Vaid A, Bingman WE, 3rd, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65(17):7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 45.Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman WE, 3rd, Erdem H, Frolov A, Smith CL, Ayala GE, Ittmann MM, Weigel NL. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res. 2006;66(21):10594–10602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]
- 46.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Exome Variant Server. NHLBI Exome Sequencing Project (ESP) Seattle, WA: (URL: http://snp.gs.washington.edu/EVS/). [Google Scholar]