Abstract
Background
Prior studies suggested that glycans were differentially expressed in patients with ovarian cancer and controls. Wehypothesized that glycan-based biomarkers might serve as a diagnostic test for ovarian cancer and evaluated the ability of glycans to distinguish ovarian cancer cases from matched controls.
Methods
Serum samples were obtained from the tissue-banking repository of the Gynecologic Oncology Group, and included healthy female controls (n = 100), women diagnosed with low malignant potential (LMP) tumors (n = 52), and epithelial ovarian cancers (EOC) cases (n = 147). Cases and controls were matched on age at enrollment within ±5 years. Serum samples were analyzed by glycomics analysis to detect abundance differences in glycan expression levels. A two-stage procedure was carried out for biomarker discovery and validation. Candidate classifiers of glycans that separated cases from controls were developed using a training set in the discovery phase and the classification performance of the candidate classifiers was assessed using independent test samples that were not used in discovery.
Results
The patterns of glycans showed discriminatory power for distinguishing EOC and LMP cases from controls. Candidate glycan-based biomarkers developed on a training set (sensitivity, 86% and specificity, 95.8% for distinguishing EOC from controls through leave-one-out cross-validation) confirmed their potential use as a detection test using an independent test set (sensitivity, 70% and specificity, 86.5%).
Conclusion
Formal investigations of glycan biomarkers that distinguish cases and controls show great promise for an ovarian cancer diagnostic test. Further validation of a glycan-based test for detection of ovarian cancer is warranted.
Impact
An emerging diagnostic test based on the knowledge gained from understanding the glycobiology should lead to an assay that improves sensitivity and specificity and allows for early detection of ovarian cancer.
Introduction
Ovarian cancer is the fifth leading cause of cancer-related deaths among women in the United States. It is speculated that early detection of ovarian cancer would be greatly enhanced with the development of improved tumor markers that are sensitive, specific, and detectable in early-stage disease when survival is the highest. The current generation of ovarian cancer tumor markers is protein based, for example, CA125, HE4, and Ova1. These tumor markers are commonly used to either monitor disease status in patients with known treated ovarian cancer, or to assess risk of malignancy in patients with a detected ovarian mass. However, there are significant limitations due to lack of sensitivity in early-stage disease and nonspecific elevations in nonmalignant states (especially CA125; refs. 1, 2).
We and other authors have studied the use of glycomics analysis of patient serum to see whether the pattern of glycan expression might discriminate between patients with and without ovarian cancer. Glycans are highly branched oligosaccharides that decorate larger parent molecules such as glycoproteins and glycolipids. The presence of the various glycans has significant influence over protein folding, receptor binding, protein clearance (3), and cell to cell recognition and signaling (4). Alterations in the glycosylation of glycoproteins are a very common post-translational event in the pathogenesis of cancer, including ovarian cancer (5). The analysis of glycans involves the determination of both their composition and isomer structures. This requires specialized mass spectroscopy techniques, among others, that our group has developed (3,4, 6).
Earlier "glycomic profiling" studies demonstrated a differential glycan expression pattern in the serum of patients with ovarian cancer compared with nondiseased controls (7–11). This present study focused on biomarker discovery and validation in ovarian cancer. We used serum samples obtained from the Gynecologic Oncology Group (GOG) cohort studies in a two-stage procedure that first identified candidate glycans (in a training set) and then tested the performance of each candidate and multiplex classifiers developed in the discovery phase in independent test samples (test set).
Materials and Methods
Sample cohorts
The Institutional Review Board (IRB) approval was obtained for this project through the University of California, Davis Medical Center (Sacramento, CA; IRB #251975) to use serum samples obtained from the GOG tissue-banking repository. The GOG collected whole blood specimens from patients with epithelial ovarian cancer (EOC), serous low malignant potential (LMP) tumors, and healthy female controls from multiple participating institutions as described by the GOG #136 protocol (revised August 2003), along with clinical information that included demographics and tumor characteristics, including stage, grade, and histology. Controls were healthy female volunteers without a history of malignancy and no family history of breast or ovarian cancer. Control samples were not obtained in conjunction with surgery. All serum samples, including controls, were uniformly prepared from the whole blood samples by the GOG per their protocol. The subjects selected for our study included healthy female volunteers (controls), and women diagnosed with LMP tumors, and EOCs. Serum samples were matched and balanced by a 5-year-age block (range, 40–65 years), as well as a balanced representation of stages I through IV EOC cases and controls. Preoperative, nonfasting blood samples were collected and de-identified before release to University of California (Davis, CA). Clinical information was provided for the patients with ovarian tumors, including age at collection, and tumor characteristics such as stage, grade, and histology. Two separate sets of serum samples were subjected to glycomics analysis independently at different times. The first set was a training set (OC1), which included control samples and patients with stages III–IV EOC. A separate second set that was selected and analyzed independently was used as a test set (OC2), which did not include any samples from the training set (OC1). The testing set (OC2) included controls subjects, patients with LMP tumors, and patients with EOC stages I–IV. Age was balanced across different cohorts (Table 1).
Table 1.
Summary statistics of characteristics of samples used in the study
| Variable | OC1 (Training set) | OC2 (Test set) |
|---|---|---|
| Total sample size, N | 91 | 208 |
| Healthy controls, n (%) | 48 (52.7%) | 52 (25%) |
| LMP, n (%) | 0 | 52 (25%) |
| Cancer cases, n (%) | 43 (47.3%) | 104 (50%) |
| By stage, n (%) | ||
| I | 0 | 34 (32.7%) |
| II | 0 | 18 (17.3%) |
| III | 34 (79.1%) | 47 (45.2%) |
| IV | 9 (2.1%) | 5 (4.8%) |
| By histology subtype, n (%) | ||
| High-grade serous adenocarcinoma, including papillary types | 43 (100%) | 66 (63.5%) |
| Endometrioid adenocarcinoma | – | 21 (20.2%) |
| Clear cell adenocarcinoma | – | 10 (9.6%) |
| Epithelial adenocarcinoma, not otherwise specified | – | 7 (6.7%) |
| Agea, mean ± SD | 52.44 ± 6.73 | 52.75 ± 5.84 |
| CA125a, mean ± SD | 261.64 ± 542.71 | 211.35 ± 425.85 |
| By disease category, mean ± SD | ||
| Healthy control | 17.26 ± 11.79 | 19.33 ± 3.95 |
| LMP | – | 119.47 ± 217.01 |
| EOC stage I–II | – | 242.82 ± 428.66 |
| EOC stage III–IV | 541.78 ± 699.88 | 463.77 ± 626.93 |
Age and CA125 level did not significantly differ between the OC1 and OC2 sets at the 0.05 significance level.
Serum processing and handling
N-glycan release of serum samples was performed as described previously (12). Briefly, proteins in 50 µL of serum were denatured in 50 µL of 200 mmol/L ammonium bicarbonate (Sigma-Aldrich) solution with 10 mnol/L dithiothreitol (Promega) using six cycles alternating between 100°C and room temperature for 10 seconds each. Two microliters of PNGaseF (New England Biolabs) was added to the samples, and enzymatic glycan release was performed in a CEM microwave. Deglycosylated proteins were precipitated using 400 µL of ice-cold ethanol, and upon centrifugation the supernatant was transferred to new Eppendorf tubes, and brought to dryness in vacuo.
Oligosaccharides released by PNGaseF were purified using graphitized carbon SPE cartridges (Grace) using a Gilson liquid handler. Briefly, cartridges were conditioned using 4 mL of 80% acetonitrile (ACN) containing 0.05% TFA (trifluoroacetic acid; EMD Chemicals), followed by 4 mL of water containing 0.05% TFA. Oligosaccharide samples were reconstituted in 500 µL of water and subsequently loaded onto the cartridges, which were washed using 3 × 4 mL of water. N-glycans were eluted using 8 mL of 40% ACN containing 0.05% TFA. All eluates were dried in vacuo before analysis.
Glycomics analysis
N-glycans were analyzed using an Agilent 6200 series nanoHPLC-chip-TOF-MS (time-of-flight mass spectrometer), consisting of an autosampler, which was maintained at 8°C, a capillary loading pump, a nanopump, HPLC-chip-MS interface, and an Agilent 6210 TOF-MS. The microfluidic chip (glycan chip II; Agilent) contained a 9 × 0.075 mm inner diameter (i.d.) enrichment column coupled to a 43 × 0.075 mm i.d. analytic column; both packed with 5-µm porous graphitized carbon. N-glycans were reconstituted in 45 µL of water and diluted 1:5 with water before analysis; 1 µL of sample was used for injection. Upon injection, the sample was loaded onto the enrichment column using 3% ACN containing 0.1% formic acid (Fluka). After the analytic column was switched in-line, the nanopump delivered a gradient of 3% ACN with 0.1 % formic acid (solvent A) and 90% ACN with 0.1 % formic acid (solvent B). Mass spectra were acquired in the positive mode over a mass range from 100 m/z to 3,000 m/z.
Data processing was performed using Masshunter qualitative analysis (version B.03.01; Agilent) and Microsoft Excel for Mac 2011 (version 14.1.3; Microsoft), according to Hua and colleagues (13) with modifications. Data were loaded into Masshunter qualitative analysis, and glycan features were identified and integrated using the molecular feature extractor algorithm. First, signals above a signal to noise threshold of 5.0 were considered. Then, signals were deconvoluted using a tolerance of 0.0025 m/z ± 10 ppm. The resulting deconvoluted masses were subsequently annotated using a retrosynthetic theoretical glycan library (14), in which a 20-ppm mass error was allowed. Glycan compositions and peak areas were exported to comma-separated values format for further statistical evaluation.
Statistical analysis
Before statistical analysis, raw peak areas were total quantity normalized on the basis of the underlying assumption that the total amount of ionized glycans that reach the detector is similar for different samples and glycan profiles for each dataset. Glycans detected in fewer than 70% of samples were discarded from downstream analysis to reduce the bias that could be induced by imputation for missing not at random. Unobserved values for any remaining undetected glycans below the predefined detection limit were imputed as one-half of the glycan-specific minimum of the observed values. Because the objective of this study was to assess the classification performance of glycan markers individually and classifiers developed with a training set when applied to an independent test set, it was necessary for the intensity values of glycans detected in these datasets to be on a comparable scale. We, therefore, normalized the intensities of the OC2 test set to the same total quantity used for the OC1 training set using a centering normalization method (15). Basically, we scaled each sample of the test set to the median sum of the samples in the training set so that the sum of the intensities is the same for all samples. Finally, the normalized data were log2 transformed to reduce the influence of extreme values and to meet homogeneity of variance assumptions. All statistical analyses were conducted in R 2.14.0 language and environment.
A differential analysis was used to identify specific glycans in which intensities differ between controls and tumor groups. For the training set (OC1), we used a t test to identify differentially expressed glycans between stages III–IV EOC cases and controls. For the test set (OC2), we used an ANOVA to test for differences among the four groups (controls, LMP, stages I–II EOC, and stages III–IV EOC). Where significant group differences were found (false discovery rate, FDR < 0.05), we applied the Dunnett test to compare the EOC and LMP groups with the healthy controls. All differential analyses were adjusted for age. Significance was determined on the basis of a permutation null distribution consisting of 10,000 permutations. FDRs were calculated to account for multiple testing.
We then used the training set to develop voting classifiers, as a multiplex panel, for classifying samples as tumor case (LMP or EOC) or control (16). Voting classifiers consist of one or more glycans. To classify a sample, each glycan in the classifier "votes" for a group membership for an unknown sample determined by whether the intensity value of the sample is higher or lower than a threshold value. For each glycan, the intensity value that yielded the highest value for the Youden index was used as the classification threshold. Alternatively, we also determined the classification thresholds under the requirement of a minimum specificity of 70%, 80%, or 90%. Then, for glycans that were upregulated in patients with cancer, samples with glycan values greater than the threshold were classified as EOC case, or as control by that glycan. In downregulated glycans, this classification was reversed. With multiple glycans in the classifier, an unknown sample is classified as tumor case or control by each glycan and classified according to the majority "vote" of the individual glycans composed of the classifier. To construct and evaluate multiplex classifiers, the glycan with the highest individual accuracy was added to the classifier first. Then, we systematically added the two glycans with the next highest accuracies and so on until the performance of the classifier improved no further. If several glycans yielded the same accuracy, they entered the classifier in order of their area under the curve (AUC) values. At each step, we constructed and assessed the accuracy of the classifier through leave-one-out cross-validation (LOOCV). Classifiers developed with the training set were "locked in place," and then applied to the independent test set samples not used during training to estimate the performance accuracy of the classifiers.
Results
Subject characteristics
All subjects selected for this study were 40 or older. The characteristics of the two independent sample sets are summarized in Table 1. The first set (OC1) consisted of serum samples obtained from 43 patients with stages III– IV serous EOC and 48 healthy controls, age matched within ±5 years. This set was regarded as the training set for biomarker discovery. The second set (test set, OC2) consisted of serum samples from 52 subjects per group in each of the following groups: healthy controls, LMP tumors, EOC stages I–II, and EOC stages III–IV. The ovarian cancer specimens had various nonmucinous epithelial histologies. All of the LMP tumors were serous histology. All four groups had similar age (40–65 years) distributions between groups as well as the training set.
Identification of informative glycans separating cancers and controls
Mass spectrometry–based glycomics analysis was performed on each of the samples in the training set (OC1) and 60 glycan compositions were detected consistently in at least 70% of the samples. Using differential analysis (EOC cases vs. controls), the 60 glycans were individually screened for association with ovarian cancer. Thirty-six glycan compositions significantly (FDR < 0.05) differed between stages III–IV EOC cases and control subjects. Using the OC2 test set, we conducted a differential analysis to confirm whether glycan compositions in which expression was found to be significantly differential between stages III–IV EOC cases and control subjects in the training set also differed significantly in the test set and further to identify glycan compositions that differed significantly across the four groups of control, stages I–II EOC, stages III–IV EOC, and LMP. For the 25 compositions that differed significantly (FDR < 0.05) across the four groups, we used the Dunnett test to compare each tumor group with the healthy control group (data not shown). More glycan compositions differed significantly between stages I–II EOC versus healthy controls and stages III–IV versus controls than LMP versus controls. In the comparison of stages III–IV EOC cases versus controls, 33 compositions were significantly different (FDR < 0.05) in the OC2 test set. Twenty-two glycan compositions, shown in Table 2, were significantly different in both datasets, suggesting consistency among them. Analysis of the expression of these glycans showed that only two glycans were overexpressed in cases compared with controls (H4N3S1, mass 1,566.56 and H7N6F1S2, mass 3,099.11), whereas the other 20 were underexpressed.
Table 2.
Glycan compositions with statistical difference in both the OC1 training and OC2 test sample sets, together with their mass, putative structure, a raw P value, FDR, and fold change
| Composition | Mass | Putative structure | OC1 P |
OC1 FDR |
OC1 Fold change |
OC2 P |
OC2 FDR |
OC2 Fold change |
|---|---|---|---|---|---|---|---|---|
| H3N2 | 910.33 | <0.001 | <0.001 | 0.57 | 0.011 | 0.025 | 0.76 | |
| H4N3 | 1,275.46 | 0.006 | 0.012 | 0.93 | 0.016 | 0.037 | 0.88 | |
| H5N3 | 1,437.51 | <0.001 | <0.001 | 0.68 | 0.007 | 0.019 | 0.53 | |
| H4N4 | 1,478.54 | <0.001 | <0.001 | 0.66 | <0.001 | 0.001 | 0.70 | |
| H7N2 | 1,558.54 | <0.001 | <0.001 | 0.78 | 0.008 | 0.019 | 0.74 | |
| H4N3S1 | 1,566.56 | <0.001 | 0.001 | 1.11 | 0.004 | 0.01 | 1.16 | |
| H5N3F1 | 1,583.57 | <0.001 | 0.001 | 0.62 | 0.007 | 0.019 | 0.58 | |
| H6N3 | 1,599.57 | <0.001 | <0.001 | 0.61 | <0.001 | 0.001 | 0.60 | |
| H4N4F1 | 1,624.60 | <0.001 | <0.001 | 0.73 | <0.001 | 0.002 | 0.88 | |
| H5N4 | 1,640.59 | 0.003 | 0.006 | 0.89 | <0.001 | 0.001 | 0.86 | |
| H4N4S1 | 1,769.63 | 0.001 | 0.002 | 0.87 | 0.004 | 0.012 | 0.88 | |
| H5N4F1 | 1,786.65 | <0.001 | <0.001 | 0.57 | <0.001 | 0.001 | 0.72 | |
| H4N5F1 | 1,827.68 | <0.001 | <0.001 | 0.73 | <0.001 | 0.001 | 0.75 | |
| H5N5 | 1,843.67 | <0.001 | <0.001 | 0.70 | <0.001 | 0.001 | 0.56 | |
| H5N3F1S1 | 1,874.67 | <0.001 | 0.001 | 0.69 | <0.001 | 0.001 | 0.56 | |
| H6N3S1 | 1,890.66 | <0.001 | <0.001 | 0.78 | 0.002 | 0.008 | 0.81 | |
| H5N5F1 | 1,989.73 | <0.001 | <0.001 | 0.72 | <0.001 | 0.001 | 0.63 | |
| H5N4F1S1 | 2,077.75 | <0.001 | <0.001 | 0.81 | <0.001 | 0.001 | 0.83 | |
| H6N5F1 | 2,151.78 | 0.014 | 0.026 | 0.71 | <0.001 | 0.001 | 0.48 | |
| H5N5F2S1 | 2,426.88 | <0.001 | <0.001 | 0.35 | 0.001 | 0.004 | 0.70 | |
| H6N5F1S1 | 2,442.88 | 0.026 | 0.044 | 0.74 | <0.001 | 0.002 | 0.71 | |
| H7N6F1S2 | 3,099.11 | 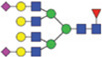 |
<0.001 | <0.001 | 3.78 | 0.001 | 0.004 | 2.17 |
NOTE: Glycans are represented as mannose (green circle), galactose (yellow circle), GlcNAc (blue square), fucose (red triangle), and sialic acid (purple diamond).
Developing multimarker classifiers using the OC1 training set
Through LOOCV of the training set, we determined the classification thresholds individually for each glycan and of CA125 based on providing the desired minimum specificity and then calculated the classification accuracy, sensitivity, and specificity of the left out samples (Fig. 1; Supplementary Table S1). The highest accuracy achieved by a single glycan composition was 87% (AUC = 0.9) by Hex6HexNAc3 (mass 1,599.57) followed by Hex5HexNAc5-Fuc2Sia1 [mass 2,426.88,accuracy 84% (AUC = 0.9)].CA125 performed better than all of the glycan compositions [accuracy of 93% (AUC = 0.94)]. We then investigated whether combining multiple compositions could yield better classifications. Accuracy increased with up to nine additional compositions and then declined (Table 3). The highest accuracy achieved was 91.2% (sensitivity, 86%; specificity, 95.8%) based on the thresholds determined by the Youden index; the nine compositions in this classifier were Hex6HexNAc3, Hex5HexNAc5Fuc2Sia1, Hex5Hex-NAc4Fuc1, Hex7HexNAc6Fuc1Sia2, Hex5HexNAc4Fuc1-Sia1, Hex8HexNAc2, Hex6HexNAc5Fuc1Sia2, Hex5Hex-NAc5Fuc1, and Hex4HexNAc4 (masses 1,599.57; 2,426.39; 1,786.65; 3,099.11; 2,077.75; 1,720.59; 2,733.97; 1,989.73; and 1,478.54; Fig. 2, middle). The order each composition entered the classifier is shown in Supplementary Table S1. This preliminary result suggests that several glycan compositions used in combination can improve classification relative to using a single glycan composition and can closely approach the accuracy of CA125.
Figure 1.
Receiver operating characteristic (ROC) curves for the top three glycans singly and CA125 that demonstrate diagnostic ability for separating healthy controls to stages III–IV EOC cases (blue line) of the OC1 training set and healthy controls to LMP tumors (green line), stages I–II EOC cases (red line), and stages III–IV EOC cases (black line) of the OC2 test set.
Table 3.
Confirmation of candidate multiplex classifiers using (a) a subset of the OC1 training set not used during training through LOOCV and (b) an independent OC2 test set
| a. LOOCV using OC1 training
set |
b. Independent test
validation using OC2 test set |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control vs. EOC (stage
III–IV) |
Control vs. EOC (stage
III–IV) |
Control vs. EOC (stage
l–ll) |
Control vs. LMP |
|||||||||||||||||
| Method determined a cutoff value |
Youden index |
Specificity level |
Youden index |
Specificity level |
Youden index |
Specificity level |
Youden index |
Specificity level |
||||||||||||
| 70% | 80% | 70% | 80% | 70% | 80% | 70% | 80% | |||||||||||||
| Number of glycans in a classifier |
Accuracy (%) |
Specificitya (%) |
Sensitivitya (%) |
Sensitivityb (%) |
Sensitivityb (%) |
Accuracy (%) |
Specificitya (%) |
Sensitivitya (%) |
Sensitivityb (%) |
Sensitivityb (%) |
Accuracy (%) |
Specificitya (%) |
Sensitivitya (%) |
Sensitivityb (%) |
Sensitivityb (%) |
Accuracy (%) |
Specificitya (%) |
Sensitivitya (%) |
Sensitivityb (%) |
Sensitivityb (%) |
| 1 | 86.8 | 91.7 | 81.4 | 84.0 | 84.0 | 70.2 | 59.6 | 80.8 | 85.0 | 85.0 | 65.4 | 59.6 | 71.2 | 81.0 | 81.0 | 58.7 | 59.6 | 57.7 | 77.0 | 73.0 |
| 3 | 90.1 | 89.6 | 90.7 | 93.0 | 88.4 | 75.0 | 84.6 | 65.4 | 73.1 | 61.5 | 69.2 | 84.6 | 53.8 | 63.5 | 53.8 | 68.3 | 84.6 | 51.9 | 65.4 | 53.8 |
| 5 | 90.1 | 93.8 | 86.0 | 88.4 | 86.0 | 62.5 | 98.1 | 26.9 | 46.2 | 38.5 | 55.8 | 98.1 | 13.5 | 25.0 | 17.3 | 55.8 | 98.1 | 13.5 | 28.8 | 21.2 |
| 7 | 87.9 | 95.8 | 79.1 | 86.0 | 83.7 | 66.3 | 94.2 | 38.5 | 51.9 | 46.2 | 56.7 | 94.2 | 19.2 | 42.3 | 26.9 | 58.7 | 94.2 | 23.1 | 42.3 | 26.9 |
| 9 | 91.2 | 95.8 | 86.0 | 88.4 | 88.4 | 73.1 | 96.2 | 50.0 | 59.6 | 48.1 | 59.6 | 96.2 | 23.1 | 46.2 | 30.8 | 58.7 | 96.2 | 21.2 | 34.6 | 25.0 |
| 11 | 87.9 | 97.9 | 76.7 | 86.0 | 81.4 | 77.9 | 86.5 | 69.2 | 76.9 | 67.3 | 71.2 | 86.5 | 55.8 | 65.4 | 55.8 | 62.5 | 86.5 | 38.5 | 51.9 | 40.4 |
| 13 | 84.6 | 97.9 | 69.8 | 83.7 | 79.1 | 76.0 | 84.6 | 67.3 | 82.7 | 75.0 | 71.2 | 84.6 | 57.7 | 75.0 | 59.6 | 64.4 | 84.6 | 44.2 | 65.4 | 50.0 |
| 15 | 83.5 | 95.8 | 69.8 | 81.4 | 76.7 | 73.1 | 65.4 | 80.8 | 92.3 | 86.5 | 70.2 | 65.4 | 75.0 | 82.7 | 80.8 | 66.3 | 65.4 | 67.3 | 76.9 | 76.9 |
| 17 | 84.6 | 95.8 | 72.1 | 79.1 | 74.4 | 72.1 | 65.4 | 78.8 | 92.3 | 86.5 | 71.2 | 65.4 | 76.9 | 82.7 | 80.8 | 67.3 | 65.4 | 69.2 | 78.8 | 76.9 |
| 19 | 83.5 | 95.8 | 69.8 | 81.4 | 72.1 | 76.0 | 82.7 | 69.2 | 86.5 | 78.8 | 74.0 | 82.7 | 65.4 | 76.9 | 65.4 | 69.2 | 82.7 | 55.8 | 69.2 | 55.8 |
NOTE: Classification performance was measured in terms of overall accuracy, sensitivity, specificity, and AUC of multiple marker classifiers for comparing healthy controls with the various tumor groups (EOC stage I–II, EOC stage III–IV, and LMP). The classifiers were voting classifiers consisting of an odd number of glycans.
Sensitivity (the true-positive rate) and specificity (the true-negative rate) have been calculated using a cutoff threshold that attained the maximal Youden index.
Sensitivity has been calculated at a classification threshold determined on the basis of providing the desired minimum specificity.
Figure 2.
ROC curves for multiplex classifiers consisting of three, nine, and 11 glycan compositions that demonstrate diagnostic ability for separating healthy controls to stages III–IV EOC cases (blue line) of the OC1 training set and healthy controls to LMP tumors (green line), stages I–II EOC cases (red line), and stages III–IV EOC cases (black line) of the OC2 test set.
Validating classifiers developed with the OC1 training set in an independent OC2 test set
We then evaluated the performance of classifiers developed with the training set in the discovery phase using the OC2 test samples. Of the top three glycan compositions identified with OC1, Hex6HexNAc3 (mass 1,599.57) retained a high AUC value of 0.84 for healthy controls versus stages III–IV EOC; however, both Hex5-HexNAc5Fuc2Sia1 and Hex5HexNAc4Fuc1 had lower AUC values with the OC2 than OC1 samples (Fig. 1; Supplementary Table S1). The highest AUC values seen for OC2 generally were for separating controls from patients with stages III–IV EOC as expected. None of the individual glycan compositions from OC2 yielded an AUC as high as CA125. Classification accuracy was lower in OC2 than estimated through LOOCV of OC1. Composition Hex6HexNAc3 (mass 1,599.57) had an accuracy of 87% in OC1 but this dropped to 70% when applied to the test set using the classification thresholds determined by the Youden index. The highest accuracy for separating patients with stages III–IV EOC from healthy controls in OC2 was 71%, achieved by Hex5-HexNAc5Fuc1 (mass 1,989.73). Considering the other group comparisons, the highest accuracy achieved by a single glycan composition was in separating controls versus stages I–IV EOC combined plus LMP tumors and was 70% to 75% for glycans Hex7HexNAc2, Hex5Hex-NAc5, Hex3HexNAc2, Hex8HexNAc2, Hex5HexNAc3, and Hex4HexNAc2. Interestingly, the classification accuracy of CA125 using the threshold determined with OC1 also became slightly lower when applied to OC2. However, for separating stages III–IV EOC from healthy controls, the accuracy of CA125 was 91%, which was still higher than any of individual glycan compositions.
Accuracy increased with multiple compositions, increasing between one and three compositions, declining to the lowest accuracy with five glycan compositions, and then again increasing in accuracy (Table 3). For classifying stages III–IV cancers versus controls, the highest accuracy of 78% (specificity, 86.5%; sensitivity, 69.2%) was achieved with the classifier consisting of the top 11 glycan compositions (Fig. 2). The decline in accuracy seen with three to seven compositions resulted from adding OC2 markers with accuracies of only about 50% (Supplementary Table S1).
Assessment of the diagnostic performance of the biomarkers
Estimated accuracies were lower with the test set than the training set. This finding suggests greater variability in OC2 (likely due to greater heterogeneity among samples in OC2 as well as some differences between the two cohorts), which would explain the lower accuracies found when the OC1 was used to develop classifiers applied to the OC2. To further assess the diagnostic performance of the candidate biomarkers, we switched the roles of the two datasets, using the OC2 (restricted to stages III–IV) as the training set and the OC1 dataset as the test set and repeated all the analyses to estimate performance accuracy of classifiers. When classifiers were developed using OC2 and tested in OC1, the estimated accuracy increased. Interestingly, all classifiers achieved accuracies greater than 85% when applied to the OC1 dataset, whereas accuracy was usually less than this value in the validation of OC2. Alternatively, we combined the two datasets and randomly split the combined dataset into training and test sets, allocating 60% of the data to a training set (117 subjects) and 40% to an independent test set. Training and test sets were comprised approximately equally of samples from the OC1 and OC2 datasets and of cancer and control samples. The new training set consisted of 57 randomly selected samples from the OC1 dataset (30 controls and 27 cases) and 60 samples from the OC2 dataset (30 cases and 30 controls). Using the new training set, we developed classifiers and estimated classification accuracy when applied to the new test set as previously described. Consistent with expectations, when classifiers developed with the new training set were applied to the new test set, accuracies were lower than found when the OC1 dataset was used as the test set but greater than the OC2 dataset used as the test set (data not shown), indicating potential bias in estimating the diagnostic performance accuracy of the candidate biomarkers depending on characteristics of a training set used for biomarker development and a test set for validation.
Discussion
We identified a set of glycan compositions that are differentially expressed, both individually and as a group, in patients with EOC and LMP tumors compared with healthy controls. By identifying and then locking down a candidate test developed on training set followed by a confirmation of the candidate test using an independent sample set, this prevalidation study aligns with the U.S. National Cancer Institute (17) guidelines for evaluating potential diagnostic cancer biomarkers. The value of following these guidelines is to avoid overstating the diagnostic accuracy of a proposed diagnostic test due to flawed clinical design methodology. The sample sets were successfully matched by age, LMP tumors, and ovarian cancer cases. We used several rigorous statistical methodologies to analyze the test performance of the glycan compositions, and demonstrated strong discriminatory power to segregate ovarian cancer cases from healthy controls. Importantly, although we found some empirical evidence that classification accuracy slightly varied depending on which data have been used during training, we still determined that the glycan compositions had strong test performance even when the two sets of samples were combined and then randomly split into two sets, one for training and the other for validation. This demonstrates that diagnostic power of the glycomics profile persists regardless of sample selection. Thus, we believe that a profile of glycans globally released from serum glycoproteins and detected by mass spectrometry, can distinguish patients with EOC from healthy controls.
When we lock down the glycan classification scheme from the training set to the test set, the test performance of the individual glycan compositions to discriminate between all stages EOC cases and healthy controls is quite good (Supplementary Table S1; Fig. 1) and even better when using a combination of multiple compositions (Table 3; Fig. 2). Specifically, the highest diagnostic accuracy came from a combination of nine or 11 glycan compositions. The expression of the glycan compositions does not distinguish as well between LMP tumors and controls when compared with EOC cases and controls (Fig. 2).
Several important characteristics of the sample sets and comparison with CA125 as a diagnostic marker should be made. First, the pattern of expression is different between the glycan profile and CA125. All except two glycans were underexpressed in the tumor cases compared with the controls (Table 2). Thus, the biology of glycan expression is different than for CA125, which is elevated in most EOCs. The biologic basis for this is discussed below. Second, because the serum samples were obtained from multiple institutions participating in the GOG tissue-banking program, the risk of overestimating the diagnostic accuracy of the glycan profile is minimized compared with using a single institution with respect to greater heterogeneity among patients and may be more comparable with real life circumstances. Third, the serum samples were selected to evaluate the diagnostic test performance of the glycan profile to distinguish between ovarian tumor cases and healthy controls, not necessarily to show that the test performance is superior to CA125. Because of inherent sample selection bias pertaining to the GOG tissue-banking program, CA125 is a near-perfect diagnostic tumor marker as it is markedly elevated in most cancer cases and normal in all controls (Table 1). Therefore, it is not surprising that CA125 has stronger diagnostic power compared with individual glycans. Nonetheless, the test performance of the multiplex glycan profile approaches that of CA125 in comparable test set groups. For example, using the independent test set OC2, the AUC for CA125 in EOC stages I–II was 0.82 (Supplementary Table S1) compared with 0.82 for a multiplex of 11 glycans (Table 3), showing that they have comparable test performance. For OC2 stages III–IV, the AUC for CA125 was 0.89, and for the 11 glycans was 0.92. Future validation studies will include ovarian cancer cases for which there are a range of CA125 values from low to high, including different histologies.
The biologic connection between aberrantly expressed glycans and ovarian cancer is a topic of intense investigation. Protein glycosylation has been recognized as a potential target for the development of biomarkers for the detection of several cancer types (18). Serum glycomic profiles have been noted to be differentially expressed in ovarian cancer cases compared with controls in a variety of studies to date (7, 8, 10, 19–21). Results from these studies, especially the studies performed previously in our group (8, 19), provide similar differential glycosylation profiles to the ones observed in this study. The pattern of over- and underexpression of glycans in tumor cases versus controls was consistent between the OC1 training and OC2 test sets especially when samples were selected and analyzed independently. This consistency of expression is critical to the development of a reproducible diagnostic test.
The glycans that were differentially expressed in EOC cases compared with controls can be subdivided in groups based on their structural properties, as depicted in Fig. 3. One group consists of high mannose– and hybrid-type glycans, of which the levels are typically decreased in cancer cases versus controls. Because the biosynthesis of N-glycans starts with the production of high mannose– type glycans, which are then converted to hybrid and later complex-type glycans using mannosidases and GlcNAc transferases (22), it is likely that the enzyme activity of these two enzymes is increased in cancer cases. Although there is limited literature related to the presence of high mannose– and hybrid-type glycans on serum proteins, a recent study using standard serum proteins suggested their presence on immunoglobulin M (IgM; ref. 23).
Figure 3.
Glycans that are differentially expressed can be subdivided into three different groups based on their structural features. Glycans are represented as mannose (green circle), galactose (yellow circle), GlcNAc (blue square), fucose (red triangle), and sialic acid (purple diamond).
Another group of glycan compositions decreased in EOC are biantennary glycans with at least one galactose residue that may or may not be decorated with fucose and bisecting GlcNAc residues. Moreover, if both antennae are galactosylated, one of them may be decorated with a sialic acid. Decreased levels of galactosylated biantennary glycans may be an indication that the activity of galactosyltransferases, which are responsible for the decoration of N-glycans with galactose residues, is hampered in EOC. Such biantennary glycan compositions have often been described on multiple serum glycoproteins, including IgG. It is widely known that decreased levels of galactosylated glycans on IgG are a hallmark for autoimmune diseases such as rheumatoid arthritis (24, 25). It is unknown which serum glycoproteins are responsible for the aberrant expression of glycans in EOC, but we suspect that an inflammatory response to the malignancy may play a role. We are currently investigating glycan expression in serum immunoglobulins of patients with ovarian cancer to understand their contribution to the glycomics profile, which we anticipate will soon provide further biologic insight into the secondary responses to the malignancy.
In this study, glycan compositions were analyzed. However, each glycan composition may potentially be subdivided into several glycan structures based on different linkages of the glycans. Initial studies using ovarian cancer profiles have shown that differentiation of cancer cases and controls may be obtained using structure-specific analysis (19). At this time, however, it is not yet feasible to perform such analyses on a routine basis (3). Further advancements in bioinformatics will facilitate such structure-specific analysis on a larger scale. This will likely provide a biomarker panel with higher accuracy, sensitivity, and specificity.
The next steps in evaluating the glycomics profile as a diagnostic biomarker for ovarian cancer involves two parallel processes. The first will be to confirm our findings on well-annotated clinical serum samples as a validation study using the "locked down" glycomics profile. It will be important to evaluate how the glycomics profile performs with various ovarian cancer histologies, and determine whether the profile is effective in both early- (particularly presymptomatic) and late-stage cancers. It is similarly important to determine the heterogeneity of serum glycan expression in general healthy female populations because this greatly affects the specificity of the diagnostic test. The second area of investigation is to establish the biologic basis for glycomics expression in ovarian cancer. Important studies will compare glycan expression in ovarian tumor specimens, ascites, and serum to see whether the patterns are similar. Other studies will focus on protein- specific glycan expression by isolating glycoproteins from patient serum.
Supplementary Material
Acknowledgments
The authors are thankful for the receipt of serum samples from the GOG tissue-banking repository.
Grant Support
This work was supported by the Ovarian Cancer Research Fund (to G.S. Leiserowitz).
Footnotes
Note: Supplementary data for this article are available at Cancer Epidemiology, Biomarkers & Prevention Online (http://cebp.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: K. Kim, S. Miyamoto, C.B. Lebrilla, G.S. Leiserowitz
Development of methodology: K. Kim, S. Ozcan, S. Miyamoto, C.B. Lebrilla, G.S. Leiserowitz
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L.R. Ruhaak, L. Dimapasoc, C. Williams, C. Stroble, S. Ozcan, S. Miyamoto, C.B. Lebrilla, G.S. Leiserowitz
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): K. Kim, L.R. Ruhaak, U.T. Nguyen, S.L. Taylor, S. Ozcan, S. Miyamoto, C.B. Lebrilla, G.S. Leiserowitz
Writing, review, and/or revision of the manuscript: K. Kim, L.R. Ruhaak, S.L. Taylor, S. Miyamoto, C.B. Lebrilla, G.S. Leiserowitz
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K. Kim, S. Ozcan, S. Miyamoto, C.B. Lebrilla, G.S. Leiserowitz
Study supervision: K. Kim, C.B. Lebrilla, G.S. Leiserowitz
References
- 1.Zgheib NB, Xiong Y, Marchion DC, Bicaku E, Chon HS, Stickles XB, et al. The O-glycan pathway is associated with in vitro sensitivity to gemcitabine and overall survival from ovarian cancer. Int J Oncol. 2012;41:179–188. doi: 10.3892/ijo.2012.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore RG, Jabre-Raughley M, Brown AK, Robison KM, Miller MC, Allard WJ, et al. Comparison of a novel multiple marker assay versus the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am J Obstet Gynecol. 2010;203:228. doi: 10.1016/j.ajog.2010.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruhaak LR, Miyamoto S, Lebrilla CB. Developments in the identification of glycan biomarkers for the detection of cancer. Mol Cell Proteomics. 2013;12:846–855. doi: 10.1074/mcp.R112.026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mechref Y, Hu Y, Garcia A, Hussein A. Identifying cancer biomarkers by mass spectrometry-based glycomics. Electrophoresis. 2012;33:1755–1767. doi: 10.1002/elps.201100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An HJ, Lebrilla CB. A glycomics approach to the discovery of potential cancer biomarkers. Methods Mol Biol. 2010;600:199–213. doi: 10.1007/978-1-60761-454-8_14. [DOI] [PubMed] [Google Scholar]
- 6.Hua S, An HJ. Glycoscience aids in biomarker discovery. BMB Rep. 2012;45:323–330. doi: 10.5483/bmbrep.2012.45.6.132. [DOI] [PubMed] [Google Scholar]
- 7.Alley WR, Jr, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, et al. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res. 2012;11:2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kronewitter SR, De Leoz ML, Strum JS, An HJ, Dimapasoc LM, Guerrero A, et al. The glycolyzer: automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics. 2012;12:2523–2538. doi: 10.1002/pmic.201100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leiserowitz GS, Lebrilla C, Miyamoto S, An HJ, Duong H, Kirmiz C, et al. Glycomics analysis of serum: a potential new biomarker for ovarian cancer? Int J Gynecol Cancer. 2008;18:470–475. doi: 10.1111/j.1525-1438.2007.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 11.Saldova R, Wormald MR, Dwek RA, Rudd PM. Glycosylation changes on serum glycoproteins in ovarian cancer may contribute to disease pathogenesis. Dis Markers. 2008;25:219–232. doi: 10.1155/2008/601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kronewitter SR, de Leoz ML, Peacock KS, McBride KR, An HJ, Miyamoto S, et al. Human serum processing and analysis methods for rapid and reproducible N-glycan mass profiling. J Proteome Res. 2010;9:4952–4959. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, et al. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kronewitter SR, An HJ, de Leoz ML, Lebrilla CB, Miyamoto S, Leiserowitz GS. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9:2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor SL, Kim K. A jackknife and voting classifier approach to feature selection and classification. Cancer Inform. 2011;10:133–147. doi: 10.4137/CIN.S7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McShane LM, Cavenagh MM, Lively TG, Eberhard DA, Bigbee WL, Williams PM, et al. Criteria for the use of omics-based predictors in clinical trials. Nature. 2013;502:317–320. doi: 10.1038/nature12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Packer NH, von der Lieth CW, Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, et al. Frontiers in glycomics: bioinformatics and biomarkers in disease. Proteomics. 2008;8:8–20. doi: 10.1002/pmic.200700917. [DOI] [PubMed] [Google Scholar]
- 19.Hua S, Williams CC, Dimapasoc LM, Ro GS, Ozcan S, Miyamoto S, et al. Isomer-specific chromatographic profiling yields highly sensitive and specific potential N-glycan biomarkers for epithelial ovarian cancer. J Chromatogr A. 2013;1279:58–67. doi: 10.1016/j.chroma.2012.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imre T, Kremmer T, Heberger K, Molnar-Szollosi E, Ludanyi K, Pocsfalvi G, et al. Mass spectrometric and linear discriminant analysis of N-glycans of human serum alpha-1-acid glycoprotein in cancer patients and healthy individuals. J Proteomics. 2008;71:186–197. doi: 10.1016/j.jprot.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Kim YG, Jeong HJ, Jang KS, Yang YH, Song YS, Chung J, et al. Rapid and high-throughput analysis of N-glycans from ovarian cancer serum using a 96-well plate platform. Anal Biochem. 2009;391:151–153. doi: 10.1016/j.ab.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 23.Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11:1958–1968. doi: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parekh RB, Roitt IM, Isenberg DA, Dwek RA, Ansell BM, Rademacher TW. Galactosylation of IgG associated oligosaccharides: reduction in patients with adult and juvenile onset rheumatoid arthritis and relation to disease activity. Lancet. 1988;1:966–969. doi: 10.1016/s0140-6736(88)91781-3. [DOI] [PubMed] [Google Scholar]
- 25.Huhn C, Selman MH, Ruhaak LR, Deelder AM, Wuhrer M. IgG glycosylation analysis. Proteomics. 2009;9:882–913. doi: 10.1002/pmic.200800715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





