Cby1 localizes to centrioles and antagonizes canonical Wnt signaling. Cby1−/− mice have cystic kidneys, and Cby1 facilitates primary cilium formation and ciliary recruitment of Arl13b. Cby1 localizes to a distal centriolar domain with Ofd1 and Ahi1, and the amount of Ahi1 at the transition zone is reduced in Cby1−/− cells.
Abstract
Defects in centrosome and cilium function are associated with phenotypically related syndromes called ciliopathies. Cby1, the mammalian orthologue of the Drosophila Chibby protein, localizes to mature centrioles, is important for ciliogenesis in multiciliated airway epithelia in mice, and antagonizes canonical Wnt signaling via direct regulation of β-catenin. We report that deletion of the mouse Cby1 gene results in cystic kidneys, a phenotype common to ciliopathies, and that Cby1 facilitates the formation of primary cilia and ciliary recruitment of the Joubert syndrome protein Arl13b. Localization of Cby1 to the distal end of mature centrioles depends on the centriole protein Ofd1. Superresolution microscopy using both three-dimensional SIM and STED reveals that Cby1 localizes to an ∼250-nm ring at the distal end of the mature centriole, in close proximity to Ofd1 and Ahi1, a component of the transition zone between centriole and cilium. The amount of centriole-localized Ahi1, but not Ofd1, is reduced in Cby1−/− cells. This suggests that Cby1 is required for efficient recruitment of Ahi1, providing a possible molecular mechanism for the ciliogenesis defect in Cby1−/− cells.
INTRODUCTION
Many mammalian cells have a single primary cilium, and some specialized cell types have one or more motile cilia. All cilia are nucleated from a centriole, or basal body—a ninefold radially symmetric organelle conserved in many eukaryotes. Most cells have a centrosome that includes a pair of centrioles, only one of which—the older, mother centriole—is competent to extend a primary cilium. Primary cilia are sensory organelles important for mechanosensation, photoreceptor function, and osmosensation and modulate a range of signaling pathways, including Hedgehog, Wnt, and platelet-derived growth factor receptor-α (Pazour et al., 2002; Huangfu et al., 2003; Nauli et al., 2003; Schneider et al., 2005; Gerdes et al., 2007; Gradilone et al., 2007; Malone et al., 2007; Corbit et al., 2008).
The centriole becomes competent to form a cilium by the recruitment of protein complexes to the distal end that enable basal body docking to the plasma membrane and define the ciliary compartment. These include proteins of the transition zone, a region at the junction of the centriole and cilium. Recruitment of these complexes is followed by extension of the axoneme, growth and modification of the specialized ciliary membrane, and localization of ciliary components such as ciliary signaling receptors (Avasthi and Marshall, 2012; Reiter et al., 2012). Genetic defects in these processes cause a group of diseases, termed ciliopathies, in which cilium formation or function is disrupted (Hildebrandt et al., 2011). Ciliopathies share subsets of common phenotypes that reflect the diversity of cilium function. Renal cystic disease is perhaps the most prevalent of phenotypes in the ciliopathies, and other phenotypes include retinal degeneration, polydactyly, situs inversus, hydrocephaly, and neurocognitive defects. The molecular basis of pathogenesis in ciliopathies is believed to be due to disruption of cilium-regulated signaling pathways such as mechanosensation, noncanonical Wnt, and Hedgehog signaling (Berbari et al., 2009).
There are three classes of proteins at the distal end of centrioles involved in cilia formation or function. Distal-end proteins, including Ofd1, localize to the distal ends of both mother and daughter centrioles (Singla et al., 2010). Appendage proteins, including Cep164, localize to the distal and/or subdistal appendages of the mother centriole (Graser et al., 2007). Both of these classes of proteins are required for cilium formation and are found on all centrioles competent to form a cilium, whether they have yet formed a cilium or not. In contrast, the third class of proteins, the transition zone proteins, including Ahi1 and Tmem237 (Hsiao et al., 2009; Huang et al., 2011; Lancaster et al., 2011), are recruited to the distal end of mother centrioles only during ciliogenesis. Some transition zone components are not required for cilium formation but instead for regulation of ciliary protein content (Reiter et al., 2012). Most transition zone components have been characterized as cytoplasmic complexes (Sang et al., 2011; Chih et al., 2012), with little information on the organization and order of assembly of the proteins at the centriole.
Cby1 was initially identified as an antagonist of canonical Wnt signaling (Takemaru et al., 2003), and subsequently shown to localize to the mother centriole (Steere et al., 2012). Deletion of Cby1 in mice resulted in chronic respiratory infection due to a reduced number of motile cilia in cells of the airway epithelium (Voronina et al., 2009). In vitro RNA interference (RNAi) experiments suggested that Cby1 is required for primary cilia formation (Steere et al., 2012). However, the majority of Cby1−/− mice typically reach birth, which is not consistent with the early embryonic lethality associated with failure to make a functional primary cilium (Marszalek et al., 1999; Murcia et al., 2000; Huangfu et al., 2003). Most Cby1−/− mice die within 3 wk of birth, likely of multiple organ dysfunction, whereas the remaining 20% are viable for >18 mo (Voronina et al., 2009). Similarly, cilia still form in Drosophila cells in which chibby, the orthologue of Cby1, has been deleted (Enjolras et al., 2012). This led us to investigate the role of Cby1 in primary cilium formation and function, using of cells and tissues from Cby1−/− mice, to clarify its contribution to ciliogenesis. We find that Cby1−/− mice have cystic kidneys and that Cby1 facilitates, but is not required for, primary cilium formation. Cby1 is needed for efficient recruitment of Arl13b to the primary cilium. Cby1 localizes to a distinct distal centriolar domain with Ofd1 and Ahi1, and the amount of Ahi1 at the transition zone is reduced in Cby1−/− cells.
RESULTS
Cby1 −/− mice have cystic kidneys and reduced primary cilium frequency
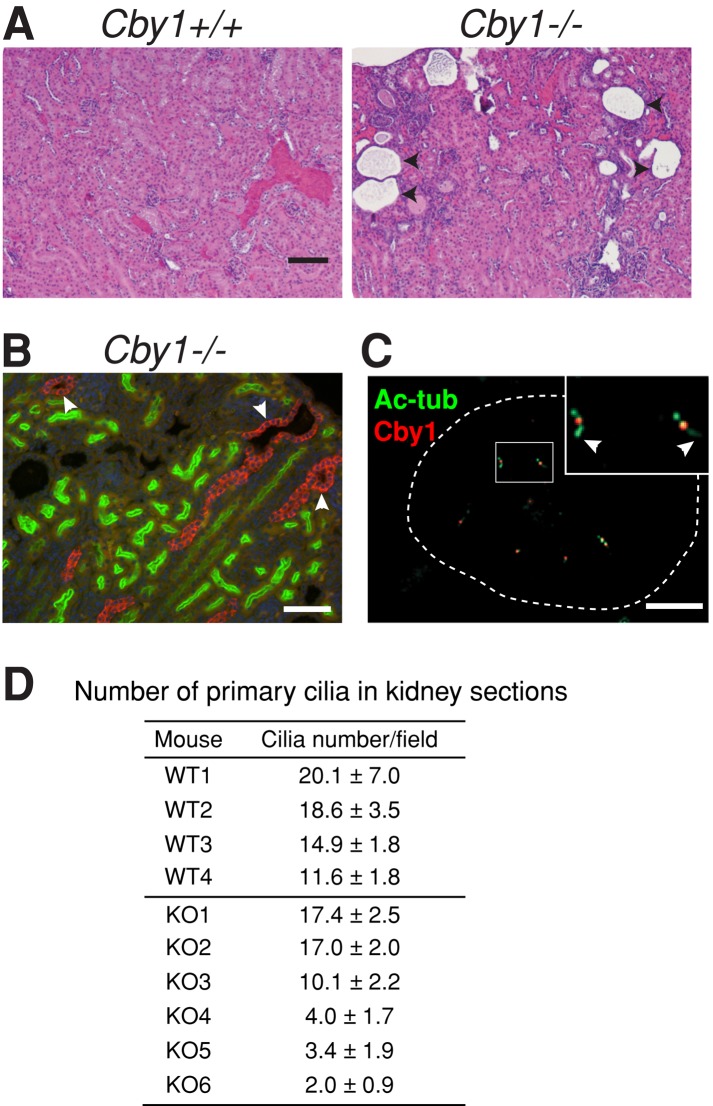
Renal disorders are a major feature of ciliopathies resulting from defects in primary cilia structure or function (Berbari et al., 2009; Gascue et al., 2011). To determine whether loss of Cby1 results in renal defects, we examined the kidneys of mutant Cby1−/− and wild-type Cby1+/+ mice. The kidneys of Cby1−/− mice were similar in size to wild-type kidneys at all ages examined. However, histological analysis revealed the presence of multiple renal cysts in Cby1−/− mice (Figure 1A). Although the extent of the cystic phenotype varied, renal cysts were consistently observed in all adult (>2 mo old) Cby1−/− animals and were detectable in some animals as early as at birth. As shown in Figure 1B, some of the cysts were positive for the collecting duct/distal tubule marker Dolichos biflorus agglutinin (DBA), but the others were negative for both DBA and the proximal tubule marker Lotus tetragonolobus lectin (LTL). LTL-positive cysts were rarely observed. Thus it appears that renal cysts predominantly derive from collecting duct/distal tubules in Cby1−/− mice. Consistent with its function in kidney development, Cby1 was detected in renal tubular cells, localizing to the centrioles at the base of primary cilia (Figure 1C). Next we determined the status of primary cilia in Cby1−/− kidneys. Assessment by immunostaining, as in Figure 1C, showed that primary cilia were present in the renal tubular cells of Cby1−/− mice but that the cilia were less abundant compared with Cby1+/+ controls. The number of primary cilia in the renal cortex was quantified at various ages, blinded to genotype for unbiased counting (Figure 1D). Only cilia that were >2 μm and protruded into the tubular lumen were scored. In Cby1+/+ kidneys from four mice, mean cilium number per microscope field varied from 11.6 to 20.1. In Cby1−/− kidneys from six mice, mean cilium number varied from 2.0 to 17.4, with greater variability than in the wild-type kidneys. Kidneys from three of the mice (KO4, 5, 6) had a significantly lower number of cilia, whereas the other three were not significantly different from wild type. Although we do not yet understand the basis of the variability of the Cby1−/− phenotype, the reduced number of cilia in the kidneys observed in some animals is consistent with Cby1 being important for primary cilium function in renal epithelia.
FIGURE 1:
Loss of Cby1 results in polycystic kidneys. (A) Kidney cyst formation in Cby1−/− mice. Kidney sections from Cby1+/+ and Cby1−/− adult mice (3 mo old) were stained with hematoxylin and eosin. Cysts are indicated with arrowheads. (B) Renal sections from postnatal day 13 Cby1−/− mice were stained with DBA (red, distal tubules), LTL (green, proximal tubules), and DAPI (blue). Note that some kidney cysts arise from collecting duct/distal tubules (arrowheads). (C) Cby1 localizes to the basal bodies of primary cilia in the renal tubular cells. Kidney sections from 3-mo-old Cby1+/+ mice were double labeled with antibodies against Cby1 (red) and the ciliary/basal body marker acetylated α-tubulin (Ac-tub, green), and the merged image is shown. Inset, high-magnification view with arrowheads pointing to primary cilia. The dotted line encircles a renal tubule. No Cby1 staining was observed in Cby1−/− kidney sections. Scale bars, 200 μm (A), 50 μm (B), 5 μm (C). (D) The number of primary cilia (>2.0 μm) per microscope field at 100× in the renal cortex of Cby1+/+ (WT) and Cby1−/− (KO) mice was counted, with 10 fields of view/mouse. Values are means ± SEM for individual mice.
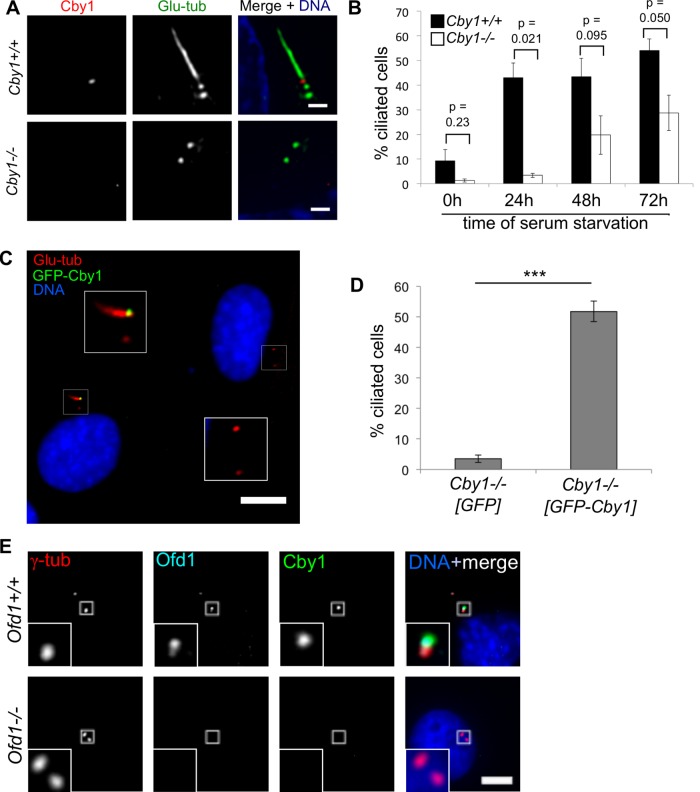
To test directly whether loss of Cby1 compromises cilium formation, we examined primary cilium formation in mouse embryonic fibroblasts (MEFs) from Cby1+/+ and Cby1−/− embryos. Cells were induced to form a cilium by serum starvation and assayed over a 72-h time course. Cby1−/− MEFs exhibited a delay in primary cilia formation relative to wild type (Figure 2, A and B). After 24 h of serum starvation, >40% of Cby1+/+ MEFs had a cilium, whereas <5% of Cby1−/− MEFs had a cilium (Figure 2B). The fraction of Cby1−/− MEFs with a cilium increased over time of serum starvation, although it remained lower than that for Cby1+/+ MEFs (Figure 2B). Complementation of the Cby1−/− deletion with a human GFP-Cby1 construct (Figure 2, C and D) rescued the defect in primary cilium formation, assessed at 24 h of serum starvation. Unlike in previous RNAi experiments (Steere et al., 2012), these data demonstrate that Cby1 is not essential for forming a primary cilium but is important for facilitating the process.
FIGURE 2:
Cby1 facilitates primary cilia formation. (A) Cby1+/+ or Cby1−/− MEFs were serum starved, fixed, and stained for polyglutamylated tubulin (Glu-tub, green), Cby1 (red), and DNA (DAPI, blue). Scale bars, 10 μm. (B) Cby1+/+ or Cby1−/− MEFs were fixed after 0, 24, 48, or 72 h of serum starvation, stained for polyglutamylated tubulin, and scored for primary cilia. Results shown are the mean of three independent experiments ± SEM (100 cells/experiment; p values shown above each time point). (C, D) Cby1−/− MEFs were transfected with GFP or GFP-Cby1, serum-starved for 24 h, fixed, and stained for GFP (green), polyglutamylated tubulin (Glu-tub, red) and DNA (DAPI, blue). A representative image of Cby1−/− MEFs transfected with GFP-Cby1 is shown in C. Insets, enlarged images of centrosomal/ciliary regions. Note in C that the cell expressing GFP-Cby1 is ciliated, whereas the neighboring cell not expressing GFP-Cby1 is not ciliated. Scale bar, 10 μm. These data are quantified in D. Results shown are the mean of four experiments ± SEM (>100 cells/experiment; ***p < 0.001). (E) Ofd1+/+ or Ofd1−/− mouse embryonic stem cells were fixed and stained for γ-tubulin (γ-tub, red), Ofd1 (cyan), Cby1 (green), and DNA (DAPI, blue). Scale bar, 5 μm.
Previous immunofluorescence studies indicated that Cby1 localizes to the distal mother centriole (Steere et al., 2012). Ofd1 localizes to a similar position and is required for primary cilium formation, likely by regulating the formation of distal centriole structures such as the distal appendages (Singla et al., 2010). To determine whether Cby1 is a component of distal centriole structures organized by Ofd1, we examined Ofd1+/+ and Ofd1−/− mouse embryonic stem (ES) cells (Singla et al., 2010) for Cby1 localization using immunofluorescence microscopy (Figure 2E). Cby1 localized to the distal end of the mother centriole in Ofd1+/+ cells but was absent from the centrosome in Ofd1−/– cells (Figure 2E), consistent with Cby1 being a component of distal centriole structures important for ciliogenesis.
Cby1 −/− MEFs have reduced ciliary Arl13b
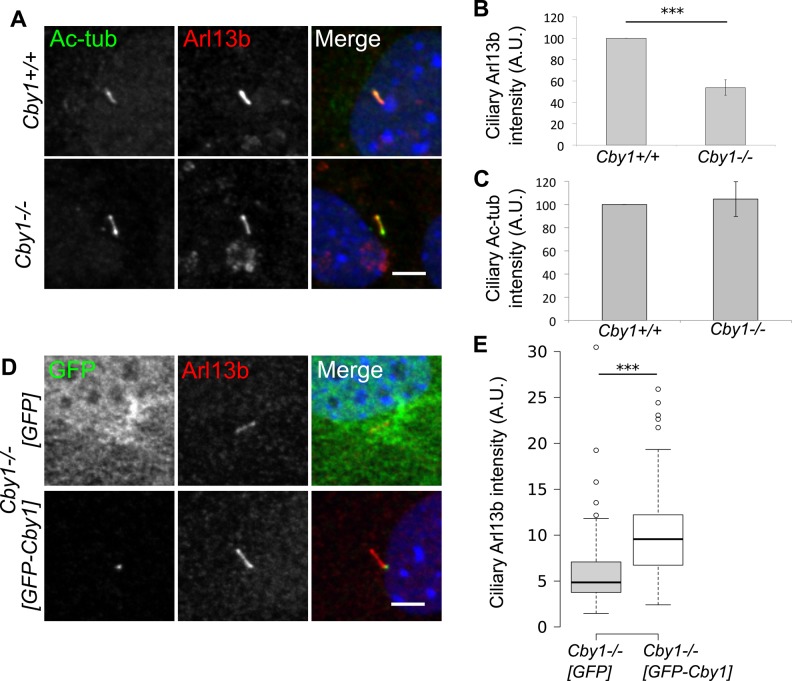
The primary cilium is a signaling center for several pathways, and concentration of signaling proteins in the cilium compartment is essential to proper signaling (Berbari et al., 2009; Mahjoub and Stearns, 2012). Arl13b is a cilium-localized small GTPase that regulates cilium morphology and ciliary trafficking (Caspary et al., 2007; Duldulao et al., 2009; Cevik et al., 2010; Larkins et al., 2011). Loss of Arl13b in the zebrafish scorpion mutant results in cystic kidneys (Sun et al., 2004; Cantagrel et al., 2008), and mutations in ARL13B are causative for the ciliopathy Joubert syndrome (Cantagrel et al., 2008). We examined the localization of Arl13b to cilia in Cby1+/+ and Cby1−/− MEFs. The amount of ciliary Arl13b in Cby1−/− MEFs was reduced twofold compared with Cby1+/+ MEFs (Figure 3, A and B), and this reduction could be complemented by a human GFP-Cby1 construct (Figure 3, D and E). The amount of ciliary acetylated α-tubulin was similar in Cby1+/+ and Cby1−/− MEFs (Figure 3, A and C). Despite the reduction of ciliary Arl13b in Cby1−/− MEFs, these cells retained the ability to control protein access to the ciliary compartment, as assessed by recruitment of the transmembrane protein Smoothened (Smo) in response to treatment with Smo agonist (SAG), an activator of the pathway (Supplemental Figure S1).
FIGURE 3:
Cby1 facilitates ciliary recruitment of Arl13b. (A) Cby1+/+ or Cby1−/− MEFs were serum starved for 48 h, fixed, and stained for acetylated α-tubulin (Ac-tub, red), Arl13b (green), and DNA (DAPI, blue). Scale bar, 5 μm. (B, C) Quantification of ciliary Arl13b and acetylated tubulin levels in Cby1+/+ or Cby1−/− MEFs. Results shown are the mean of three independent experiments ± SEM (>100 cells/experiment, *p < 0.05). (D) Cby1−/− MEFs were transduced with lentivirus expressing GFP or GFP-Cby1, serum-starved for 48 h, fixed, and stained for GFP (green), Arl13b (red), and DNA (DAPI, blue). Scale bar, 5 μm. (E) Ciliary Arl13b intensity was quantified and displayed as a Tukey boxplot. Results from duplicate independent experiments, >30 observations/experiment, ***p < 0.001 by Mann–Whitney U test.
Cby1 localizes in close proximity to the transition zone
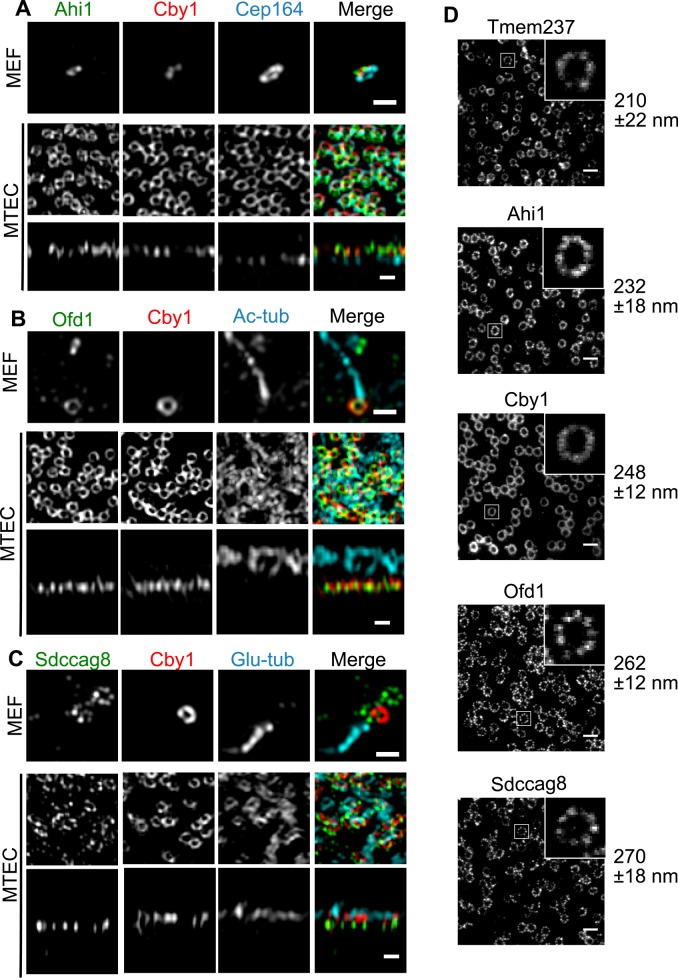
Control of ciliary protein trafficking occurs in part by the action of protein complexes at the base of the cilium. This region includes the distal end of the mother centriole and the transition zone between the centriole and cilium and is a site to which many ciliopathy proteins localize (Reiter et al., 2012). Given that Cby1 localizes to the mother centriole (Steere et al., 2012) and that our aforementioned results show that it depends on Ofd1 for centriolar localization and is involved in ciliary trafficking, we sought to examine its localization at higher resolution. We used a combination of three-dimensional structured illumination (3D-SIM) and stimulated emission depletion (STED) superresolution imaging (Klar et al., 2000; Willig et al., 2006; Gustafsson et al., 2008) to visualize Cby1 and five other centriole proteins in MEFs and multiciliated mouse tracheal epithelial cells (MTECs). These other centriole proteins are Cep164, a component of the distal appendages (Graser et al., 2007); Ahi1 and Tmem237, components of the transition zone (Hsiao et al., 2009; Huang et al., 2011; Lancaster et al., 2011); and Ofd1 and Sdccag8, which localize to the distal end of centrioles (Otto et al., 2010; Singla et al., 2010). We and others previously examined Cep164 at high resolution (Sillibourne et al., 2011; Lau et al., 2012; Sonnen et al., 2012), but no such experiments have been reported for the other proteins.
When visualized by 3D-SIM, all of the proteins appeared to be organized as rings of ∼250–300 nm, consistent with being in close apposition to the centriole microtubules, in both cell types (Figure 4, A–C). For this characterization we used MTECs, which, in contrast to MEFs, have hundreds of centrioles docked in the same orientation and plane at the apical surface, allowing for determination of the relative localization of the proteins along the longitudinal axis of the centriole. XZ-projections of 3D-SIM images of MTECs demonstrated that Cby1 is located at a similar position along the longitudinal axis as Ahi1 and Ofd1 (Figure 4, A and B). In contrast, Cep164 and Sdccag8 are located closer to the proximal end of the centriole than Cby1 (farther from the cilium; Figure 4, A and C). It was not possible to image Tmem237 under the 3D-SIM conditions.
FIGURE 4:
Superresolution imaging of Cby1 and centriole proteins. (A–C) 3D-SIM imaging. (A) MEFs or MTECs were fixed and stained for Ahi1 (green), Cby1 (red), and Cep164 (cyan). (B) MEFs or MTECs were fixed and stained for Ofd1 (green), Cby1 (red), and acetylated α-tubulin (Ac-tub, cyan). (C) MEFs or MTECs were fixed and stained for Sdccag8 (green), Cby1 (red), and polyglutamylated tubulin (Glu-tub, cyan). For A–C, MEF and MTEC top rows are maximum Z-projections; MTEC bottom rows are XZ-projections. Scale bars, 500 nm. (D) MTECs were fixed and stained for Tmem237, Ahi1, Cby1, Ofd1, or Sdccag8 and imaged using STED microscopy. Insets, enlarged centriolar regions. Mean diameters and SDs of centriolar rings for each protein (n ≥ 45) are indicated to the right. Mean diameters were derived from maximum intensity points of radial intensity profiles (Supplemental Figure S3). Ring diameters of each protein are statistically different from all other proteins with p < 0.01 (Student's two-tailed t test). Scale bars, 500 nm.
3D-SIM imaging does not have sufficient lateral resolution to clearly resolve differences in ring sizes of the various proteins, which might reveal structural relationships within centriolar domains. STED immunofluorescence microscopy (Lau et al., 2011, 2012) was used to image the same set of proteins at higher resolution (Figure 4D). We previously used STED, with a lateral resolution of ∼60 nm, to show that Cep164 forms a ring of ninefold symmetric clusters with a diameter of 310 nm (Lau et al., 2012). Under the same conditions, the proteins examined by STED were also organized as rings, with diameters ranging from 210 nm for Tmem237 to 270 nm for Sdccag8 (Figure 4D). It was possible to discern apparent ninefold symmetric organization of Sdccag8 and Ofd1, similar to our previous data for Cep164 (Lau et al., 2012). Cby1, Ahi1, and Ofd1 had similar ring diameters (248, 232, and 262 nm, respectively). Together the 3D-SIM and STED data demonstrate that Cby1, Ahi1, and Ofd1 are similarly positioned in the longitudinal and transverse axes of the centriole, suggesting possible common function at the distal end of the centriole.
Cby1 facilitates recruitment of Ahi1 to the transition zone
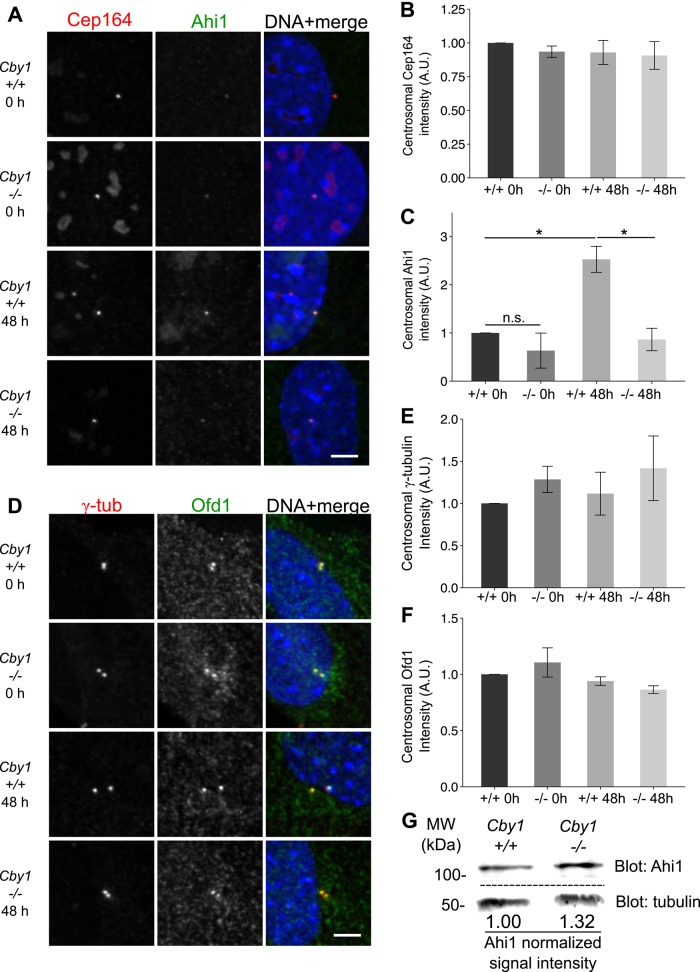
Given the close spatial relationship between Cby1, Ahi1, and Ofd1, we tested whether association of either Ahi1 or Ofd1 with the centrosome depends on Cby1. The fluorescence signal for each protein was determined in Cby1−/− MEFs and Cby1+/+ controls. The amount of Ahi1 at the centrosomes of serum-starved Cby1−/− MEFs was more than twofold lower than in Cby1+/+ controls (Figure 5, A and C), and this could be complemented with a human GFP-Cby1 construct (Figure 6, A and B). This was likely due to a defect in recruitment of Ahi1 to the centrosome rather than a reduction of total Ahi1 level, because centrosomal Ahi1 was similar in Cby1−/− and Cby1+/+ cycling cells before starvation-induced recruitment of Ahi1 (Figure 5, A and C), and total Ahi1 assessed by Western blotting of cycling cells was unchanged (Figure 5G). The reduction in centrosomal Ahi1 was apparent in both ciliated and nonciliated Cby1−/− cells (Figure 6, C and D) and thus is not due to failure of the centrioles in Cby1−/− cells to undergo the transition to forming a cilium. In contrast to Ahi1, the amounts of centrosomal Cep164 (Figure 5B), Ofd1 (Figure 5, D and F), Tmem237, and Sdccag8 (Supplemental Figure S2, A–E) were unchanged in Cby1+/+ and Cby1−/− cells, regardless of time in serum starvation. Loss of CP110 from the mother centriole precedes cilia formation (Tsang et al., 2008); this loss still occurred in Cby1−/− MEFs (Supplemental Figure S2, F and G). These data demonstrate that Cby1 is required, directly or indirectly, for efficient recruitment of the transition zone protein Ahi1 and that this defect is manifested after initiation of the centriole-basal body transition marked by CP110 loss.
FIGURE 5:
Cby1 is required for efficient recruitment of Ahi1 to the centrosome. (A) Cby1+/+ or Cby1−/− MEFs were fixed after 0 or 48 h of serum starvation and stained for Cep164 (red), Ahi1 (green), and DNA (DAPI, blue). Scale bar, 5 μm. (B, C) Centrosomal levels of Cep164 (B) or Ahi1 (C) were quantified and normalized to Cby1+/+ 0-h serum starvation condition. Results shown are the mean of three independent experiments ± SEM (>100 centrosomes/experiment, *p < 0.05). (D) Cby1+/+ or Cby1−/− MEFs were fixed after 0 or 48 h of serum starvation and stained for γ-tubulin (γ-tub, red), Ofd1 (green), and DNA (DAPI, blue). Scale bar, 5 μm. (E) Centrosomal levels of γ-tubulin were quantified and normalized to Cby1+/+ 0-h serum starvation condition. Results shown are the mean of three independent experiments ± SEM (>100 centrosomes/experiment). (F) Centrosomal levels of Ofd1 were quantified as fluorescence intensity within the region defined by γ-tubulin labeling and then normalized to Cby1+/+ 0-h serum starvation condition. Results shown are the mean of three independent experiments ± SEM (>100 centrosomes/experiment). (G) Western blot analysis of extracts from Cby1+/+ or Cby1−/−MEFs probed with anti-Ahi1 or anti–α-tubulin as a loading control. For each sample, Ahi1 signal intensity was quantified and divided by tubulin signal intensity. Values shown are normalized to Cby1+/+ signal intensity.
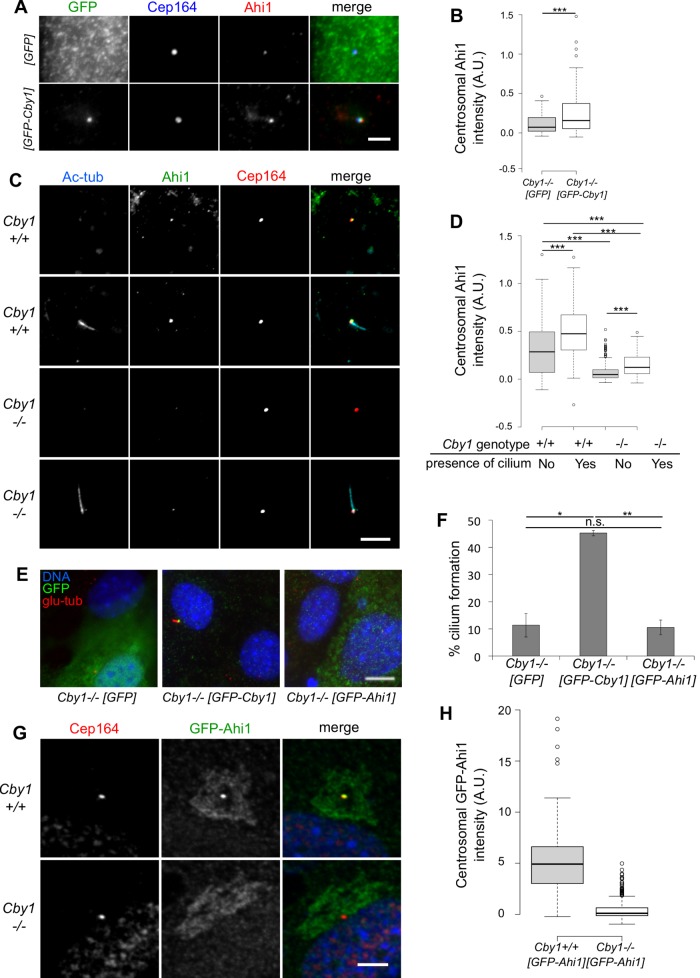
FIGURE 6:
Regulation of Ahi1 by Cby1. (A) Cby1−/− MEFs were transduced with lentivirus expressing GFP or GFP-Cby1, serum starved for 48 h, fixed, and immunostained for GFP (green), Cep164 (blue), and Ahi1 (red). Scale bar, 2.5 μm. (B) Centrosomal levels of Ahi1 in Cby1−/− MEFs expressing GFP or GFP-Cby1 quantified and displayed as a Tukey boxplot. Results from duplicate independent experiments, >50 observations/experiment, ***p < 0.001 by Mann–Whitney U test. (C) Cby1+/+ or Cby1−/− MEFs were fixed after 48 h of serum starvation and stained for Cep164 (red), Ahi1 (green), and acetylated tubulin (cyan). Scale bar, 5 μm. (D) Centrosomal levels of Ahi1 in MEFs with or without acetylated tubulin-labeled cilia quantified and displayed as a Tukey boxplot. Results from duplicate independent experiments, >60 observations/experiment, ***p < 0.001 by Mann–Whitney U test. (E) Cby1−/− MEFs were transduced with lentivirus expressing GFP or GFP-Cby1 or GFP-Ahi1, serum starved for 48 h, fixed, stained for GFP (green), polyglutamylated tubulin (glu-tub, red), and DNA (blue), and (F) scored for frequency of primary cilium formation. Results shown are the mean of three independent experiments ± SEM (>100 cells/experiment; *p < 0.05, **p < 0.01). Scale bar, 5 μm. (G) Cby1+/+ or Cby1−/− MEFs were transduced with lentivirus expressing GFP-Ahi1, serum starved for 24 h, fixed, and stained for GFP (green) and Cep164 (red). Scale bar, 5 μm. (H) Centrosomal levels of GFP quantified and displayed as a Tukey boxplot. Results from three independent experiments, >100 observations/experiment, ***p < 0.001 by Mann–Whitney U test.
If the reduction of centrosomal Ahi1 in Cby1−/− MEFs were due simply to reduced affinity of Ahi1 for the centriole, then overexpression of Ahi1 might be able to rescue the localization and ciliogenesis defects. Cby1−/− MEFs were stably transduced with lentivirus expressing green fluorescent protein (GFP), GFP-Cby1, or GFP-Ahi1, serum starved for 24 h, and stained for cilia. As described in Figure 2, GFP-Cby1 rescued the ciliogenesis defect in Cby1−/− MEFs. However, neither GFP nor GFP-Ahi1 was able to rescue the defect (Figure 6, E and F). Consistent with this, overexpression of GFP-Ahi1 was not able to overcome the reduced centrosomal amount of the protein in Cby1−/− MEFs relative to Cby1+/+ MEFs (Figure 6, G and H). These results suggest that the defect in association of Ahi1 with the centriole involves a structural change, possibly affecting other proteins, rather than simple reduction in affinity for Ahi1.
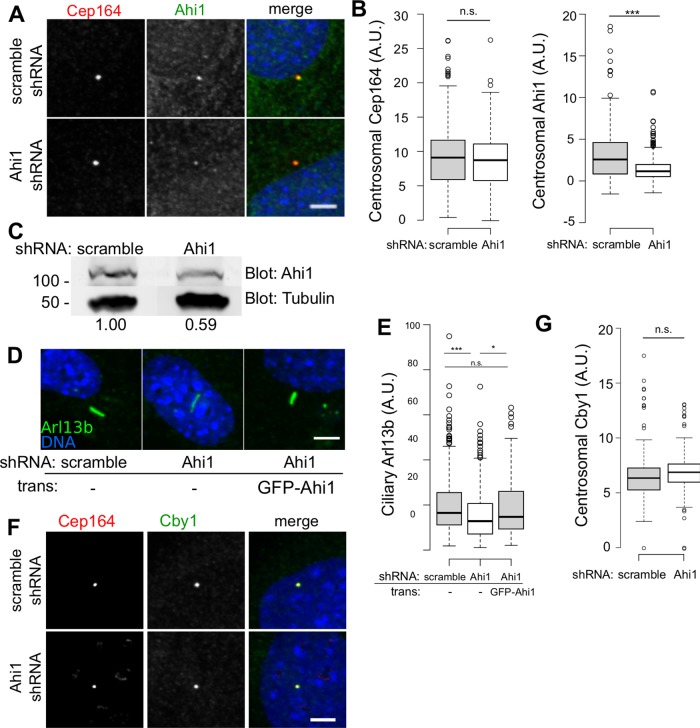
To determine whether Ahi1, like Cby1, also regulates ciliary transport, we stably transduced Cby1+/+ MEFs with lentivirus expressing a short hairpin interfering RNA (shRNA) targeting Ahi1 or a scrambled nontargeting control. MEFs were serum starved for 24 h and stained for Ahi1, with the markers indicated in Figure 7. Fluorescence intensity measurements indicated that centrosomal Ahi1 levels in cells depleted of Ahi1 were reduced to approximately half of the levels in control cells (Figure 7, A and B). Western blots of whole-cell lysates indicated similar depletion of total Ahi1 (Figure 7C). Fluorescence intensity measurements of Arl13b indicated that ciliary Arl13b in Ahi1-depleted cells was reduced to ∼75% of the levels in control cells (Figure 7, D and E), consistent with a report that Arl13b was lost from some primary cilia of Ahi1-mutant patient fibroblasts (Tuz et al., 2013). Overexpression of depletion-resistant GFP-tagged human Ahi1 compensated for the observed defect in Arl13b, confirming that the shRNA used specifically depleted Ahi1. Cby1 levels were similar in both control cells and cells depleted of Ahi1 (Figure 7, F and G). These data indicate that partial depletion of Ahi1 results in a phenotype that is similar to, but less severe than, the reduction of centrosomal Ahi1 caused by deletion of Cby1 and that Ahi1 acts downstream of Cby1. These experiments, together with previous work linking Ahi1 to cilium formation and function (Hsiao et al., 2009; Lancaster et al., 2009; Tuz et al., 2013), suggest that defective recruitment of Ahi1, and probably other transition zone proteins, in Cby1−/− MEFs might provide a molecular explanation for the observed ciliogenesis defect.
FIGURE 7:
Effects of depleting Ahi1. (A) Cby1+/+ MEFs transduced with lentivirus expressing shRNA targeting Ahi1 or scrambled control were serum starved for 24 h, fixed, and stained for Cep164 and Ahi1. Scale bar, 5 μm. (B) Centrosomal levels of Cep164 or Ahi1 quantified and displayed as Tukey boxplots. Results from duplicate experiments, >100 observations/experiment, ***p < 0.001 by Mann–Whitney U test. (C) Western blot analysis of extracts from Cby1+/+ MEFs stably transduced with lentivirus expressing shRNA targeting Ahi1 or scrambled control and probed with anti-Ahi1 or anti–α-tubulin as a loading control. For each sample, Ahi1 signal intensity, shown at bottom, was quantified and divided by tubulin signal intensity. Values shown are normalized to control shRNA signal intensity. (D) MEFs transduced with lentivirus expressing short hairpin RNA against Ahi1 or a scrambled control or with shRNA against Ahi1 followed by GFP-human-Ahi1 transgene, serum starved for 24 h, fixed, and stained for Arl13b. Scale bar, 5 μm. (E) Ciliary Arl13b intensity quantified and displayed as a Tukey boxplot. Results from three independent experiments, >40 observations/experiment. ***p < 0.001; *p < 0.05; n.s., not statistically significant; by Mann–Whitney U test. (F) Cby1+/+ MEFs transduced with lentivirus expressing shRNA targeting Ahi1 or scrambled control were fixed and stained for Cep164 and Cby1. Scale bar, 5 μm. (G) Centrosomal levels of Cby1 quantified and displayed as Tukey boxplots. Results from duplicate experiments, >100 observations/experiment.
DISCUSSION
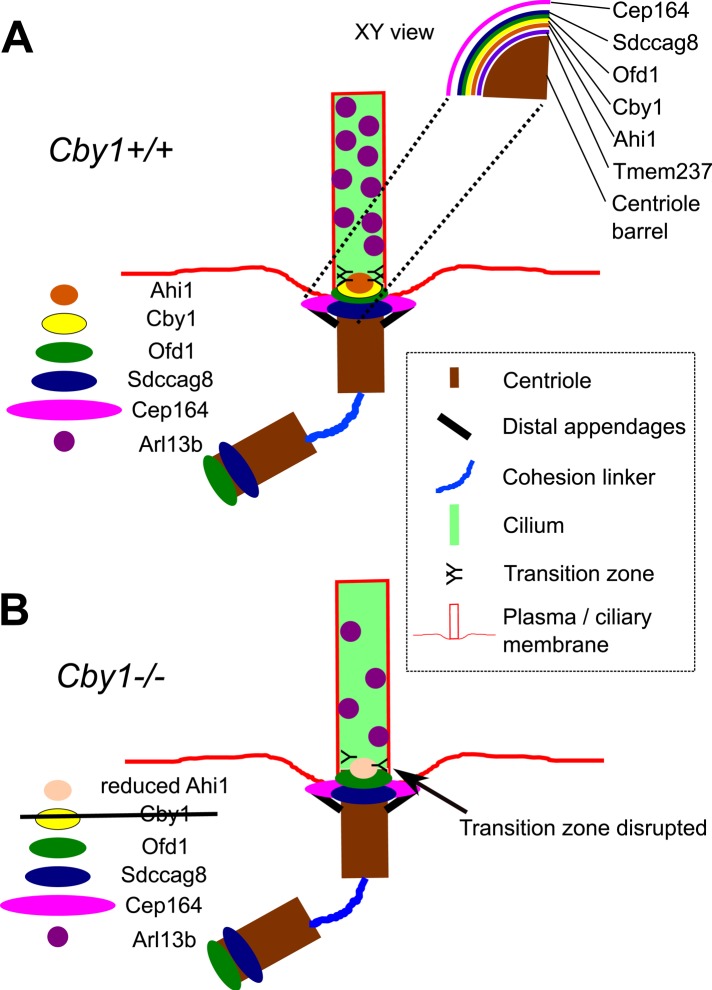
We showed that deletion of Cby1 results in reduced efficiency of primary cilium formation in culture and in cystic kidneys, a cilium-associated defect, in vivo. A possible molecular basis for these effects was revealed by our finding that Cby1 is required for full recruitment of Arl13b to the primary cilium, suggesting a role in determining ciliary protein content. In addition, we showed that the Cby1 protein localizes to a specific compartment at the distal end of the centriole with Ofd1 and Ahi1 and is important for the function of the transition zone between centriole and cilium. The function of Cby1 may be mediated in part by Ahi1, which is reduced at the centrosome of Cby1−/− cells. Given the known functions of these proteins in cilium structure and function (Hsiao et al., 2009; Singla et al., 2010; Lancaster et al., 2011), we propose that the defect in determining ciliary protein content in Cby1−/− cells is due to functional disruption of this domain. We summarize these findings in a model in Figure 8.
FIGURE 8:
Model for localization of centriole/cilium complex proteins and Cby1 function. (A) Representative localizations in Cby1+/+ cells for Ahi1, Cby1, Ofd1, Sdccag8, and Cep164 from superresolution imaging shown on the longitudinal axis of the centriole in the diagram. Localizations on the transverse axis of the centriole are shown in the cutout (XY view). (B) In Cby1−/− cells, levels of centrosomal Ahi1 and ciliary Arl13b are reduced, suggesting that the transition zone is disrupted, resulting in a defect in maintaining ciliary composition.
The cystic kidney phenotype is common to many ciliopathies. The basis for this relationship is not known but has been attributed to disrupted planar cell polarity signaling or spindle orientation defects (Gascue et al., 2011). Using superresolution microscopy, we show here that Ofd1, Cby1, and Ahi1 localize to the same subdomain of the centriole and that Ofd1 is required for Cby1 localization, whereas Cby1 promotes localization of Ahi1 to the centriole. Similar to the cystic kidneys observed in Cby1−/− mice, defects in Ofd1 (Ferrante et al., 2001; 2006; Prattichizzo et al., 2008; Zullo et al., 2010) and Ahi1 (Utsch et al., 2006; Lancaster et al., 2009) in mice and human patients can also result in cystic kidneys. Ofd1 is required for cilium formation (Singla et al., 2010), whereas Ahi1 and Cby1 are only needed to facilitate this process (Hsiao et al., 2009, 2012; Lancaster et al., 2009). The similarity of localization and phenotype of Cby1 and Ahi1 suggests that these two proteins might have related molecular functions.
The defect in recruitment of Arl13b to cilia that we observed in Cby1−/− cells is consistent with the involvement of Cby1 in function of the transition zone, a structure originally defined by electron microscopy and believed to be a barrier to free diffusion into the ciliary compartment (Musgrave et al., 1986). A similar defect in recruiting Arl13b has also been observed in cells from mice with mutations in the transition zone component Tmem67, and these mice also have kidney cysts (Garcia-Gonzalo et al., 2011). The sensory cilia of Caenorhabditis elegans mks-5, nphp-4, mksr-1, and mksr-2 transition zone mutants have defects in Arl13b recruitment as well (Cevik et al., 2013), and a defect in Arl13b recruitment was observed in patient cells with Ahi1 mutations (Tuz et al., 2013). Of importance, we did not observe a disruption in centrosomal localization of another transition zone protein, Tmem237, in Cby1−/− cells; thus Cby1 is likely required for proper assembly or function of a subset of transition zone components. In this sense the phenotype of Cby1 loss is similar to that of depletion of the ciliogenesis-promoting centriole distal-end protein Cep162, which also disrupts only a subset of transition zone proteins (Wang et al., 2013).
Cby1 directly binds and antagonizes β-catenin by competing with Tcf/Lef transcription factors for β-catenin binding and promoting β-catenin nuclear export (Takemaru et al., 2003, 2009; Li et al., 2008, 2010). Ahi1, in addition to being a transition zone component, is a Wnt agonist believed to be sequestered at the centrosome to dampen canonical Wnt signaling (Lancaster et al., 2009, 2011). It is possible that the reduced Ahi1 at the centrosome in Cby1−/− cells makes more Ahi1 available for activating Wnt signaling, contributing in part to elevated canonical Wnt signaling observed in Cby1−/− MEFs and mouse tissues (Voronina et al., 2009; Love et al., 2010).
On the basis of the depletion phenotype of Cby1 in cultured cells, Steere et al. (2012) proposed that Cby1 is required for primary cilium formation. However, Cby1−/− mice (Voronina et al., 2009) did not exhibit the complete embryonic lethality associated with loss of primary cilia (Marszalek et al., 1999; Murcia et al., 2000; Huangfu et al., 2003). Our results with null Cby1−/− cells revealed a delay in primary cilium formation upon serum starvation rather than a block to the process, which is more consistent with the mouse phenotype. One possible explanation for this disparity comes from similar experiments on the centrosome satellite protein Azi1, in which acute depletion of the protein in cultured cells causes a more severe phenotype than that observed in null cells derived from the mutant mouse (Hall et al., 2013). Another potential disparity is that the defect in primary cilium number that we observed in Cby1−/− mouse kidneys might have been expected to result in a phenotype more typical of other mutations that disrupt cilium formation and cause Hedgehog signaling defects. We have not investigated Hedgehog-associated defects such as situs inversus or polydactyly in detail in the Cby1−/− mice, but we note that any defect in cilium formation caused by loss of Cby1 might be variable by tissue and thus not as severe as in the kidney. With regard to Hedgehog signaling at the cellular level, we observed apparently normal translocation of Smo to the primary cilium upon Hedgehog pathway activation. In hennin mice with null alleles of Arl13b, Smo can translocate to the primary cilium in response to Hedgehog pathway activation, but translocation of Gli2 and Gli3 is impaired (Caspary et al., 2007; Larkins et al., 2011). Although we did not investigate this, it is possible that the reduced Arl13b levels in Cby1−/− MEFs might result in Gli translocation defects even though Smo translocation appears normal.
The nanoscale molecular organization of the centrosome has been determined in unprecedented detail by the recent application of superresolution imaging methods (Sillibourne et al., 2011; Sir et al., 2011; Fu and Glover, 2012; Lau et al., 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). Most previous work focused on the pericentriolar material, which is the largest domain of the centrosome; here, we analyze several components localizing to the distal end of the centriole, a more compact domain critical to organizing cilia. We show that Cby1, Ofd1, and Ahi1 localize as tightly apposed rings at a similar position along the long axis of the centriole. Ahi1 is considered to be a protein of the transition zone (Reiter et al., 2012), and, along with other transition zone proteins, it is assembled into the structure during ciliogenesis. In contrast, Ofd1 and Cby1 are constitutive centriole components. The colocalization of the three proteins suggests that Ofd1 and Cby1 link the centriole distal end and the transition zone proper. In contrast, Sdccag8, although also at the distal end of the centriole, is localized in a separate domain from Ofd1 and Cby1, closer to the proximal end of the centriole. In the case of Ofd1, the protein with the largest-diameter ring among these three, it was possible to resolve ninefold symmetric clusters, as shown for the distal appendage protein Cep164 (Lau et al., 2012), the pericentriolar protein PLP (Mennella et al., 2012), and the cartwheel protein SAS-6 (Keller et al., 2014). This suggests that Ofd1, and perhaps the other proteins at the interface of the centriole and cilium, is linked to the centriole/axoneme microtubule doublets that impart that ninefold symmetry. Further application of superresolution methods to the centriole/cilium complex will be crucial to understanding the relationship between the unique structure of this organelle and its function.
MATERIALS AND METHODS
Cby1 −/− mice, histology, and immunohistochemistry
The generation of Cby1−/− mice has been described previously (Voronina et al., 2009), and mice were maintained on a mixed genetic background (87.5% C57BL/6J and 12.5% 129SvJ). Animals were housed in pathogen-free conditions, and all experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee of Stony Brook University. Mice were killed by CO2 asphyxiation, and the kidneys were fixed in 4% paraformaldehyde and processed for paraffin sectioning. The paraffin sections were subjected to hematoxylin and eosin staining and lectin staining with rhodamine-DBA and fluorescein isothiocyanate–LTL (Vector Laboratories, Burlingame, CA). For immunostaining for Cby and acetylated α-tubulin, kidney samples were fresh-frozen in Cryo-Gel medium (Instrumedics, Ann Arbor, MI), and frozen sections were postfixed with methanol/acetone (1:1), processed for immunostaining as described previously, and mounted using Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL). Images of representative fields were acquired using a BX61 microscope (Olympus, Center Valley, PA) equipped with a Cooke Sensicam QE charge-coupled device camera. For quantification of the number of renal cilia, 5-μm paraffin sections of kidneys were processed for immunostaining for acetylated α-tubulin and 4′,6-diamidino-2-phenylindole (DAPI). Ten nonoverlapping images in cortex regions devoid of noticeable cysts were captured at 100× objective magnification for each kidney. Primary cilia >2.0 μm that protruded into the tubular lumen were counted, blind to genotype.
Plasmids, recombinant proteins, and antibodies
GFP-Cby1 plasmid was described previously (Li et al., 2010). pEGFP-N1 (Clontech, Mountain View, CA) was used for transient expression of enhanced GFP (EGFP). A full-length cDNA clone of Cep164 was obtained from Kazusa DNA Research Institute (Chiba, Japan). To generate a hexahistidine (6xHis)-tagged N-terminal fragment spanning positions 1–298 of Cep164 (pTS2775), the relevant codons were amplified from full-length cDNA and cloned into pDONR221 using Gateway recombination technology (Invitrogen, Carlsbad, CA) to obtain pENTR-Cep164-1-298 (pTS2701). pTS2775 was then obtained by Gateway recombination between pTS2701 and pDEST 17 (Invitrogen). A human Cby1 Gateway entry clone, pTS3506, was obtained by PCR amplification from GFP-Cby1 (Li et al., 2010) and Gateway recombination into pDONR221. To obtain EGFP-Cby1 lentiviral transfer vector (pTS3617), Gateway recombination was carried out between pTS3506 and pTS3517, a Gateway lentiviral destination plasmid that after recombination results in an EGFP tag at the N-terminus of the gene of interest. A human Ahi1 Gateway entry clone, pTS3601, was obtained by PCR amplification of EGFP-Ahi1 (Lancaster et al., 2009), obtained from Addgene (Cambridge, MA; plasmid 30494), followed by Gateway recombination into pDONR221. An EGFP-Ahi1 lentiviral transfer vector (pTS3591) was obtained by Gateway recombination between pTS3601 and pTS3517. pSicoR (Ventura et al., 2004) was used for lentivirus-mediated expression of EGFP. Lentiviral transfer vectors expressing a nontargeting (SHC002) shRNA or an shRNA targeting mouse Ahi1 (TRCN0000190390) were obtained from Sigma-Aldrich (St. Louis, MO). 6xHis-Cep164-1-298 protein was produced and purified as described (Graser et al., 2007). Rabbit anti-Cep164 antisera were raised against this antigen (Cocalico, Reamstown, PA), affinity purified on nitrocellulose blots with the antigen, and used at 1 ng/μl for immunofluorescence. Other antibodies used in this study for immunofluorescence were mouse anti–polyglutamylated tubulin (GT335; Adipogen, San Diego, CA) at 1:500, mouse anti–acetylated α-tubulin (6-11B-1; Abcam, Cambridge, MA) at 1:5000, mouse anti–γ-tubulin (GTU-88; Sigma-Aldrich) at 1:1000, mouse anti-Ahi1 (ab93386; Abcam, Cambridge, MA) at 1:1000, mouse-anti Arl13b (75-287; Antibodies Incorporated, Davis, CA) at 1:200, mouse anti-Cby1 (8-2; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:100, mouse anti-centrin3 (clone 3E6, H00001070-M01; Abnova, Taipei, Taiwan), rabbit anti-CP110 (Chen et al., 2002) at 1:500, mouse anti-GFP (clone 3e6, A-11120; Invitrogen, Carlsbad, CA) at 1:500, rabbit anti-GFP (Hatch et al., 2010) at 1.5 μg/ml, rabbit anti-Ofd1 (Singla et al., 2010) at 1:1000, rabbit anti-Sdccag8 (13471-1-AP; Proteintech, Chicago, IL) at 1:500, rabbit anti-Smoothened (ab38686; Abcam) at 1:1000, and rabbit anti-Tmem237 (Zuniga and Craft, 2010) at 1:500. For Western blots, the following antibodies were used: mouse anti–α-tubulin (DM1α; Sigma-Aldrich) at 1:10,000 and rabbit anti-Ahi1 (22045-1-AP; Proteintech) at 1:500. For immunohistochemistry, the foregoing antibodies were used, but at higher concentrations: mouse anti acetylated α-tubulin at 1:500 and mouse anti-Cby1 at 1:100.
Cell culture, transfections, and lentivirus transduction
Cby1+/+ and Cby1−/− MEFs were derived from mice on a mixed genetic background (87.5% C57BL/6J and 12.5% 129SvJ) as described previously (Voronina et al., 2009) and grown in DMEM (Cellgro, Manassas, VA) with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA). Mouse tracheal epithelial cells were cultured as previously described (Vladar and Stearns, 2007; Vladar and Brody, 2013). Ofd1+/+ and Ofd1−/− ES cells were described previously (Singla et al., 2010) and grown in Knockout DMEM (Invitrogen) with 15% ES cell–qualified fetal bovine serum (Omega Sciences, Tarzana, CA), recombinant leukemia inhibitory factor, 1 mM sodium pyruvate, 4 mM l-glutamine, 1 mM nonessential amino acids, 0.1 mM 2-β-mercaptoethanol, and 1× penicillin/streptomycin (all cell culture additives were from Thermo Fisher Scientific, Waltham, MA). For primary cilium formation experiments, cells were cultured with reduced (0.5%) serum. Plasmid transfections were performed using Lipofectamine 2000 or Lipofectamine LTX according to the manufacturer's protocol (Invitrogen). For Smoothened translocation experiments, MEFs were grown to near confluence (80–100%) and switched to reduced serum medium for 24 h. The medium was then changed to reduced-serum medium with 200 nM SAG (Enzo, Farmingdale, NY) and fixed for immunofluorescence after incubation for a further 24 h. Lentivirus constructs were generated and cells were infected as previously described (Schaub and Stearns, 2013), except that lentivirus was concentrated with PEG 6000 (Kutner et al., 2009). Pools of MEFs stably expressing short hairpin RNA were obtained by selecting infected cells in 1 μg/ml puromycin (Invivogen, San Diego, CA) for at least 48 h.
Immunofluorescence and microscopy
For immunofluorescence of tissue culture cells, cells were grown on poly-l-lysine–coated #1.5 glass coverslips (Electron Microscopy Sciences, Hatfield, PA) or high-tolerance #1.5 coverslips (Marienfeld, Lauda Konigshofen, Germany) for 3D-SIM, washed with PBS, and fixed in either 4% paraformaldehyde in PBS at room temperature or −20°C methanol for 5–10 min. Paraformaldehyde-fixed samples were quenched in 1 mg/ml NaBH4. After fixation, cells were washed with PBS, followed by extraction and blocking with PBS containing 3% bovine serum albumin (Sigma-Aldrich), 0.1% Triton X-100, and 0.02% sodium azide (PBS-BT). Coverslips were incubated with primary antibodies diluted in PBS-BT for 1–3 h at room temperature or overnight at 4°C. Alexa Fluor dye–conjugated secondary antibodies (Invitrogen) were diluted in PBS-BT 1:500 and incubated at room temperature for 45 min. In cases in which cells were labeled with two mouse monoclonal antibodies, appropriate isotype-specific secondary antibodies were used to distinguish the antibodies. Nuclei were stained by brief incubation with DAPI (1 μg/ml). Coverslips were mounted using Mowiol (Polysciences, Warrington, PA) in glycerol containing 1,4,-diazobicycli-[2.2.2]-octane (Sigma-Aldrich) as antifade or with SlowFade Gold (Invitrogen) for 3D-SIM. Mouse tracheal epithelial cells grown on Transwell filters were fixed and stained as previously described (Vladar and Stearns, 2007; Vladar and Brody, 2013) and mounted on conventional coverslips or high-tolerance coverslips for 3D-SIM.
For standard immunofluorescence, images were acquired using an Axiovert 200M microscope (Carl Zeiss, Jena, Germany) with Plan-Neofluar 100×/1.3 numerical aperture (NA) and PlanApoChromat 63×/1.4 NA objectives and a cooled, charge-coupled device (CCD) camera (Orca ER; Hamamatsu Photonics, Hamamatsu, Japan). For quantitative immunofluorescence, Z-stacks encompassing the entire thickness of the centrosome or cilium were collected at 0.3-μm intervals on a Zeiss Axio Observer microscope with a confocal spinning-disk head (Yokogawa, Tokyo, Japan), PlanApoChromat 63×/1.4 NA objective, and a Cascade II:512 electron-multiplying (EM) CCD camera (Photometrics, Tucson, AZ). For some quantitative immunofluorescence experiments involving four-color imaging, Z-stacks encompassing the entire thickness of the centrosome were collected at 0.3-μm intervals on the Axiovert 200M.
3D-SIM imaging was performed on a DeltaVision OMX V4 BLAZE system (Applied Precision, Issaquah, WA) equipped with a 100×/1.42 NA U-PLANAPO SIM oil immersion objective (Olympus), 405-, 488-, 568-, and 642-nm lasers, and three EMCCD cameras. Images were acquired in the sequential imaging mode. SI patterns were generated using an electro-optical high-speed SI diffraction grating engine. Image stacks 3–6 μm in height with 15 images per plane (five phases, three angles) and a z-distance of 0.125 μm were acquired and computationally reconstructed to generate superresolution optical serial sections with twofold extended resolution in all three axes. Color channels were computationally aligned based on control measurements with 0.1-μm multispectral fluorescent beads (Tetraspeck beads; Invitrogen). SI reconstruction and image processing were performed with the SoftWoRx 3.7 imaging software package (Applied Precision).
STED microscopy was performed with a home-built pulsed STED microscope optimized for imaging Atto647N labels. The inverted fluorescence microscope (Olympus IX71; Plan Fluor 100×/1.3 NA oil immersion; Nikon, Melville, NY) used a pulsed pump source at 640 nm with ∼100-ps pulse width (PDL 800-B; PicoQuant, Berlin, Germany) and a pulsed, donut-shaped STED beam at 750 nm produced by an 80-MHz mode-locked Ti:sapphire laser (Mira 900D; Coherent, Santa Clara, CA) pumped by a 10-W, 532-nm laser (Verdi-10; Coherent). The STED beam pulse was stretched by propagation in dispersive glass and 100 m of silica fiber and phase modulated by a 2π spiral phase plate (RPC Photonics, Rochester, NY). Typical pump powers at the sample are as follows: for 640 nm, 10–40 kW cm−2, and for 750 nm, 140–150 MW cm−2. Emitted fluorescence was filtered with a confocal pinhole and detected by a Si avalanche photodiode (SPCM-ARQH-13; PerkinElmer, Waltham, MA). For each image, the sample was moved by stage scanning at a rate of 0.5 ms/pixel of size 18.5 × 18.5 nm. Further details of the experimental setup are provided in previous publications (Lau et al., 2011, 2012). Images were processed using ImageJ (National Institutes of Health, Bethesda, MD).
Quantitation and statistical analysis of diffraction-limited immunofluorescence images
Quantitative immunofluorescence was performed on cells by acquiring confocal stacks of control and mutant cells using identical settings. For some experiments involving four-color imaging, stacks were acquired with a widefield microscope. Maximum intensity projections were used for all quantification. The region of interest (ROI) in the images was defined using a centrosome (γ-tubulin staining), centriole (Cep164 or centrin staining), or cilium (acetylated tubulin or Arl13b) marker in each cell. ImageJ or Cellprofiler (Carpenter et al., 2006) was used to quantify signal intensity. In Cellprofiler, background subtraction was performed by subtracting the mean intensity from a 5-pixel region immediately surrounding the ROI from the mean intensity of the ROI. The background-subtracted integrated intensity of the ROIs was obtained by multiplying background-subtracted mean intensity by area. In ImageJ, background subtraction was performed by subtracting the integrated intensity of a neighboring area of identical dimensions to the ROI. Primary cilium formation was assessed by counting the total number of cells and the number of cells with primary cilia, as detected by acetylated or polyglutamylated tubulin staining. Statistical analysis was performed using Student's t test or Mann–Whitney U-test in Excel (Microsoft, Redmond, WA) or R (www.r-project.org/) and RStudio (www.rstudio.com/about/). Error bars reflect SEM. Bar graphs were drawn in Excel or R, and Tukey boxplots were drawn using R or BoxPlotR (Spitzer et al., 2014).
Quantitation and statistical analysis of STED images
Ring radii were determined using a bespoke MATLAB (MathWorks, Natick, MA) program (outlined in Supplemental Figure 3A). First, ∼1 × 1 μm2 square ROIs containing well-isolated centrioles were identified from STED images. The cell edges and obvious regions above and below the focal plane were avoided to minimize underestimating the radii due to projection effects. An annulus with fixed radii (inner/outer radii in nanometers: Tmem237, 35/180; Ahi1, 50/180; Cby1, 55/205; Ofd1, 55/210; Sdccag8, 65/210) around a user-determined center further isolated the centrioles in order to fit an ellipse. The maximum intensity pixels in seven radially equal sections of the annulus were used to fit an ellipse using a linear least squares approach, and the fits were verified by eye (Supplemental Figure S3B). A fraction of centrioles (∼15%) were discarded due to poor ellipse fits. The raw data from the entire ROI were then interpolated 10 times using a cubic convolution (Matlab function interp2) and the interpolated image used to determine the radial intensity profile by calculating the average pixel intensity in 5-nm-wide annuli about the centriole center determined via the ellipse fit (Supplemental Figure S3C, top). The ring radius for each centriole was defined as the maximum intensity point of the radial intensity profile. Ring radii distributions were analyzed using a two-sample t test (Supplemental Figure S3C, bottom). As expected, the ring radii and the average of the major and minor axes from the ellipse fit are well correlated, with a Pearson's r of 0.88 (Supplemental Figure S3D).
Cell extracts and Western blots
Whole-cell extracts for Western blotting were prepared by lysing cells in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40 substitute, 1 μg/ml each leupeptin, pepstatin, and chymostatin). Protein extracts were resolved by SDS–PAGE and transferred to nitrocellulose membrane (Bio-Rad, Hercules, CA). Membranes were blocked with 5% milk, washed with Tris-buffered saline containing 0.5% Tween-20 (Sigma-Aldrich), and probed with primary antibodies. Bound primary antibodies were detected using secondary antibodies conjugated to IRDye 800CW or IRDye 680RD on an Odyssey CLx fluorescence imaging system according to manufacturer's recommendations (LI-COR, Lincoln, NE). Relative protein concentrations were quantified using ImageJ.
Supplementary Material
Acknowledgments
We thank Daniel Van de Mark and Christian Hoerner for critical reading of the manuscript, Christian Hoerner for generating pTS3517, Johanna Schaub for recombinant LIF, Jeremy Reiter (University of California, San Francisco, San Francisco, CA) for the gift of Ofd1+/+ and Ofd1−/− ES cells, and Cheryl M. Craft (Mary D. Allen Laboratory for Vision Research, Doheny Eye Institute, University of Southern California, Los Angeles, CA), Brian Dynlacht (New York University, New York, NY), and Jeremy Reiter (University of California, San Francisco) for gifts of antibodies. This work was supported in part by National Institutes of Health Grants R01GM52022 (T.S.), R01HL107493 (K.T.), and R01GM086196 (W.E.M.), the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (ZIA DK075042; G.G.G.), a graduate fellowship from the Singapore Agency for Science, Technology and Research (Y.L.L.), and a National Science Foundation Graduate Fellowship (C.J.C.). The 3D-SIM superresolution experiments were supported, in part, by Award 1S10OD01227601 from the National Center for Research Resources.
Abbreviations used:
- BBS
Bardet–Biedl syndrome
- DBA
Dolichos biflorus agglutinin
- 3D-SIM
three-dimensional structured illumination
- IFT
intraflagellar transport
- LTL
Lotus tetragonolobus lectin
- MEFs
mouse embryonic fibroblasts
- MKS
Mekel–Gruber syndrome
- MTECs
mouse tracheal epithelial cells
- ROI
region of interest
- Smo
Smoothened
- STED
stimulated emission depletion
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-02-0735) on August 7, 2014.
REFERENCES
- Avasthi P, Marshall WF. Stages of ciliogenesis and regulation of ciliary length. Differentiation. 2012;83:S30–S42. doi: 10.1016/j.diff.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O'Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attie-Bitach T, Holden KR, Dobyns WB, et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley-Fisher C, Cottell D, Katada T, Kontani K, Blacque OE. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol. 2010;188:953–969. doi: 10.1083/jcb.200908133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S, Sanders AAWM, van Wijk E, Boldt K, Clarke L, van Reeuwijk J, Hori Y, Horn N, Hetterschijt L, Wdowicz A, et al. Active transport and diffusion barriers restrict Joubert syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. 2013;PLoS Genet 9, e1003977 doi: 10.1371/journal.pgen.1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen M-H, Chuang P-T, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 2009;136:4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjolras C, Thomas J, Chhin B, Cortier E, Duteyrat J-L, Soulavie F, Kernan MJ, Laurençon A, Durand B. Drosophila chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J Cell Biol. 2012;197:313–325. doi: 10.1083/jcb.201109148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Giorgio G, Feather SA, Bulfone A, Wright V, Ghiani M, Selicorni A, Gammaro L, Scolari F, Woolf AS, et al. Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet. 2001;68:569–576. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, Dollé P, Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascue C, Katsanis N, Badano JL. Cystic diseases of the kidney: ciliary dysfunction and cystogenic mechanisms. Pediatr Nephrol. 2011;26:1181–1195. doi: 10.1007/s00467-010-1697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Liu Y, Zaghloul NA, Leitch CC, Lawson SS, Kato M, Beachy PA, Beales PL, DeMartino GN, Fisher S, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- Gradilone SA, Masyuk AI, Splinter PL, Banales JM, Huang BQ, Tietz PS, Masyuk TV, Larusso NF. Cholangiocyte cilia express TRPV4 and detect changes in luminal tonicity inducing bicarbonate secretion. Proc Natl Acad Sci USA. 2007;104:19138–19143. doi: 10.1073/pnas.0705964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL, Shao L, Carlton PM, Wang CJR, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall EA, Keighren M, Ford MJ, Davey T, Jarman AP, Smith LB, Jackson IJ, Mill P. Acute versus chronic loss of mammalian azi1/cep131 results in distinct ciliary phenotypes. PLoS Genet. 2013;9:e1003928. doi: 10.1371/journal.pgen.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y-C, Tong ZJ, Westfall JE, Ault JG, Page-McCaw PS, Ferland RJ. Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking. Hum Mol Genet. 2009;18:3926–3941. doi: 10.1093/hmg/ddp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y-C., Tuz K, Ferland RJ. Trafficking in and to the primary cilium. Cilia. 2012;1:4. doi: 10.1186/2046-2530-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Szymanska K, Jensen VL, Janecke AR, Innes AM, Davis EE, Frosk P, Li C, Willer JR, Chodirker BN, et al. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am J Hum Genet. 2011;89:713–730. doi: 10.1016/j.ajhg.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Keller D, Orpinell M, Olivier N, Wachsmuth M, Mahen R, Wyss R, Hachet V, Ellenberg J, Manley S, Gonczy P. Mechanisms of HsSAS-6 assembly promoting centriole formation in human cells. J Cell Biol. 2014;204:697–712. doi: 10.1083/jcb.201307049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner RH, Zhang X-Y, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Louie CM, Silhavy JL, Sintasath L, Decambre M, Nigam SK, Willert K, Gleeson JG. Impaired Wnt-beta-catenin signaling disrupts adult renal homeostasis and leads to cystic kidney ciliopathy. Nat Med. 2009;15:1046–1054. doi: 10.1038/nm.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Schroth J, Gleeson JG. Subcellular spatial regulation of canonical Wnt signalling at the primary cilium. Nat Cell Biol. 2011;13:700–707. doi: 10.1038/ncb2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins CE, Aviles GDG, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Lee YL, Matis M, Axelrod J, Stearns T, Moerner WE. STED Super-resolution microscopy in Drosophila tissue and in mammalian cells. Proc Soc Photo Opt Instrum Eng. 2011:2011(Feb 11), 79101N. doi: 10.1117/12.881221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Lee YL, Sahl SJ, Stearns T, Moerner WE. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys J. 2012;102:2926–2935. doi: 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Li F-Q, Mofunanya A, Fischer V, Hall J, Takemaru K-I. Nuclear-cytoplasmic shuttling of Chibby controls beta-catenin signaling. Mol Biol Cell. 2010;21:311–322. doi: 10.1091/mbc.E09-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F-Q, Mofunanya A, Harris K, Takemaru K-I. Chibby cooperates with 14-3-3 to regulate beta-catenin subcellular distribution and signaling activity. J Cell Biol. 2008;181:1141–1154. doi: 10.1083/jcb.200709091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love D, Li F-Q, Burke MC, Cyge B, Ohmitsu M, Cabello J, Larson JE, Brody SL, Cohen JC, Takemaru K-I. Altered lung morphogenesis, epithelial cell differentiation and mechanics in mice deficient in the Wnt/β-catenin antagonist Chibby. PLoS One. 2010;5:e13600. doi: 10.1371/journal.pone.0013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr Biol. 2012;22:1628–1634. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone AMD, Anderson CT, Tummala P, Kwon RY, Johnston TR, Stearns T, Jacobs CR. Primary cilia mediate mechanosensing in bone cells by a calcium-independent mechanism. Proc Natl Acad Sci USA. 2007;104:13325–13330. doi: 10.1073/pnas.0700636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, Woychik RP. The Oak Ridge Polycystic Kidney (ORPK) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- Musgrave A, de Wildt P, van Etten I, Pijst H, Scholma C, Kooyman R, Homan W, van den Ende H. Evidence for a functional membrane barrier in the transition zone between the flagellum and cell body of Chlamydomonas eugametos gametes. Planta. 1986;167:544–553. doi: 10.1007/BF00391231. [DOI] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AEH, Lu W, Brown EM, Quinn SJ, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattichizzo C, Macca M, Novelli V, Giorgio G, Barra A, Franco B, Oral-Facial-Digital Type I (OFDI) Collaborative Group Mutational spectrum of the oral-facial-digital type I syndrome: a study on a large collection of patients. Hum Mutat. 2008;29:1237–1246. doi: 10.1002/humu.20792. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 2012;13:608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Stearns T. The Rilp-like proteins Rilpl1 and Rilpl2 regulate ciliary membrane content. Mol Biol Cell. 2013;24:453–464. doi: 10.1091/mbc.E12-08-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Sillibourne JE, Specht CG, Izeddin I, Hurbain I, Tran P, Triller A, Darzacq X, Dahan M, Bornens M. Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton. 2011;68:619–627. doi: 10.1002/cm.20536. [DOI] [PubMed] [Google Scholar]
- Singla V, Romaguera-Ros M, Garcia-Verdugo J-M, Reiter JF. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev Cell. 2010;18:410–424. doi: 10.1016/j.devcel.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J-H, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D'Santos C, Woods CG, Gergely F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet. 2011;43:1147–1153. doi: 10.1038/ng.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M, Wildenhain J, Rappsilber J, Tyers M. BoxPlotR: a web tool for generation of box plots. Nat Methods. 2014;11:121–122. doi: 10.1038/nmeth.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere N, Chae V, Burke M, Li F-Q, Takemaru K-I, Kuriyama R. A Wnt/beta-catenin pathway antagonist Chibby binds Cenexin at the distal end of mother centrioles and functions in primary cilia formation. PLoS One. 2012;7:e41077. doi: 10.1371/journal.pone.0041077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- Takemaru K-I, Fischer V, Li F-Q. Fine-tuning of nuclear-catenin by Chibby and 14-3-3. Cell Cycle. 2009;8:210–213. doi: 10.4161/cc.8.2.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K-I, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peränen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuz K, Hsiao YC, Juarez O, Shi B, Harmon EY, Phelps IG, Lennartz MR, Glass IA, Doherty D, Ferland RJ. The Joubert syndrome-associated missense mutation (V443D) in the Abelson-helper integration site 1 (AHI1) protein alters its localization and protein-protein interactions. J Biol Chem. 2013;288:13676–13694. doi: 10.1074/jbc.M112.420786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsch B, Sayer JA, Attanasio M, Pereira RR, Eccles M, Hennies H-C, Otto EA, Hildebrandt F. Identification of the first AHI1 gene mutations in nephronophthisis-associated Joubert syndrome. Pediatr Nephrol. 2006;21:32–35. doi: 10.1007/s00467-005-2054-y. [DOI] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Brody SL. Analysis of ciliogenesis in primary culture mouse tracheal epithelial cells. Methods Enzymol. 2013;525:285–309. doi: 10.1016/B978-0-12-397944-5.00014-6. [DOI] [PubMed] [Google Scholar]
- Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina VA, Takemaru K-I, Treuting P, Love D, Grubb BR, Hajjar AM, Adams A, Li F-Q, Moon RT. Inactivation of Chibby affects function of motile airway cilia. J Cell Biol. 2009;185:225–233. doi: 10.1083/jcb.200809144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-J, Tay HG, Soni R, Perumal GS, Goll MG, Macaluso FP, Asara JM, Amack JD, Tsou M-FB. CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat Cell Biol. 2013;15:591–601. doi: 10.1038/ncb2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nat Cell Biol. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- Zullo A, Iaconis D, Barra A, Cantone A, Messaddeq N, Capasso G, Dollé P, Igarashi P, Franco B. Kidney-specific inactivation of Ofd1 leads to renal cystic disease associated with upregulation of the mTOR pathway. Hum Mol Genet. 2010;19:2792–2803. doi: 10.1093/hmg/ddq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga FI, Craft CM. Deciphering the structure and function of Als2cr4 in the mouse retina. Invest Ophthalmol Vis Sci. 2010;51:4407–4415. doi: 10.1167/iovs.10-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.