Although cytoplasmic interactions between Caenorhabditis elegans centrosome proteins SPD-5, RSA-1, and RSA-2 can be detected in vivo, the key proteins required for centrosome assembly, SPD-2 and SPD-5, exist mostly as monomers and are separated from the centrosome-related kinases PLK-1 and AIR-1 in the cytoplasm.
Abstract
Centrosomes are the main microtubule-organizing centers in animal cells. Centrosomes consist of a pair of centrioles surrounded by a matrix of pericentriolar material (PCM) that assembles from cytoplasmic components. In Caenorhabditis elegans embryos, interactions between the coiled-coil proteins SPD-5 and SPD-2 and the kinase PLK-1 are critical for PCM assembly. However, it is not known whether these interactions promote the formation of cytoplasmic complexes that are added to the PCM or whether the components interact only during incorporation into the PCM matrix. Here we address this problem by using a combination of live-cell fluorescence correlation spectroscopy, mass spectrometry, and hydrodynamic techniques to investigate the native state of PCM components in the cytoplasm. We show that SPD-2 is monomeric, and neither SPD-2 nor SPD-5 exists in complex with PLK-1. SPD-5 exists mostly as a monomer but also forms complexes with the PP2A-regulatory proteins RSA-1 and RSA-2, which are required for microtubule organization at centrosomes. These results suggest that the interactions between SPD-2, SPD-5, and PLK-1 do not result in formation of cytoplasmic complexes, but instead occur in the context of PCM assembly.
INTRODUCTION
Centrosomes consist of a pair of barrel-shaped centrioles surrounded by an amorphous protein matrix called the pericentriolar material (PCM). During the cell cycle the PCM assembles from a pool of cytoplasmic PCM proteins (for a more in-depth overview of PCM assembly see Woodruff et al., 2014; Zwicker et al., 2014). Thus identification of the cytoplasmic state of PCM proteins is essential to understanding the underlying mechanism of PCM assembly. This is also of interest because the formation of aberrant centrosomes can severely impair cell viability, indicating that cells need to strictly control the cytoplasmic state of its PCM proteins and prevent ectopic interactions (Sluder et al., 1997; Sluder and Nordberg, 2004; Godinho et al., 2009; Krämer et al., 2011). Surprisingly, little is known about the native complexes formed by centrosome components before their incorporation into the PCM (Habermann and Lange, 2012). A recent study in Drosophila melanogaster suggested that a subset of centriole and PCM components, including SAS-4, CNN, D-PLP, and ASL, interact in the cytoplasm to form preassembled complexes that could serve as intermediates in PCM assembly (Gopalakrishnan et al., 2011). However, it is not yet clear whether formation and incorporation of preassembled complexes is a general principle of PCM assembly across eukaryotes.
In the nematode Caenorhabditis elegans, the key proteins required for centriole and PCM assembly have been defined. SAS-4, SAS-5, SAS-6, and the Plk4 homologue ZYG-1 are critical for centriole assembly (O'Connell et al., 2001; Kirkham et al., 2003; Leidel and Gönczy, 2003; Dammermann et al., 2004; Delattre et al., 2004; Leidel et al., 2005; Sönnichsen et al., 2005; Lettman et al., 2013), whereas SPD-5 and SPD-2, along with the mitotic kinases PLK-1 and Aurora A, control assembly of the PCM (Chase et al., 2000; Hannak et al., 2001; Hamill et al., 2002; Kemp et al., 2004; Pelletier et al., 2004; Sönnichsen et al., 2005; Decker et al., 2011). All known PCM proteins require SPD-2 and SPD-5 to localize to centrosomes, suggesting that these two proteins are core building blocks of PCM in C. elegans (Sluder et al., 1997; Sluder and Nordberg, 2004; Hamill et al., 2002; Kemp et al., 2004; Pelletier et al., 2004; Godinho et al., 2009; Krämer et al., 2011). Protein–protein interaction mapping has identified potential interactions between SPD-2 and PLK-1, as well as between SPD-2 and SPD-5 (Boxem et al., 2008), and subsequent in vitro work confirmed that PLK-1 interacts with a region of SPD-2 predicted to be a polo box–binding site (Decker et al., 2011). Aurora A is not required for the recruitment of the initial shell of PCM that forms after the introduction of centrioles into the cytoplasm during fertilization but does play a role in the subsequent expansion of the PCM that accompanies centrosome maturation after mitotic entry (Hannak et al., 2001; Hamill et al., 2002). On the basis of this information, we define SPD-2 and SPD-5 as the core structural components of the PCM and consider PLK-1 as the major kinase driving PCM assembly in C. elegans. To determine the cytoplasmic state of the core C. elegans PCM proteins, we combined mass spectrometry, hydrodynamic protein analysis, and fluorescence correlation spectroscopy to acquire a detailed picture about the cytoplasmic state of SPD-2 and SPD-5.
RESULTS
Identification of cytoplasmic interactions of SPD-5 and SPD-2 in embryo extracts using label-free quantitative mass spectrometry
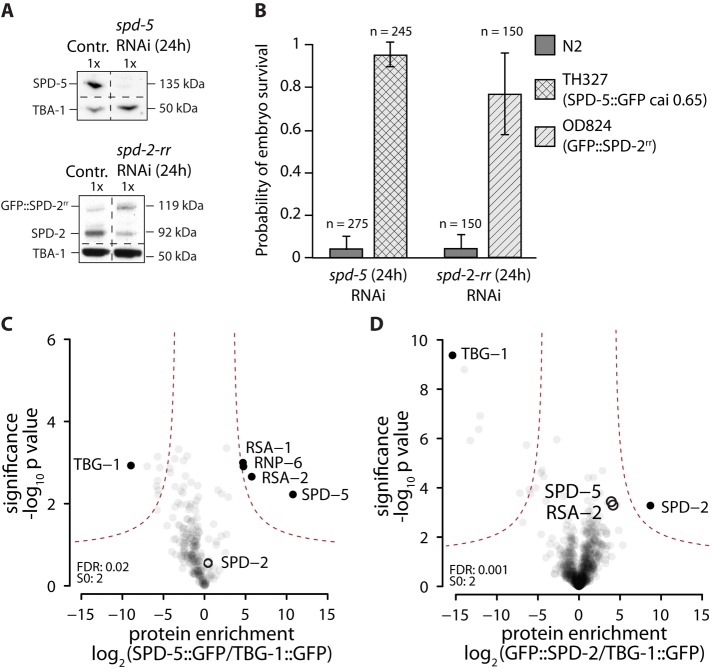
To identify potential cytoplasmic interactions of the PCM proteins SPD-2 and SPD-5, we used label-free-quantitative mass spectrometry. For proper quantification of protein amounts using this technique, the immunoprecipitated proteins must carry the same tag. We therefore used green fluorescent protein (GFP)–tagged versions of SPD-2 and SPD-5 (GFP::SPD-2 and SPD-5::GFP, respectively), which rescue depletion of the corresponding endogenous proteins and therefore are functional (Figure 1, A and B). First, we used ultracentrifugation to generate centrosome-free cytoplasmic extracts from transgenic C. elegans embryos expressing GFP::SPD-2 and SPD-5::GFP, similar to what was described by Gopalakrishnan et al. (2011). GFP::SPD-2 or SPD-5::GFP was then immunoprecipitated from these extracts using bead-coupled GFP antibodies and eluted, and then the coprecipitating proteins were identified by mass spectrometry; each experiment was repeated three times. The enrichment and significance of the enrichment of an identified interactor were assessed via label-free quantification using control protein (TBG-1::GFP) immunoprecipitates as described by Hubner et al. (2010). Analysis of SPD-5::GFP immunoprecipitates revealed that, in addition to SPD-5 (2022-fold enriched in comparison to the control), the most enriched proteins were RSA-2 (63-fold enriched) and RSA-1 (29-fold enriched) (Figure 1C), two known regulatory subunits of the protein phosphatase 2A (PP2A) complex (Schlaitz et al., 2007). This result is consistent with previous LAP::RSA-1 immunoprecipitations from centrosome-containing whole-worm extracts by Schlaitz et al. (2007). A novel interaction was found between SPD-5 and the RNA-binding protein RNP-6 (25-fold enriched above simulated noise level; see Materials and Methods), which shows a postembryonic lethal phenotype upon depletion (Sönnichsen et al., 2005). However, since RNP-6 depletion did not affect centrosomes during the first cell division (unpublished data), its role was not investigated further. Of importance, SPD-2, the centrosome-related kinases PLK-1 and AIR-1, and the centriole proteins SAS-4/-5/-6 were not detected in the immunoprecipitates. This data suggest that SPD-5 molecules interact with RSA-1, RSA-2, and RNP-6 in cytoplasmic embryo extracts.
FIGURE 1:
Identification of cytoplasmic interactors of GFP::SPD-2 and SPD-5::GFP using label-free quantification mass spectrometry. (A) Western blot showing depletion of endogenous SPD-5 or SPD-2 after 24 h of RNAi treatment. SPD-5 is fully removed from N2 worms after 24 h of RNAi. Endogenous SPD-2 levels in OD824 (GFP::SPD-2-rr) are reduced to 25% after 24 h of spd-2-rr RNAi, whereas GFP::SPD-2-rr levels increase threefold. (B) Embryonic viability of N2, OD824 (GFP::SPD-2rr), and TH327 (SPD-5::GFP codon optimized to cai 0.65) embryos after 24 h of treatment with spd-5 or spd-2-rr RNAi. Both transgenes were found to restore embryo viability when endogenous protein was removed. (C, D) Identification of cytoplasmic interactors of SPD-5::GFP and GFP::SPD-2. SPD-5::GFP or GFP::SPD-2 was immunopurified from C. elegans embryo extracts, and the coimmunoprecipitated proteins were identified via label-free quantification mass spectrometry. TBG-1::GFP immunoprecipitates were used as a control. Experiments were repeated in triplicate. Results are plotted as volcano plots, with the enrichment of identified peptides in sample vs. control plotted on the x-axis and the significance of enrichment on the y-axis. p values and enrichments were calculated as described previously (Hubner et al., 2010). Black circles indicate nonenriched centrosome proteins; black dots indicate the control protein, as well as significant interactors enriched in the sample.
In the GFP::SPD-2 immunoprecipitates, only SPD-2 was significantly enriched (488-fold; Figure 1D). SPD-5 and RSA-2 were detected, but their levels of enrichment and significance were below our threshold in a range similar to that of many other nonspecific interactors. Therefore we do not consider SPD-5 and RSA-2 to be true interaction partners of cytoplasmic SPD-2. The kinase PLK-1, which has been shown to interact with SPD-2 in vitro (Decker et al., 2011), and SAS-4/-5/-6 and AIR-1 were also not detected. These results suggest that SPD-2 might not form stable complexes with other proteins in cytoplasmic embryo extracts. Taken together, our data suggest that in cytoplasmic extracts, SPD-5 forms complexes with RSA-1, RSA-2, and RNP-6; however, no evidence was obtained for stable interactions between SPD-2, SPD-5, and PLK-1. Because an interaction between SPD-2 and SPD-5 was suggested previously (Boxem et al., 2008), this result suggests that this interaction might only occur in the context of PCM assembly around centrioles. Consistent with this idea, SPD-2 was detected when SPD-5::GFP was immunoprecipitated from centrosome-containing low-spin extracts but not from cytoplasmic high-spin extracts (Supplemental Figure S1).
Stoichiometry and shape of SPD-2, SPD-5, and the SPD-5/RSA-1/RSA-2 complex in cytoplasmic extracts
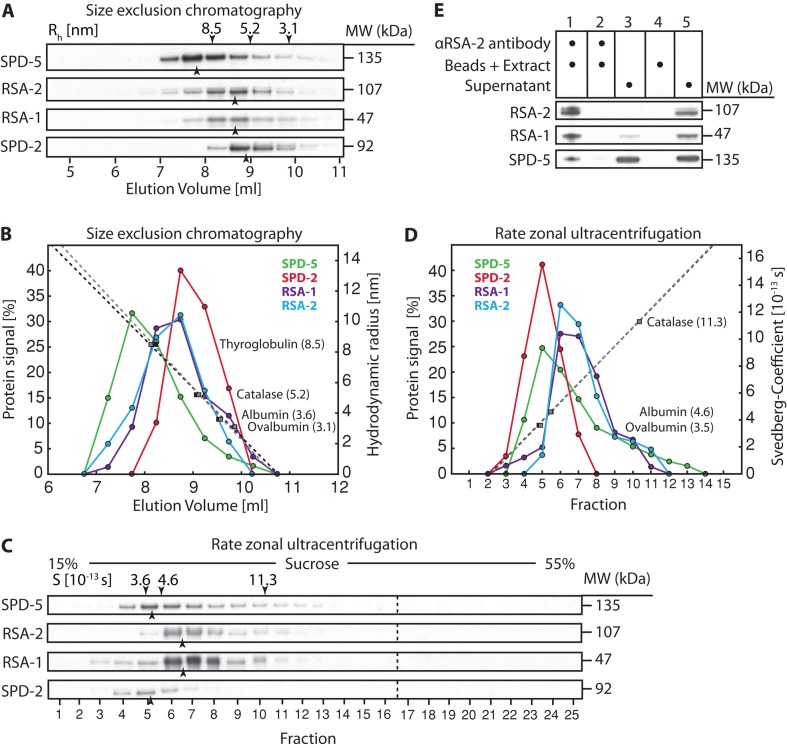
Our immunoprecipitation experiments suggest that SPD-5 forms complexes with RSA-1 and RSA-2 but that SPD-5 and SPD-2 do not interact with each other in cytoplasmic extracts. To confirm these observations and determine whether SPD-2 and SPD-5 form homo-oligomers, which cannot be identified by mass spectrometry, we conducted size-exclusion chromatography and rate zonal ultracentrifugation on cytoplasmic extracts isolated from wild-type embryos.
Size-exclusion chromatography revealed that SPD-2 migrated with a hydrodynamic radius (Rh) of ∼6.0 nm (Figure 2, A and B), and rate-zonal ultracentrifugation showed a peak fraction for SPD-2 with a Svedberg coefficient (Sexp) of 3.8 S (Figure 2, C and D). These values yield a calculated molecular weight of 95 kDa (monomeric molecular weight, 92 kDa) and a shape factor of 1.94 (Table 1), indicating that SPD-2 exists as an elongated monomer (Erickson et al., 2009).
FIGURE 2:
Stoichiometry and shape analysis of SPD-2, SPD-5, and the SPD-5/RSA-1/RSA-2 complex. (A) Cytoplasmic extracts prepared from C. elegans embryos were fractionated by size exclusion chromatography, and an immunoblot of the fractions (collected in 0.5-ml steps) probed for SPD-5, RSA-2, RSA-1, and SPD-2 is shown. Black arrowheads mark the center of the peak for each protein. The centers of the peaks for each standard protein are indicated, along with their hydrodynamic radius, by the black arrowheads above the top lane. (B) Graph blotting the measured signal for each protein vs. elution volume. Standard calibration curves were repeated twice (dotted lines) and calculated from the position of the standard proteins (filled rectangles). (C) Svedberg coefficients for SPD-5, RSA-1, RSA-2, and SPD-2 were determined by rate zonal ultracentrifugation. An immunoblot of the 25 × 0.2 ml fractions obtained from a 5-ml 15–55% sucrose gradient is shown. Black arrowheads mark the center of the peak for each protein. The centers of the peaks for each standard protein are indicated, along with their Svedberg coefficients, by the black arrowheads above the top lane. (D) Graph plotting the measured signal for each protein vs. fraction. The standard calibration curve (dotted line) was calculated from the position of the standard proteins (filled rectangles). (E) Immunoblot of RSA-2 coimmunoprecipitation experiment using RSA-2–specific antibodies on cytoplasmic extract, showing that SPD-5 and RSA-1 coimmunoprecipitate with RSA-2. 1) αRSA-2 antibody–coupled beads were incubated with fresh cytoplasmic extract; 2) the remaining extract from step 1 was reincubated with fresh αRSA-2 antibody–coupled beads to control for full depletion of RSA-2; 3) remaining supernatant from step 2; 4) blank beads were incubated with fresh cytoplasmic extract to control for unspecific binding of beads; 5) remaining supernatant from step 4.
TABLE 1:
Experimental hydrodynamic radii, Svedberg coefficients, and calculated molecular weight and Smax/Sexp values.
| Rh (nm) | Sexp (S) | Smax (S) | MWexpect (kDa) | MWcalc (kDa) | Shape factor | |
|---|---|---|---|---|---|---|
| Ovalbumin | 3.1 | 3.6 | 4.5 | 43 | 46 | 1.25 |
| Albumin | 3.6 | 4.6 | 5.9 | 66 | 68 | 1.28 |
| Catalase | 5.2 | 11.3 | 13.6 | 244 | 247 | 1.20 |
| Thyroglobulin | 8.5 | 19 | 27.4 | 660 | 679 | 1.44 |
| SPD-5 | 9.5 | 4.0 | 9.5 | 135 | 160 | 2.36 |
| RSA-2 | 6.8 | 5.6 | 8.1 | 107 | 160 | 1.44 |
| RSA-1 | 6.9 | 5.7 | 4.6 | 47 | 163 | 0.81 |
| SPD-2 | 6.0 | 3.8 | 7.3 | 92 | 95 | 1.94 |
| RSA-1/RSA-2 | 6.8 | 5.6 | 10.3 | 153 | 160 | 1.80 |
Rh, hydrodynamic radius; Sexp, Svedberg value determined in experiment; Smax, maximum Svedberg value estimated for a spherical particle of a given molecular weight; MWexpect, molecular weight estimated from sequence; MWcalc, molecular weight as calculated from Rh and Sexp; Shape factor, ratio Smax/Sexp, which provides information about the shape of a molecule (Erickson et al., 2009).
SPD-5 had Rh ≈ ∼9.5 nm and a Svedberg value of 4.0 S (Figure 2, A–D). These values yield a calculated molecular weight of 160 kDa (SPD-5 monomeric molecular weight, 135 kDa) and a shape factor of 2.36, which is in the range typical for highly elongated proteins such as tropomyosin or fibrinogen (Erickson, 2009). Considering that the estimated molecular weight of a SPD-5 homodimer is 270 kDa, we conclude that the majority of SPD-5 does not form homodimers or large homo-oligomers in cytoplasmic extracts. Nevertheless, the molecular weight of 160 kDa is larger than expected for monomeric SPD-5 (135 kDa), even when considering an expected error of 10% in this calculation (Erickson, 2009). This relative 25-kDa shift in expected molecular weight could represent transient dynamic interactions with other extract components.
We considered the possibility that the difference between the calculated and predicted molecular weights of SPD-5 results from its interactions with RSA-1 and RSA-2. In size exclusion chromatography and rate zonal ultracentrifugation analysis RSA-1 and RSA-2 migrated in an essentially identical fashion (Figure 2, A–D). The calculated molecular weight of RSA-1 and RSA-2 was ∼160 kDa each (Table 1), similar to the 153 kDa expected for an RSA-1/RSA-2 heterodimer. Consistent with the idea that RSA-1 and RSA-2 form heterodimers, ∼80% of total RSA-1 coimmunoprecipitated with RSA-2 from cytoplasmic extracts (Figure 2E). SPD-5 ran as a single peak that did not comigrate with the RSA-1/RSA-2 peak in either experiment (Figure 2, B and D), and only 20% of total SPD-5 coimmunoprecipitated with RSA-2 from cytoplasmic extracts (Figure 2E). Cumulatively these results suggest that SPD-5 does not form a tight heterotrimeric complex with RSA-1/RSA-2 (which would have an expected mass of 288 kDa), but rather that RSA-1/RSA-2 heterodimers might exhibit a lower-affinity dynamic interaction with SPD-5 that shifts the SPD-5 peak toward the observed higher molecular weight.
Investigation of the cytoplasmic interactions of SPD-2 and SPD-5 in vivo using fluorescence correlation spectroscopy
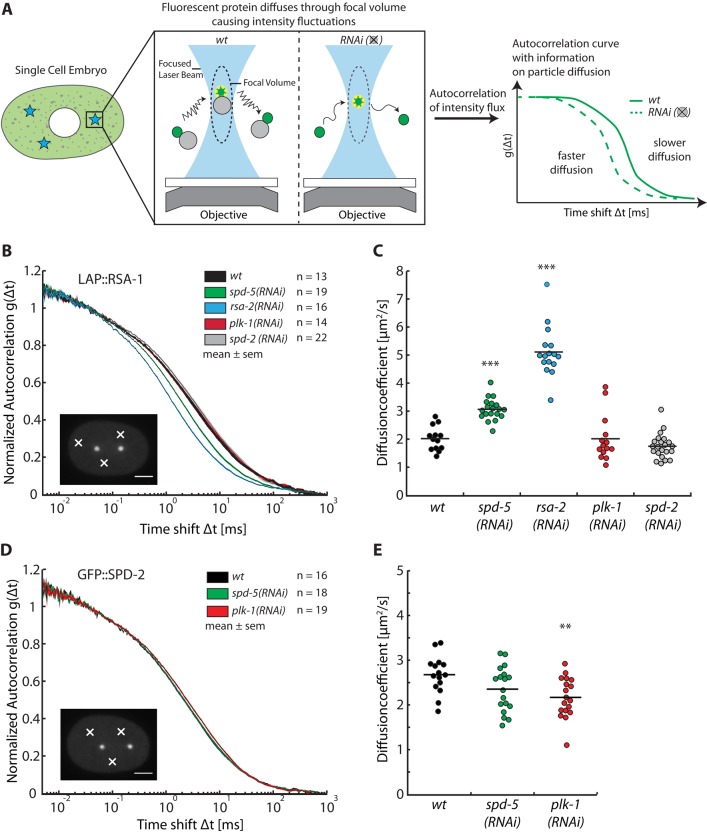
To confirm that the protein interactions we observed in cytoplasmic extracts also occur in living embryos, we employed fluorescence correlation spectroscopy (FCS) to assess the diffusion of fluorescently tagged proteins in vivo. Protein–protein interactions involving fluorescently tagged proteins can be deduced by combining FCS with RNA interference (RNAi)–mediated depletion of candidate binding partners because depletion of a binding partner is expected to decrease the hydrodynamic radius of complexes containing the tagged protein, thereby increasing its diffusion (Figure 3A). Such differences in diffusion can be observed as time shifts of the autocorrelation curve of the fluorescent protein or by changes in the diffusion coefficient obtained from fitting the autocorrelation curve with a diffusion model. Changes in diffusion strongly depend on the relative change in hydrodynamic radius of the particle and thus are best tested by observing a small molecule under depletion of its larger interaction partner.
FIGURE 3:
Fluorescence correlation spectroscopy reveals cytoplasmic interactions in vivo. (A) Schematic overview of the experimental procedure. FCS measurements were conducted in the cytoplasm of single-cell embryos at three randomly chosen cytoplasmic positions away from the centrosomes and nucleus. Intensity fluctuations caused by GFP-tagged proteins diffusing through the focal volume were recorded over time. (B) Diffusion of LAP::RSA-1 is affected by depletion or RSA-2 and SPD-5. Autocorrelation curves are depicted as mean ± SEM as calculated from all embryos for each condition. The number of embryos analyzed is indicated next to each condition. Scale bar, 10 μm; white crosses point to exemplary locations of measurements. (C) Diffusion coefficient plot showing diffusion coefficients of LAP::RSA-1. LAP::RSA-1 diffusion changes from 2.0 ± 0.4 to 3.1 ± 0.4 μm2/s (p < 0.001, n = 19) under spd-5 RNAi and 5.1 ± 0.9 μm2/s (p < 0.001, n = 16) under rsa-2 RNAi. Diffusion coefficients were obtained from fitting the autocorrelation curve of LAP::RSA-1 from each embryo. Asterisks indicate significance with p < 0.001. The results show a clear interaction between LAP::RSA-1 and RSA-2, as well as SPD-5. An interaction with PLK-1 or SPD-2 could not be detected. (D) GFP::SPD-2 diffusion is unaffected by depletion of SPD-5 but slightly decreases under PLK-1 depletion. Representation as in B, showing GFP::SPD-2 autocorrelation curves under various conditions. (E) Diffusion coefficient plot of GFP::SPD-2. GFP::SPD-2 diffuses indistinguishably, with ∼2.5 μm2/s in wild- type and spd-5 RNAi conditions. Its diffusion is significantly reduced to ∼2.2 μm2/s under plk-1 RNAi conditions.
We investigated the cytoplasmic interaction of SPD-5, RSA-2, and RSA-1 by measuring the diffusion of RSA-1 tagged with a GFP-containing LAP tag (LAP::RSA-1) in the cytoplasm of C. elegans embryos. We compared the diffusion of LAP::RSA-1 in wild-type embryos to that in embryos depleted of SPD-5, RSA-2, SPD-2, and PLK-1. Protein knockdown was confirmed by Western blotting (Supplemental Figure S2A). Autocorrelations from the LAP::RSA-1 measurements were shifted from larger (wt) to smaller time scales, indicating that LAP::RSA-1 changed its diffusion from slower to faster speeds when RSA-2 or SPD-5 was depleted from the cytoplasm (Figure 3B). We then fitted the autocorrelation curves using a one-component anomalous diffusion model to obtain diffusion coefficients for each respective experimental condition. A representative fit, along with R2 values and χr2 values for the fits, are shown in Supplemental Figure S2, B–D. The diffusion coefficient of LAP::RSA-1 in nonmanipulated cytoplasm was 2.0 ± 0.4 μm2/s. Depletion of SPD-2 or PLK-1 did not affect the diffusion of LAP::RSA-1 (1.8 ± 0.4 and 2.0 ± 0.8 μm2/s, respectively). LAP::RSA-1 diffusion changed to 5.1 ± 0.9 μm2/s (p < 0.001, n = 16) when the embryonic cytoplasm was depleted of RSA-2 (Figure 3C), in support of the idea that RSA-1 and RSA-2 heterodimerize in vivo. Upon removal of SPD-5, the diffusion of LAP::RSA-1 increased significantly, from 2.0 ± 0.4 to 3.1 ± 0.4 μm2/s (p < 0.001, n = 19). Similar shifts could not be detected when SPD-5::GFP was observed and RSA-2 was depleted from the cytoplasm (Supplemental Figure S3, A and B). We confirmed this observation using size exclusion chromatography and rate zonal ultracentrifugation on RSA-2–depleted extract. We found that the hydrodynamic radius of SPD-5 was 9.5 nm and the sedimentation coefficient was 3.7 S in RSA-2–depleted extracts, values very similar to those in undepleted extracts (Supplemental Figure S3, C and D), resulting in a calculated molecular weight of SPD-5 of 148 kDa. We believe that our ability to detect a shift for RSA-1 but not SPD-5 when RSA-2 is depleted is due to the fact that the RSA-1/RSA-2/SPD-5 complex has a hydrodynamic radius distinct from the 6.8-nm radius of the RSA-1/RSA-2 heterodimer but quite similar to the 9.5-nm radius of the SPD-5 monomer. Taken together, our data clearly show interactions between LAP::RSA-1, RSA-2, and SPD-5 in the cytoplasm of living embryos, confirming our previous observations.
Next we compared the diffusion of GFP::SPD-2 in the cytoplasm of wild-type embryos to that in embryos depleted of SPD-5 and PLK-1. A time shift in the autocorrelation functions could not be detected with absolute certainty (Figure 3D). The diffusion coefficient of SPD-2 in wild-type embryo cytoplasm was 2.7 ± 0.4 μm2/s (Figure 3E). In embryos depleted of SPD-5, GFP::SPD-2 diffusion was 2.4 ± 0.5 μm2/s. A significant result of 2.2 ± 0.5 μm2/s (p = 0.0036) was obtained for GFP::SPD-2 diffusion in PLK-1–depleted cytoplasm. Contrary to a disruption of a cytoplasmic SPD-2/PLK-1 complex that would increase the diffusion of GFP::SPD-2, the diffusion of GFP::SPD-2 was reduced by PLK-1 depletion. Combined with the observation that SPD-2 exists as a monomer in the cytoplasm, the result indicates that PLK-1 likely does not form a stable complex with SPD-2 but instead directly or indirectly influences its cytoplasmic conformation. A similar effect was not observed for GFP::SPD-2 or SPD-5::GFP when AIR-1 was depleted (Supplemental Figure S4). The fact that depletion of SPD-5 does not lead to a detectable effect on the diffusion of GFP::SPD-2 is consistent with our previous biochemical analysis, enforcing the notion that SPD-2 does not form stable complexes with SPD-5 in the cytoplasm of living C. elegans embryos but instead exists as a monomer.
DISCUSSION
Here we present an analysis of the cytoplasmic state of the C. elegans PCM proteins SPD-2 and SPD-5, their interactions, protein characteristics, and diffusion rates. By combining various techniques, we were able to draw a precise picture of the stable interactions of these proteins occurring in the cytoplasm of C. elegans embryos. The diffusion rates and detailed complex characteristics determined here (Tables 1 and 2) will help in building models for centrosome assembly.
TABLE 2:
Values for free parameters obtained from fitting single-component anomalous diffusion model to the autocorrelation curves obtained for transgenes in wild-type conditions.
| D (μm2/s) | τblinking (μs) | Fraction τ (%) | α | n | χ2corr of fit | |
|---|---|---|---|---|---|---|
| GFP::SPD-2 | 2.7 ± 0.4 | 41 ± 45 | 10 ± 4 | 0.75 ± 0.05 | 5 ± 1 | 2.02 ± 0.35 |
| LAP::RSA-1 | 2.0 ± 0.4 | 38 ± 17 | 11 ± 2 | 0.65 ± 0.06 | 19 ± 6 | 2.58 ± 0.56 |
D, diffusion coefficient; τblinking, triplet blinking decay time; fraction τ, percentage of molecules predicted to be in blinking state; α, anomalous α to correct for subdiffusive or superdiffusive behavior of particles; n, number of particles calculated to be in the focal volume at any given time; χ2corr, corrected χ2 as a measure for the goodness of the fit.
The methods used in this study mainly focus on stable cytoplasmic associations of proteins. Immunoprecipitation and mass spectrometry, as well as size exclusion chromatography and sucrose gradient rate zonal ultracentrifugation, allow for detection and analysis of protein associations that are stable throughout biochemical analyses. On the other hand, FCS is not limited by these constraints, as it analyzes molecular diffusion inside unperturbed living cells. However, the sensitivity of the FCS-RNAi assay strongly depends on the relative concentrations of the proteins, the amount of interaction occurring, and the relative differences in hydrodynamic radii of the different complexes (for more information see Schwille, 2001; Heinze and Schwille, 2007). Thus we cannot exclude that we might have missed transient or low-affinity interactions. Future studies implementing fluorescence cross-correlation spectroscopy, BioID, and/or S-Cross may be necessary to identify such interactions (Baudendistel et al., 2005; Bacia et al., 2006; Bacia and Schwille, 2007; Gautier et al., 2009; Roux et al., 2012).
The evidence presented here suggests that the C. elegans core PCM proteins SPD-2 and SPD-5 do not form stably associated complexes inside the cytoplasm of living C. elegans embryos. Instead, SPD-2 was found to be an elongated monomer, and SPD-5 was mostly monomeric and highly elongated in shape and can associate with RSA-1 and RSA-2. Published data show that PLK-1 does bind to a SPD-2 fragment via a region containing a polo box binding site in vitro (Decker et al., 2011). Furthermore, it was shown that the polo box domain of human polo kinase associates with the targeted phosphopeptide with a binding constant (Kd) of ∼280 nM (Elia et al., 2003), a value similar to the strong and biochemically detectable association of spindle checkpoint protein Bub3 and the phosphorylated MELT peptide (Primorac et al., 2013). Thus the interaction between SPD-2 and PLK-1 is likely to be strong enough to be detected even by our biochemical analyses. However, none of our methods did detect such an interaction in the cytoplasm. Therefore it seems likely that SPD-2 and PLK-1 interact exclusively at centrosomes. The evidence for the interaction between SPD-2 and SPD-5 in the literature is less clear. Yeast two-hybrid data analyzing protein fragments suggested an interaction between SPD-2 and SPD-5 (Boxem et al., 2008). Although not definitive, the existence of the two-hybrid interaction combined with the coimmunoprecipitation of SPD-2 and SPD-5::GFP from centrosome-containing extracts, as well as the fact that SPD-2 and SPD-5 are the only two proteins genome wide whose loss has been shown to lead to a nearly complete loss of PCM assembly (Sönnichsen et al., 2005), support the idea that these proteins do interact at centrosomes.
It was previously reported that SPD-5 can interact with the PP2A regulatory subunit RSA-2 in yeast-two hybrid assays and biochemical analysis (Schlaitz et al., 2007; Boxem et al., 2008). Our data suggest that SPD-5 interacts with RSA-1/RSA-2 heterodimers in the cytoplasm, although this may be a relatively low affinity interaction. We know RSA-1 and RSA-2 are not required for incorporation of SPD-5 at centrosomes, as centrosomes still form in the absence of RSA-1 or RSA-2 (Schlaitz et al., 2007). However, it is possible that RSA-1 and RSA-2 preserve cytoplasmic SPD-5 in a dephosphorylated state, possibly by promoting an association between SPD-5 and PP2A. Future work will be needed to investigate the potential roles of this interaction and how the interactions between PCM proteins are regulated.
Finally, neither SPD-2 nor SPD-5 was found to form stable complexes with the centriolar protein SAS-4. Recent work in D. melanogaster shows that SAS-4 forms cytoplasmic S-CAP complexes with the key pericentriolar proteins CNN, ASL, D-PLP, CP-190, and β‑tubulin (Gopalakrishnan et al., 2011). On the basis of this work, Gopalakrishnan et al. (2011) proposed a mechanism for PCM assembly in which cytoplasmic S-CAP complexes are recruited to the centrosome in an SAS-4–dependent manner. In contrast, our data indicate that PCM components are monomeric in the cytoplasm, suggesting that they are recruited independently to the centrosome, where they are incorporated into the PCM matrix. Additional work is needed to analyze the similarities and differences in the cytoplasmic state of PCM components and the process of PCM assembly across species.
MATERIALS AND METHODS
Worm strains
C. elegans strains were maintained according to standard protocols (Stiernagle, 2006). The C. elegans strains used in this work are listed in Table S1. All strains except OD824 were described previously. OD824 expresses GFP::SPD-2. The gfp::spd-2 transgene was rendered RNAi resistant by reencoding a 622–base pair sequence (Supplemental Figure S5). The engineered spd-2 locus was cloned into pCFJ151 (Frøkjaer-Jensen et al., 2008), which contains homology arms that direct transposase-mediated insertion of intervening sequence into the ttTi5605 Mos I insertion site on chromosome II. Single-copy integrants were generated by injecting the transgene-containing plasmid (50 ng/μl) along with a plasmid encoding the Mos transposase (Pglh-2::transposase, pJL43.1, 50 ng/μl) and three plasmids encoding different fluorescent markers—Pmyo-2::mCherry (pCFJ90, 2.5 ng/μl), Pmyo-3::mCherry (pCFJ104, 5 ng/μl), and Prab-3::mCherry (pGH8, 10 ng/μl)—into the strain EG4322. Injected worms were singled and incubated at room temperature for two or three generations until all worms were starved. Moving worms lacking the fluorescent coinjection markers were identified among the progeny by visual inspection. Transgene integration was confirmed by PCR of regions spanning each side of the insertion.
RNA interference
Worms were treated with feeding RNAi as previously described (Timmons and Fire, 1998; Kamath et al., 2001). Regions targeted by the feeding clones are flanked by the following primers: spd-2-fw (GAATGCATCAATGGCAGATG), spd-2-rev (AATTTTGTGCCGGTACTTCG); spd-2-rr-fw (ACATTCTGAAACAATGATGACAA), spd-2-rr-rev (GACAGTGAAACATTGCCATT); spd-5-fw (AACCATTACTGATATGGGGA), spd-5-rev (CAAATGAACAATTCTTTCAAGCA); plk-1-fw (TCAACAACAAGCTGCAGAGG), plk-1-rev (TGGGACTAAAAGGGTCGATG); and rsa-2-fw (AAAATAGGCGGATGAAACCC), rsa-2-rev (GTATCTGTCACTGCTGCCGA).
Scoring of embryo viability
Worms were exposed to feeding RNAi for 24 h. Worms were then dissected, and 50 embryos were collected and transferred to fresh OP50 plate for hatching. The number of hatched larva was counted 48 h post transfer. The embryo viability was then scored by normalizing the number of hatched embryos from the RNAi condition against the non-RNAi condition.
Worm culture and cytoplasmic embryo extracts
All equipment used for centrifugations was obtained from Beckmann Coulter (Brea, CA). Worms were cultured on 3.5% agar peptone media plates seeded with C600 bacteria. To extract embryos from the worm culture, worm cultures were mixed with 1× volume bleach solution (400 mM NaOH, 1.6 M NaClO solution with 6–14% active chlorine base (Sigma-Aldrich, St. Louis, MO) and vortexed for 5 min. The bleached worm culture was repeatedly washed via centrifugation–resuspension cycles to remove any active bleach solution. Cell Strain 40-μm nylon filters (BD, Franklin Lakes, NJ) were used to retrieve the embryos and separate them from remaining worm carcasses. Pure embryo fractions were weighed, frozen in liquid nitrogen, and stored at −80°C for later use.
Extracts were generated in the following way, Embryos were thawed in 1× volume of extract buffer (1× phosphate-buffered saline [PBS] 100 mM KCl, 1 mM ethylene glycol tetraacetic acid [EGTA], 1 mM MgCl2, 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS], 1× Complete Protease Inhibitor cocktail [Roche, Basel, Switzerland]). Thawed embryos were sonicated and centrifuged at 20,000 × g for 10 min at 4°C. The resulting supernatant was centrifuged for 30 min using a TLA120.2 rotor in an Optima Ultracentrifuge at 97,000 RPM (κ-factor = 12.4). The supernatant was filtered using 0.22-μm Cellulose Acetate Spin Filters (Agilent Technologies, Santa Clara, CA) at 16,000 × g for 1 min. Protein concentration in the supernatant was determined by ultraviolet absorption using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Waltham, MA) in preparation for subsequent experiments.
Immunoprecipitation and label-free quantitative mass spectrometry
Transgenic cytoplasmic embryo extracts were incubated with magnetic Dynabeads (Life Technologies, Carlsbad, CA) coupled to goat anti-GFP antibodies (affinity purified against GFP by the protein expression and purification facility of the Max Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany) for >30 min at 4°C. Proteins were washed in extract buffer as described and eluted from the antibodies in SDT buffer (1% SDS, 10 mM Tris-HCl, pH 7.6) for mass spectrometry analysis by boiling and subsequent denaturing by adding 0.1 M dithiothreitol (DTT) and reboiling. Each sample was filter-aided sample preparation processed (Wic´niewski et al., 2009), diluted in 300 μl of UA buffer (8 M urea), and transferred to a 30-kDa MWCO Vivacon (Sartorius, Goettingen, Germany) 500 filter, followed by three washes with UA buffer with intermittent spins to remove the SDS and alkylated with iodoacetamide; the samples were digested with trypsin overnight. The trypsinized samples were spun through the filter, acidified with 0.1% trifluoroacetic acid (TFA), and desalted using stage tips (C18 activated with 50 μl of methanol and then equilibrated with 50 μl of 0.1% TFA) with two washes of 0.1% TFA. Peptides were eluted with 50 μl of solvent B (80% acetonitrile, 0.5% acetic acid), and then acetonitrile was removed by vacuum centrifugation and peptides were acidified to 0.1% TFA.
Mass spectrometric (MS) analysis was performed essentially as described (Hubner et al., 2010) on an LTQ Orbitrap or a QExactive (Thermo Scientific) equipped with a nano-electrospray ion source (Thermo Scientific) coupled to a Thermo EASY-nLC II system. The acidified peptides were separated on a nanoscale reversed-phase column (C18 ReproSil-Pur, 3μm) packed in a fused silica capillary emitter with 75-μm inner diameter and a length of 20 cm. Peptides were eluted during a 120-min linear gradient of solvents A (0.5% acetic acid) and B (80% acetonitrile and 0.5% acetic acid) with a flow of 250 nl/min. Upon elution, the peptides were ionized using a voltage of 2.3 kV and no sheath and auxiliary gas flow. Spectra were recorded in positive-ion mode with data-dependent acquisition automatically switching between recording an MS survey scan of the precursor ions and MS/MS of the most intense ions.
Label-free data processing using MaxQuant, version 1.3.0.3 or 1.5.0.6 (Cox and Mann, 2008), and data analysis were performed as described (Hubner et al., 2010) with the Andromeda database search against the C. elegans Uniprot database and the MaxLFQ module for label-free quantification (Cox et al., 2014). In cases in which a protein was detected only in the sample and not in the control, the enrichment was calculated over a simulated noise level instead. For the determination of statistically significant protein interactions, the false discovery rate was set to 0.02 for SPD-5 and 0.001 for SPD-2, with the SO value (=curve bend) of 2 in both cases (Hubner et al., 2010).
Size exclusion chromatography and rate zonal ultracentrifugation analysis
The following purified proteins (Sigma-Aldrich) were used as standards for the calibration: thyroglobulin (8.5 nm, 19 S), catalase (5.2 nm, 11.3 S), albumin (3.6 nm, 4.6 S), and ovalbumin (3.1 nm, 3.5 S)
Size exclusion chromatography was carried out with N2 cytoplasmic embryo extracts using the Akta Ettan LC system (GE Healthcare, Chalfont St Giles, UK) equipped with a Tosoh G5000PWxl column (Tosoh, Tokyo, Japan). Samples were loaded according to specification of the system and the column and run in storage buffer (2× PBS, 0.1% CHAPS, 1 mM DTT, 0.1× Complete Protease Inhibitor cocktail [Roche], 1 mM EGTA, 1 mM MgCl2). Samples were collected in 500-μl fractions.
Sucrose solutions were prepared from storage buffer and D-Sucrose (AppliChem, Gatersleben, Germany). Sucrose gradients, 15–55%, were prepared using an Auto Densi-Flow II C gradient mixer (Buchler Instruments, Fort Lee, NJ). Total protein N2 extracts (2.4 mg) were loaded onto 4.8-ml gradients. Gradients were then spun using a SW-Ti 55 rotor in an Optima LE‑80K Ultracentrifuge at 192,000 × g at 4°C for 13 h. After the spin, the gradient was separated into 25 fractions using the removal function of the Auto Densi-Flow II C gradient mixer.
Twenty microliters of all fractions was used for immunoblot analysis to identify the peak fraction. The hydrodynamic radius and Svedberg value for the proteins were estimated using calibration curves from the standard proteins.
Molecular weight and shape analysis
The hydrodynamic radii and Svedberg values were used to estimate the molecular weight (MW) and the shape as previously described (Erickson, 2009; see note to Table 1 for definitions):
MW = 4204SRh
Shape-factor = Smax/Sexp
Immunoblot analysis
All immunoblot analyses were performed using protein denatured in 1× sample buffer (2% SDS, 10% glycerol, 10% 2-mercaptoethanol, 0.01% bromophenol blue, and 0.0625 M Tris-HCl). Proteins were loaded onto NuPAGE Novex 4-12% Bis-Tris gels (Life Technologies). Proteins were detected using primary rabbit antibodies at 1 μg/ml as previously described (Schlaitz et al., 2007; Decker et al., 2011). Secondary detection was achieved using a 1:30,000 dilution of horseradish peroxidase–coupled secondary antibodies (Bio-Rad, Hercules, CA).
Fluorescence correlation spectroscopy and diffusion analysis
FCS measurements were made using a LSM 780 microscope equipped with a 40×/1.2 numerical aperture water immersion objective and an avalanche photodiode (Zeiss, Jena, Germany) at room temperature. The focal volume was calibrated using Alexa 488 dye (Life Technologies), resulting in the following parameters: beam diameter (ωxy) = 0.19 μm, structural parameter (S) = 5 and confocal volume (V) = 0.19 fl. Worms were dissected to obtain single embryos mounted in M9 on 2% (wt/vol) agarose pads (Life Technologies). Excitation of GFP was achieved using a 488-nm argon laser. Three measurements with a total of 72 s (24 s each) were taken in each embryo at random positions in the cytoplasm (excluding cell membrane, pronuclei, and centrosomes). Each measurement was preceded by a 1-s prebleach to quench immobile fluorescent particles. Autocorrelation curves were calculated from the intensity profiles obtained from the measurements and then averaged for each embryo. To obtain diffusion coefficients, each averaged autocorrelation curve was fitted within a time range of 500 ns to 1 s with a single component three-dimensional anomalous diffusion model including a free triplet component to account for fluorophore blinking (Banks and Fradin, 2005; Heinze and Schwille, 2007). Statistical analysis on diffusion coefficients was done using Wilcoxon rank sum test. Autocorrelation analysis and data plotting were carried out using a MATLAB script developed in our lab.
Supplementary Material
Acknowledgments
We thank Markus Decker for essential discussions on centrosome biology, David Zwicker and Zdenek Petrasek for comments on protein diffusion and fluorescence correlation spectroscopy, and David Drechsel and Barbara Borogonov for their support with regard to biochemical techniques. We also thank David Drechsel and Mark Leaver for critical reading of the manuscript. We thank the Protein Expression and Purification Facility and the Sequencing Facility of the Max Planck Institute for Molecular Cell Biology and Genetics for their services and the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN; funded by National Institutes of Health Office of Research Infrastructure Program P40 OD010440) for providing some of the strains used in this study, as well as the Max Planck Society and the European Commission's 7th Framework Programme Grant, Systems Biology of Mitosis (FP7-HEALTH-2009-241548/MitoSys), for funding.
Abbreviations used:
- FCS
fluorescence correlation spectroscopy
- PCM
pericentriolar material
- RNAi
RNA interference
- wt
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-09-0514) on August 7, 2014.
*These authors contributed equally to this work.
REFERENCES
- Bacia K, Kim SA, Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat Methods. 2006;3:83–89. doi: 10.1038/nmeth822. [DOI] [PubMed] [Google Scholar]
- Bacia K, Schwille P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat Protoc. 2007;2:2842–2856. doi: 10.1038/nprot.2007.410. [DOI] [PubMed] [Google Scholar]
- Banks DS, Fradin C. Anomalous diffusion of proteins due to molecular crowding. Biophys J. 2005;89:2960–2971. doi: 10.1529/biophysj.104.051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudendistel N, Müller G, Waldeck W. 2005. Two-hybrid fluorescence cross-correlation spectroscopy detects protein–protein interactions in vivo Chemphyschem 6, 984–990. [DOI] [PubMed]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul J, van de Peut D, Devos M. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D, Serafinas C, Ashcroft N, Kosinski M, Longo D, Ferris DK, Golden A. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis. 2000;26:26–41. doi: 10.1002/(sici)1526-968x(200001)26:1<26::aid-gene6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Müller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Decker M, Jaensch S, Pozniakovsky A, Zinke A, O'Connell KF, Zachariae W, Myers E, Hyman AA. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol 21, 1259–1267. 2011 doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gönczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- Elia AEH, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proc Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Nakata E, Lukinavicˇius G, Tan K-T, Johnsson K. Selective cross-linking of interacting proteins using self-labeling tags. J Am Chem Soc. 2009;131:17954–17962. doi: 10.1021/ja907818q. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J, Mennella V, Blachon S, Zhai B, Smith AH, Megraw TL, Nicastro D, Gygi SP, Agard DA, Avidor-Reiss T. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun. 2011;2:359. doi: 10.1038/ncomms1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann K, Lange BM. New insights into subcomplex assembly and modifications of centrosomal proteins. Cell Division. 2012;7:17. doi: 10.1186/1747-1028-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Hannak E, Kirkham M, Hyman A, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol. 2001;155:1109–1115. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze K, Schwille P. Fluorescence correlation spectroscopy in living cells. Nat Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kirkham M, Müller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Krämer A, Maier B, Bartek J. Centrosome clustering and chromosomal (in)stability: a matter of life and death. Mol Oncol. 2011;5:324–335. doi: 10.1016/j.molonc.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leidel S, Gönczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Lettman MM, Wong YL, Viscardi V, Niessen S, Chen S-H, Shiau AK, Zhou H, Desai A, Oegema K. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev Cell. 2013;25:284–298. doi: 10.1016/j.devcel.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Pelletier L, Ozlü N, Hannak E, Cowan C, Habermann B, Ruer M, Müller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, Musacchio A. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2013;2:e01030. doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz A-L, Srayko M, Dammermann A, Quintin S, Wielsch N, MacLeod I, de Robillard Q, Zinke A, Yates JR, 3rd, Müller-Reichert T, et al. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P. Fluorescence correlation spectroscopy and its potential for intracellular applications. Cell Biochem Biophys. 2001;34:383–408. doi: 10.1385/CBB:34:3:383. [DOI] [PubMed] [Google Scholar]
- Sluder G, Nordberg JJ. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Sluder G, Thompson EA, Miller FJ, Hayes J, Rieder CL. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J Cell Sci. 1997;110:421–429. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook 2006(Feb 11) 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Wis´niewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Phil Trans R Soc B. 2014:1–10. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker D, Decker M, Jaensch S, Hyman AA, Julicher F. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc Natl Acad Sci USA. 2014;111:E2636–E2645. doi: 10.1073/pnas.1404855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.