Rab10 can increase insulin-stimulated activation of RalA in 3T3-L1 adipocytes. Rlf is a new effector of Rab10 and is required for maximal glucose uptake in adipocytes. Rab10 recruits Rlf to activate RalA.
Abstract
Insulin-stimulated glucose uptake in fat and muscle is mediated by the major facilitative glucose transporter Glut4. Insulin controls the trafficking of Glut4 to the plasma membrane via regulation of a series of small G proteins, including RalA and Rab10. We demonstrate here that Rab10 is a bona fide target of the GTPase-activating protein AS160, which is inhibited after phosphorylation by the protein kinase Akt. Once activated, Rab10 can increase the GTP binding of RalA by recruiting the Ral guanyl nucleotide exchange factor, Rlf/Rgl2. Rab10 and RalA reside in the same pool of Glut4-storage vesicles in untreated cells, and, together with Rlf, they ensure maximal glucose transport. Overexpression of membrane-tethered Rlf compensates for the loss of Rab10 in Glut4 translocation, suggesting that Rab10 recruits Rlf to membrane compartments for RalA activation and that RalA is downstream of Rab10. Together these studies identify a new G protein cascade in the regulation of insulin-stimulated Glut4 trafficking and glucose uptake.
INTRODUCTION
Insulin stimulates glucose uptake in adipocytes and skeletal muscle by promoting the translocation of the facilitative glucose transporter Glut4 to the plasma membrane (Pessin and Saltiel, 2000). In the basal state, Glut4 is contained in endosomes and postendosomal Glut4 storage vesicles (GSVs), which traffic to the cell surface in response to insulin via a process that involves the activation of small G proteins (Leto and Saltiel, 2012). Although our understanding of the signaling cascades that govern these GTPases is incomplete, activation of the protein kinase Akt downstream of phosphoinositide 3-kinase is known to be a key event (Whiteman et al., 2002; Hou and Pessin, 2007; Sakamoto and Holman, 2008). Several studies indicate that Akt can phosphorylate different GTPase-activating proteins, in the process reducing the inactivation of their cognate GTPases and thus producing increased G protein activity (Kane et al., 2002; Chen et al., 2011b).
One Akt substrate that has been implicated in regulating Glut4 translocation is AS160 (Sano et al., 2003; Thong et al., 2007), which contains multiple Akt phosphorylation sites and a TBC1 domain with putative GTPase-activating protein (GAP) activity toward certain Rab GTPases (Kane et al., 2002). Small interfering RNA (siRNA)–mediated depletion of AS160 in 3T3-L1 adipocytes increases basal Glut4 translocation and glucose uptake, an effect that can be reversed by reintroducing the wild-type AS160, but not the GAP-deficient AS160 RK mutant, suggesting that AS160 retains Glut4 in intracellular compartments in a GAP activity–dependent manner (Eguez et al., 2005; Larance et al., 2005). In vitro GTP hydrolysis assays revealed that AS160 can catalyze GTP hydrolysis of Rab2, Rab8A, Rab10, and Rab14 (Miinea et al., 2005). Among these, only Rab10 is required for insulin-stimulated Glut4 translocation in 3T3-L1 adipocytes (Sano et al., 2007, 2008). However, there is no direct evidence that any of these Rab GTPases are the functional targets of AS160 and whether phosphorylation of AS160 by Akt is related to Rab target activation.
Another Akt substrate that has been invoked in insulin-stimulated glucose uptake is the RalA GAP complex RGC1/2 (Chen et al., 2011b). This complex bears some homology to the tuberous sclerosis complex, which targets the G protein Rheb upstream of mTORC1 activation by growth factors (Inoki et al., 2003; Shirakawa et al., 2009). RGC2 has multiple Akt phosphorylation sites and contains a RalGAP domain that inactivates RalA. The complex also requires a regulatory protein, RGC1, which stabilizes RGC2. Akt2 phosphorylation of RGC2 leads to its inactivation, in the process producing increased GTP-bound RalA. RalA, in turn, resides in Glut4 vesicles and directs GSVs to the plasma membrane by interacting with Sec5 and Exo84 of the exocyst complex, an evolutionarily conserved, eight-subunit structure that assembles at the plasma membrane (Moskalenko et al., 2002; Chen et al., 2007, 2011a). After this interaction, phosphorylation of Sec5 by protein kinase C releases RalA from the exocyst, leading to disengagement of Glut4 vesicles, thus permitting their fusion with the plasma membrane (Chen et al., 2011a).
Although Rab10 and RalA are believed to be controlled mainly by inactivation of their cognate GAP proteins, the role of exchange factors in the hormonal regulation of their activities is not well understood. Moreover, the relative roles of these two G proteins and their relationship to each other in the Glut4 trafficking process remain uncertain. In this study, we establish Rab10 as a functional target of AS160 and demonstrate that the GAP activity of AS160 toward Rab10 is negatively regulated by Akt phosphorylation. We show that Rab10 and RalA are both required for insulin-stimulated glucose uptake in 3T3-L1 adipocytes. We also investigate the relationship of Rab10 and RalA and reveal that Rab10 activity is required for, and lies upstream of, RalA in a crucial insulin signaling cascade, due to the recruitment by Rab10 of its effector, the Ral guanine nucleotide exchange factor (GEF) Rlf, leading to the activation of RalA. These findings establish the important role of a G protein cascade in the control of Glut4 trafficking.
RESULTS
AS160 inhibits Rab10 activity
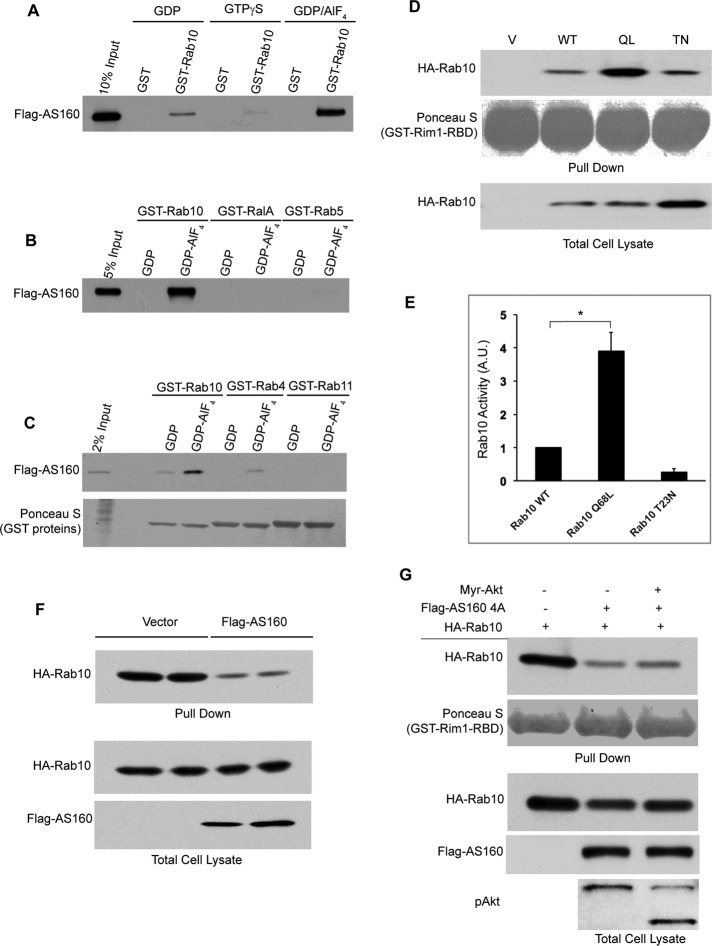
Previous studies demonstrated that GAP proteins preferentially interact with their cognate small GTPases during the transition state of GTP hydrolysis (Scheffzek et al., 1997; Bos et al., 2007). To identify a cellular target of AS160, we loaded glutathione S-transferase (GST)–Rab proteins with guanosine diphosphate (GDP)/AlF4 to mimic this transition state, incubating the fusion proteins with lysates from COS-1 cells overexpressing FLAG-AS160. AS160 specifically interacted with GST-Rab10 bound to GDP/AIFx but not with GST-Rab10 bound to GDP or GTPγS (Figure 1A), indicating that the interaction between AS160 and Rab10 is transition state dependent. Meanwhile, AS160 did not interact, or interacted only modestly, with other GDP/AIFx-loaded small GTPases, including the Ras subfamily member RalA and the Rab subfamily members Rab4, Rab5, and Rab11 (Figure 1, B and C). These results suggest that the transition state–dependent binding between AS160 and Rab10 is specific and characteristic of a GAP-substrate GTPase interaction, consistent with the notion that AS160 possesses GAP activity toward Rab10 in vitro.
FIGURE 1:
Rab10 is an in vivo target of AS160. (A–C) COS-1 cell lysates overexpressing FLAG-AS160 was incubated with immobilized GST or GST-Rab4, 5, 10, 11, or RalA bound to GDP, GTPγS, or GDP/AIFx as indicated. Pull downs were subjected to SDS–PAGE and Western blot with anti-FLAG antibody. GST proteins were visualized with Ponceau S. (D) COS-1 cells were transiently transfected with HA-Rab10 WT, QL, or TN constructs as indicated, and cell lysates were incubated with immobilized GST-Rim1-RBD. Pull downs were resolved on SDS–PAGE and visualized by Western blot with anti-HA antibody. (E) Quantification of Rab10 activity in D from three independent experiments. Rab10 activity was quantified by normalizing the amount of pull down with the total cell lysate. The value of the activity of wild-type Rab10 was then set as 1 arbitrary unit. *p < 0.05. (F) 3T3-L1 adipocytes were transfected with FLAG-AS160 and HA-Rab10 (QL), and the activity of Rab10 was determined by GST-Rim1-RBD pull-down assay as described previously. (G) COS-1 cells were transiently transfected with HA-Rab10 alone or with FLAG-AS160 4A and/or Myr-Akt constructs as indicated. Cell lysates were subjected to GST-Rim1-RBD pull-down assay as described previously.
To test whether AS160 can catalytically inactivate Rab10 in vivo, we developed an effector pull-down assay to evaluate the activation state of Rab10 (Supplemental Figure S1A). Rab3-interacting molecule 1 (Rim1) was reported to interact with Rab10 through the Rab-binding domain (RBD) at its N-terminus (Fukuda, 2003). Lysates from COS-1 cells expressing wild-type, constitutively active (Q68L/QL), or a GDP-locked version (T23N/TN) of Rab10 were subjected to the GST-Rim1-RBD pull-down assay, validating the assay for activity of Rab10 (Figure 1, D and E). Similar interactions were detected by immunoprecipitation in which Myc-Rim1-RBD preferably formed a complex with the active mutant of Rab10 (QL) (Supplemental Figure S1B) and by pull-down experiments with GDP/GTPγS-loaded Rab10 in vitro (Supplemental Figure S1C). We then used the Rim1-RBD pull-down assay to test whether AS160 can inactivate Rab10 in vivo. As shown in Figure 1F, <40% of Rab10 was pulled down by GST-Rim1-RBD in the presence of overexpressed AS160 in 3T3-L1 adipocytes, indicating that AS160 can inactivate Rab10 in cells. AS160 phosphorylation was enhanced in COS-1 cells expressing constitutively active Myr-Akt compared with control cells. Moreover, the inhibitory effect of AS160 on Rab10 activity in vivo was almost completely abolished by Myr-Akt phosphorylation (Supplemental Figure S1, D and E). Furthermore, overexpression of the nonphosphorylable AS160 4A mutant, in which four (Ser-318, Ser-588, Thr-642, Ser-751) of the five Akt phosphorylation sites are mutated to alanine (Sano et al., 2003), significantly inhibited Rab10 activity. Myr-Akt expression did not influence the GAP activity of the nonphosphorylable mutant (Figure 1G). Together these data establish that Rab10 is a functional target of AS160 in vivo.
Rab10 functions upstream of RalA
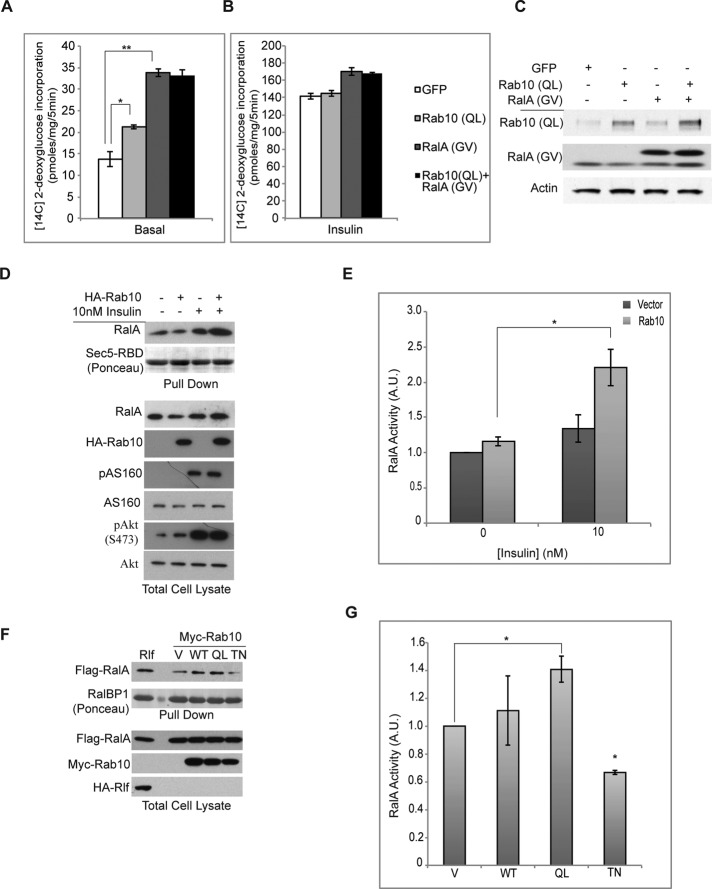
Recent evidence on the sequential action of G proteins in vesicular trafficking (Mizuno-Yamasaki et al., 2012) suggests that G protein cascades could be a general mechanism in the regulation of vesicle recycling. Of interest, upon insulin stimulation in fat cells, Akt1/2 phosphorylates and inactivates both AS160 (Kane et al., 2002) and the RalGAP complex RGC1/2 (Chen et al., 2011b), leading to the activation of Rab10 and RalA. However, the relative roles of these G proteins and their relationship to each other remain unknown. Whereas the overexpression of constitutively active RalA (G23V/GV) or Rab10 (Q68L/QL) (Figure 2C) independently increased basal glucose uptake in adipocytes (Figure 2A), no additive effects were observed with co-overexpression of the two active G proteins in the same cells. Expression of active RalA modestly increased insulin-stimulated glucose uptake, without further effect from coexpression of Rab10 (QL) (Figure 2B). These data suggest the possibility that RalA and Rab10 might reside in a linear pathway, and we thus determined whether either of the two G proteins might influence the activity of the other. Although overexpression of constitutively active RalA did not alter cellular Rab10 activity, measured as described (Supplemental Figure S2), Rab10 overexpression produced an increase in RalA activity under insulin-stimulated conditions (Figure 2, D and E) as measured by GST-Sec5-RBD pull-down assay. Activation of RalA was dependent on the activation status of Rab10. Overexpression of the constitutively active Rab10 (QL) allele caused hyperactivation of RalA, whereas a GDP-locked version of Rab10 (TN) decreased RalA activity (Figure 2, F and G). These data suggest that Rab10 functions upstream of RalA, and further that its activation can stimulate RalA activity.
FIGURE 2:
Rab10 activates RalA. (A) 3T3-L1 adipocytes expressing the indicated active G proteins using lentiviral vectors were subjected to 2-deoxyglucose uptake assay. Glucose uptake under basal conditions are shown. *p < 0.05; **p < 0.01. (B) Glucose uptake under conditions in A upon insulin stimulation. (C) Level of indicated proteins in 3T3-L1 adipocytes under conditions in A and B were assessed by immunoblots. (D) 3T3-L1 adipocytes were electroporated with 50 μg of the indicated plasmids. Two days after electroporation, cells were starved for 16 h and treated with 10 nM insulin for 5 min, and pull-down assay was performed using GST-Sec5-RBD beads. (E) Immunoblots from three independent experiments as in D were quantified using ImageJ (National Institutes of Health, Bethesda, MD) and graphed. *p < 0.05. (F) Indicated plasmids were expressed in Cos-1 cells, and cells were lysed after 24 h. Pull down with GST-RalBP1 beads was performed, and samples were run on SDS–PAGE and immunoblotted. (G) RalA activity as measured by quantification of three independent experiments as in F. Statistical tests were performed by comparing each condition to that of the vector. *p < 0.01.
Rab10 activates RalA through Rlf
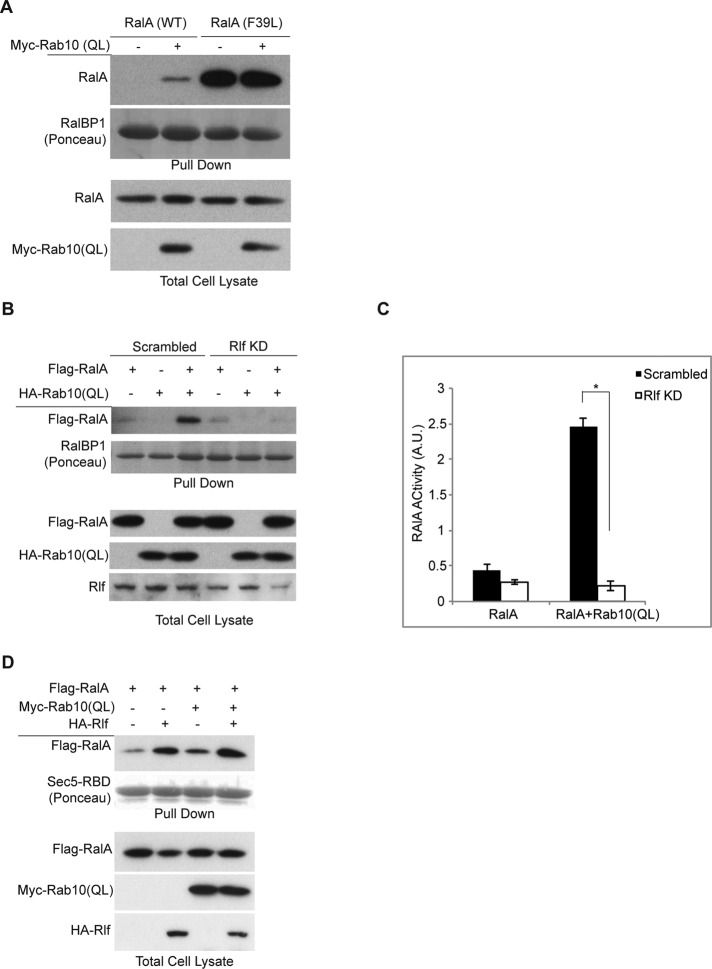
The activation of RalA by Rab10 could be due to inactivation of the RalGAP complex, activation of a RalGEF, or both. RGC1/2 is inactivated upon insulin stimulation through Akt phosphorylation of the catalytic subunit RGC2 (Chen et al., 2011b). Rab10 overexpression did not accelerate or affect the phosphorylation status of RGC2 (unpublished data). Moreover, a point mutation in RalA (F39L/FL) with increased nucleotide dissociation rates that quickly cycles between GDP and GTP (Reinstein et al., 1991; Lim et al., 2005), and thus does not require a GEF for activation, was not influenced by overexpression of Rab10 (QL), unlike what was observed for RalA (wild type [WT]; Figure 3A).
FIGURE 3:
Rab10 activates RalA through Rlf. (A) Rab10 activates RalA through a GEF. Wild-type and F39L mutant of RalA were expressed with or without HA-Rab10 in Cos-1 cells. Pull-down assay was performed with GST-RalBP1 beads, and samples were run on SDS–PAGE and Western blotted. (B) Stealth siRNA knockdown of Rlf in 293T cells. Two days posttransfection, cells were lysed, and RalA pull down with GST-RalBP1 beads was performed. Levels of indicated proteins in total cell lysates and pull down are shown. (C) Images from experiment in B were quantified by ImageJ and plotted. *p = 0.05. (D) Cos-1 cells were transfected with FLAG-RalA together with Myc-Rab10 (QL), HA-Rlf, or both. At 24 h later, cells were lysed and subjected to pull-down assay using GST-Sec5-RBD beads. Lysates and pull-down samples were run on SDS–PAGE and immunoblotted.
Together these data suggest that Rab10 might activate RalA through recruitment or activation of a specific GEF. There are several known GEFs for RalA, including Rgl1, Rgl2/Rlf, Rgl3, RalGDS, and others (Neel et al., 2011). Among these, Rlf, the mouse homologue of Rgl2, is most abundant in 3T3-L1 adipocytes (unpublished data). To evaluate the role of Rlf in Rab10-stimulated RalA activation, we knocked down Rlf in 293T cells, since this cell line has high basal Akt phosphorylation and as a result reduced RalGAP activity. Knockdown of Rlf decreased basal RalA activity. Whereas overexpression of Rab10 (QL) caused a twofold increase in RalA activity, depletion of Rlf blocked this effect (Figure 3, B and C). We also introduced Rab10 (QL) in the presence of ectopic expression of hemagglutinin (HA)-Rlf. Whereas Rab10 (QL) and Rlf increased RalA activity independently, coexpression of the two proteins was synergistic (Figure 3D), suggesting the possibility that Rab10 can recruit Rlf to activate RalA.
Rlf is an effector of Rab10
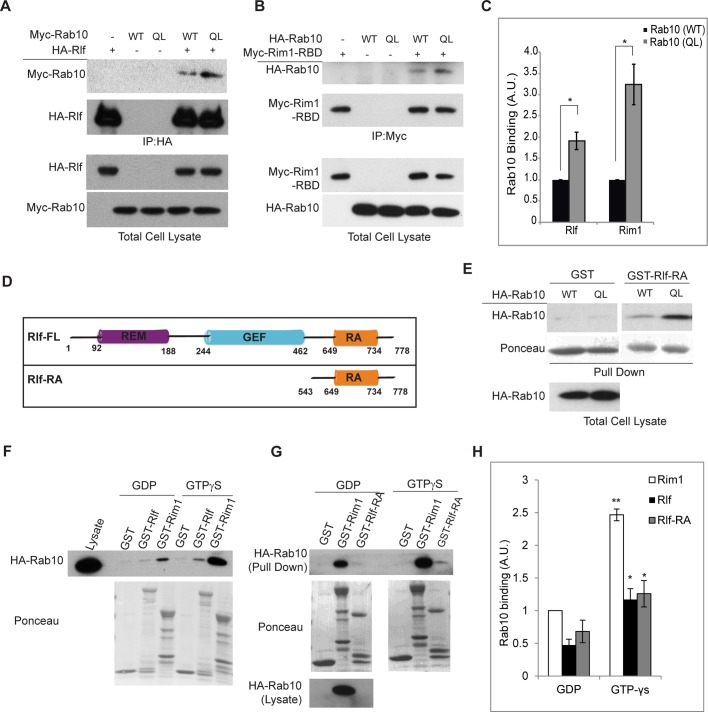
Because Rlf is required for Rab10-stimulated activation of RalA, we tested whether Rlf might be a direct effector for Rab10. Immunoprecipitation of HA-Rlf from Cos-1 cells overexpressing Myc-Rab10 (WT or QL) revealed that Rab10 specifically binds to Rlf, and further that this binding depends on the activation state of Rab10 (Figure 4A). Constitutively active Rab10 (QL) was twofold more effective in binding to Rlf than with the wild-type G protein (Figure 4C). Rlf binding to Rab10 (QL) was comparable to that observed with Rim1-RBD, a known effector of Rab10 (Figure 4, B and 4C). In addition, reciprocal immunoprecipitation of Myc-Rab10 also showed specific binding of Rlf to active Rab10 (QL) (Supplemental Figure S3).
FIGURE 4:
Rab10 interacts with Rlf. (A) Indicated proteins were ectopically expressed in Cos-1 cells. Immunoprecipitation with HA-Rlf shows specific interaction with Rab10 in immunoblots. (B) Cos-1 cells were transfected with indicated plasmids. At 24 h later, cells were lysed and immunoprecipitated with antibodies against Myc. Level of proteins in the total cell lysate and the immunoprecipitates are shown. (C) Quantification of blots in A and B. Images from three independent experiments were used to generate error bars. *p < 0.05. (D) Schematic representation of various domains in full-length and truncated Rlf. (E) Pull-down assay with GST-Rlf-RA. Indicated plasmids were expressed in Cos-1 cells, and GST or GST-Rlf-RA beads were used for pull down. Specific and increased binding to Rab10 (QL) are shown. (F) Lysates from Cos-1 cells expressing HA-Rab10 were treated with 1.0 mM GDP or 0.5 mM GTPγS. Pull-down experiments were performed with indicated GST beads. Protein levels in the total cell lysate, as well as pull down, are shown. (G) Pull-down experiment was performed as in F, except that GST-Rlf-RA beads were used instead of full-length Rlf. (H) Quantification of blots in F and G. Binding of Rim1 to GDP-bound Rab10 was set to 1, and fold changes in binding of Rlf and Rlf-RA to Rab10 in GDP/GTP-bound states were calculated. Error bars were acquired from at least three independent experiments. **p < 0.01, *p < 0.05.
Together these data suggested that Rlf could be a downstream effector of Rab10. Rlf is a known effector of another small G protein, Ras; constitutively active Ras (RasV12) binds to Rlf in the Ras activation (RA) domain of the latter protein (Figure 4D) and functions in cellular transformation (Wolthuis et al., 1997). Using a version of Rlf that contained only the RA domain (amino acids 543–778; Wolthuis et al., 1996), we pulled down HA-Rab10 overexpressed in Cos-1 cells (Figure 4E). To further confirm that this binding is dependent on the activation status of Rab10, we performed a pull-down assay using lysates from Cos-1 cells overexpressing HA-Rab10 after in vitro addition of GDP or nonhydrolyzable GTP (GTPγS). This assay demonstrated that upon binding to GTPγS, Rab10 interacted with a Rim1-RBD fusion protein significantly better than did Rab10-GDP (Figure 4, F and H). Similarly, both the full-length and RA domain of Rlf were equally effective in specifically binding to Rab10-GTPγS (Figure 4, F–H). These data indicate that Rlf is a downstream effector of Rab10 and that the RA domain of Rlf is sufficient to interact with Rab10, as is the case for its interaction with Ras (Peterson et al., 1996; Wolthuis et al., 1996).
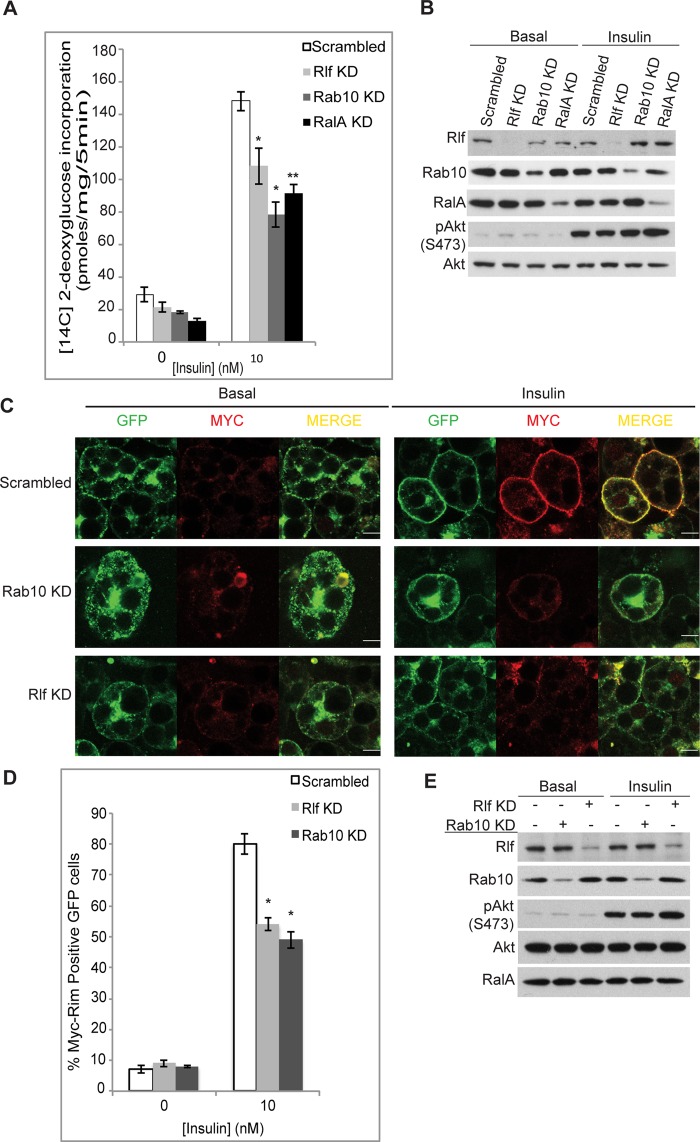
Rab10, RalA, and Rlf are required for insulin-stimulated glucose uptake in 3T3-L1 adipocytes
Rab10 and RalA are established components of the insulin regulatory network and are required for Glut4 translocation and glucose uptake in adipocytes (Chen et al., 2007; Sano et al., 2007, 2008). Because Rlf appears to be an intermediate in this activation cascade, we hypothesized that the GEF may also be essential. We depleted Rlf using siRNA in 3T3-L1 adipocytes before assaying 2-deoxyglucose uptake. The expression of Rlf was reduced by ∼70% after incubation with siRNA oligos (Figure 5B). Knockdown of Rlf caused a 15–20% loss in insulin-stimulated glucose uptake, comparable to the effect achieved by knockdown of RalA (Chen et al., 2007) and Rab10 (Figure 5A). Because RalA and Rab10 are required for translocation of Glut4 storage vesicles to the plasma membrane upon insulin stimulation (Chen et al., 2007; Sano et al., 2007), we also tested the effect of Rlf knockdown on Glut4 translocation. Knockdown of Rlf in 3T3-L1 adipocytes stably expressing Myc-Glut4–enhanced green fluorescent protein (eGFP; Bogan et al., 2001; Lodhi et al., 2007) caused a significant reduction (∼25%) in surface Glut4 levels similar to the effect of knockdown of RalA and Rab10 (Figure 5, C–E). Together these data suggest that Rlf is a new component required for Glut4 translocation and is essential for maximal glucose uptake in adipocytes.
FIGURE 5:
Rlf is required for maximal glucose uptake in 3T3-L1 adipocytes. (A) siRNA-mediated depletion of indicated genes in 3T3-L1 adipocytes. Six days after knockdown, cells were starved, and 2-deoxyglucose uptake was measured. A representative assay from five independent repetitions is shown. *p < 0.05, **p < 0.01. (B) Knockdown efficiency and insulin signaling in conditions in A. (C) Glut4 translocation assay. 3T3-L1 adipocytes stably expressing Myc-Glut4-eGFP were used for knockdown of indicated genes. Scrambled siRNA was used as control. Immunostaining for Myc (red) in nonpermeabilized cells indicates Glut4 translocation to plasma membrane. GFP levels in cells indicate total Glut4 expressed. A merge of GFP and Myc is shown. Bar, 10 μm. (D) Number of cells that show Myc staining at the rim in the total number of GFP-positive cells was counted for control and Rab10 and Rlf knockdown. Percentage of cells that undergo Glut4 translocation is graphed. Error bars are indicative of at least three independent experiments. *p < 0.05. (E) Knockdown efficiency at protein level is shown by immunoblots. Intact insulin signaling under conditions in D is shown by Western blotting.
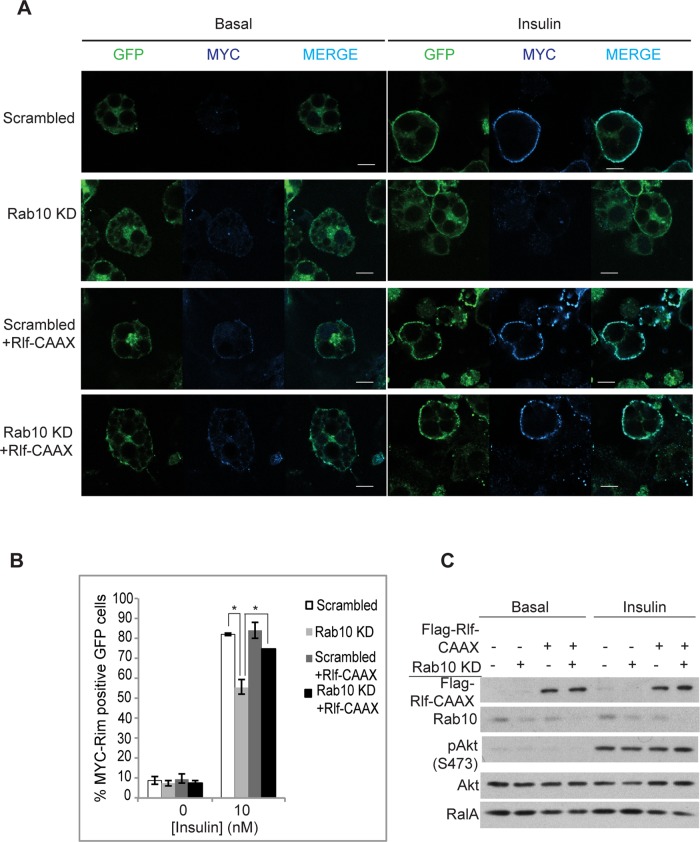
Rab10 affects RalA activity through Rlf in adipocytes
As we showed, active Rab10 increases RalA activity through Rlf (Figure 3, B– D). Because Rab10 can increase RalA activity via recruitment of Rlf (Figures 3 and 4), we attempted to rescue the effect of Rab10 knockdown by expression of an activated version of Rlf. We used a tagged version of Rlf (Rlf-CAAX) through addition of a CAAX motif to its C-terminus, thus tethering the protein to membranous compartments (Vigil et al., 2010). We confirmed that Rlf-CAAX partitioned more in the membrane fraction than in the cytosolic fraction, unlike the wild-type protein (Supplemental Figure S4A). Overexpression of Rlf-CAAX also produced a larger increase in RalA activity compared with the wild-type Rlf (Supplemental Figure S4B; Wolthuis et al., 1997). Whereas Rab10 knockdown caused a 25–30% loss in surface Glut4 levels (Figure 6, A and B), overexpression of Rlf-CAAX in these cells (Figure 6C) brought the surface Glut4 levels closer to that seen in control insulin-stimulated cells (Figure 6B). These data indicate that Rab10 affects Glut4 translocation at least partly by recruiting Rlf to activate RalA.
FIGURE 6:
Rab10 acts on RalA through Rlf in 3T3-L1 adipocytes. (A) 3T3-L1 adipocytes stably expressing Myc-Glut4-eGFP were electroporated with scrambled or Rab10 siRNA with or without FLAG-Rlf-CAAX. Two days later, cells were starved and treated with or without 10 nM insulin for 15 min. Nonpermeabilized cells were fixed and immunostained with anti-Myc antibodies. GFP indicates total Glut4 levels, and Myc staining in the rim (blue) represents presence or absence of Glut4 at the plasma membrane. Bar, 10 μm. (B) Percentage of cells that show Glut4 translocation upon insulin stimulation under various conditions in A was calculated by counting at least 100 cells in three independent experiments. *p < 0.05. (C) Knockdown efficiency and level of ectopically expressed FLAG-Rlf-CAAX in A are shown using Western blots. Insulin signaling was unaffected as detected by immunoblotting for pAkt (S473).
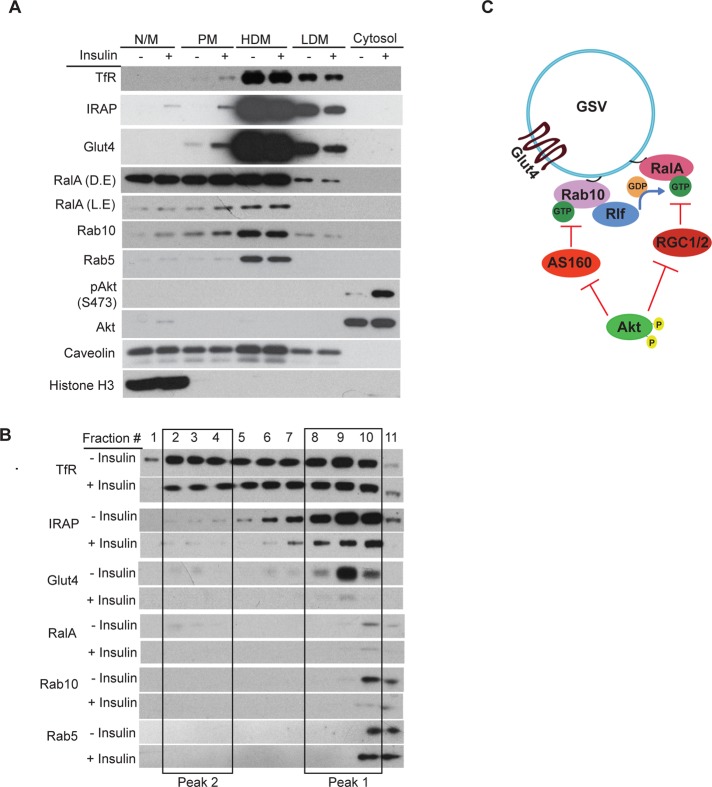
Rab10 and RalA function in the same subcellular compartment
The foregoing results suggest that Rab10 might coexist in the same cellular compartment as RalA. Furthermore, ectopic expression of a version of AS160 that is enriched at sites of Rab10 action through a C-terminal Rab10 membrane attachment motif (AS160GAP-Rab10 tail) decreased RalA activity (Supplemental Figure S4C), indicating a localized effect of Rab10 on RalA.
We performed subcellular fractionation of 3T3-L1 adipocytes to assess the insulin-dependent changes in the cellular localization of RalA and Rab10. As expected, Glut4, IRAP, and RalA were enriched in the plasma membrane fractions upon insulin stimulation, with a concurrent loss of these proteins in the low-density microsome (LDM) fractions. Of interest, Rab10 localization mirrored that of RalA (Figure 7A).
FIGURE 7:
Rab10 and RalA reside in insulin-sensitive GSVs. (A) Subcellular fractionation of 3T3-L1 adipocytes. Indicated fractions were run on SDS–PAGE and immunoblotted with antibodies as specified. HDM, high-density microsomes; LDM, low-density microsomes; N/M, nuclear/mitochondrial; PM, plasma membrane. L.E and D.E. denote light and dark exposures of the RalA blot, respectively. (B) LDM fractions from several independent experiments as in A were pooled to obtain an enriched fraction, which was then run on 14% iodixanol gradient. Fractions 1–11 represent lighter to denser peaks in the gradient. Insulin-sensitive pool of GSVs (fractions 8–10) is indicated by a box (peak 1). Peak 2 represents the second-less-responsive recycling endosomes, containing Glut4 (fractions 2–4). (C) Proposed model for the role of Rab10 in Glut4 translocation. Rab10 recruits Rlf to insulin-sensitive GSVs to promote GEF activity on RalA. Activated RalA allows Glut4 translocation through its interactions with the myosin motor and the exocyst.
These data suggest that RalA and Rab10 might be found together in some insulin-responsive Glut4 vesicles. LDM fractions were thus further fractionated on an iodixanol density gradient to distinguish the insulin-responsive Glut4 vesicles (peak 1) from the recycling endosomal fractions (peak 2; Hashiramoto and James, 2000). Optiprep fractions corresponding to insulin-responsive GSVs (peak 1) contained Glut4 and IRAP as well as RalA and Rab10 (Figure 7B). The appearance of RalA and Rab10 peaks in fraction 10, but less so in fraction 9, which is enriched in Glut4 and IRAP, indicates that these small G proteins are associated with only a subpopulation of the insulin-sensitive Glut4 vesicles. Taken together, our studies show that Rab10 recruits Rlf to activate RalA, which plays a role in the translocation of Glut4 to the plasma membrane (Figure 7C).
DISCUSSION
Studies indicated that Rab10 may be a crucial target of the GAP protein AS160 (Sano et al., 2007). Although data presented here confirm that Rab10 is indeed a target of AS160 in adipocytes, the molecular events that occur as a consequence of changes in Rab10 activity, and how they in turn contribute to the control of Glut4 trafficking, have remained a mystery. We reveal here the first novel effector for Rab10 in adipocytes, the guanyl nucleotide exchange factor Rlf, and demonstrate its role as an intermediate in a G protein cascade that mediates insulin-stimulated glucose transport. We show that Rab10 is upstream of RalA and plays an important role in RalA activation by insulin, leading to its interaction with the exocyst. Furthermore, this activation cascade is required for insulin-stimulated glucose transport and Glut4 translocation to the plasma membrane.
Many studies illustrated the importance of G protein activation cascades in control of vesicular trafficking in both mammalian and yeast cells, including TC10 to Rab5 (Lodhi et al., 2008), Rab11 to Rab8 (Bryant et al., 2010; Knodler et al., 2010), Rab5 to Rab7 (Bottanelli et al., 2012), Ras to RalA (Wolthuis et al., 1997), and others (Hutagalung and Novick, 2011; Mizuno-Yamasaki et al., 2012). A key feature of such cascades is the colocalization of the relevant G proteins. In this scenario, the G proteins are at least transiently found in the same population of vesicles, and the upstream protein recruits a GEF for activation of the downstream G protein. We show here that Rab10 and RalA are found in the same insulin-sensitive vesicle pool, and the cross-talk between them appears to be mediated by the RalA GEF, Rlf. Moreover, whereas Rab10, RalA, and Rlf are all required for full insulin-stimulated Glut4 translocation and glucose transport, expression of a membrane-tagged Rlf can rescue cells in which Rab10 has been depleted, strongly suggesting that it is the localization of the exchange factor that leads to RalA activation. This finding is particularly interesting in light of previous studies indicating that Rab10 may be upstream of the exocyst in certain instances (Jiang et al., 2008).
Although these studies provide the first hint of a relevant effector for Rab10 in the regulation of glucose uptake, several questions remain, including the relative roles of GEFs and GAPs in the regulation of these two G proteins. Both AS160 and RGC are specific GAPs for their cognate G proteins and are coordinately regulated by Akt-catalyzed phosphorylation, leading to inactivation of the GAPs and subsequent increased activity of the G proteins (Sano et al., 2003; Chen et al., 2011b). However, blockade of GAP activity alone is not expected to produce G protein activation, since there must be a mechanism to increase GTP binding once GDP is bound to the protein. This insight, plus the data presented here, suggests that RalA activation requires negative regulation of the GAP and positive regulation of its GEF. Previous studies suggest a similar scenario for Rab10 via its GEF Dennd4C (Sano et al., 2011).
Although numerous studies demonstrated that G proteins play important roles in vesicle trafficking, it is also known that their activation must be transient, so that vesicles can move into the next compartment (Mizuno-Yamasaki et al., 2012). However, if their GAPs are used for G protein activation, how are the signals terminated? One solution to this perplexing question might lie in changes to their effectors. Our previous studies indicated that the RalA effector Sec5 undergoes phosphorylation in adipocytes to permit the release of the Glut4 vesicle from the exocyst, thus permitting fusion with the plasma membrane (Chen et al., 2011a). One possibility is that Rlf undergoes a similar modification, ensuring that RalA is not constitutively activated. These and other possibilities are under investigation.
MATERIALS AND METHODS
Cell culture and transfections
COS-1 cells and 293T cells were cultured in DMEM containing 10% fetal bovine serum (FBS). Transfections were carried out using Lipofectamine 2000 following the manufacturer's protocol (Life Technologies, Grand Island, NY). For overexpression experiments, cells were collected 18–24 h after transfection. Knockdown experiments in 293T cells were conducted for 48 h from the time of transfection. 3T3-L1 preadipocytes were differentiated into mature adipocytes as described before (Liu et al., 2005). Electroporation into mature adipocytes was done within 1–3 d postdifferentiation at 160 kV, 950 μF (Inoue et al., 2006). Knockdown was continued for 4–6 d after electroporation unless specified otherwise. Viral work was carried out as described previously (Chen et al., 2011a). 3T3-L1 preadipocytes stably expressing Myc-Glut4-eGFP were maintained and differentiated as described previously (Lodhi et al., 2007). Cells were starved in reduced serum (0.5% FBS) medium for 16 h except indicated. Concentration of insulin used was 10 nM for specified times, unless stated otherwise.
Plasmids, siRNAs, and purification
Stealth siRNAs were purchased from Life Technologies, and oligo sequences are listed in the Supplemental Materials. Rab10 was subcloned into pKH3 and pRK5-Myc vectors. Site-Directed Mutagenesis (QuikChange; Agilent Technologies, Santa Clara, CA) was used to generate active (Q68L) and inactive (T23N) mutants of Rab10. FLAG-RalA constructs (wild type, G23V, S28N, F39L) have been described before (Chen et al., 2007). The N-terminal RBD of Rim1 (residues 11–399) was cloned from Marathon-Ready adult brain cDNA (Clontech, Mountain View, CA) by PCR and subcloned into pKCmyc (myc tag) vector and pGEX (GST tag) vector. Rab5 and RalA cDNA were obtained as described (Chen et al., 2007; Lodhi et al., 2007). Both were subcloned into pGEX vector. The Myr-Akt construct was kindly provided by Anne Vojtek (University of Michigan, Ann Arbor, MI). To make the AS160GAP-Rab10tail chimera, the GAP domain of AS160 was amplified by PCR with a 3′ primer containing the last 132 base pairs of Rab10 cDNA and subcloned into the red fluorescent protein (RFP) vector, which was kindly provided by Roger Tsien (University of California, San Diego, La Jolla, CA). Full-length Rlf and Rlf-RA (amino acids 543–778) were subcloned from HA-Rlf (Chen et al., 2011a) into pGEX4T1 vector (GE Healthcare, Piscataway, NJ). pBABE-HAII-Rlf-CAAX was described previously (McFall et al., 2001) and purchased from Addgene, Cambridge, MA (plasmid 12592). Rlf and Rlf-CAAX were subcloned into pKFLAG and pCS2-MT vectors. All constructs were verified by sequencing at the University of Michigan DNA sequencing core.
GST-RalBP1 beads were purchased from EMD Millipore (Billerica, MA). Purification of GST-Sec5-Ral binding domain has been previously described (Chen et al., 2011a). Purification of Rim1-RBD using high-performance Glutathione Sepharose Beads was carried out according to the manufacturer's instructions (GE Healthcare). GST-Rlf and GST-Rlf-RA were expressed in Rosetta (DE3 pLysS) cells (EMD Millipore, Billerica, MA), and purification protocol was adapted from Wolthuis et al. (1997). GST alone or GST-Rim1 RBD, RalA, Rab4, 5, 10, and 11 were expressed in BL21(DE3)pLysS Escherichia coli strain and purified as described (Smith and Johnson, 1988).
GDP-AIFx loading assay
GST alone or GST-fusion proteins bound to glutathione beads were incubated for 30 min at 25°C in loading buffer (20 mM Tris, pH 7.5, 1 mM dithiothreitol [DTT], 50 mM NaCl) with 2 mM EDTA and Complete, EDTA-free protease inhibitor tablets (Roche, San Francisco, CA). GST-Rab10 was then loaded with nucleotide by incubating in loading buffer supplemented with 2 mM GDP or 200 mM GTPγS for 1 h at 25°C. To stop loading, 10 mM MgCl2 was added for 5 min at 25°C. To produce GDP/AlF4-bound Rab10, 30 mM AlCl3 and 10 mM NaF were included in the GDP loading buffer. Loaded GST-Rab10 beads were then added to COS-1 cells that were lysed in buffer (25 mM Tris, pH 7.5, 137 mM NaCl, 10% glycerol, 1% NP-40, 5 mM MgCl2, and 1 mM DTT) supplemented with 10 μM GDP, 10 μM GTPγS, or 10 μM GDP, 30 μM AlCl3, and 10 mM NaF, and Complete, EDTA-free protease inhibitor tablets. The reaction was incubated for 2 h at 4°C, and then beads were washed three times in wash buffer (25 mM Tris, pH 7.5, 40 mM NaCl, 30 mM MgCl2, 1% NP-40, and 1 mM DTT) and once in rinse buffer (25 mM Tris, pH 7.5, 40 mM NaCl, 30 mM MgCl2, and 1 mM DTT) supplemented with 10 μM GDP, 10 μM GTPγS, or 10 μM GDP, 30 mM AlCl3, and 10 mM NaF, and Complete EDTA-free protease inhibitor tablets. The pull downs were solubilized in 2× SDS sample buffer and subjected to SDS–PAGE, and Western blotting was performed using the indicated antibodies.
Statistical analysis
All error bars are depicted as SEM. Statistical significance was determined via Student's t test.
Pull-down and coimmunoprecipitation experiments
Rab10-GTP was pulled down using GST-Rim1-RBD beads as a measure of active Rab10. GST-RalBP1 or GST-Sec5-RBD beads were used interchangeably to quantify RalA activity. Pull-down experiments were carried out in a buffer containing 100 mM Tris, pH 7.5, 10% glycerol, 1% NP-40, 1 mM DTT, 129 mM NaCl, 10 mM NaF, and 5 mM MgCl2 with protease inhibitor tablet (Roche) for 40 min at 4°C. Proteins were extracted from beads in SDS-sample buffer and analyzed. Immunoprecipitation experiments were carried out with indicated antibodies in a lysis buffer containing 100 mM Tris, pH 7.5, 129 mM NaCl, 1% Triton X-100, 10 mM NaF, 5 mM MgCl2, 1 mM EDTA, and 1 mM NaVO3 with protease inhibitor tablet for 2 h to overnight at 4°C. Immunoprecipitates were adsorbed on Protein A/G plus agarose (Santa Cruz Biotechnology, Dallas, TX) for 1 h, washed three times in lysis buffer, and eluted in 2× SDS-sample buffer.
Reagents, antibodies, and immunoblotting
Samples were run on 4–20% precast SDS–PAGE gels (Novex, Life Technologies) and transferred to nitrocellulose membrane. Antibodies to HA and Myc (monoclonal mouse and polyclonal rabbit) and human Rlf were purchased from Santa Cruz Biotechnology. Mouse-FLAG antibodies were from Agilent Technologies. Anti-RFP antibodies were from Abcam (Cambridge, MA). AS160 and pAS160 (T642) antibodies were from EMD Millipore. Rabbit anti-Rlf serum described previously (Wolthuis et al., 1997) was affinity purified (Pacific Immunologies, Ramona, CA) and used for immunoblotting. RalA and caveolin3 antibodies were from BD Biosciences (San Jose, CA). Transferrin receptor antibody was from Zymed Labs. Glut4 and Sec8 antibodies were purchased from Alpha Diagnostics (San Antonio, TX) and Enzo LifeSciences (Farmingdale, NY). Rab10, Akt, pAkt (S473), pAkt-substrate, histone H3, and IRAP antibodies were from Cell Signaling (Danvers, MA). Horseradish peroxidase–conjugated secondary antibodies were obtained from Thermo Scientific (Waltham, MA), and Alexa Flour secondary antibodies were from Molecular Probes, Life Technologies. Insulin, GDP, GTPγS, and Optiprep gradient solution were purchased from Sigma-Aldrich (St. Louis, MO).
Glucose uptake assay
The 2-deoxyglucose uptake assay was performed as previously described (Inoue et al., 2006). Briefly, adipocytes that are 7–10 d postdifferentiation were starved for 6 h and treated with 10 nM insulin for 30 min. The 14C-labeled glucose incorporation in 5 min was measured to calculate glucose uptake by 3T3-L1 adipocytes.
Glut4 translocation assay
Glut4 translocation in response to 10 nM insulin treatment for 15 min was measured as described before (Inoue et al., 2006). Differentiated adipocytes stably expressing Myc-Glut4-eGFP were used 4–6 d after knockdown except when proteins were overexpressed, during which cells were analyzed after 2 d. Immunostaining was performed using indicated antibodies. Glut4 translocation to the plasma membrane was measured by staining with Myc antibodies in nonpermeabilized cells. Percentage of cells with insulin-stimulated Glut4 translocation was calculated as the percentage of cells with Myc staining in the GFP-positive pool.
Subcellular fractionation and iodixanol gradient
3T3-L1 adipocytes that were 9–10 d postdifferentiation were lysed in HES buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 1 mM EDTA, 250 mM sucrose) and homogenized in a Dounce homogenizer. Fractionation was performed by ultracentrifugation using established protocols (Hashiramoto and James, 2000; Chen et al., 2007). LDMs were further fractionated on a 14% iodixanol (Optiprep) gradient at 56,000 rpm in a VTi65.2 rotor, and fractions were collected from lighter to denser layers (peaks 1–11; Chen et al., 2007).
Supplementary Material
Acknowledgments
Rlf antiserum was a generous gift from Johannes L. Bos (Utrecht University, Utrecht, Netherlands). Rab10 and AS160 cDNA were provided by Gustav Lienhard (Dartmouth Medical School, Hanover, NH). This study was supported by National Institutes of Health Grants R01DK076906 and DK061618 to A.R.S.
Abbreviations used:
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1060) on August 7, 2014.
S.K. and A.R.S developed the research design and prepared the manuscript. S.K. designed and performed the experiments and analyzed data. T.X. designed and performed experiments. M.U. collaborated on the fractionation experiments. D.L. performed some of the initial experiments and contributed to the development of the project. J.S. generated some of the expression vectors and performed the experiment with RalA (FL). X.W.C. carried out the glucose uptake assay with active G proteins and commented on the manuscript.
No conflicts of interest are associated with this article.
REFERENCES
- Bogan JS, McKee AE, Lodish HF. Insulin-responsive compartments containing GLUT4 in 3T3-L1 and CHO cells: regulation by amino acid concentrations. Mol Cell Biol. 2001;21:4785–4806. doi: 10.1128/MCB.21.14.4785-4806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Bottanelli F, Gershlick DC, Denecke J. Evidence for sequential action of Rab5 and Rab7 GTPases in prevacuolar organelle partitioning. Traffic. 2012;13:338–354. doi: 10.1111/j.1600-0854.2011.01303.x. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XW, Leto D, Chiang SH, Wang Q, Saltiel AR. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev Cell. 2007;13:391–404. doi: 10.1016/j.devcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Chen XW, Leto D, Xiao J, Goss J, Wang Q, Shavit JA, Xiong T, Yu G, Ginsburg D, Toomre D, et al. Exocyst function is regulated by effector phosphorylation. Nat Cell Biol. 2011a;13:580–588. doi: 10.1038/ncb2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XW, Leto D, Xiong T, Yu G, Cheng A, Decker S, Saltiel AR. A Ral GAP complex links PI 3-kinase/Akt signaling to RalA activation in insulin action. Mol Biol Cell. 2011b;22:141–152. doi: 10.1091/mbc.E10-08-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguez L, Lee A, Chavez JA, Miinea CP, Kane S, Lienhard GE, McGraw TE. Full intracellular retention of GLUT4 requires AS160 Rab GTPase activating protein. Cell Metab. 2005;2:263–272. doi: 10.1016/j.cmet.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Fukuda M. Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2. Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J Biol Chem. 2003;278:15373–15380. doi: 10.1074/jbc.M212341200. [DOI] [PubMed] [Google Scholar]
- Hashiramoto M, James DE. Characterization of insulin-responsive GLUT4 storage vesicles isolated from 3T3-L1 adipocytes. Mol Cell Biol. 2000;20:416–427. doi: 10.1128/mcb.20.1.416-427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JC, Pessin JE. Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Curr Opin Cell Biol. 2007;19:466–473. doi: 10.1016/j.ceb.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Chiang SH, Chang L, Chen XW, Saltiel AR. Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol Biol Cell. 2006;17:2303–2311. doi: 10.1091/mbc.E06-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Fan J, Bai L, Wang Y, Chen Y, Yang L, Chen L, Xu T. Direct quantification of fusion rate reveals a distal role for AS160 in insulin-stimulated fusion of GLUT4 storage vesicles. J Biol Chem. 2008;283:8508–8516. doi: 10.1074/jbc.M708688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S, Sano H, Liu SC, Asara JM, Lane WS, Garner CC, Lienhard GE. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J Biol Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larance M, Ramm G, Stockli J, van Dam EM, Winata S, Wasinger V, Simpson F, Graham M, Junutula JR, Guilhaus M, et al. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J Biol Chem. 2005;280:37803–37813. doi: 10.1074/jbc.M503897200. [DOI] [PubMed] [Google Scholar]
- Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Liu J, DeYoung SM, Zhang M, Cheng A, Saltiel AR. Changes in integrin expression during adipocyte differentiation. Cell Metab. 2005;2:165–177. doi: 10.1016/j.cmet.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Lodhi IJ, Bridges D, Chiang SH, Zhang Y, Cheng A, Geletka LM, Weisman LS, Saltiel AR. Insulin stimulates phosphatidylinositol 3-phosphate production via the activation of Rab5. Mol Biol Cell. 2008;19:2718–2728. doi: 10.1091/mbc.E08-01-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi IJ, Chiang SH, Chang L, Vollenweider D, Watson RT, Inoue M, Pessin JE, Saltiel AR. Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 2007;5:59–72. doi: 10.1016/j.cmet.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall A, Ulku A, Lambert QT, Kusa A, Rogers-Graham K, Der CJ. Oncogenic Ras blocks anoikis by activation of a novel effector pathway independent of phosphatidylinositol 3-kinase. Mol Cell Biol. 2001;21:5488–5499. doi: 10.1128/MCB.21.16.5488-5499.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miinea CP, Sano H, Kane S, Sano E, Fukuda M, Peranen J, Lane WS, Lienhard GE. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- Neel NF, Martin TD, Stratford JK, Zand TP, Reiner DJ, Der CJ. The RalGEF-Ral effector signaling network: the road less traveled for anti-Ras drug discovery. Genes Cancer. 2011;2:275–287. doi: 10.1177/1947601911407329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessin JE, Saltiel AR. Signaling pathways in insulin action: molecular targets of insulin resistance. J Clin Invest. 2000;106:165–169. doi: 10.1172/JCI10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Trabalzini L, Brtva TR, Fischer T, Altschuler DL, Martelli P, Lapetina EG, Der CJ, White GC., 2nd Identification of a novel RalGDS-related protein as a candidate effector for Ras and Rap1. J Biol Chem. 1996;271:29903–29908. doi: 10.1074/jbc.271.47.29903. [DOI] [PubMed] [Google Scholar]
- Reinstein J, Schlichting I, Frech M, Goody RS, Wittinghofer A. p21 with a phenylalanine 28–leucine mutation reacts normally with the GTPase activating protein GAP but nevertheless has transforming properties. J Biol Chem. 1991;266:17700–17706. [PubMed] [Google Scholar]
- Sakamoto K, Holman GD. Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am J Physiol Endocrinol Metab. 2008;295:E29–E37. doi: 10.1152/ajpendo.90331.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Eguez L, Teruel MN, Fukuda M, Chuang TD, Chavez JA, Lienhard GE, McGraw TE. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Sano H, Kane S, Sano E, Miinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- Sano H, Peck GR, Kettenbach AN, Gerber SA, Lienhard GE. Insulin-stimulated GLUT4 protein translocation in adipocytes requires the Rab10 guanine nucleotide exchange factor Dennd4C. J Biol Chem. 2011;286:16541–16545. doi: 10.1074/jbc.C111.228908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Roach WG, Peck GR, Fukuda M, Lienhard GE. Rab10 in insulin-stimulated GLUT4 translocation. Biochem J. 2008;411:89–95. doi: 10.1042/BJ20071318. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Shirakawa R, Fukai S, Kawato M, Higashi T, Kondo H, Ikeda T, Nakayama E, Okawa K, Nureki O, Kimura T, et al. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem. 2009;284:21580–21588. doi: 10.1074/jbc.M109.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Thong FS, Bilan PJ, Klip A. The Rab GTPase-activating protein AS160 integrates Akt, protein kinase C, and AMP-activated protein kinase signals regulating GLUT4 traffic. Diabetes. 2007;56:414–423. doi: 10.2337/db06-0900. [DOI] [PubMed] [Google Scholar]
- Vigil D, Martin TD, Williams F, Yeh JJ, Campbell SL, Der CJ. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem. 2010;285:34729–34740. doi: 10.1074/jbc.M110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- Wolthuis RM, Bauer B, van ‘t Veer LJ, de Vries-Smits AM, Cool RH, Spaargaren M, Wittinghofer A, Burgering BM, Bos JL. RalGDS-like factor (Rlf) is a novel Ras and Rap 1A-associating protein. Oncogene. 1996;13:353–362. [PubMed] [Google Scholar]
- Wolthuis RM, Ruiter ND de, Cool RH, Bos JL. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 1997;16:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.