Period 2 forms a trimeric complex with p53 and Mdm2. As a result, p53’s transcriptional activity and stability are modulated in unstressed cells, ensuring that basal levels are present if a p53-mediated response is needed. These data provide evidence of cross-talk between circadian and checkpoint components, adding a level of regulation to the checkpoint.
Abstract
Human Period 2 (hPer2) is a transcriptional regulator at the core of the circadian clock mechanism that is responsible for generating the negative feedback loop that sustains the clock. Its relevance to human disease is underlined by alterations in its function that affect numerous biochemical and physiological processes. When absent, it results in the development of various cancers and an increase in the cell's susceptibility to genotoxic stress. Thus we sought to define a yet-uncharacterized checkpoint node in which circadian components integrate environmental stress signals to the DNA-damage response. We found that hPer2 binds the C-terminal half of human p53 (hp53) and forms a stable trimeric complex with hp53’s negative regulator, Mdm2. We determined that hPer2 binding to hp53 prevents Mdm2 from being ubiquitinated and targeting hp53 by the proteasome. Down-regulation of hPer2 expression directly affects hp53 levels, whereas its overexpression influences both hp53 protein stability and transcription of targeted genes. Overall our findings place hPer2 directly at the heart of the hp53-mediated response by ensuring that basal levels of hp53 are available to precondition the cell when a rapid, hp53-mediated, transcriptional response is needed.
INTRODUCTION
Circadian rhythms are conserved mechanisms of disparate phylogenetic origin and complexity that measure time on a scale of about 24 h and adjust the organism's physiology to external environmental signals (for review, see Bell-Pedersen et al., 2005). Accordingly, core circadian clock genes are genes whose protein products are necessary components for the generation and regulation of circadian rhythms. In addition to defining the core of the molecular circadian clock, high-throughput studies permitted identification of a large number of circadian molecular outputs that control many aspects of an organism's physiology, ranging from organ function and cognitive performance to systems-level behavior (Duffield et al., 2002; Duffield, 2003; Panda et al., 2002; Storch et al., 2002). Several of those studies showed that an estimated 10% of genes in a given tissue exhibit a pattern of circadian expression and ∼7% of all circadian-controlled genes regulate either cell-cycle progression or cell death processes (Duffield et al., 2002; Panda et al., 2002; Storch et al., 2002; Lowrey and Takahashi, 2004). These observations led to the hypothesis that the circadian and cell cycle systems operating within an individual cell are interlocked through the sharing of some critical elements. An emerging relationship among these mechanisms arises from the observation that cell-cycle progression is impaired in circadian gene–deficient animals, providing the first direct link between circadian gene regulation and cell division (Matsuo et al., 2003). A less predictable discovery was that mutation of one of the circadian clock genes, period 2, resulted in a cancer-prone animal that developed spontaneous tumors more rapidly than normal animals. Moreover, this tumor-rate difference increased when animals were exposed to γ-irradiation (γ-IR), arguing for a role of the per2 gene in tumor suppression and DNA damage response through control of cell proliferation and death (Fu et al., 2002).
We focus our studies on Per2, a molecule whose function and structural organization is viewed as pivotal for its role as a “molecular integrator” of the organism's physiology and biochemical network (Albrecht et al., 2007). However, Per2’s potential pleiotropic functions remain to be established. Not surprisingly, regulation of per2 gene expression is tightly controlled by a plethora of transcription factors that either interact directly within regulatory regions of per2, such as p53 (Miki et al., 2013), or modulate the activity of other components of the transcription machinery (Horikawa et al., 2000; Fukuhara et al., 2001; Cajochen et al., 2006; Segall et al., 2006). Accordingly, fine-tuning of Per2 stability, localization, and posttranscriptional and posttranslational modifications is relevant for Per2 interaction with various proteins and ligands, all of which have an effect on numerous regulatory signaling pathways (for review, see Albrecht et al., 2007, and references therein).
To understand fully how Per2’s function and oscillatory behavior modulate signaling events that influence critical aspects of disease development, we first looked for novel partners of human Period2 (hPer2) that were essential components of cell proliferation and death pathways. We then investigated the regulatory consequences of their association in cell signaling. In the present study, we report the identification of the human tumor suppressor p53 (hp53 hereafter) as a novel direct interactor of hPer2, a finding supported by two-hybrid screening studies and further validated in cells. We hypothesize that hPer2, a core circadian component and tumor suppressor protein, is a novel downstream effector of the DNA-damage checkpoint pathway and is an important regulatory factor that selectively modulates p53 function. Remarkably, hPer2 association with hp53 occurs within p53’s DNA-binding and C-terminal end. This region has a critical regulatory role for its tumor suppression function by virtue of containing the tetramerization domain required for hp53 oligomerization, the nuclear localization signals needed for shuttling, and various residues targeted for posttranslational modification, including those that are polyubiquitinylated (Kruse and Gu, 2009). Moreover, our findings establish the presence of a trimeric complex in cells in which the murine double minute-2 (Mdm2) E3 ubiquitin ligase, a negative regulator of hp53 that acts by modifying the hp53’s C-terminus and promoting its proteasomal degradation, binds to the N-terminus region of hp53. Conversely, hPer2 modulates the extent of Mdm2-mediated ubiquitination by binding to the C-terminus portion of hp53. Furthermore, our studies show that hPer2 binding to hp53 directly controls hp53-mediated transcriptional activity, suggesting a novel multilevel mechanism for regulating hp53 function to include circadian components. These findings not only provide important insights into the already complex mechanism of hp53 regulation, but also shed light on the emerging role of circadian components as modulators of checkpoint molecules, thus supporting a potential role for circadian factors in tumorigenesis.
RESULTS
Mice without functional mouse Period2 (mPer2) show increased sensitivity to γ-IR exposure and reduced apoptotic response, all of which are phenotypically manifested by premature hair graying and hair loss, early onset of hyperplastic growth, increased tumor occurrence, and severe morbidity (Fu et al., 2002). Despite the lack of a detailed mechanistic foundation for these observations, a number of cell cycle and tumor suppressor genes known to be under circadian control (CCND1, CCNA2, and MYC, which encode cyclin D1, A, and c-myc, respectively) were found to be deregulated in per2-deficient animals (Fu et al., 2002). This suggests a role of Per2 in tumor suppression by modulating DNA-damage-responsive pathways. Therefore we focused our initial studies on identifying hPer2 partners acting on key nodes of the protein–protein interaction network whose circadian deregulation can directly affect cellular homeostasis.
The hPer2 transcription factor interacts with the tumor suppressor protein p53
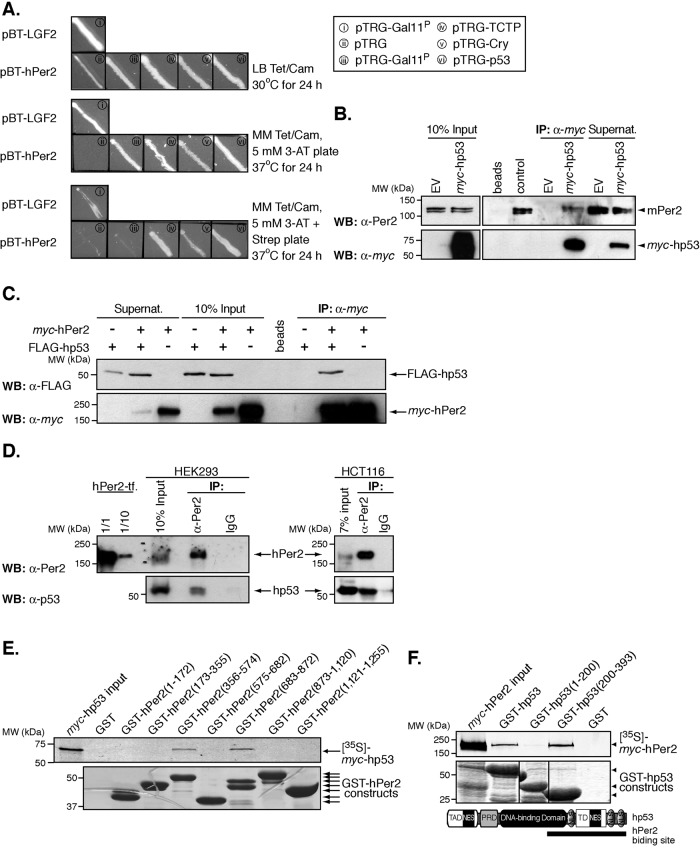
As part of an effort to define novel factors important for hPer2 regulation, we used a bacterial two-hybrid system to screen a human liver cDNA library to search for interacting partners. We chose this library because liver is known to be a peripheral oscillator tissue, and there are comprehensive studies on how circadian components are interlocked and operate in liver tissue (Lamia et al., 2008). Three baits were independently used for screening: full-length hPer2 and two fragments of cDNA encoding the N-terminal (residues 1–821) and C-terminal (residues 822–1255) regions of hPer2. These regions were chosen because of their relevance to Per2 function in various cellular processes and the presence of functional and structural domains known to bind protein counterparts (Griffin et al., 1999; Kume et al., 1999) or small ligand molecules (Yang et al., 2008). The human liver cDNA library (primary size, 6.9 × 106) was screened with the generated pBT recombinant plasmids. Approximately 4 × 106 clones were screened, and 120 were identified as putative positive interactors (67 strong and 53 weak interactors). These clones were maintained in nonselective media containing antibiotics and later patched on selective screening medium containing 5 mM 3-amino-1,2,4-triazole (3-AT) (Figure 1A). All putative clones were then subjected to further screening on a dual selective screening medium (5 mM 3-AT and streptomycin) and confirmed positive. To validate protein–protein interactions, we performed retransformation of the reporter strain using the pTRG-positive clones and recombinant pBT baits. Our results show that 17 clones, which include proteins involved in cellular metabolic processes, RNA binding, regulation of programmed cell death, transcriptional activation, response to stress, and cell cycle progression, reproducibly grow on selective screening medium when cotransformed with the bait plasmid but failed to grow under the same conditions when cotransformed with the empty pBT vector. Among the clones identified in the screening were hp53, the translationally controlled tumor protein (TCTP), and various fragments encoding open reading frame regions of the circadian factor crytochrome (Cry), a known direct interactor of hPer2 (Figure 1A). Remarkably, hp53 was also identified as a positive interactor when screenings were carried out using the sequences encompassing the N- and C-terminal fragments of hPer2, suggesting that more than one interaction site for hp53 might exist within the circadian factor.
FIGURE 1:
The circadian factor hPer2 interacts with hp53. (A) Two-hybrid protein–protein interaction between hPer2, hp53, TCTP, and Cry. Each pair of plasmids—i) pBT-LGF2 + pTRG-Gal11P, ii) pBT-hPer2 + pTRG, iii) pBT-hPer2 + pTRG-Gal11P, iv) pBT-hPer2 + pTRG-TCTP, v) pBT-hPer2 + pTRG-Cry, and vi) pTRG-hp53—was grown on nonselective medium plus antibiotics (LB tetracycline [Tet]/chloramphenicol [Cam]) and later patched on both selective screening minimum medium (MM Tet/Cam/5 mM 3-AT) and dual-selective minimum medium containing MM Tet/Cam/5 mM 3-AT/streptomycin (Strep). Positive controls were i and v, whereas ii and iii were negative. (B) Pellets from CHO cells transfected with pCS2+myc-hp53 were lysed in 25 mM Tris-phosphate pH 7.8, 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, and 1% Triton X-100, and extracts (∼300 μg) were incubated with α-myc beads. Endogenous mPer2 and recombinantly expressed hp53 were detected by using either α-Per2 (top) or -myc antibodies (bottom). Control indicates 20 μg of total extract. (C) Samples from CHO cells cotransfected with pCS2+myc-hPer2 and pCS2+FLAG-hp53 were immunoprecipitated using α-myc beads and immunoblotted using α-FLAG (top) and -myc antibodies (bottom). (D) One milligram of HEK293 and HCT116 extracts was incubated with either α-Per2 or IgG. Complexes were immunoprecipitated using protein A beads and immunoblotted for endogenous proteins using α-Per2 (top) and -p53 antibodies (bottom). For the positive control, HEK293 cells were transfected with pCS2+hPer2 (hPer2-tf), and total cell extracts (20 μg [1/1] and 2 μg [1/10]) were loaded. (E) Recombinant GST-tagged fragments of hPer2 were purified using affinity chromatography, and bound beads were incubated with 35S-labeled myc-hp53 and assayed for binding as described in the Supplemental Material. Bound complexes were visualized by Coomassie staining (bottom) and the radiolabeled protein detected by autoradiography (top). (F) Mapping of hPer2-binding regions in hp53. Schematic representation of hp53 architecture (393 residues), including the transactivation (TAD; residues 1–42), proline-rich (PRD; residues 61–92), DNA-binding (residues 101–300), and tetramerization domains (TD; residues 326–356). The binding site in hPer2 is indicated with a solid line. Bead-bound GST-hp53 and recombinant proteins were incubated with [35S]myc-hPer2 and analyzed for complex formation as described. In all cases, GST beads were used as a negative control; beads, matrix sample with no antibody bound; EV, empty vector; Supernat, supernatant fraction after immunoprecipitation (IP); transf, transfected cells; B–F show immunoblot data from a single experiment that was repeated three times with similar results.
To investigate whether Per2 forms a complex with p53 in cells, we examined various scenarios in which recombinantly expressed proteins and their endogenous counterparts were monitored for direct interaction in mammalian cells and extracts (Figure 1, B–D, and Supplemental Figure S1). We analyzed diverse cell lines (CHO cells, human embryonic kidney 293 [HEK293], and human colon carcinoma [HCT116] cells) to probe the prevalence of the complex in different backgrounds. These cell lines, besides being relatively easy to manipulate, were chosen because 1) CHO cells can be circadian synchronized, and mPer2 has been shown to be functional in this system (Yang et al., 2008), 2) human HEK293 cells have been an effective model to study the dynamic process of shuttling among hPer proteins as a result of posttranslational modifications (Vielhaber et al., 2001), the transcriptional regulation of genes by Clock/NPAS2 and Bmal dimers (Shi et al., 2010), and the validation of neurotransmitters regulating circadian locomotor rhythms (Mertens et al., 2005), and, finally, 3) human carcinoma HCT116 cells were used to study the role of Per1 in cell growth (Gery et al., 2006), the contribution of specific SCF ubiquitin E3 ligases for circadian protein turnover (Yang et al., 2009a), and the role of hPer2 for expression of cell cycle genes (Yang et al., 2009b). In addition, all cell lines express the wild-type forms of p53 and Per2.
To examine the interaction between p53 and Per2, we first transfected CHO cells with myc-hp53 and evaluated the presence of endogenous mPer2 within the complex by immunoprecipitation (Figure 1B). Results show mPer2 associates with the recombinantly expressed protein, further supporting our two-hybrid data, whereas unbound proteins remained available in the soluble fraction (Figure 1B, top). Endogenous mPer2 is usually detected as a doublet by its specific antibody in CHO cells, as previously described (Yang et al., 2008). We also performed reciprocal coimmunoprecipitation assays using CHO extracts from cells cotransfected with myc-hPer2, FLAG-hp53, or both plasmids. As shown in Figure 1C, immunoprecipitation of recombinant hPer2 specifically associates with hp53, a result of relevance for validating further in vitro experiments. The prevalence of this interaction was then assessed in both HEK293 and HCT116 cells by immunoprecipitating both endogenous components of the hp53/hPer2 complex (Figure 1D). Thus cell extracts were immunoprecipitated with either α-Per2 antibody or immunoglobulin G (IgG). As expected, immunoblot analysis revealed that hp53 is only detected in immunoprecipitates obtained using the α-Per2 antibody but not control IgG (Figure 1D). In sum, these results indicate that p53 and Per2 can be coimmunoprecipitated from extracts of either transfected cells or endogenous pools and are consistent with direct interaction between both proteins. To further test for this latter possibility directly, we incubated recombinant glutathione S-transferase (GST)-hp53–bound beads with labeled myc-hPer2 in the presence of increasing concentrations of untagged hp53 (Supplemental Figure S1A). Our results indicate that hPer2 is efficiently displaced from the complex as a result of adding increasing amounts of hp53, which support a model of direct interaction among these two protein components.
The hPer2 protein associates with the C-terminal half of hp53
To define the regions in hp53 and hPer2 responsible for their interaction, we first generated a number of recombinant constructs based on sequence homology, secondary structure prediction, and molecular modeling for each protein and determined their binding capacity to its full-length radiolabeled counterpart. Each of the seven recombinant fragments of hPer2—GST-hPer2(1-172), GST-hPer2(173-355), GST-hPer2(356-574), GST-hPer2(575-682), GST-hPer2(683-872), GST-hPer2(873-1120), and GST-hPer2(1121-1255)—was incubated with 35S-labeled myc-hp53 and protein association evaluated by pull-down assays (Figure 1E and Supplemental Figure S1B). Results showed that hp53 interacts within the central region of hPer2 comprising residues 356–574 (C-terminus of the PAS domain) and 683–872, a stretch of sequence predicted to be structurally flexible and strongly posttranslationally processed (www.expasy.org/structural_bioinformatics and www.expasy.org/proteomics/post-translational_modification). Similar studies were carried out in the presence of 35S-labeled myc-hPer2 using various constructs of hp53, including the recombinant GST-hp53(1-200), GST-hp53(1-296), GST-hp53(1-325), GST-hp53Δ30, GST-hp53(100-310), GST-hp53(200-393), and GST-hp53(300-393) fragments (Figure 1F and Supplemental Figure S1C). Pull-down assays showed that hPer2 binds hp53 on its C-terminal half, comprising most of the DNA-binding domain and oligomerization regions. Current models establish the DNA-binding functionality of hp53 to be dependent on both the core DNA-binding domain to provide for sequence specificity and the C-terminal region to recognize topological structural features of the target DNA (Kim et al., 1999; Ayed et al., 2001). Thus our results support a model of direct association between hPer2 and hp53 and predict a functional regulatory role for these proteins when acting as a complex.
The hPer2 protein forms a ternary complex with hp53 and its negative regulator, the Mdm2 oncogene protein
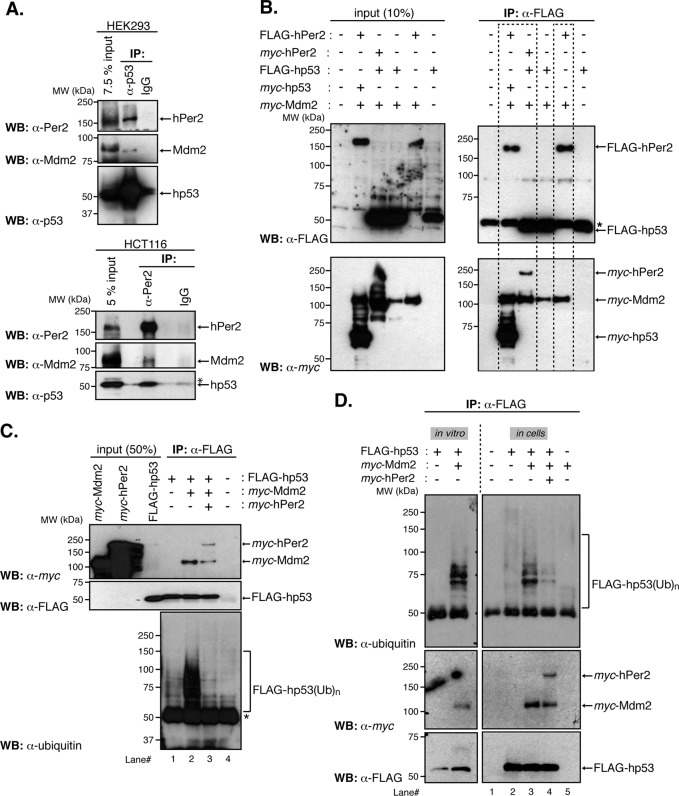
The Mdm2 protein acts as a bona fide E3-ubiquitin ligase for p53 by binding to its N-terminus and promoting either p53 monoubiquitination and nuclear export or p53 polyubiquitination and degradation through the proteasomal pathway (Honda et al., 1997; Li et al., 2003). Our results show that hPer2 primarily interacts within the C-terminal half of hp53. Thus it is possible that Mdm2 would also be sequestered into the complex. Extracts from HEK293 and HCT116 cells were immunoprecipitated with α-p53 and α-Per2 antibodies, respectively, and complex molecules were detected by immunoblotting (Figure 2A). Consistent with this hypothesis, coimmunoprecipitation and immunoblotting showed that, in both cases and under physiological conditions, Mdm2 associates with hp53/hPer2. This reveals the presence of the hp53/hPer2/Mdm2 endogenous complex in cells (Figure 2A). This result is in line with observations that all three proteins colocalize in similar cellular compartments and thus their interaction could readily occur (Supplemental Figure S2A; Giannakakou et al., 2000; Lohrum et al., 2000; Miyazaki et al., 2001).
FIGURE 2:
The hPer2 protein forms a ternary complex with hp53 and Mdm2, controlling the extent of hp53 ubiquitination. (A) HEK293 and HCT116 protein extracts (1 and 2 mg, respectively) were incubated with either α-p53 or α-Per2, respectively, and protein A beads. Rabbit IgG was used as a negative control. Immunoprecipitated complexes were analyzed for the presence of hPer2, hp53, and Mdm2 using specific antibodies (top for HEK293 and bottom for HCT116). Asterisk indicates a nonspecific signal. (B) HEK293 cells were transfected with pCS2+myc-Mdm2, pCS2+myc-hp53, pCS2+FLAG-hp53, pCS2+FLAG-hPer2, pCS2+myc-hPer2, or a combination of plasmids and complexes immunoprecipitated using α-FLAG–coupled beads. Complex components were identified by immunoblotting using α-FLAG and α-myc antibodies (right top and bottom). Input amounts were monitored in cell lysates (20 μg) and shown in the left top and bottom. Results similar to those presented were observed in two independent experiments. (C) In vitro–synthesized myc-hPer2 and FLAG-hp53 proteins were preincubated before the addition of myc-Mdm2 (ratio 1:2:5: FLAG-hp53:myc-Mdm2:myc-hPer2). Samples were then subjected to in vitro ubiquitination, followed by immunoprecipitation of hp53-bound proteins using α-FLAG antibody and protein A beads. Bound proteins were detected by immunoblotting and are indicated with arrows (top and middle). FLAG-hp53(Ub)n forms of hp53 were detected using α-ubiquitin antibody (bottom). Asterisk indicates IgG heavy chain. (D) HEK293 cells were transfected with pCS2+myc-Mdm2, pCS2+myc-hPer2, pCS2+FLAG-hp53, or a combination of plasmids and collected 12 h after treatment with 10 μM MG132. Cell lysates (100 μg) were incubated with α-FLAG and protein A beads and hp53-ubiquitinated complexes (FLAG-hp53(Ub)n) detected using α-ubiquitin antibody (top right). Bound proteins were visualized by immunoblotting using α-myc and α-FLAG antibodies (middle and lower right). An in vitro ubiquitination reaction was performed as described in C and is shown as control and for comparison purposes with the “in cells” result (left). Brackets denote ubiquitinated hp53. A, C, and D show immunoblot data from a single experiment that was repeated three times with similar results.
To study the functional relevance of interactions of these molecules, we devoted further experiments to establishing an “in-cell” system that allowed us to manipulate the complex components. HEK293 cells were transfected with various tagged forms of hPer2, hp53, and Mdm2 and complexes were monitored by immunoprecipitation and blotting (Figure 2B). Results show ternary complexes that included Mdm2 were detected using reciprocally tagged forms of both hp53 and hPer2 proteins (Figure 2B, left dashed box). Furthermore, an additional finding established that hPer2 is able to interact and form a stable complex with Mdm2 (Figure 2B, right dashed box). To rule out the contribution of endogenous hp53 in bridging hPer2/Mdm2 association, we transfected human non–small cell lung carcinoma cells (H1299 cells, p53-null; they contain a homozygous partial deletion of the p53 protein and lack expression of p53) with FLAG-hPer2 and myc-Mdm2 and evaluated their interaction by coimmunoprecipitation (Supplemental Figure S2B). Interaction between hPer2 and its E3-ubiquitin ligase β-TrCP was used as a positive control (Supplemental Figure S2B; Ohsaki et al., 2008). Results indicate that the hPer2/Mdm2 complex exists even in an hp53-null background, providing evidence for the physical association of hPer2 to Mdm2 in a p53-independent manner. Further studies established that the formation of the hp53/hPer2/Mdm2 complex is independent of the order of association of its components, as shown in Supplemental Figure S2C. In vitro binding of transcribed and translated recombinant hp53, hPer2, and Mdm2 into the trimeric complex occurred when hp53 was first associated with either Mdm2 or hPer2. Of note is the level of hPer2 associated with the preformed hp53/Mdm2 complex versus the one detected associated with hp53. This level seems to be lower, suggesting that binding of hPer2 precedes Mdm2 association with hp53 (Supplemental Figure S2C, lane 1 vs. lane 3). Overall these results suggest a dynamic scenario in which hPer2 might influence the activity and/or function of the Mdm2/hp53 complex or any of its components in a model in which Mdm2 acts as an E3-ubiquitin ligase for hp53, further linking components of the checkpoint machinery to circadian factors.
Binding of hPer2 modulates the extent of Mdm2-mediated hp53 ubiquitination
Next we asked whether binding of hPer2 to the C-terminus of hp53 modulates the extent of its ubiquitination when complexed to Mdm2. To test this possibility, we used a modified in vitro ubiquitination assay in which hp53, Mdm2, and hPer2 proteins were incorporated stepwise into a cell-free system. First, we allowed the hp53/hPer2 complex to form, followed by addition of Mdm2 in the presence of inhibitors of both proteasome and ubiquitin deconjugating enzymes, thus favoring the accumulation of ubiquitinated hp53 complexes (FLAG-hp53(Ub)n) before immunoprecipitation (Figure 2C and Supplemental Figure S4A). Results show that hPer2 efficiently prevented Mdm2-mediated hp53 ubiquitination when prebound. This is in contrast to what was observed when hp53 was solely incubated with Mdm2 (Figure 2C, lane 2 vs. lane 3, bottom, and Supplemental Figure S4A). A similar result was obtained using proteasome inhibitor–treated HEK293 cells cotransfected with hp53 and Mdm2 in the presence or absence of hPer2-expressing plasmid (Figure 2D, lane 3 vs. lane 4, top, and Supplemental Figure S4B). Comparative ratios of all proteins were detected in cells (Supplemental Figure S2D), complexes were efficiently formed (Figure 2D), and hp53 ubiquitinated forms were detected by specific immunoprecipitation (Figure 2D, top) and quantified (Supplemental Figure S4B). Although the data presented here suggest a role for hPer2 in controlling hp53 modifications mediated by Mdm2, they do not rule out the possibility of a deubiquitination activity associated with hPer2 that might counteract Mdm2 action. However, in vitro preliminary studies using hp53(Ub)n as a substrate suggest that this might not be the case (unpublished data). Consequently, the evidence suggests that binding of hPer2 to hp53 prevents Mdm2-mediated ubiquitination, allowing for the formation of a stable trimeric complex.
Binding of hPer2 influences hp53 stability
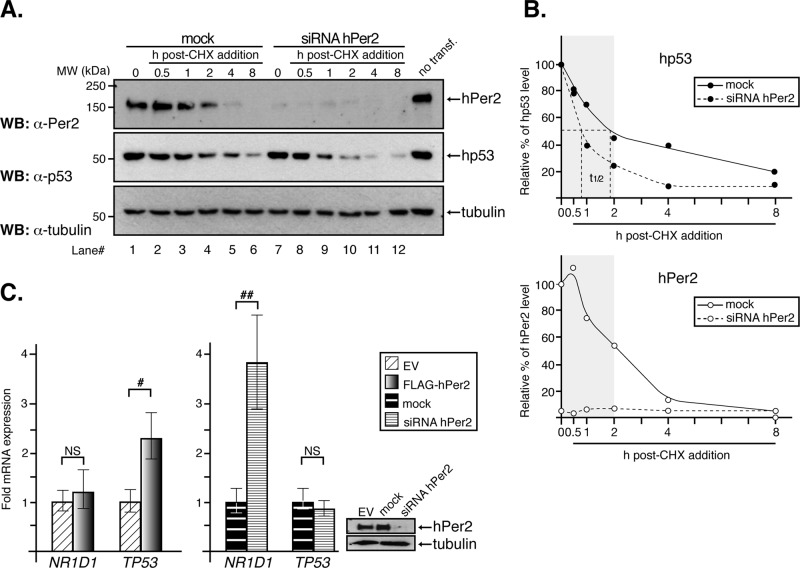
Rhythmic expression of p53 protein, but not its mRNA, has been observed in human tissues and correlates with those of the circadian oscillators Per and Bmal1 (Bjarnason et al., 1999, 2001). Remarkably, hp53 oscillations are not due to rhythmic expression of Mdm2, since its mRNA and protein levels exhibit a very modest oscillation throughout a 24-h cycle (Fu et al., 2002; Panda et al., 2002). Consequently, we hypothesized that, when available, hPer2 binds to the C-terminal half of hp53 and inhibits its Mdm2-mediated ubiquitination, thus stabilizing hp53. To test this possibility, we knocked down the endogenous expression of hPer2 using small interfering RNA (siRNA) in HCT116 cells and monitored hp53 stability in the presence of cycloheximide (CHX), an inhibitor of protein translation (Figure 3A). Effective down-regulation of hPer2 was achieved 48 h after siRNA transfection (Figure 3A, top, lane 1 vs. lane 7) and was sustained over the time course analyzed (Figure 3A, top, lanes 7–12). Expression of hp53 was then evaluated at different times after CHX addition (t = 0, Figure 3A, middle). Our findings show that endogenous hp53 levels dramatically dropped 1 h after CHX treatment (from 100 to 70 and 40% in mock- vs. siRNA-treated samples, respectively (Figure 3A, middle, lane 3 vs. lane 9) and decreased thereafter (Figure 3B, top). Thus hp53 half-life (t1/2) decreased ∼60% in the absence of hPer2 expression, supporting a role for hPer2 in hp53 stability. Accordingly, we observed a direct correlation between the down-regulation of hPer2 after CHX addition in mock samples and the progressive decrease of hp53 (Figure 3A, lanes 1–6) to the point in which hPer2 was undetectable and only trace levels of hp53 were identified (Figure 3, A, lane 6, and B). It is worth noting both the role of de novo transcription/translation in keeping hp53 levels detectable in the cell even in the absence of hPer2 as evidenced in Figure 3A (lane 7) and the relevance of hPer2 presence in sustaining hp53 levels (Figure 3A, middle). A clearer picture of the effect of hPer2 in hp53 stability arises from the quantitative analysis of the experiments shown in Figure 3A and summarized in Figure 3B. Here we represent the remaining levels of hp53 (Figure 3B, top) and hPer2 (Figure 3B, bottom) detected in both mock- and siRNA hPer2 CHX-treated cells. The drop in hp53 levels is evident within the first 2 h post CHX treatment (Figure 3B, top, shaded box) and is concomitant with a decrease in hPer2 levels in mock samples (Figure 3B, bottom, shaded box). Further support for our observations came from transfection studies in which hPer2 was overexpressed in HCT116 and endogenous hp53 levels were monitored after CHX addition (Supplemental Figure S3). As a result, we observed a significant increase (∼60%) in endogenous hp53 levels in samples transfected with hPer2 (t = 0; Supplemental Figure S3). As expected, this effect was sustained while hPer2 was overexpressed in cells but dropped dramatically once CHX affected hPer2 translation, and its own stability was compromised by 4 h (Supplemental Figure S3A, middle, and graph, shaded box).
FIGURE 3:
Binding of hPer2 to hp53 modulates its stability. (A) HCT116 cells were transfected with siRNA hPer2 (25 nM) or not (mock), and a sample equivalent to t = 0 h was collected 72 h later. Samples (t = 0.5–8 h) were harvested from cells maintained in complete medium plus cycloheximide (CHX, 100 μg/ml). Extracts were analyzed by immunoblotting using specific antibodies. A nontransfected control sample was loaded onto lane 12 and indicates the position of endogenous proteins. Immunoblot data from a single experiment that was repeated three times with similar results. (B) Protein levels (hPer2 and hp53) from mock- and siRNA hPer2-treated samples were quantified using ImageJ, version 1.45 (Schneider et al., 2012), and values were normalized to tubulin levels. Bar graphs indicate the percentage of protein remaining plotted as a function of time. The curve was fitted and hp53 half-life calculated using GraphPad Prism software (top). The gray box indicates the window of time while endogenous hPer2 was readily detected in mock samples. Data from a single experiment that was repeated three times with similar results. In all cases, mock and siRNA are represented by solid and dashed lines, respectively, and hp53 and hPer2 proteins are symbolized by ● and ◯, respectively. (C) HCT116 cells were transfected with either pCS2+FLAG-hPer2 (left bar graph) or siRNA hPer2 (right bar graph) and samples subjected to qRT-PCR as described in Materials and Methods. Data are presented as mean ± SEM from three independent experiments performed in triplicate. Statistical comparisons were done by two-tailed unpaired t test and analyses performed using SPSS. NS, not significant; #p ≤ 0.05; ##p ≤ 0.01. In all samples, endogenous hPer2 levels were monitored by immunoblotting and before performing quantitative RNA analyses. Tubulin was a loading control (right).
Although our results establish a role for hPer2 in promoting hp53 stability, its influence on total hp53 cellular levels includes an additional transcriptional component when hPer2 is overexpressed. This scenario became clear from experiments performed in HCT116 cells transfected with either FLAG-hPer2 or hPer2 siRNA and where hp53 mRNA levels were measured in real time (Figure 3C). It also became clear from analyses of lung cancer cells, in which overexpression of hPer2 led to an increase in p53 mRNA and p53-mediated apoptosis (Hua et al., 2006). As shown in Figure 3C (left), overexpression of hPer2 resulted in a significant increase in TP53 transcription (encodes p53), an effect that was overturned in cells transfected with siRNA hPer2 (Figure 3C, right). Transcription of NR1D1 (encodes orphan nuclear receptor Rev-erbα) was used as an internal control, as its level is influenced by the presence of hPer2 in the cell, with repression taking place when hPer2 is at its highest (Ko and Takahashi, 2006). In summary, our data place hPer2 as a modulator of hp53 cellular levels by acting through different mechanisms; a canonical signaling pathway via TP53 gene transcription and, independently, a noncanonical pathway that involves hPer2 binding to hp53, modulation of Mdm2-mediated hp53 ubiquitination and therefore hp53 stability.
The hPer2 factor influences the expression of hp53-target genes
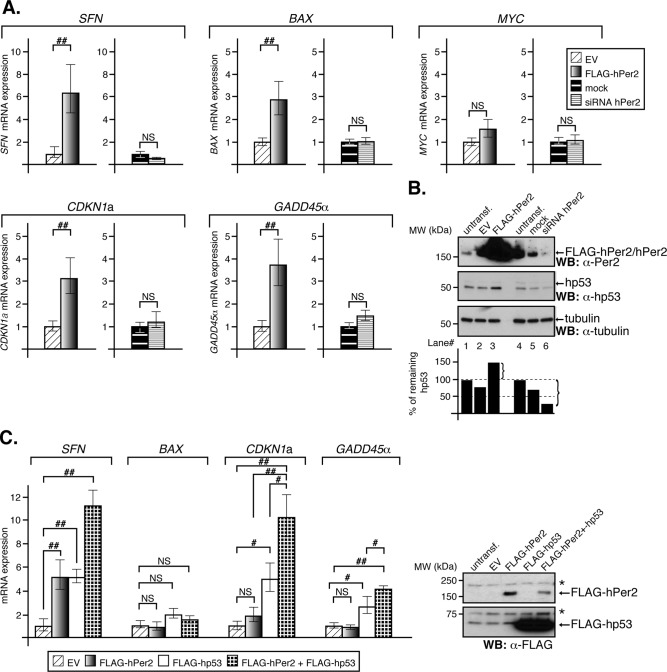
We then asked whether increased stability of hp53 as a result of hPer2 transfection (Supplemental Figure S3) affects activation of hp53-mediated gene transcription, thus functionally linking hPer2 to the hp53 pathway. First, we monitored the expression of p53-targeted genes in HCT116 cells that were either overexpressing hPer2 (Figure 4A, left, for each set of genes) or silenced for hPer2 expression (Figure 4A, right, for each set of genes). We chose to analyze the expression of those genes because they represent the diversity of cellular pathways controlled by p53 and the various forms of regulation, ranging from transcriptional repression (i.e., MYC) to activation (i.e., SFN [encodes 14-3-3σ ], BAX [encodes Bcl-2–associated X protein (Bax)], CDKN1a [encodes cyclin-dependent kinase inhibitor human p21 (hp21WAF1/CIP1)], and GADD45α [encodes the growth arrest and DNA-damage-inducible protein 45 α (Gadd45α)]). Accordingly, hPer2-transfected cells showed a significant increase in expression of CDKN1a, SFN, GADD45α, and the proapoptotic component BAX, whereas the expression of the MYC oncogene remained invariable (Figure 4A, left, for each set of genes). Specificity of response toward hPer2 was assessed by effectively abrogating its expression using siRNA in HCT116 and monitoring gene transcription in real time (Figure 4A, right, for each set of genes). As shown, down-regulation of hPer2 counteracts the effect of the transcription of p53-target genes triggered by hPer2 overexpression, supporting a role for this circadian modulator in their transcriptional control. Consistent with our previous observations, increased hp53 levels were solely observed in hPer2-transfected samples (Figure 4B), supporting a model in which hPer2 action on hp53-mediated transcription might be the result of its stabilization.
FIGURE 4:
The hPer2 protein influences the expression of hp53 target genes. (A) HCT116 cells were transfected with either FLAG-hPer2 or siRNA hPer2 and collected at 24 and 48 h posttransfection, respectively. Empty vector (EV) and mock samples were controls. qRT-PCR data are presented as the mean ± SEM from three independent experiments performed in triplicate. Statistical comparisons were done by two-tailed unpaired t test and analyses performed using SPSS. NS, not significant; ##p ≤ 0.01. (B) HCT116 cell extracts (20 μg) from the various samples in A were analyzed for hPer2 (top), endogenous hp53 (middle), and tubulin (bottom) by immunoblotting. Bands were quantified using ImageJ, version 1.45, and values normalized to tubulin levels (loading control). Bar graphs indicate the percentage of the remaining hp53 protein. Immunoblot data from a single experiment that was repeated three times with similar results. (C) H1299 cells were transfected with pCS2+FLAG-hPer2, pCS2+FLAG-hp53, empty vector (EV), or a combination of plasmids. Cells were harvested 24 h after transfection and samples prepared for qRT-PCR. Aliquots of cell extracts were analyzed by immunoblotting (right). Asterisks indicate nonspecific signal. Data are presented as the mean ± SEM from three independent experiments performed in triplicate. Statistical comparisons were evaluated by ANOVA using Bonferroni or Games–Howell post hoc analyses when needed (SPSS). NS, not significant; #p ≤ 0.05; ##p ≤ 0.01. SFN encodes 14-3-3σ; BAX encodes the proapoptotic factor Bax; MYC encodes the oncogene c-myc; CDKN1a encodes hp21WAF1/CIP1; GADD45α encodes the gadd45α protein.
Next we wanted to ascertain hp53 as the chief mediator of the hPer2 transcriptional effect shown in Figure 4A. We reasoned that if hPer2 induces CDKN1a, SFN, GADD45α, and BAX transcription via a p53-dependent pathway, transfection with hPer2 should not restore their expression in an hp53-null background unless either a p53-independent mechanism exists or hp53 is cotransfected along with hPer2 and the p53-mediated signaling pathway is restored. To test this possibility, H1299 cells (p53-null) were transfected with FLAG-tagged forms of hp53, hPer2, or both plasmids together (Figure 4C, inset) and analyzed for transcription of specific genes in real time (Figure 4C). Results show FLAG-hPer2 by itself was unable to significantly induce the expression of CDKN1a, GADD45α, or BAX in a p53-null background. Although the contribution of other mediators and additional signaling mechanisms cannot be completely ruled out, hPer2 seems to act primarily through hp53 and not vice versa, as hp53 does not seem to have a direct effect on hPer2 mRNA and protein expression in HCT116 cells (Supplemental Figure S5). Of interest, SFN expression seemed to be enhanced by just hPer2 transfection, suggesting the existence of at least an hp53-independent but hPer2-dependent mechanism involved in SFN expression. As expected, FLAG-hp53 transfection led to increased transcriptional expression of SFN, CDKN1a, and GADD45α, whereas BAX levels remained largely unchanged. Of note, cotransfection of FLAG-hp53 and -hPer2 resulted in a synergistic effect that led to overexpression of SFN, CDKN1a, and GADD45α but not BAX.
Overall these results establish that 1) hp53 is a mediator of the hPer2 transcriptional response, 2) hp53 target genes such as SFN and BAX might have at least an additional mode of regulation by hPer2 that is independent of hp53, and 3) hPer2 most likely influences the ability of hp53 to function as a sequence-specific transcription factor indirectly by influencing hp53 levels. Thus our data substantiate the biological relevance of hPer2 in modulating hp53 target gene expression, and we speculate that the hPer2-hp53 axis might act on multiple hp53 canonical and noncanonical response elements to varying extents.
DISCUSSION
The emerging view of the role of core circadian clock components is that their function is no longer restricted to the generation of circadian rhythms but that they also intervene in other cellular pathways believed to be under circadian regulation. Examples are many and growing and include signaling pathways involved in cell growth, division, death regulation, metabolism, behavioral disorders, immunity, and xenobiotic responses (for review, see Takahashi et al., 2008, and references therein). These findings highlight the complexity of the circadian-controlled network and emphasize its physiological relevance for human health and new therapeutic interventions.
Our findings provide the first evidence of direct protein–protein interaction between the clock factor hPer2 and the tumor suppressor hp53 (Figures 1 and 2). They are further validated by converse studies performed in human cells in search of new modulators of p53-mediated proliferation arrest using large-scale RNAi screening (Berns et al., 2004). Although it is unclear how Per2 is involved in p53-mediated cell cycle arrest from the high-throughput studies, those results support Per2 as a tumor suppressor relevant to the p53 pathway. Other forms of validation of the p53–clock connection result from the identification of modifiers of the clock's amplitude and period. These include molecules that physically interact with p53 (CDK9, NCL, ABL1, BRCA2; Zhang et al., 2009), down-regulation of which causes either short- or long-period changes, as well as high-amplitude phenotypes in mammalian cells (Zhang et al., 2009). Furthermore, knockdown of hp53 itself generates low-amplitude circadian rhythms in U2OS cells, further linking the tumor suppressor to the clock (Zhang et al., 2009).
Binding of the E3 ligase Mdm2 to the N-terminus domain of hp53 allows for the formation of a trimeric complex in which hPer2 binding to p53’s C-terminus prevents its ubiquitination and promotes hp53 stabilization (Figures 2, C and D, 3, and 5). The stability of p53 has been the subject of numerous studies and is known to be largely influenced by posttranslational modifications, intracellular distribution, and binding to other interacting proteins (for review, see Lavin and Gueven, 2006). As a result, different scenarios should be considered when evaluating the mechanism by which hPer2 leads to hp53 stabilization, including 1) intracellular localization of the hPer2/hp53 complex (unpublished data), 2) inhibition of Mdm2 E3 ligase activity when in contact with hPer2 in the trimeric complex (Supplemental Figure S2C, lane 6), and, alternatively, 3) blockage of Mdm2 access to its substrate region within hp53. In support of these scenarios is the existence of multiple proteins that influence the stabilization of p53, although the major control mechanism remains its interaction with and ubiquitination by Mdm2 (Harris and Levine, 2005). However, proteins that reverse this modification and others that either enhance translation of p53 mRNA or alter its subcellular localization influence the total level of p53 present in the cell at a given time. The herpes virus–associated ubiquitin-specific protease (HAUSP) is a p53-interacting protein that stabilizes p53 by deubiquitination even in the presence of an excess of Mdm2 (Li et al., 2002). More recently, the ovarian tumor domain-containing ubiquitin aldehyde-binding protein 1 (Otub1) was found to directly suppress Mdm2-mediated p53 ubiquitination and drastically stabilize p53 in response to DNA damage (Sun et al., 2012). An additional ubiquitin-independent but proteasome-dependent p53 degradation process is linked to its association to NAD(P)H quinone oxidoreductase (NQO1; Asher and Shaul, 2005). Both p53 and p73 interact with NQO1, which is largely associated with the 20S proteasome, unless an excess of NADH is present and competes off their interaction, preventing p53 degradation. Unlike in unstressed cells, in which Mdm2 prevents p300 from associating with p53, both CBP/p300 transcriptional coactivators and Strap, a partner protein of p300, associate with p53 in response to DNA damage, increasing its level and half-life through a mechanism that involves p53 acetylation (Lambert et al., 1998; Barlev et al., 2001). Methylation of p53 by overexpression of various methyltransferases has also been linked to its hyperstabilization (Chuikov et al., 2004). Finally, protein–protein interaction has been proven effective in increasing the stability of various forms of mutant p53. For example, heat shock protein 90 (hsp90) binds to the C-terminus of mutant p53 when in complex with Mdm2; however, in this case, hsp90 acts by inhibiting the ubiquitin ligase activity of Mdm2, blocking the ubiquitination of both Mdm2 itself and mutant p53 (Peng et al., 2001). An additional mode of regulation results from association of the central and C-terminus regions of p53 to poly(ADP-ribose) polymerase-1 (PARP-1; Wesierska-Gadek et al., 2003). In this case, inactivation of PARP-1 resulted in decreased levels of p53, whereas inhibition of nuclear export by leptomycin B prevented accelerated degradation of p53 in PARP-1–knockout cells, favoring p53 accumulation. As a result, p53 stability results from its nuclear accumulation, as PARP-1 blocks p53’s nuclear export signal located in its carboxy-terminal area (Wesierska-Gadek et al., 2003). Overall the mechanisms for controlling p53 stabilization are many and varied, but they all serve the purpose of monitoring different aspects of cellular homeostasis.
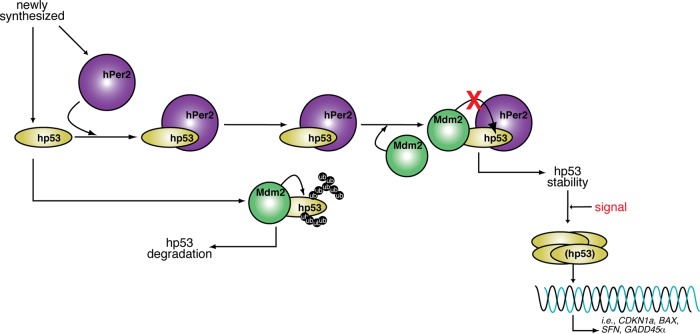
FIGURE 5:
Proposed model of hPer2 and hp53 interaction and function. The hPer2 protein associates with cytosolic hp53, forming a stable complex that keeps hp53 in a stable state and ensuring that basal levels of hp53 exist (“priming state”). This heterodimer eventually incorporates Mdm2, forming a trimeric and stable Mdm2/hp53/hPer2 complex. In this scenario, Mdm2 is not able to ubiquitinate hp53 unless binding between these two proteins occurs in the absence of hPer2. We hypothesize that existence of the trimeric complex is compromised under, for example, stress signals, leading to release and activation of the hp53 downstream pathway.
What makes hPer2’s interaction with hp53 particularly attractive is the possibility of building a new level of regulation by understanding how cell homeostasis harmonizes with its environment. Accordingly, we asked about the relevance of the hp53/hPer2 regulatory mechanism in a physiological context. Here we embrace the concept of “biological anticipation” first introduced to explain the function of the cardiomyocyte circadian clock in both myocardial health and disease (for review, see Durgan and Young, 2010). Basically, the concept proposes that a biological system is able to prepare for an event before it happens, thus ensuring rapid response in a temporally appropriate manner. Consequently, we hypothesize that the presence of hPer2 allows for the formation of the hp53/hPer2 complex and hp53 stabilization. Thus the existence of basal levels of the hp53 tumor suppressor helps “prime” the signaling pathway to rapidly respond to a stress condition (metabolic, genotoxic). In this scenario, induction of TP53 transcription via hPer2 (Figure 3C) or by any additional factor is a secondary step and would most likely help to sustain the response. Of interest, Miki et al. (2013) showed that transcriptional regulation may be reciprocal and that p53 targets the Clock/Bmal1 complex, inhibiting the transcriptional expression of Per2 in the suprachiasmatic nuclei, all of which result in p53−/− animals that exhibit altered circadian behavior, further supporting various levels of regulation in the Per2-p53 axis.
The role of hPer2 as mediator of the hp53 transcriptional response was established in a p53-knockout background (H1299, p53-null cells) in which the hPer2-hp53 axis was reconstituted in a stepwise manner (Figure 4). First, we showed that manipulation of hPer2 levels influences both the transcription of a broad range of hp53 target genes and the stability of endogenous hp53 (Figure 4, A and B). In fact, knocking down hPer2 expression counteracts any effect in transcription of hp53 target genes caused by hPer2 overexpression (Figure 4A). Whereas this result establishes a correlation between the presence of hPer2 and the expression of genes whose transcription is mediated by hp53, a direct connection is further established when coexpression of hPer2 and hp53 synergistically enhances the transcription of SFN, CDKN1a, and GADD45α in an hp53-null background (Figures 4C and 5). Overall our results pose more relevant questions that refer to how both selectivity in terms of gene expression and diversity in terms of regulation are accomplished in the hPer2/hp53 axis and whether posttranslational modifications play a role in fine-tuning the strength of their interaction for a wider range of control.
MATERIALS AND METHODS
Bacterial two-hybrid screening
To assay for two-hybrid interactions between a specific bait (pBT-hPer2) and target plasmid pair from a library (pTRG cDNA library), we performed a two-hybrid screening analysis using the BacterioMatch II system (Stratagene, La Jolla, CA) following the manufacturer's instructions. Briefly, BacterioMatch II validation reporter–competent cells were cotransformed with ∼200 ng each of the bait vector plus pTRG target vector. Aliquots of each of the cotransformation mixtures were plated in both nonselective screening medium (no 3-AT; Sigma-Aldrich, St. Louis, MO) and selective screening medium (5 mM 3-AT). Strong interactors were isolated from selective media within the first 24 h of incubation, and positive clones were maintained in lysogeny broth (LB) tetracycline/chloramphenicol (Tet/Cam) agar plates. Activation of a second reporter gene, aadA, encoding streptomycin resistance, was used to verify the specificity of the interaction between bait and target proteins. In this case, putative positive colonies were patched from selective screening medium onto a dual-selective screening medium plate containing both 3-AT and streptomycin. The pBT-LFG2/pTRG-Gal11P cotransformant was used as a positive control in the experiment and taken from a selective screening medium plate. Negative controls were the recombinant pBT-hPer2 cotransformed with either empty pTRG or pTRG-Gal11P vectors taken from a nonselective screening medium plate. Purification of plasmid DNA from the 3-AT–resistant colonies isolated during library screening was performed from cultures grown in LB supplemented with tetracycline. All cDNAs were sequenced to confirm the identity of the clones.
RNA extraction and quantitative reverse-transcription PCR
Total RNA was extracted from cell pellets using the TRIzol reagent (Life Technologies, Grand Island, NY) following the manufacturer's instructions. RNA was quantified by spectrophotometric reading at 260 nm and analyzed for quality assurance using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) at the Virginia Bioinformatic Institute Proteomics Core Facility. Quantitative reverse-transcription PCR (qRT-PCR) was conducted essentially as previously described (Yang et al., 2008). Briefly, total RNA was pretreated with DNaseI (Promega, Madison, WI) at 37°C for 30 min, and a 1-μg sample was used as a template for first-strand cDNA synthesis using the iScript cDNA Synthesis system (Bio-Rad, Hercules, CA). qRT-PCR assay was performed using IQ SYBR Green Supermix (Bio-Rad) as follows: 10 ng of cDNA (50 ng for the 14-3-3σ gene) was added to a 20-μl reaction volume containing the indicated primers for amplification (see Supplemental Materials and Methods and Supplemental Table S1). Real-time assays were performed in triplicate on a MyIQ single color Real-Time PCR Detection instrument (Bio-Rad). Data were collected and analyzed with Optical System software, version 1.0. The glyceraldehyde-3-phosphate dehydrogenase and β-actin genes were used as internal controls to compute the relative expression level (ΔCt) for each sample. The fold change of gene expression in each sample was calculated as 2−ΔΔCt.
Immunoprecipitation and immunoblot assays
For (co)immunoprecipitation experiments, transfected cells were harvested in lysis buffer, and extracts (∼100 μg) were incubated with either α-FLAG M2 agarose beads (Sigma-Aldrich) or α-myc (9E10) beads (Santa Cruz Biotechnology, Dallas, TX) either for 2 h or overnight at 4°C with rotation before washing. Where indicated, immunoprecipitations were carried out in a two-step procedure, with extracts being incubated with the antibody (α-myc, α-FLAG, or α-p53) overnight at 4°C before the addition of protein A beads (50% slurry; Sigma-Aldrich). Sample beads were then washed four times with lysis buffer, resolved by SDS–PAGE, and analyzed by immunoblotting using specific primary antibodies (α-FLAG [Sigma-Aldrich], α-myc [Santa Cruz Biotechnology], α-Per2 [Sigma-Aldrich, Farmingdale, NY], α-p53 [Santa Cruz Biotechnology], α-ubiquitin [Enzo Biomol]). For (co)immunoprecipitation experiments of endogenous proteins, cells were harvested in lysis buffer, and extracts (∼1 mg) were incubated with α-Per2 (Santa Cruz Biotechnology) or α-p53 (Santa Cruz Biotechnology) overnight at 4°C before the addition of protein A beads (50% slurry; Sigma-Aldrich). Samples were then kept for an additional 2 h at 4°C with rotation before washing and then processed as described, with the exception that in some of the experiments, blots were incubated with either α-Mdm2 (Santa Cruz Biotechnology and Calbiochem, Billerica, MA) primary antibody. In all cases, horseradish peroxidase–conjugated α-rabbit or α-mouse IgG secondary antibodies (GE Healthcare Life Sciences, Buckinghamshire, UK; Cell Signaling, Danvers, MA) were used for immunoblotting following standard procedures. Chemiluminescence reactions were performed using the SuperSignal West Pico substrate (Pierce, Rockford, IL).
In vitro binding assays
In vitro transcription and translation of pCS2+ myc-hPer2, myc-Mdm2, FLAG-hPer2, and FLAG-hp53 were carried out using the SP6 high-yield TNT system (Promega) following the manufacturer's instructions, although, unlike the standard procedure, the reaction was cold. Aliquots (1-4 μl) of indicated recombinant proteins were preincubated for 20 min at room temperature to allow the complex to form before adding NP40 lysis buffer. Immunoprecipitation of the various complexes was carried out essentially as described.
Protein pull-down assays
GST fusion proteins were expressed in Escherichia coli strain Rosetta (Novagen) and purified by glutathione-Sepharose chromatography based on the manufacturer's instructions (GE Healthcare Life Sciences). For pull-down assays, a total of 5 μg of GST-hp53-bound beads, its recombinant fragments, or an equivalent amount of glutathione beads (GST control) were washed in binding buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, and 0.1% Triton X-100) and incubated with 2 μl of in vitro–transcribed and translated [35S]myc-hPer2 at 4°C for 1 h. After washing of the beads with low- and high-salt binding buffer (100 mM and 1 M NaCl, respectively), bound proteins were eluted by boiling in Laemmli sample buffer and analyzed by SDS–PAGE and autoradiography. A similar procedure was followed to evaluate binding of [35S]myc-hp53 to various tagged recombinant fragments of hPer2. For competition assays, untagged hp53 was generated by digestion of the fusion protein with thrombin, followed by concentration and buffer exchange (10 mM Tris-HCl, pH 8.0). Reactions were set using 5 μg of GST-hp53 and increasing amounts of untagged p53 (0–20 μg) in the presence of a constant radiolabeled hPer2. Reactions were incubated at 4°C for 1 h, beads were washed, and samples analyzed by SDS–PAGE as described. Densitometric quantitation was carried out using a FluoChem digital imaging system (Alpha Innotech, Santa Clara, CA).
Ubiquitination assays
For in vitro assays, pCS2+ constructs of myc-hPer2, myc-Mdm2, and FLAG-hp53 were transcribed and translated in vitro as described. Aliquots of each tagged protein (1–4 μl), or a combination of them, were incubated at room temperature for 30 min to allow complex formation to happen before adding the reaction buffer containing 1× ubiquitination buffer (Enzo Biomol), 2 mM dithiothreitol, 20 μg/ml ubiquitin-aldehyde, 100 μg/ml ubiquitin, 1xATP-energy regeneration system (5 mM ATP/Mg2+; Enzo Biomol), 40 μM MG132, and 1 mg/ml HeLa S100 lysate fraction (Enzo Biomol) to a final volume of 10 μl. Reactions were further incubated at 37°C for 30 min before being terminated by the addition of lysis buffer. Diluted samples were incubated twice at 4°C for 1 h, first after addition of 3 μg of α-FLAG antibody (Sigma-Aldrich) and then after the addition of 8 μl of protein A–Sepharose 4B (50% slurry; Sigma-Aldrich). Bound complex were washed four times with NP40 lysis buffer, resuspended in Laemmli buffer, and resolved by SDS–PAGE and immunoblotting.
For detection of in-cell ubiquitination, HCT116 cells were transfected with pCS2+myc-hp53 and either pCS2+FLAG-hPer2 or empty vector and maintained in complete medium for 20 h before the addition or not (–MG132) of 50 μM MG132 and ubiquitin aldehyde (5 nM). Cells were harvested 4 h later and subjected to subcellular fractionation as previously described (Schreiber et al., 1989). Lysates were immunoprecipitated with α-FLAG or α-myc antibodies as described, and the complexes were resolved in SDS–PAGE. Ubiquitinated complexes were detected by immunoblotting using an α-ubiquitin antibody.
Analysis of hp53 half-life
Extracts were from HCT116 cells treated with 100 μg/ml cycloheximide. Protein levels were quantitated by immunoblot analysis using ImageJ, version 1.45 (National Institutes of Health software package; Schneider et al., 2012), and values normalized to tubulin levels. The percentage of protein remaining was calculated based on t = 0, and the data were fitted using Prism software (GraphPad Software, La Jolla, CA).
Immunofluorescence microscopy
HCT116 was used for detection of endogenous hp53 and hPer2 proteins. Cells were fixed (3.7% formaldehyde/phosphate-buffered saline [PBS]/0.5% Triton X-100) 2 h after exposure at room temperature, washed with PBS/0.5% Triton X-100 and then 0.1% Triton X-100, and blocked with goat serum at room temperature for 30 min. Subcellular localization was detected using commercially available antibodies. Nuclei were visualized by incubating fixed cells with SYTO 60 (Life Technologies). Fluorescence was visualized using a Nikon Eclipse TE2000-E microscope (Nikon, Melville, NY) equipped with a Cascade II E2V CCD97 camera (Photometrics, Tucson, AZ) at 488, 568, and 647 nm. Images were processed using the NIS-Elements AR 3.0 Nikon software.
Gene reporter activity
H1299 cells were seeded onto 12-well plates and cotransfected with ∼200 ng of hp21WAF1/CIP1-luciferase (Luc), pCS2+FLAG-hp53, -hp53(ch)GST, -hp53(ch)hPer2, or empty vector (pCS2+FLAG) each. The pCMV-β-gal (∼200 ng) plasmid was included as an internal control and harvested 2 h later. Reporter activity was measured using the Bright-Glo Luciferase Assay System (Promega) according to the manufacturer's instructions. Readings were recorded from a Glomax Luminometer, and results were normalized for expression of β-gal, which was determined separately using the Galacto-Light Plus System (Bio-Rad). Experiments were done in triplicate and repeated at least twice.
Statistical analyses
To assess the overall significance of our results, data were processed using either a two-tailed unpaired Student's t test or analysis of variance (ANOVA), followed by either Bonferroni or, when necessary, Games–Howell post hoc test (SPSS statistical software; IBM, Armonk, NY). Levene's test was used to determine homogeneity of variances, whereas data normality was examined using the Shapiro–Wilk W test and the Box–Cox Y transformation (JMP 9 statistical software; SAS, Cary, NC) and applied when necessary. Values of p ≤ 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank John Tyson, Jill Sible, Daniel Capelluto, and James Maller for critical reading of the manuscript and all members of the Finkielstein laboratory for help and discussions. We also thank J. Webster for comments and manuscript editing. We are grateful to Steven L. McKnight (University of Texas Southwestern Medical Center, Dallas, TX), Daiqing Liao (University of Florida, Gainesville, FL), and Bert Volgestein (Johns Hopkins University, Baltimore, MD) for providing us with the hPer2 cDNA, pWAF1-Luc constructs, and HCT116 p53−/− cells, respectively. This work was supported by a National Science Foundation CAREER Award (MCB-0844491), the Avon Foundation (02-2009-033), the Fralin Life Science Institute (F441598), and the Susan G. Komen Foundation (BCTR0706931) to C.V.F.
Abbreviations used:
- BAX
encodes Bcl-2–associated X protein (Bax)
- CDKN1a
encodes cyclin-dependent kinase inhibitor human p21 (hp21WAF1/CIP1)
- GADD45α
encodes the growth arrest and DNA-damage-inducible protein 45 α (Gadd45α)
- H1299
human non–small cell lung carcinoma cells
- HCT116
human colon carcinoma cells
- HEK293
human embryonic kidney 293 cells
- hp53
human p53 transcription factor
- hPer2
human Period 2
- Mdm2
murine (human) double minute-2
- mPer2
mouse Period 2
- NR1D1
encodes orphan nuclear receptor Rev-erbα
- SFN
encodes 14-3-3σ
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-05-0993) on August 7, 2014.
*These authors contributed equally.
Present addresses: †Department of Biology, University of Padova, Padova 35121, Italy.
‡Eosinophil Pathology Unit, National Institute of Allergy and Infectious Diseases, Bethesda, MD 20814.
§Dupont Pioneer, Johnston, IA 50131.
T.G. performed all experiments except for those mentioned in what follows. M.V.-C. provided data in Figures 3C, S2A, and 4, A and C, and performed all statistical analyses. J.J.L. provided data in Figure 4B and Supplemental Figure S2B and blots in Figure 4C and contributed to many stages of this research. C.S.S. performed the research included in Figure 1, A, E, and F, and Supplemental Figure S1. J.Y. contributed data shown in Figure 1, B and C. C.S.S. and J.Y. generated reagents used throughout this research. C.V.F. and C.S.S. conceived this project. C.V.F., T.G., and M.V.-C. analyzed the overall data, refined the hypothesis, and proposed the model. C.V.F. supervised and coordinated all investigators for the project and wrote the manuscript.
REFERENCES
- Albrecht U, Bordon A, Schmutz I, Ripperger J. The multiple facets of Per2. Cold Spring Harb Symp Quant Biol. 2007;72:95–104. doi: 10.1101/sqb.2007.72.001. [DOI] [PubMed] [Google Scholar]
- Asher G, Shaul Y. p53 proteasomal degradation: poly-ubiquitination is not the whole story. Cell Cycle. 2005;4:1015–1018. doi: 10.4161/cc.4.8.1900. [DOI] [PubMed] [Google Scholar]
- Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Bjarnason GA, Jordan RC, Sothern RB. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am J Pathol. 1999;154:613–622. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnason GA, Jordan RC, Wood PA, Li Q, Lincoln DW, Sothern RB, Hrushesky WJ, Ben-David Y. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001;158:1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajochen C, Jud C, Munch M, Kobialka S, Wirz-Justice A, Albrecht U. Evening exposure to blue light stimulates the expression of the clock gene PER2 in humans. Eur J Neurosci. 2006;23:1082–1086. doi: 10.1111/j.1460-9568.2006.04613.x. [DOI] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol. 2003;15:991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Durgan DJ, Young ME. The cardiomyocyte circadian clock: emerging roles in health and disease. Circ Res. 2010;106:647–658. doi: 10.1161/CIRCRESAHA.109.209957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Fukuhara C, Brewer JM, Dirden JC, Bittman EL, Tosini G, Harrington ME. Neuropeptide Y rapidly reduces Period 1 and Period 2 mRNA levels in the hamster suprachiasmatic nucleus. Neurosci Lett. 2001;314:119–122. doi: 10.1016/s0304-3940(01)02304-7. [DOI] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Giannakakou P, Sackett DL, Ward Y, Webster KR, Blagosklonny MV, Fojo T. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat Cell Biol. 2000;2:709–717. doi: 10.1038/35036335. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Horikawa K, Yokota S, Fuji K, Akiyama M, Moriya T, Okamura H, Shibata S. Nonphotic entrainment by 5-HT1A/7 receptor agonists accompanied by reduced Per1 and Per2 mRNA levels in the suprachiasmatic nuclei. J Neurosci. 2000;20:5867–5873. doi: 10.1523/JNEUROSCI.20-15-05867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, Wang X, Wang Z, Cornelissen-Guillaume G, Halberg F. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97:589–596. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Rohaly G, Heinrichs S, Gimnopoulos D, Meissner H, Deppert W. Influence of promoter DNA topology on sequence-specific DNA binding and transactivation by tumor suppressor p53. Oncogene. 1999;18:7310–7318. doi: 10.1038/sj.onc.1203139. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. (Spec No 2) [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Miki T, Matsumoto T, Zhao Z, Lee CC. p53 regulates Period2 expression and the circadian clock. Nat Commun. 2013;4:2444. doi: 10.1038/ncomms3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Mesaki M, Ishida N. Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein. Mol Cell Biol. 2001;21:6651–6659. doi: 10.1128/MCB.21.19.6651-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki K, Oishi K, Kozono Y, Nakayama K, Nakayama KI, Ishida N. The role of β-TrCP1 and β-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J Biochem. 2008;144:609–618. doi: 10.1093/jb/mvn112. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Peng Y, Chen L, Li C, Lu W, Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem. 2001;276:40583–40590. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall LA, Perrin JS, Walker CD, Stewart J, Amir S. Glucocorticoid rhythms control the rhythm of expression of the clock protein, Period2, in oval nucleus of the bed nucleus of the stria terminalis and central nucleus of the amygdala in rats. Neuroscience. 2006;140:753–757. doi: 10.1016/j.neuroscience.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol. 2010;20:316–321. doi: 10.1016/j.cub.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhaber EL, Duricka D, Ullman KS, Virshup DM. Nuclear export of mammalian PERIOD proteins. J Biol Chem. 2001;276:45921–45927. doi: 10.1074/jbc.M107726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesierska-Gadek J, Wojciechowski J, Schmid G. Central and carboxy-terminal regions of human p53 protein are essential for interaction and complex formation with PARP-1. J Cell Biochem. 2003;89:220–232. doi: 10.1002/jcb.10521. [DOI] [PubMed] [Google Scholar]
- Yang J, Kim KD, Lucas A, Drahos KE, Santos CS, Mury SP, Capelluto DG, Finkielstein CV. A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol Cell Biol. 2008;28:4697–4711. doi: 10.1128/MCB.00236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wood PA, Ansell CM, Ohmori M, Oh EY, Xiong Y, Berger FG, Pena MM, Hrushesky WJ. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009a;145:289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJ. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. 2009b;117:423–431. doi: 10.1007/s10549-008-0133-z. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.