On the basis of previously published biophysical and biochemical parameters of mRNA export, a three-dimensional, coarse-grained, agent-based model is developed for the study and characterization of mRNA nucleocytoplasmic export.
Abstract
mRNA export from the nucleus is an essential step in the expression of every protein- coding gene in eukaryotes, but many aspects of this process remain poorly understood. The density of export receptors that must bind an mRNA to ensure export, as well as how receptor distribution affects transport dynamics, is not known. It is also unclear whether the rate-limiting step for transport occurs at the nuclear basket, in the central channel, or on the cytoplasmic face of the nuclear pore complex. Using previously published biophysical and biochemical parameters of mRNA export, we implemented a three-dimensional, coarse-grained, agent-based model of mRNA export in the nanosecond regime to gain insight into these issues. On running the model, we observed that mRNA export is sensitive to the number and distribution of transport receptors coating the mRNA and that there is a rate-limiting step in the nuclear basket that is potentially associated with the mRNA reconfiguring itself to thread into the central channel. Of note, our results also suggest that using a single location-monitoring mRNA label may be insufficient to correctly capture the time regime of mRNA threading through the pore and subsequent transport. This has implications for future experimental design to study mRNA transport dynamics.
INTRODUCTION
In eukaryotes, a large quantity of RNAs, such as tRNA, microRNA (miRNA), small nuclear RNA (snRNA), rRNA, and mRNA, has to be transported across the nuclear envelope. tRNA, miRNA, snRNA, and rRNA all follow a karyopherin-mediated nuclear export pathway that is similar to that of nuclear import, relying on the Ran GTPase cycle for directionality. For example, tRNAs recruit Exportin-t (Los1) of the karyopherin superfamily in complex with RanGTP to facilitate export, whereas miRNAs recruit Exportin-5 (Msn5) in complex with RanGTP. snRNAs, as well as many viral RNAs such as HIV-1 RNA, use Exportin-1 (Xpo1; also known as CRM1) in complex with RanGTP to achieve export (Rodriguez et al., 2004; Kohler and Hurt, 2007). These exportins rely on the energy liberated by the RanGAP/RanBP2-mediated hydrolysis of RanGTP to provide RNAs with export directionality.
In contrast, the bulk of mRNA is exported from the nucleus via the NXF1/NXT1-mediated pathway, which does not directly depend on the RanGTPase system. During and after transcription, a nascent pre-mRNA undergoes multiple cotranscriptional changes before it can be successfully recruited to the nuclear pore complex (NPC) and exported. In addition to splicing, as well as 5′- and 3′-end processing, a maturing mRNA binds a number of factors to become a messenger ribonucleoprotein particle (mRNP). In this regard, the transcription elongation-mRNA export (TREX) complex appears to play a critical role in the recruitment of the NXF1/NXT1 (Mex67/Mtr2) heterodimeric export factor via a set of core proteins termed the transcription elongation (THO) complex and the associated protein Aly/REF (Yra1). Once the mRNA is processed and the NXF1/NXT1 export receptors are recruited, components of THO/TREX are removed from the mRNP (Natalizio and Wente, 2013), and the mRNP is considered to be export competent. The mRNP is then recruited to the NPC, where it translocates to the cytoplasm via a series of binding and unbinding events between NXF1/NXT1 and phenylalanine-glycine (FG)-rich repeats within specific nuclear pore proteins (Nups). Unlike RNA export pathways in which exportins of the karyopherin superfamily are recruited, bulk mRNA export via the NXF1/NXT1-mediated pathway appears to rely on the superfamily 2 DEAD-box ATPase DDX19 (Dbp5) for directionality. DDX19 was previously proposed to remodel the mRNP as it translocates through the NPC and reaches the cytoplasm-facing Nups. DDX19 is localized to the cytoplasmic Nup214 (Nup159) and is activated by the export factor Gle1 along with its cofactor, inositol hexakisphosphate (IP6). Remodeling of the mRNP by DDX19 frees transport receptors to return to the nucleus for another round of transport and prevents the return of the mRNP into the nucleus (Hodge et al., 2011; Montpetit et al., 2011; Noble et al., 2011). With multiple mRNP/export factors bound on an mRNA, stepwise remodeling by DDX19 via a virtual “Brownian ratchet” mechanism has been proposed as a way to ensure one-way transport of the mRNP through the NPC (Stewart, 2007).
Although the mRNA-ratcheting model appears feasible, the number and distribution of transport receptors required to bind an mRNP for efficient transport remain unknown. Binding of the TREX complex has been observed at the 5′-terminal end of mRNA (Cheng et al., 2006). Coincidentally, it has been reported that transport of large Balbiani ring mRNAs occurs with the 5′ end leading through the pore, as seen in electron microscopy (EM) experiments (Mehlin et al., 1992, 1995; Visa et al., 1996). As a result, having transport receptors bound at the 5′ end may be necessary for the observed mRNA export orientation. Other groups observed the binding of transport receptor recruiting factors such as REF along the length of the mRNA at the site of exon junction complexes (Kataoka et al., 2000; Le Hir et al., 2000, 2001), suggesting that transport receptors are distributed at multiple sites along an mRNA and lending support to the Brownian ratchet model. However, if transport receptors are distributed along the mRNA, it is unclear how a preference for the 5′ end could be accomplished. In addition, it is not known how the density and distribution of transport receptors affect mRNP export competence.
In recent years, new molecular techniques have been developed that have allowed the tracking of single mRNA molecules, providing insight into mRNA export kinetics. Grunwald and Singer (2010) used the MS2-GFP system (Querido and Chartrand, 2008) to track the movement of β-actin mRNA (∼3.3 kb) through the NPC, with a temporal resolution of 20 ms and a spatial precision of 26 nm. They observed that it is not transport through the central pore (5–20 ms in duration), but docking to and release from the NPC, that appears to be the rate-limiting step in export (∼160 ms in duration). The Kubitscheck group (Siebrasse et al., 2012) used light sheet fluorescence microscopy to track single export events of mRNPs with temporal and spatial resolutions of 20 ms and 10 nm, respectively. Briefly, they used a fluorescent hrp36 protein that natively binds mRNA before export, and, by adding it at sufficiently low concentrations, ensured stoichiometric tagging of mRNAs within the imaging plane. An average transport time of 65 ms was observed for the mixture of mRNAs present within the nucleus, but with transport times of up to several seconds for what are likely very large mRNAs. They further observed an arrest of the mRNAs at the nuclear face of the pore that typically lasted ∼50 ms.
The discrepancy between the transport times of these two methods (180 vs. 65 ms) could be due to the fact that in the MS2-GFP system, a specific mRNA of known length was tracked (β-actin mRNA, ∼3.3 kb), whereas the hrp36 protein can bind mRNAs of varying lengths (with an average length reported as ∼2.2 kb). On the other hand, longer mRNAs have a greater number of hrp36-binding sites, suggesting that larger mRNAs are more likely to be tagged. However, the typical length of mRNAs tracked in the fluorescent hrp36 system is also expression dependent. Nevertheless, the fluorescent hrp36 system allows for the tracking of native mRNAs without introducing multimers of RNA stem loops that bind MS2 coat proteins, which might alter the size and dynamics of the mRNA molecule (Querido and Chartrand, 2008).
Despite the discrepancy in transport kinetics between these works, both single-molecule tracking studies did observe a rate-limiting step at the nuclear basket during transport. Based on previous sequential electron microscopy work (Mehlin et al., 1995), this was suggested to reflect a 5′-end leading orientation requirement for export—the large polymer required a considerable amount of time to achieve an appropriate orientation. Another potential source of mRNA arrest in the nuclear basket might be the mRNA quality control mechanism present there (Tutucci and Stutz, 2011). This would be of more significance for smaller mRNAs, since they do not need much time to reach a favorable export orientation.
On the other hand, evidence for mRNA threading not being the rate-limiting step has also been proposed. The Yang group (Ma et al., 2013) used single-point edge-excitation subdiffraction microscopy to measure the kinetics and transport pathways of mCherry-tagged firefly luciferase mRNA and yellow fluorescent protein–tagged β-actin mRNA with 8-nm and 2- ms spatiotemporal accuracy They observed much faster dwell times of ∼12 ms. During analysis, they calculated spatial density (NPC volume-normalized) of mRNP signals in cumulative transport paths and found that it is highest within the central channel. From spatial densities, they also arrived at normalized diffusion coefficients for single cargo and found it to be approximately two to three times lower in the central channel than in either the nucleoplasm or cytoplasm. Successful and aborted cargo transport events appeared to have the largest differences in transport path in the cytoplasmic region of the NPC. These findings were used to support the claim that the rate-limiting step occurs in the central channel rather than in the nuclear basket during threading.
In this study, we sought to use a computational, agent-based modeling (ABM) approach to further investigate the dynamics of mRNA export. We built upon our ABM framework (Azimi et al., 2011) to understand how binding, unbinding, and diffusive events enable the efficient transport of long polymeric molecules across the nuclear envelope. The ABM allowed us to address the following questions: 1) How do the number and distribution of transport receptors across an mRNA molecule affect export efficiency? 2) Is tracking a single site of a polymeric chain transporting through a narrow pore sufficient to accurately quantify transport times? We observed the following in our model: 1) mRNA export is very sensitive to the number of transport receptors coating the mRNA; 2) a single transport receptor at the 5′ end is insufficient for facilitating export; 3) increasing transport receptor coverage along the length of the mRNA improves the chances of successful export; 4) the presence of a transport receptor near either the 5′ or 3′ terminus is required for successful export; and 5) there is a rate-limiting step in the nuclear basket that is potentially associated with the mRNA finding an optimal configuration for threading through the central channel. Of interest, our results also suggest that using a single location-monitoring probe to label mRNA might be insufficient both to capture correctly the time regime of mRNA threading through a pore and its subsequent transport and fully distinguish between partial and failed transport.
RESULTS
Model validation
We developed a three-dimensional (3D), computationally efficient, and spatiotemporally detailed agent-based model (ABM) to simulate molecular diffusion, binding, and unbinding events with consideration for physical factors such as molecular crowding and steric repulsion (Azimi et al., 2011; Azimi and Mofrad, 2013). We performed in silico experiments to determine the ability of our ABM model to recapitulate experimentally determined, size-dependent permeabilities for passive cargoes as well as for Impβ (Riddick and Macara, 2005; Mohr et al., 2009). After the microinjection of noninteracting species agents in the cytoplasm, the NPC was observed to inhibit the influx of larger species while allowing smaller species to diffuse through the pore (Figure 1). Influx rates of noninteracting species with Stokes radii of ∼1 nm are on the order of 0.1 s−1, whereas larger species with Stokes radii of >2.5 nm have influx rates of <0.001 s−1. As expected, a reduced influx rate was not observed for larger species that had affinity for the FG Nups. To test this behavior, we repeated experiments similar to those performed for noninteracting species, replacing the noninteracting species with 2.5 μM labeled Impβ in addition to the steady-state concentration of unlabeled Impβ (unpublished data). The influx rate of Impβ into the nucleus was observed to be 0.367 s−1. This value is comparable with an experimentally measured influx rate of 0.4 s−1 for Impβ (Riddick and Macara, 2005).
FIGURE 1:
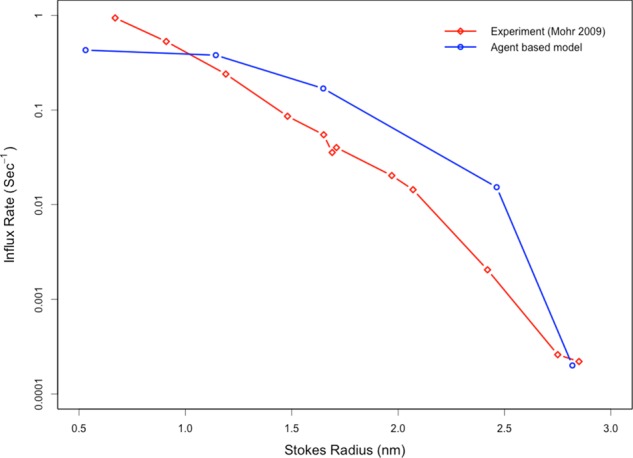
The ABM recapitulates the experimentally observed, size-dependent permeabilities of passive cargo3s through the nuclear pore. After a simulated microinjection of noninteracting species in the cytoplasm, the in silico pore is observed to inhibit the influx of larger species while allowing smaller species to diffuse through the pore. This is in agreement with previous experimental observations.
Effect of Nup-NXF1 affinity on mRNA export rate and efficacy
Recent studies reported an in vitro avidity for the NXF1 yeast homologue Mex67 to the nucleoporin Nsp1 to be on the order of 100 nM (Hobeika et al., 2009). However, it has been demonstrated experimentally that the typically reported in vitro affinities between Nups and transport receptors are much higher (∼1000-fold) than actual in vivo affinities (Tetenbaum-Novatt et al., 2012). We were interested in testing our model's sensitivity to changes in NTR/FG Nup affinity.
To obtain the default NTR number and affinity configuration settings that are in line with mRNA export efficiencies observed in vitro and in vivo, we tested configurations of seven, nine, and 13 NTRs along the mRNA for a select set of affinities ranging from 2 to 200 μM (Supplemental Figures S1 and S2). We arrived at these choices of NTR configurations by considering that a typical mRNA of the length studied would be expected to have nine NTRs and selecting close alternative configurations that entail either less dense or more dense NTR placement. Our export efficiency and transport time results validated the default choice of nine NTRs and an NTR/FG Nup affinity of 100 μM; this configuration yielded a transport efficiency and time that best recapitulated those measured in vitro (just >20% successful transport, and export time within tens of nanoseconds).
Keeping the nine-NTR configuration constant, we then varied the affinity between NXF1 and FG Nups more meticulously (Figure 2), observing nonmonotonic behavior in the export efficiency. When mRNA-bound NXF1 had a dissociation constant of 200 μM, no transport was observed. Increasing the affinity incrementally toward 20 μM led to an increase in the percentage of simulations in which successful transport was observed. When affinity was increased further to 2 μM, however, this percentage plummeted (Figure 2, blue). For successful transport events, average residence times in the basket and central channel increased more or less monotonically with increasing affinities (Figure 3).
FIGURE 2:
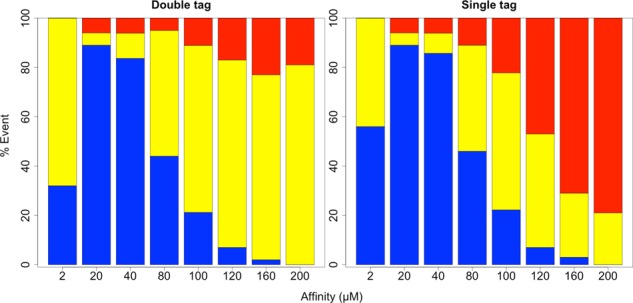
Bar graphs showing the cumulative percentage of successful (blue), partial (yellow), and unsuccessful (red) transport events observed for an mRNA with nine NTRs across different binding affinities between NXF1 and FG Nups. The bar graph on the left corresponds to observations captured by monitoring both the 5′ and 3′ ends, and that on the right corresponds to observations captured by monitoring a single, randomly placed probe.
FIGURE 3:
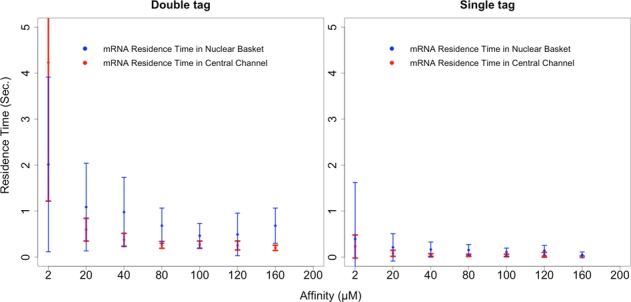
Two graphs that capture the effect of NXF1/FG Nup affinity on the mRNA's nuclear basket and central channel residence times during successful export events. The graph on the left corresponds to what is observed through probes placed at 5′ and 3′ ends of the mRNA, and the graph on the right corresponds to what is observed through a single probe located randomly along the length of the mRNA. Error bars represent 1 SD.
With the affinity between NXF1 and FG Nups in the ABM set to 100 μM and under single-tag observation conditions (consistent with experiments), we measured the mean nuclear basket and central channel residence times as 107.3 ± 87.8 and 34.5 ± 27.0 ms, respectively. These are comparable to residence times measured in vivo (Siebrasse et al., 2012). As mentioned previously, the fraction of mRNA observed successfully trafficking out of the nucleus at 9 NTRs per mRNA and 100 μM NTR/FG Nup affinity is also in agreement with experimental observations (Siebrasse et al., 2012).
Comparison of single-and double-tag mRNA export efficiency and times
In addition to using the experimentally consistent single-tag tracking approach when analyzing mRNA export efficiency and time, we also observed the positioning of the mRNA's 5′ and 3′ termini in the simulations. This equated to using a double-tag approach to track mRNA export. Although single probes placed at either the 3′ end or a random site along the length of the mRNA were previously used in experimental studies of mRNA export, we hypothesized that they may inaccurately measure residence time in the pore, as well as the fraction of successful transport. A single probe might less precisely indicate the location of the mRNA, potentially leading to overestimation or underestimation of true pore residence time. In addition, the fraction of successful transports may be overestimated if a probe on a partially transported mRNA reaches far enough ahead of the rest of the molecule within a particular time cutoff.
As indicated in Figures 2 and 3, the choice to use single or double tagging had notable bearing on both mRNA export efficiency and time. With regard to transport efficiency (Figure 2), whereas successful mRNA transport remained approximately the same under the default NTR configuration, partial and unsuccessful transport events were portrayed differently. Of note, in the double-tag case, the fraction of unsuccessful transport events did not appear to significantly change with affinity. Increasing NXF1's affinity to FG Nups appears to have caused more partial transport events to become successful ones. Meanwhile, this trend was not found with the single-tag approach. With regard to transport time (Figure 3), slightly higher central channel residence times were recorded across NTR/FG Nup affinities using the double-tag method than the single-tag method. Meanwhile, the differences in nuclear basket residence times were more profound.
Considering the default configuration of nine NTRs per mRNA and an NTR/FG Nup affinity of 100 μM, using a double-tag approach yielded mean nuclear basket and central channel residence times of 462.9 ± 265.4 and 269.5 ± 78.6 ms, respectively. As mentioned earlier, the corresponding residence times using the single-tag approach are 107.3 ± 87.8 and 34.5 ± 27.0 ms, respectively. With the double-tag approach, the residence times observed in our ABM appear to be significantly longer than those reported in in vivo studies.
More-granular representations of single-tag and double-tag mRNA residence times are shown in Figure 4 for the nuclear basket and Figure 5 for the central channel. A nonparametric Wilcoxon rank-sum test comparing residence times obtained using the two different mRNA- tracking approaches confirmed that residence times were significantly different for all affinities, with the exception of 120 μM for the nuclear basket and 160 μM for the central channel. We suspect that there was a lack of significance at those affinities partly due to sparse sampling of successful transport events.
FIGURE 4:
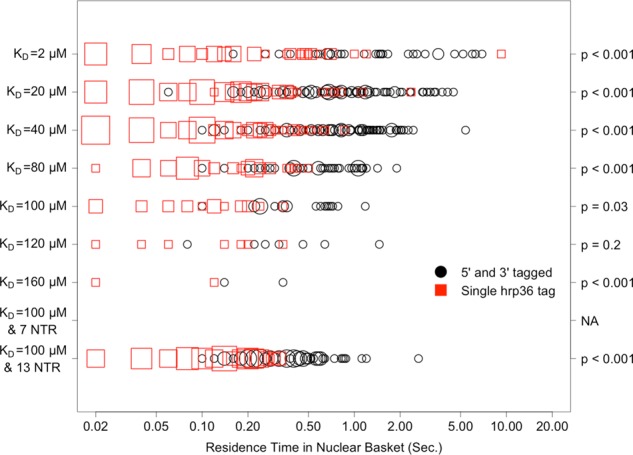
Distribution, by affinity, of mRNA residence times in the nuclear basket for successful export events as calculated using a single, randomly placed hrp36 probe, compared with using two probes that are placed at both 5′ and 3′ ends of the mRNA. The x-axis is on a log10 scale. Larger points indicate higher frequency of the specified measurement. The p values on the right correspond to a nonparametric (Wilcoxon rank sum) test of significance in the difference between the residence-time distributions obtained with each probe method.
FIGURE 5:
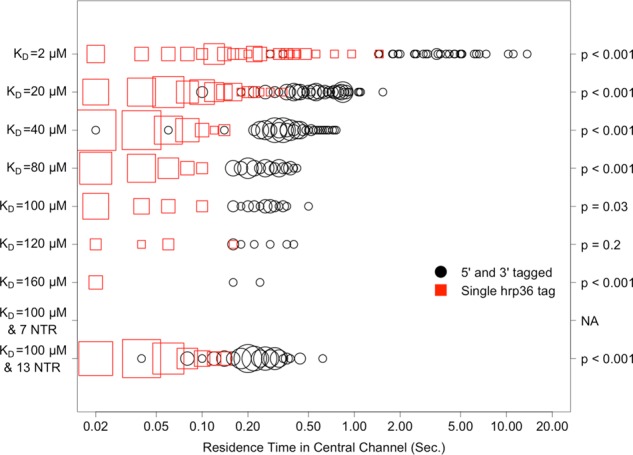
Distribution, by affinity, of mRNA residence times in the central channel for successful export events as calculated using a single, randomly placed hrp36 probe, compared with using two probes that are placed at both 5′ and 3′ ends of the mRNA. The x-axis is on a log10 scale. Larger points indicate higher frequency of the specified measurement. The p values on the right correspond to a nonparametric (Wilcoxon rank sum) test of significance in the difference between the residence-time distributions obtained with each probe method.
Effect of number and distribution of mRNA-bound NXF1 transport receptors
In the aforementioned experiments, the default number of NXF1 transport receptors bound to the mRNA was set to nine. This configuration turns out to represent the number of exon junction complexes present in a typical mRNA of this length. Under this configuration, an NTR/FG Nup affinity of 100 μM yielded a frequency of successful transport events that was in agreement with experimentally reported values (Figure 2). To assess the sensitivity of mRNA export to the variation in the number of transport receptors for an mRNA of fixed length (i.e., receptor density), we increased the number of transport receptors from nine to 13. This led to a significant increase in the number of successful transport events (Figure 6). Meanwhile, the nuclear basket and central channel residence times did not appear to change significantly (Figure 7). A corresponding decrease in the number of transport receptors from nine to seven resulted in no successful transport events observed. Of interest, with the double-tag tracking, we continued to observe the maintenance of a relatively constant fraction of unsuccessful transport events; however, with single-tag tracking, this consistency disappeared (Figure 6). Furthermore, the residence times captured with double-tag tracking remained higher than those captured with single-tag tracking (Figure 7).
FIGURE 6:
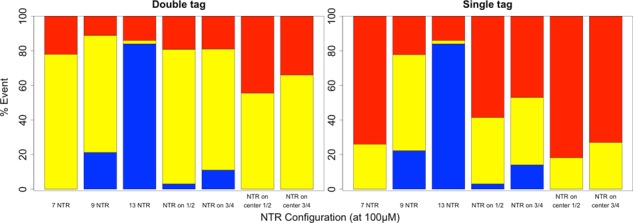
Bar graphs showing the relative percentage of successful (blue), partial (yellow), and unsuccessful (red) transport events observed for different distributions of NTRs on an export-competent mRNA. Note that all configurations used the baseline NXF1 to FG Nup affinity of 100 μM. “NTR on ½” and “NTR on ¾” represent configurations where transport receptors were placed on the terminal one-half and three-fourths length of the mRNA, respectively, with the same spacing as was used in the baseline configuration for a total of five transport receptors in the one-half configuration and seven transport receptors in the three-fourths configuration. “NTR on center ½” and “NTR on center ¾” represent configurations where transport receptors were placed in the center one-half and three-fourths length of the mRNA, respectively, with the same spacing as used in the baseline configuration for a total of five transport receptors in the one-half configuration and seven transport receptors in the three-fourths configuration (i.e., these configurations lacked transport receptors near the 5′ and 3′ termini). The graph on the left captures observations recorded with a double tag (5′ and 3′ end) system, and the graph on the right captures those recorded with a single tag.
FIGURE 7:
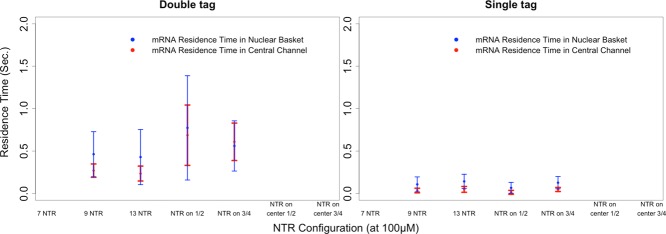
Graphs showing the effect of varying NTR distribution along the length of an mRNA on nuclear basket and central channel residence times for successful export events (error bars represent 1 SD). Note that all configurations used the baseline NXF1 to FG Nup affinity of 100 μM. “NTR on ½” and “NTR on ¾” represent configurations where transport receptors were placed on the terminal one-half and three-fourths length of the mRNA, respectively, with the same spacing as used in the baseline configuration for a total of five transport receptors in the one-half configuration and seven transport receptors in the three-fourths configuration. The graph on the left captures observations recorded with a double tag (5′ and 3′ end) system, and the graph on the right captures those recorded with a single tag.
We also analyzed the effect of transport receptor localization on transport kinetics. As shown in Figure 6, the placement of transport receptors along the entire length of the mRNA resulted in more robust transport than under partial coverage. Keeping transport receptor spacing fixed between trials, configurations of full-length coverage, three-fourths–length coverage, and one-half–length coverage yielded successful transport rates of 21, 11, and 3%, respectively. Of interest, whereas no successful transport occurred when seven transport receptors were uniformly distributed along the length of the mRNA, 11% successful export was obtained when the seven transport receptors were distributed along three-fourths of the length (starting at the 5′ end). This suggests that transport receptor spacing (or density), rather than absolute receptor quantity, plays a greater role in determining export likelihood. When we positioned the five and seven transport receptors in the central one-half and three-fourths length of the mRNA polymer, we observed no transport. The number of partial transports was reduced, whereas the number of failed transports increased for both configurations compared with all others. This may indicate that having transport receptors on at least one of the termini is necessary for successful transport.
Dynamics of an mRNA polymer undergoing export
Previous studies reported lengthening of the mRNA as it approaches the central channel (Daneholt, 1997). This observation was for the Balbiani ring mRNA, which is much larger than the mRNA modeled in our simulations (∼37 vs. ∼2.2 kb). Nevertheless, we wanted to explore whether such lengthening would be observed in our simulations. As shown in Figure 8, the average end-to-end length of mRNA approaching the basket dropped initially from that of the globular conformation taken on when diffusing freely throughout the nucleoplasm. This reduced end-to-end length appears to be the result of the mRNA taking on a more compact conformation as NXF1 proteins bind multiple Nups in the basket. Average end-to-end length then increased as the polymer translocated through the central channel. This behavior is consistent with what has been observed in EM studies of mRNA export. Of note, increased affinity and number of transport receptors both resulted in increased compaction of the mRNA polymer in the basket (Figure 8).
FIGURE 8:
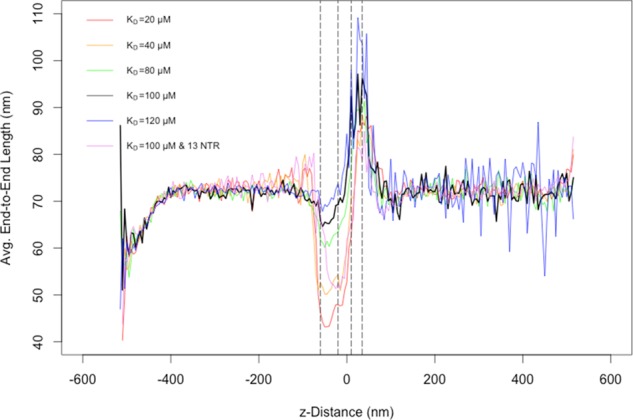
A combined plot of end-to-end length averaged over 100 simulations for multiple affinity and nuclear transport receptor configurations in the ABM. Note that, where unspecified, the number of transport receptors was set to the baseline value of nine, distributed uniformly along the length of the mRNA. The x-axis represents the position along the axis perpendicular to the nuclear envelope (z-distance), with x = 0 set at the center of the central channel of the NPC. Left to right, the dashed lines represent the distal edge of the nuclear basket, the nuclear edge of the central channel, the cytoplasmic edge of the central channel, and the distal edge of the cytoplasmic filaments, respectively. Note that low sampling of successful transport events accounts for the increased observed variability in average end-to-end lengths of low Nup-NXF1– affinity transports on the cytoplasmic side.
DISCUSSION
Although accurate experimental techniques are ultimately essential for enhancing our understanding of mRNA export dynamics, computational models can aid in constructing hypotheses and experimental design while also revealing potential systematic limitations. Using the ABM modality, we constructed a rudimentary model of mRNA export through the NPC that allowed us to test how several different parameters affect this process. The model was designed to capture only coarse-grained aspects of mRNA export, and, for example, it assumes homogeneous affinity between FG Nups and mRNPs, whereas the affinities between NTR-cargo complexes and FG Nups is very likely to vary within the NPC. As a result, all of the quantitative results produced have to be taken with a grain of salt. Nonetheless, the model does reliably recapitulate previously observed coarse-grained facets of mRNA export dynamics.
We first assessed the effect of affinity between mRNA-binding nuclear transport receptor (NTR) NXF1 and the NPC's FG Nups. As shown in Figure 2, having nine NTRs on the mRNA and an affinity of 100 μM yielded mRNA export efficiency that is most consistent with previous experimental observations. Furthermore, residence time within both the nuclear basket and the central channel was minimized under this affinity (Figure 3). These results suggest a balance between export likelihood and pore residence time of an mRNA. Taken in the context of bulk export, this balance may improve transport efficiency for other cargoes as well.
In addition, the effect of density of NTR binding to the mRNA was also examined. Raising the density of NTRs bound to the mRNA led to increased transport efficiency, whereas lowering it dramatically ablated transport. These observations hint at the degree of stringency behind the process of NTR-mediated export that cells would have to evolve to leverage the biomechanics that are relevant to this process. In further support of this idea, whereas full coverage of the mRNA by NTRs appears to favor optimal transport conditions, coverage of at least one mRNA terminus may be necessary to ensure any successful export. To speculate on a potential explanation for this requirement, having an NTR-bound terminus may ensure that the mRNA can begin threading through the pore after sweeping the conformational space. Because similar trends in NTR distribution-dependent export efficacy were observed for the case of 20 μM NXF1-Nup affinity (unpublished data), they may be independent of affinity. Of interest, NTR configuration on the mRNA did not appear to affect the nuclear basket or central channel residence times of successfully transported mRNA as much as we had suspected.
Our ABM also reveals potential sources of systematic error that may be present in previously used experimental approaches aimed at studying this process. The data show that, regardless of affinity or NTR distribution, using a single mRNA-tracking probe may lead to an overestimation of transport rate, presumably because a single probe provides only a local snapshot for the region along the mRNA that contains the tag. However, the choice of a single- or double-tag probing system appears not to substantially alter the observed fraction of successful transport events. We did observe that the use of a single tracking probe may overestimate the number of successful export events under some circumstances (unpublished data). However, this overestimation was in the range of 0–5%, depending on affinity and transport receptor distribution. We can therefore conclude that the use of a single tracking probe is sufficient for determining the fraction of successful mRNA export events. This is likely due to the relative duration of a typical mRNA export event being much shorter than the duration of observation—in this case, ∼1.0 s of pore residence compared with 20 s of observation. As the difference between these durations is reduced, the confidence in single-probe measurements of transport likelihood becomes questionable.
With our ABM, we observed that the coarse elongation of the mRNA as it threaded through the NPC (Figure 8) is consistent with previous experimental findings. In addition, a collapse in configuration as the mRNA contacted the nuclear basket was observed. Both our simulations and some of the previous experimental findings suggest that successful threading of the mRNA terminus is the rate-limiting step in mRNA transport. As such, it could be speculated that by forcing the mRNA into a more compact conformation, the basket reduces the size of the potential configuration space that the mRNA would sweep in order to find one that is optimal for export through the narrow central channel, thus accelerating the rate of transport. Crucially, such behavior of the mRNA cannot be observed in experimental studies that use a single probe per mRNA molecule for labeling purposes.
Our findings are in contrast with those of the Yang group (Ma et al., 2013), which suggest that the rate-limiting step in transport occurs in the central channel. However, the relevance of the group using spatial signal density and region-specific diffusion coefficients for distinguishing which phase of NCT is the rate-limiting step is unclear. The calculations performed assumed homogeneous, continuum-based diffusion coefficients, which, we would argue, oversimplifies the transport process. Finally, it is unclear how the differences in cytoplasmic distribution between successful and aborted transport events indicate anything related to the rate-limiting step of transport. Instead, such differences may simply reflect the researcher's method of judging which transport events are successful versus which are not, given the experimental constraints.
As with other studies, ours also has its limitations, which we wish to reiterate. The ABM that we produced recapitulates only coarse-grained behavior of an mRNA-like molecule as it transports through the NPC. Certainly, the detailed numbers behind our findings should be taken with a grain of salt. For example, although it has been strongly suggested that different FG Nups within the NPC exhibit varying affinities to NTR-cargo complexes, our model uses homogeneous affinities and thus potentially fails to capture some of the intricacies in the export process. Furthermore, the detailed organization of FG repeats within the NPC remains unclear and thus cannot be accurately reflected in our model. Despite these and other limitations, however, the model does faithfully recapitulate many of the coarse-grained experimentally observed processes and therefore has potential for further refinement in the future; it is hoped that it will stimulate further research in this area.
MATERIALS AND METHODS
Agent-based modeling
ABM is a robust computational technique used to simulate the spatiotemporal actions and interactions of real-world entities or “agents” in an effort to extract their combined effect on the system as a whole. Both space and time can be discretized in an ABM. The autonomous agents can move and interact with other agents and their environment at each time step. Simple behavioral rules govern the movement and interaction of each individual agent; added together, the emergent collective behavior of all the agents can simulate complex phenomena that may not be apparent when simply considering individual agents.
In their simplest form, on-lattice ABMs consist of a mesh of “cells” that assemble the discretized space that agents occupy (Figure 9). The agents occupy these cells and are typically aware only of other agents within their “neighborhood” of cells, with the simplest neighborhood consisting of adjacent cells. Agents can be given the ability to move into adjacent cells and interact with other agents with some probability in conjunction with governing rules that define what movement and interactions are possible. On-lattice ABMs have been applied to biological systems involving diffusion, binding, and unbinding (Bonchev et al., 2010; Devillers et al., 2010; Dong et al., 2010).
FIGURE 9:
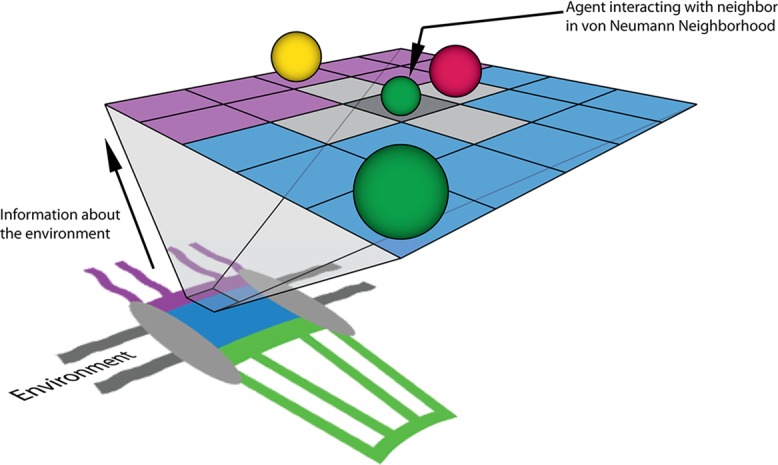
A cartoon representation of the NPC's environment (not to scale) projected onto a two-dimensional, on-lattice ABM with agents (spheres) representing protein factors. These agents move within the system and interact with other agents within their von Neumann neighborhood. One such neighborhood of cells is highlighted in gray for the smaller green agent that is pointed to by an arrow. The actual model consists of a 3D representation of the NPC structure and physiologically relevant concentrations of biochemical factors and channel dimensions. In our model, the purple region representing the cytoplasmic periphery is treated as a compartmentalized volume containing both noninteracting Nup and transport receptor–interacting FG Nup agents. The central channel (blue) and nuclear basket (green) regions are analogously represented by compartmentalized volumes and functionalized with Nup and FG Nup agents. Gray regions of the NPC diagram represent the scaffold and nuclear envelope regions of the model that are impermeable to diffusing species.
Our 3D, computationally efficient, and spatiotemporally detailed ABM was developed specifically for modeling molecular diffusion, binding, and unbinding events with consideration for physical factors such as molecular crowding and steric repulsion. Where possible, we used diffusion, binding, and unbinding rates derived from in vitro or in vivo experiments. In addition to movement and interaction rules, probabilities for movement and interaction events govern system dynamics in the ABM. Establishing methods for accurate selection of movement, binding, and unbinding probabilities to best represent actual diffusion coefficients and kinetic rate constants would improve model accuracy and thus build confidence in both the output from the ABM and any deductions made (Azimi et al., 2011).
Probability selection for molecular movement and binding/unbinding on an ABM lattice
In our previous work, we proposed a method for movement probability selection based on the following molecular diffusion coefficient, along with algorithms for realistic consideration of crowding and steric repulsion (Azimi et al., 2011; Azimi and Mofrad, 2013; Jamali et al., 2013) that were also used in the current model:
|
|
Here movement probability of an agent is determined by its diffusion coefficient (D), simulation time step (Δt), and lattice discretization length (ΔL). We implemented the reduced probability method to account for the steric effects of multiple agents occupying individual lattice sites (Azimi et al., 2011).
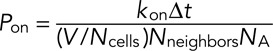
In our more recent work (Azimi and Mofrad, 2013), we proposed and validated a method for probability selection of binding and unbinding events in the ABM that was also used in the current model. Probability selection of unbinding and binding, respectively, can be determined from kinetic rate constants as follows:
|
|
 (3) (3)
|
Here event likelihoods are determined from koff and kon (which represent the real-world kinetic rate constants), simulation time step, Δt; system volume, V; number of lattice cells, Ncells; number of lattice neighbors that each cell has, Nneighbors; and Avogadro's number, NA.
mRNA model structure and dynamics
In the ABM, the mRNA is represented as a collection of bound agents that are limited in their movements through the constraint of maintaining connection with their nearest neighbors. Further, movements are only permitted in the diagonal direction into a nearest neighbor's von Neumann neighborhood. This ensures that all movements are of fixed length and ensures that a single movement probability can accommodate all movement events of a specific agent type.
The probability of movement for agents belonging to the mRNA polymer was determined through successive simulation of a representative 2.2-kb ABM polymer composed of 130 agents in a 5 × 5 × 5 nm3 cubic lattice over 5 million time steps (2.5 μs/step, 12.5 s total). Movement probability was varied, and the corresponding effective diffusion coefficient was determined by calculating the mean squared displacement with a minimum sampling of 1000 time steps. A movement probability of 0.5 was determined to yield an effective diffusion coefficient of ∼0.01 μm2/s, which is in agreement with previous in vivo measurements of mRNA diffusion (Mor et al., 2010). Analysis of mean square displacement as a function of time also confirmed that diffusion of the ABM polymer followed a normal diffusion regime, which is expected from previous reports and our own comparison with Brownian dynamics simulations. Finally, to ensure that the ABM correctly recapitulated the behavior of a freely jointed chain, we simulated ABM polymers of varying lengths to confirm that their average end-to-end length was in agreement with the length-dependent prediction postulated by the freely jointed chain model:
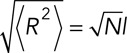 (4) (4)
|
Here 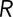 represents the total end-to-end vector of an ideal chain, N represents the number of monomers in the chain, which is equivalent to the number of agents that compose a polymer chain in the ABM, and l represents the Kuhn segment length of one of the monomers (for mRNA, ranging from ∼0.5 to 3 Å, as previously reported; Magee and Warwicker, 2005; Vanzi et al., 2005). Then l is equivalent to the discretization of the lattice in the ABM. We observed that, for very short length polymer chains of five agents, there was ∼10% discrepancy between the predicted average end-to-end length (Eq. 4) and the average end-to-end length from 10,000 randomly sampled ABM configurations. This error was reduced to <1% when the number of monomers/agents was increased to 50. The error was determined to be negligible for the configuration of 130 monomers/agents that was used to simulate a typical mRNA.
represents the total end-to-end vector of an ideal chain, N represents the number of monomers in the chain, which is equivalent to the number of agents that compose a polymer chain in the ABM, and l represents the Kuhn segment length of one of the monomers (for mRNA, ranging from ∼0.5 to 3 Å, as previously reported; Magee and Warwicker, 2005; Vanzi et al., 2005). Then l is equivalent to the discretization of the lattice in the ABM. We observed that, for very short length polymer chains of five agents, there was ∼10% discrepancy between the predicted average end-to-end length (Eq. 4) and the average end-to-end length from 10,000 randomly sampled ABM configurations. This error was reduced to <1% when the number of monomers/agents was increased to 50. The error was determined to be negligible for the configuration of 130 monomers/agents that was used to simulate a typical mRNA.
ABM system and simulation details
The model environment consists of a 42,108-element, 3D lattice composed of elements with dimensions of 5 × 5 × 5 nm3. The lattice size was selected to accommodate the volume associated with the Stokes radius of the largest single-agent species in the system—in this case a collection of nucleotides representing twice the persistence length of the mRNP, or Kuhn length. In addition, the model allowed for multiple agents of the same or different species type to occupy the same lattice element at any given time, so long as the available volume of a lattice element was not exceeded by agents diffusing into it. Discrete lattice elements belong to one of six region types: cytoplasmic, nuclear membrane, nucleoplasm, cytoplasmic filament periphery, central channel, or nuclear basket. The cytoplasmic region contains a high concentration of Dbp5 in complex with Gle1 and IP6, whereas the nucleoplasm in each simulation contained a single 2.2-kb mRNP discretized into 130 agents. The 35-nm-thick nuclear membrane, which partitions the two compartments, is impermeable to all agent types and contains a single nuclear pore with a diameter of 30 nm at the center and 50 nm at the peripheries. The cytoplasmic filament periphery consists of a 50-nm-diameter region that extends 30 nm into the cytoplasm, and the nuclear basket is composed of a basket-shaped region that extends 55 nm into the nucleoplasm (Adam, 2001; Jamali et al., 2011; Loschberger et al., 2012). The cytoplasmic periphery, central channel, and nuclear basket contain 24, 80, and 32 agents, respectively, representing the distribution of FG Nups (Yamada et al., 2010). In addition to these FG agents, non-FG agents are added to the channel to represent regions of the Nups that lack affinity for transport receptors but play a role in sterically repelling molecules, with the sum of the volume of these Nups corresponding to experimentally reported volumes (Yamada et al., 2010). The collection of agents representing the mRNP are free to diffuse throughout the system, whereas FG agents and non-FG agents are restricted to movement within their respective pore regions in order to maintain the permeability barrier.
Distribution of transport receptors on the mRNA and their affinity for FG Nups was varied across simulations. For each configuration investigated, 100 replicate simulations were generated and analyzed. Each simulation contained a single mRNA with a random initial configuration in the nucleoplasm. The 5′ and 3′ termini of the mRNA were tracked in each simulation. In addition, a randomized site along the length of the mRNA was also tracked in each replicate to simulate the use of a fluorescently labeled hrp36 molecule for comparison with experiments carried out by Siebrasse et al. (2012). Each simulation was carried out for the duration of 20 s using a time step of 2.5 μs.
The locations of the 5′ and 3′ termini, along with the location of the randomly placed “hrp36 tag” agent, were tracked over the course of the simulation. Throughout the rest of the article, we referred to the data captured through these tracking methods as double- and single-tag data, respectively. To analyze these trajectories, kymographs were generated illustrating the location of the 5′ and 3′ termini of the mRNA over time (Figure 10). The trajectories were further analyzed to determine the fraction of partial and successful transports per configuration, along with mRNA residence times in the nuclear basket and central channel, for comparison with previous in vivo observations. In this context, partial transport was defined as the initiation of transport from either the 5′ or 3′ end of the mRNA but a failure in the export of the other end.
FIGURE 10:
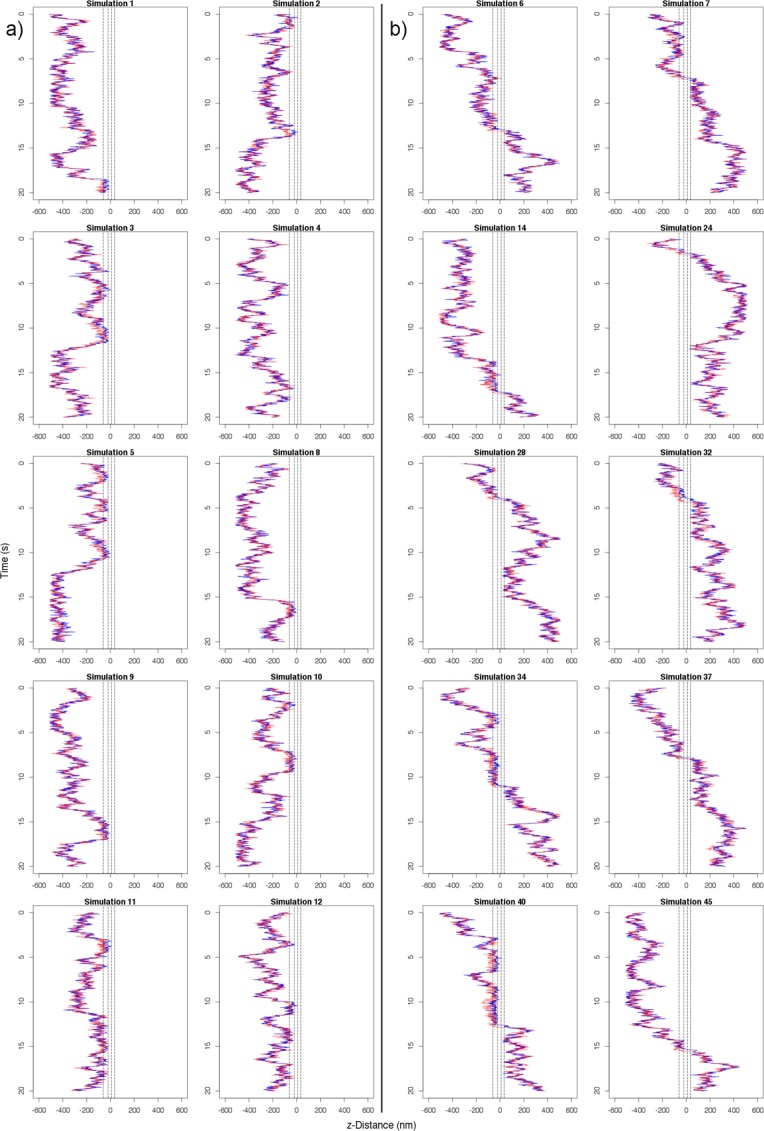
Kymographs of 20 replicate simulations of mRNA export using the default configuration of transport receptor/FG Nup affinity of 100 μM and nine transport receptors along the length of the mRNA. The blue lines represent the position of the 5′ end, and the red lines represent the position of the 3′ end. The NPC central channel is centered at z = 0 nm in each plot. Plots are distributed to show (A) 10 failed exports and (B) 10 successful exports.
Supplementary Material
Acknowledgments
We gratefully acknowledge discussions with and technical assistance by Ruhollah Moussavi-Baygi, Hanif Mahboobi, Alex Javanpour, and the rest of Molecular Cell Biomechanics Laboratory. In addition, we are grateful for numerous fruitful discussions with Ulrich Kubitscheck, which helped identify computational experiments of interest.
Abbreviations used:
- ABM
agent-based model
- NPC
nuclear pore complex
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1065) on September 24, 2014.
*These authors contributed equally to this work.
REFERENCES
- Adam SA. The nuclear pore complex. Genome Biol. 2001;2:REVIEWS0007. doi: 10.1186/gb-2001-2-9-reviews0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi M, Jamali Y, Mofrad MR. Accounting for diffusion in agent based models of reaction-diffusion systems with application to cytoskeletal diffusion. PLoS One. 2011;6:e25306. doi: 10.1371/journal.pone.0025306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi M, Mofrad MR. Higher nucleoporin-Importinbeta affinity at the nuclear basket increases nucleocytoplasmic import. PLoS One. 2013;8:e81741. doi: 10.1371/journal.pone.0081741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchev D, Thomas S, Apte A, Kier LB. Cellular automata modelling of biomolecular networks dynamics. SAR QSAR Environ Res. 2010;21:77–102. doi: 10.1080/10629360903568580. [DOI] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- Devillers J, Devillers H, Decourtye A, Aupinel P. Internet resources for agent-based modelling. SAR QSAR Environ Res. 2010;21:337–350. doi: 10.1080/10629361003773963. [DOI] [PubMed] [Google Scholar]
- Dong X, Foteinou PT, Calvano SE, Lowry SF, Androulakis IP. Agent-based modeling of endotoxin-induced acute inflammatory response in human blood leukocytes. PLoS One. 2010;5:e9249. doi: 10.1371/journal.pone.0009249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Singer RH. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika M, Brockmann C, Gruessing F, Neuhaus D, Divita G, Stewart M, Dargemont C. Structural requirements for the ubiquitin-associated domain of the mRNA export factor Mex67 to bind its specific targets, the transcription elongation THO complex component Hpr1 and nucleoporin FXFG repeats. J Biol Chem. 2009;284:17575–17583. doi: 10.1074/jbc.M109.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25:1052–1064. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamali T, Jamali Y, Mehrbod M, Mofrad MRK. Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int Rev Cell Mol Biol. 2011;287:233–286. doi: 10.1016/B978-0-12-386043-9.00006-2. [DOI] [PubMed] [Google Scholar]
- Jamali Y, Jamali T, Mofrad MRK. An agent based model of integrin clustering: Exploring the role of ligand clustering, integrin homo-oligomerization, integrin–ligand affinity, membrane crowdedness and ligand mobility. J Comput Phys. 2013;244:264–278. [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschberger A, van de Linde S, Dabauvalle MC, Rieger B, Heilemann M, Krohne G, Sauer M. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J Cell Sci. 2012;125:570–575. doi: 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]
- Ma J, Liu Z, Michelotti N, Pitchiaya S, Veerapaneni R, Androsavich JR, Walter NG, Yang W. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat Commun. 2013;4:2414. doi: 10.1038/ncomms3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee J, Warwicker J. Simulation of non-specific protein–mRNA interactions. Nucleic Acids Res. 2005;33:6694–6699. doi: 10.1093/nar/gki981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U. Structural interaction between the nuclear pore complex and a specific translocating RNP particle. J Cell Biol. 1995;129:1205–1216. doi: 10.1083/jcb.129.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- Natalizio BJ, Wente SR. Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol. 2013;23:365–373. doi: 10.1016/j.tcb.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble KN, Tran EJ, Alcazar-Roman AR, Hodge CA, Cole CN, Wente SR. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25:1065–1077. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Chartrand P. Using fluorescent proteins to study mRNA trafficking in living cells. Methods Cell Biol. 2008;85:273–292. doi: 10.1016/S0091-679X(08)85012-1. [DOI] [PubMed] [Google Scholar]
- Riddick G, Macara IG. A systems analysis of importin-α-β mediated nuclear protein import. J Cell Biol. 2005;168:1027–1038. doi: 10.1083/jcb.200409024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Stutz F. Nuclear export of RNA. Biol Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Siebrasse JP, Kaminski T, Kubitscheck U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc Natl Acad Sci USA. 2012;109:9426–9431. doi: 10.1073/pnas.1201781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Tetenbaum-Novatt J, Hough LE, Mironska R, McKenney AS, Rout MP. Nucleocytoplasmic transport: a role for nonspecific competition in karyopherin-nucleoporin interactions. Mol Cell Proteomics. 2012;11:31–46. doi: 10.1074/mcp.M111.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol. 2011;12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- Vanzi F, Takagi Y, Shuman H, Cooperman BS, Goldman YE. Mechanical studies of single ribosome/mRNA complexes. Biophys J. 2005;89:1909–1919. doi: 10.1529/biophysj.104.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


