STIM1 and Orai1 move to endoplasmic reticulum–plasma membrane (ER-PM) junctions to trigger store-operated Ca2+ entry. Single-molecule tracking reveals pure diffusion of STIM1 in the ER, subdiffusion of Orai1 in the PM, and trapping at junctions via STIM-Orai binding. STIM-Orai binding is loose, generating free proteins that can exchange freely with extrajunctional pools.
Abstract
Following endoplasmic reticulum (ER) Ca2+ depletion, STIM1 and Orai1 complexes assemble autonomously at ER–plasma membrane (PM) junctions to trigger store-operated Ca2+ influx. One hypothesis to explain this process is a diffusion trap in which activated STIM1 diffusing in the ER becomes trapped at junctions through interactions with the PM, and STIM1 then traps Orai1 in the PM through binding of its calcium release-activated calcium activation domain. We tested this model by analyzing STIM1 and Orai1 diffusion using single-particle tracking, photoactivation of protein ensembles, and Monte Carlo simulations. In resting cells, STIM1 diffusion is Brownian, while Orai1 is slightly subdiffusive. After store depletion, both proteins slow to the same speeds, consistent with complex formation, and are confined to a corral similar in size to ER–PM junctions. While the escape probability at high STIM:Orai expression ratios is <1%, it is significantly increased by reducing the affinity of STIM1 for Orai1 or by expressing the two proteins at comparable levels. Our results provide direct evidence that STIM-Orai complexes are trapped by their physical connections across the junctional gap, but also reveal that the complexes are surprisingly dynamic, suggesting that readily reversible binding reactions generate free STIM1 and Orai1, which engage in constant diffusional exchange with extrajunctional pools.
INTRODUCTION
Store-operated calcium channels carry out essential signaling functions throughout the body, in particular in the immune system, where they are required for the adaptive immune response to antigens (Parekh and Putney, 2005; Shaw and Feske, 2012). Store-operated Ca2+ channels such as the well-characterized Ca2+ release–activated Ca2+ (CRAC) channel are activated through a process initiated by the receptor-triggered depletion of Ca2+ from the endoplasmic reticulum (ER), which triggers the coordinated redistribution and interaction of stromal interaction protein 1 (STIM1) and Orai1 proteins (Lewis, 2011). In resting cells, the ER Ca2+ sensor STIM1 is diffusely distributed throughout the ER, while the CRAC channel Orai1 resides throughout the plasma membrane (PM). After Ca2+ in the ER ([Ca2+]ER) declines following a receptor-triggered increase in inositol 1,4,5-trisphosphate, STIM1 and Orai1 redistribute to ER–PM junctions, where they interact to open the CRAC channel (Liou et al., 2005; Zhang et al., 2005; Luik et al., 2006; Wu et al., 2006; Xu et al., 2006). This process is reversible upon refilling of the ER Ca2+ store (Várnai et al., 2007; Smyth et al., 2008).
The assembly of the functional CRAC channel from STIM1 and Orai1 after store depletion is an autonomous process triggered by the oligomerization of STIM1 (Luik et al., 2008). Release of Ca2+ from the EF hand allows dimerization of luminal domains (Stathopulos et al., 2006), which triggers a rearrangement of the cytosolic domains of STIM1 and likely induces oligomerization of multiple STIM1 dimers (Liou et al., 2007; Muik et al., 2009; Covington et al., 2010; Korzeniowski et al., 2010; Zhou et al., 2013). Artificial dimerization of the luminal domains of STIM1 is sufficient to elicit STIM1 redistribution, clustering, and activation of CRAC channels in cells with full stores, suggesting that once STIM1 has adopted its active conformation, all further events in assembly and opening of the CRAC channel occur independently of [Ca2+]ER (Luik et al., 2008).
What is the basis of this self-organizing behavior? A common mechanism for self-organization in biological systems is the diffusion trap, in which passive, thermally driven motion brings molecules together, and binding interactions maintain their colocalization. The diffusion trap model for store-operated calcium entry (SOCE) posits that resting STIM1 and Orai1 passively diffuse, but once activated, STIM1 binds via exposed polybasic domains to phosphatidylinositol 4,5-bisphosphate (PIP2) or phosphatidylinositol 3,4,5-trisphosphate in the PM at ER–PM junctions (Ercan et al., 2009; Walsh et al., 2010). Trapped STIM1 in turn binds to Orai1 in the overlying PM through a direct binding of its CRAC activation domain (CAD; also known as SOAR or Ccb9) to the C-terminus of Orai1 (Kawasaki et al., 2009; Park et al., 2009; Yuan et al., 2009; Zhou et al., 2010).
Several studies have reported passive movements of STIM1 and Orai1 before and after store depletion. Fluorescence recovery after photobleaching (FRAP) results suggest STIM1 and Orai1 diffuse freely (Liou et al., 2007; Park et al., 2009; Covington et al., 2010; Madl et al., 2010), and STIM1-Orai1 redistribution and clustering persists in ATP-depleted cells (Chvanov et al., 2008). Although FRAP measurements at the cellular level have been interpreted in terms of pure diffusion of a mobile population mixed with immobile fraction, this approach cannot reveal the full range of molecular behaviors in the population, possibly obscuring a combination of active and passive transport mechanisms. While the ATP depletion study argues against active transport, the high ATP sensitivity of motors like myosin II (Murphy et al., 2001) may make it difficult to reduce ATP to a level that fully prevents motor function. Several studies have reported microtubule-associated STIM1 transport (Baba et al., 2006; Wu et al., 2007; Grigoriev et al., 2008; Smyth et al., 2008) mediated by binding to EB1 at the tips of growing microtubules in cells overexpressing STIM1 (Grigoriev et al., 2008). Although SOCE is apparently not dependent on EB1 or microtubule extension (Grigoriev et al., 2008), it is unclear whether these types of active mechanisms contribute to the movement of endogenous levels of STIM1 proteins, particularly in resting cells.
Our goals in this study were to test the diffusion trap model and answer a number of questions related to the nanoscale dynamics of STIM1 and Orai1 before and after ER Ca2+ store depletion. Do STIM1 and Orai1 move purely by random diffusion in resting cells, or is an active component involved? After store depletion, to what extent does STIM1 by itself slow or stop once it reaches the junction, and how tightly is it held there by its polybasic domain? How does the presence of Orai1 alter the mobility of STIM1 at junctions? Can we detect the trapping of single STIM1 and Orai1 molecules at ER–PM junctions, and once trapped, are they immobile or do they move as a complex and sample the area within junctional boundaries? Do STIM1 and Orai1 move across the junctional boundaries after store depletion, or is trapping absolute if stores are maintained in a Ca2+-depleted state? The dynamics of exchange across junctional boundaries may provide information about the kinetics of STIM1-Orai1 binding reactions that ultimately control the Ca2+ flux through individual CRAC channels.
To accomplish these goals and achieve a more complete quantitative view of STIM1 and Orai1 mobility, we used superresolution techniques to track single green fluorescent protein (GFP)-labeled STIM1 and Orai1 molecules and photoactivation to track ensembles of photoactivatable GFP (PAGFP)-labeled STIM1 and Orai1 in intact HEK cells. Single-particle tracking (SPT) can reveal heterogeneous rates of diffusion within an ensemble of molecules, as well as provide sensitive assays for active transport, pure versus hindered diffusion, and diffusion within a boundary. Our results provide direct support for the diffusion trap model for STIM1-Orai1 complex formation, with no indication of active transport. In store-depleted cells, while STIM1 diffusion is slowed progressively by interaction with the PM and binding to Orai1, and Orai1 diffusion is slowed and restricted by STIM1 at ER–PM junctions, a large proportion of STIM1 and Orai1 continue to move throughout the junctions. Using photoactivation techniques to track the escape of STIM1 and Orai1 from junctions at a population level, we also find that both proteins exchange across junctional boundaries with surrounding pools of free STIM1 and Orai1 at a rate that depends on the STIM:Orai expression ratio. Thus, despite the apparent constancy of STIM1/Orai1 puncta at a population level, our results reveal a surprising degree of heterogeneity and dynamics in STIM1 and Orai1 molecular mobility after Ca2+ store depletion.
RESULTS
Diffusive behavior of STIM1 and Orai1 in resting cells with replete Ca2+ stores
To enable single-particle tracking, we imaged fluorescently labeled STIM1 and Orai1 proteins under conditions in which the separation between adjacent proteins exceeded the diffraction limit (∼250 nm). We tested several labeling methods, including PAGFP, mEos2, and GFP. While all three methods gave qualitatively similar results, we found that GFP-labeled proteins produced the longest tracks before bleaching. Trajectories of single GFP-STIM1 and Orai1-GFP particles in resting HEK cells are shown in Figure 1 and Supplemental Videos S1 and S2. Several observations indicate that these particles represent single molecules of STIM1 in the ER and Orai1 in the PM. The fluorescence intensities of the particles were narrowly distributed and single- or double-step photobleaching events were observed, indicating that most particles were labeled with a single GFP (Supplemental Figure S1). Furthermore, after bleaching by prolonged total internal reflection fluorescence (TIRF) illumination, new Orai1-GFP particles entered the cell footprint from the edges of the cell (unpublished data), confirming that Orai1 molecules being tracked were in the PM, not in intracellular vesicles, which would be expected to insert throughout the cell footprint.
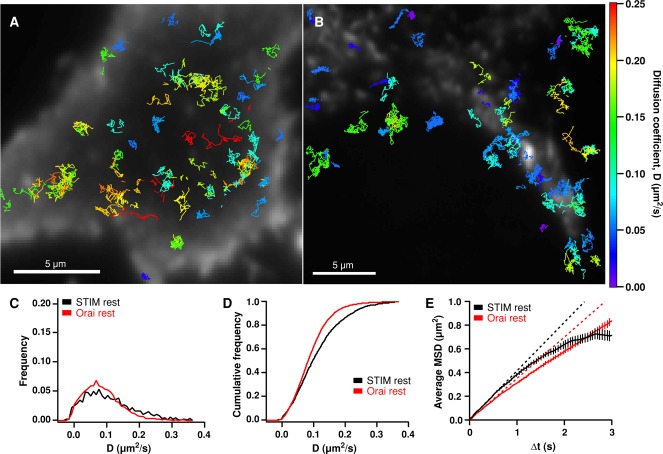
FIGURE 1:
Diffusion of STIM1 and Orai1 in resting cells. (A) A resting HEK cell in 2 mM Ca2+ Ringer's expressing a low level of GFP-STIM1 and a moderate level of mCherry (mCh)-myc-Orai1. Colored GFP-STIM1 tracks are overlaid on the mCh-myc-Orai1 TIRF image. (B) A resting HEK cell expressing a low level of Orai1-GFP and a moderate level of mCh-STIM1. Orai1-GFP tracks are overlaid on the mCh-STIM1 TIRF image. Tracks in A and B are color-coded by their average diffusion coefficient according to the color scale at far right. (C) Diffusion coefficient histograms for STIM1 (black) and Orai1 (red) in resting cells. STIM1 mean D = 0.116 μm2/s (1744 tracks, 3 cells); Orai1 mean D = 0.090 μm2/s (4994 tracks, 6 cells). (D) Cumulative histograms of the diffusion coefficients shown in C. (E) Average MSD vs. ∆t for all STIM1 and Orai1 tracks in resting cells (weighted average ± weighted SD; see Materials and Methods). The dashed lines indicate linear least-squares fits to points 2–5 (∆t = 50–200 ms) of the MSD vs. ∆t curves for STIM1 and Orai1.
We analyzed the mean square displacement (MSD) of single particles over increasing time intervals (∆t) to estimate diffusion coefficients (D) and characterize mobility behavior. For each particle, the MSD versus ∆t was calculated, and the initial slope of the plot (from time intervals of 50–200 ms) was used to estimate the mean D over the duration of the particle's track (Kusumi et al., 1993). The distribution of D values for STIM1 particles in resting cells is shown in Figure 1C, and the cumulative distribution is shown in Figure 1D. The mean D for STIM1 was 0.116 ± 0.002 μm2/s (mean ± SEM, n = 1744 tracks), similar to values derived from FRAP experiments (0.15 μm2/s [ Covington et al., 2010]; 0.1 μm2/s [ Liou et al., 2007]). SPT also reveals the distribution of diffusion coefficients, which is not obtainable by ensemble methods such as FRAP. The distribution of STIM1 diffusion coefficients extends from 0 to >0.4 μm2/s. Some of this variation results from the statistical uncertainty of estimating D given the limited duration of tracks we observed; however, the observed range of Ds is much greater than predicted by statistical uncertainty of a single D, indicating that particles actually do move with a wide range of speeds (Figure S2A). The least and most mobile particles displayed the same narrow distribution of fluorescence intensities, consistent with their being single molecules of STIM1. A defining threshold for “immobility” was estimated from the apparent diffusion coefficients of Orai1-GFP in fixed cells (Figure S3); based on this definition for immobility (D < 0.007 μm2/s), ∼5% of GFP-STIM1 particles in resting cells were considered to be immobile.
The two-dimensional MSD versus ∆t is related to the area a particle explores over time, and the shape of the curve offers additional information about the mode of particle mobility—for example, anomalous diffusion or directed transport—which cannot be readily obtained from FRAP measurements (Kusumi et al., 1993; Saxton and Jacobson, 1997). The MSD versus ∆t curve for STIM1 in resting cells is slightly sublinear (Figure 1E, compare with dashed linear fit). The MSD versus ∆t relation for particles undergoing free diffusion should be linear, as long as the diffusion coefficients are independent of track length. However, under our experimental conditions, slower STIM1 particles tended to yield longer tracks, while faster-moving particles generally had shorter tracks, probably due to movement out of the evanescent field (Figure S4 caption). To assess how this might affect MSD versus ∆t linearity, we performed Monte Carlo simulations to measure the MSD versus ∆t from particles having our experimentally measured D values and track lengths but undergoing purely Brownian motion (Figure S4A). The simulations show that our experimental STIM1 data fall within the sublinearity predicted for free diffusion, suggesting that STIM1 moves by Brownian diffusion in resting cells. To test whether a small subset of particles might move by directed transport (e.g., by attachment via EB1 to growing microtubules), we computed the MSD versus ∆t for the fastest STIM1 particles (D ≥ 0.2 μm2/s). Rather than curving upward as expected from directed transport, the curve remained sublinear (Figure S5), confirming that STIM1 in resting cells moves primarily by free diffusion.
The mean D for Orai1 (0.090 ± 0.001 μm2/s; mean ± SEM, n = 4994 tracks) was slightly lower than that of STIM1, but the D values were widely distributed (Figure 1, C and D). The mean D falls within the range of previous measurements from FRAP (0.07 μm2/s; Park et al., 2009) and SPT (0.13 μm2/s; Madl et al., 2010). Orai1-GFP in resting cells was predominantly mobile (mobile fraction, 96%), somewhat more so than estimated previously by FRAP (83% mobile [ Park et al., 2009]; 91% [ Madl et al., 2010]). The MSD versus ∆t plot for Orai1-GFP is slightly sublinear (Figure 1E). In this case, the sublinearity did not fall within the variation range predicted for free diffusion (Figure S4B), suggesting that Orai1 diffusion is slightly restricted in resting cells.
STIM1 and Orai1 are confined after store depletion
We next studied the behavior of single STIM1 and Orai1 particles in store-depleted cells. After Ca2+ store depletion by treatment with thapsigargin (TG), labeled STIM1 and Orai1 accumulate at ER–PM junctions, generating fluorescent puncta at the light microscope level. To visualize puncta in SPT experiments, we expressed moderate levels of mCherry (mCh)-labeled STIM1 or Orai1 and applied local thresholding to define puncta boundaries (see Materials and Methods). Very low levels of GFP-STIM1 or Orai1-GFP were coexpressed for SPT, and we analyzed the trajectories of particles in relation to puncta.
In store-depleted cells, trajectories of STIM1 and Orai1 were found mostly in puncta and were much more spatially restricted than in resting cells (compare Figure 2, A and B, with Figure 1, A and B). Diffusion coefficients of STIM1 and Orai1 within puncta of TG-treated cells were significantly reduced compared with D values in resting cells (STIM1 mean D = 0.031 ± 0.001 μm2/s, n = 1288 tracks; Orai1 mean D = 0.030 ± 0.001 μm2/s, n = 832 tracks). The mean D value for Orai1 in depleted cells agrees well with the average D measured by FRAP (0.036 ± 0.006 μm2/s; Park et al., 2009). Interestingly, the distributions of D values for STIM1 and Orai1 in store-depleted cells were virtually identical, unlike in resting cells (Figure 2, C and D), consistent with STIM1 and Orai1 moving together as a complex within the ER–PM junction. The size of the immobile fraction also increased significantly, from 5 to 24% for STIM1 particles and from 4 to 25% for Orai1. Possible contributors to the slowing of STIM1 and Orai1 in puncta include complex formation between the two proteins, interactions of STIM1 with the PM, molecular crowding, and diffusion within compartments whose sizes are comparable to the distance traveled within the sampling interval (see Discussion).
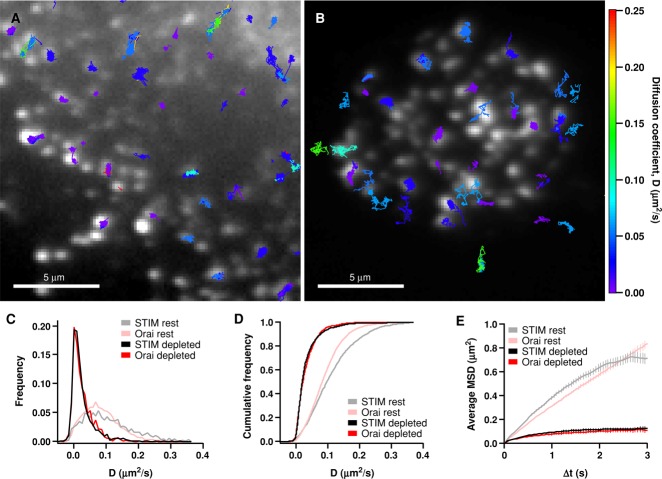
FIGURE 2:
Restricted mobility of STIM1 and Orai1 after store depletion. (A) A store-depleted HEK cell (in 0 Ca2+ Ringer's + 1 μM TG) expressing a low level of GFP-STIM1 and a moderate level of mCh-myc-Orai1. GFP-STIM1 tracks are overlaid on the mCh-myc-Orai1 TIRF image. (B) A TG-treated HEK cell expressing a low level of Orai1-GFP and a moderate level of mCh-STIM1; Orai1-GFP tracks are overlaid on the mCh-STIM1 TIRF image. Tracks in A and B are color-coded by their average diffusion coefficient according to the color scale at right. (C) Diffusion coefficient histograms for STIM1 (1288 tracks, 7 cells) and Orai1 (832 tracks, 6 cells) in store-depleted cells. D values were calculated from junctional sojourns (see Materials and Methods). Histograms from resting cells in Figure 1C are overlaid for comparison. (D) Cumulative histograms of data shown in C. (E) Average MSD vs. ∆t for STIM1 (1225 tracks, 6 cells) and Orai1 (439 tracks, 5 cells) particle trajectories that started in puncta. MSD vs. ∆t graphs from resting cells in Figure 1E are overlaid for comparison.
The slowing of STIM1 and Orai1 diffusion at ER–PM junctions by itself could cause local accumulation at these sites. In a region of reduced mobility but lacking actual diffusion barriers, slowed particles would be expected to accumulate until they reach a concentration at which the rates of particle exit and entry are equal; in this way, particles should accumulate in proportion to the reduction of the diffusion coefficient. The degree of slowing we measured in puncta (approximately threefold for Orai1 and STIM1) was significantly less than the observed accumulation of fluorescent Orai1 and STIM1 (expressed at moderate levels) in puncta after store depletion (∼10-fold; Figure S6). In addition, as described below, the MSD versus ∆t approaches a plateau rather than being linear as would result from an accumulation mechanism based solely on slowing. For these reasons, slowing alone cannot account for the accumulation of STIM1 and Orai1 in puncta.
On the other hand, accumulation is also expected from a diffusion trap mechanism in which the escape of STIM1 and Orai1 from junctions is restricted by a one-way barrier and/or by tethering. A general hallmark of particles diffusing within a restricted space (“corral”) is that the MSD versus ∆t is sublinear and approaches a plateau (Kusumi et al., 1993). Consistent with such a confinement mechanism, the MSD versus ∆t curves for STIM1 and Orai1 were highly nonlinear in depleted cells, in both cases reaching a similar asymptote within several seconds (Figure 2E). These results suggest that STIM1 and Orai1 encounter a common barrier that confines their movement within junctions. The dimensions of the barrier were estimated from the plateau MSD value (Saxton and Jacobson, 1997). For Orai1, the MSD plateau was ∼0.115 μm2 (Figure 2E), which can be approximated by a circular corral with a diameter of 0.68 μm (Saxton and Jacobson, 1997). For STIM1, the estimated corral diameter was 0.76 μm (estimated from an MSD plateau of ∼0.14 μm2). We also used local thresholding to measure the areas of all fluorescent mCh-STIM1 puncta that contained tracked Orai1 particles (see Materials and Methods). In depleted cells, Orai1 particles were located in mCh-STIM1 puncta with mean area of 0.95 μm2 (Figure 4D), equivalent to a circle with a diameter of 1.1 μm. Considering the varying size and shape of puncta, the 0.7- to 0.8-μm corral diameter approximated from the MSD plateaus corresponds reasonably well to the average puncta diameter of ∼1.1 μm, suggesting the boundaries of the diffusion trap correspond to the edges of ER–PM junctions.
FIGURE 4:
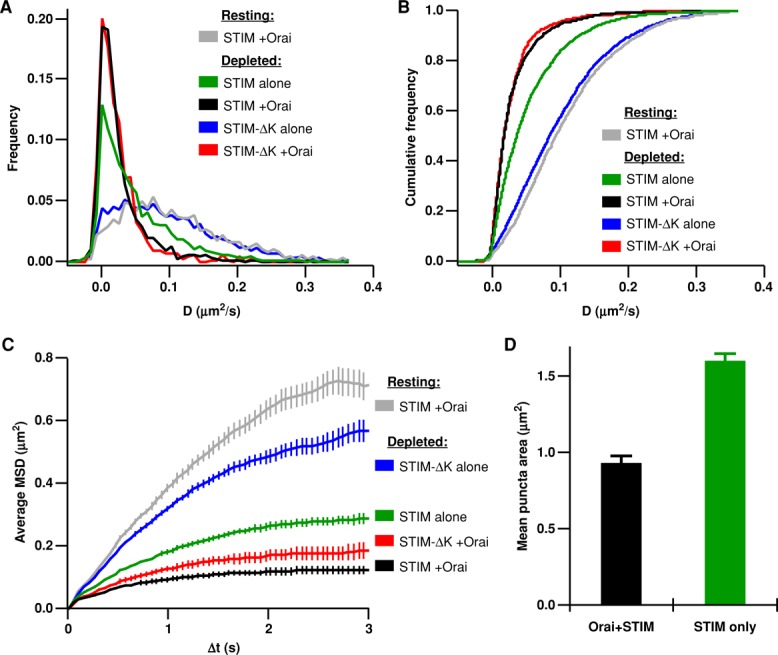
Binding to the PM and Orai1 slows and confines STIM1 diffusion in store-depleted cells. All HEK cells were store depleted by treatment with 1 μM TG + 0 Ca2+ Ringer's. (A) Histograms of diffusion coefficients for STIM1 alone (green, 4077 tracks, 7 cells), STIM1 particles with Orai1 (black, 1288 tracks, 7 cells), STIM1-∆K alone (blue, 3305 tracks, 6 cells), and STIM1-∆K with Orai1 (red, 729 tracks, 6 cells). (B) Cumulative histograms of data shown in A. (C) Average MSD vs. ∆t for STIM1-∆K alone (blue, 3305 tracks, 6 cells), STIM1 alone (green, 3574 tracks, 6 cells), and STIM1-∆K with Orai1 (red, 753 tracks, 6 cells). STIM1 with Orai1 resting (gray) and depleted (black) D histograms and MSD plots are reproduced from Figures 1 and 2. (D) Average area of all puncta containing tracked Orai1 or STIM1 particles, measured from thresholded mCh-STIM1 images (for Orai1+STIM1, n = 439 puncta; for STIM1 only, n = 3574 puncta).
Importantly, SPT offers the unique opportunity to observe the trapping event itself. Figure 3 and Videos S3–S6 show examples of single STIM1 and Orai1 particles undergoing free diffusion before being captured and escaping from a diffusion trap at the ER–PM junction, clearly illustrating the distinction between free diffusion outside junctions and confined diffusion within junctions. As indicated by the D histograms (Figure 2C), the most common behavior within puncta was very low mobility, and these particles bleached without escaping. Escaping particles generally showed a higher mobility within the punctum before escaping.
FIGURE 3:
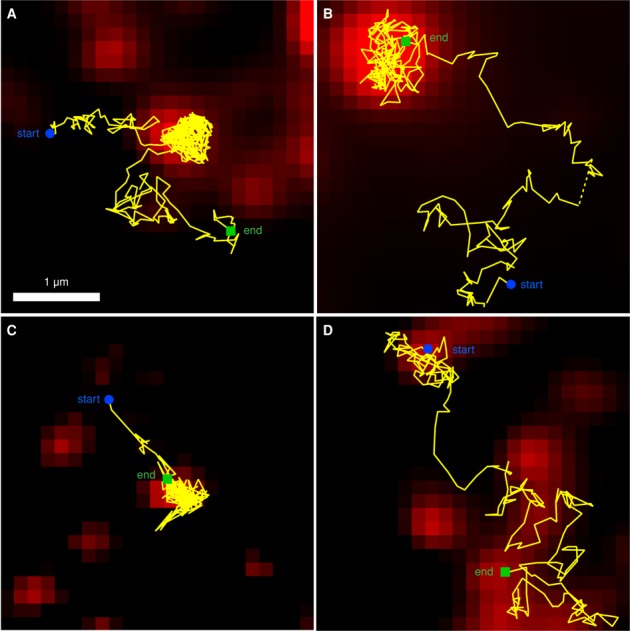
Trapping and escape of single Orai1 and STIM1 particles at ER–PM junctions. All images are from HEK cells treated with 1 μM TG + 0 Ca2+ Ringer's for 3–6 min. (A and B) Orai1-GFP tracks (yellow) overlaid on the corresponding mCh-STIM1 TIRF image (red). (C) GFP-STIM1 track (yellow) overlaid on the mCh-myc-Orai1 TIRF image (red). (D) Orai1-L273D-GFP track (yellow) overlaid on the mCh-STIM1 TIRF image (red). Starting and ending points of tracks are indicated, and dashed yellow lines indicate tracking gaps identified and linked by u-track (see Materials and Methods).
Trapping of STIM1 at ER–PM junctions by interactions with the PM
How do STIM1 and Orai1 become trapped at ER–PM junctions? STIM1 is known to interact with PIP2 in the PM via its C-terminal polybasic domain, providing a means of trapping STIM1 at ER–PM junctions independently of its binding to Orai1 (Liou et al., 2007; Ercan et al., 2009; Park et al., 2009; Walsh et al., 2010). To test this notion, we compared the mobilities of STIM1 and STIM1-ΔK (which lacks the polybasic domain) in the absence of added Orai1.
After store depletion, single GFP-STIM1 particles (without Orai1) were observed in ER–PM junctions, which were identified as fluorescent puncta of coexpressed mCh-STIM1. STIM1 in puncta slowed down after store depletion, but not to the extent seen when Orai1 was present (Figure 4, A and B, green vs. black; STIM1 only, mean D = 0.056 ± 0.001 μm2/s; STIM1+Orai1, mean D = 0.031 ± 0.001 μm2/s). Previous studies have ascribed slowing of STIM1 diffusion after store depletion to oligomerization (Liou et al., 2007; Covington et al., 2010). To distinguish the effects of oligomerization from PM binding, we analyzed the mobility of STIM1-ΔK in store-depleted cells. STIM1-ΔK oligomerizes after store depletion (Figure S7; Liou et al., 2007) but cannot bind the PM and does not redistribute to ER–PM junctions in the absence of Orai1 (Liou et al., 2007; Park et al., 2009). Surprisingly, STIM1-ΔK (without Orai1) did not slow down detectably after store depletion, appearing similar to wild-type (wt)-STIM1 in resting cells (Figure 4, A and B, blue vs. gray; STIM1-ΔK only, depleted mean D = 0.105 ± 0.001 μm2/s, n = 3305 tracks). In depleted cells, STIM1-ΔK molecules only slow down when coexpressed with Orai1, and under these conditions, STIM1-ΔK slows to the same extent as STIM1 + Orai1 (Figure 4, A and B, red vs. black; STIM1-ΔK+Orai, depleted, mean D = 0.029 ± 0.001 μm2/s, n = 729 tracks). These data indicate that binding to the PM (in the absence of Orai1) slows STIM1 mobility, but binding to Orai1 (in the absence of PM binding) slows STIM1 even more (Figure 4B, compare green vs. red), and may account for the overall speed of coexpressed STIM1 and Orai1 in the junctions.
In depleted cells, the MSD versus ∆t plot for STIM1 alone is not linear, but as with STIM1 + Orai1, it approaches an asymptote (Figure 4C, green), showing that interactions with the PM are sufficient to trap STIM1. When PM interactions are disrupted, as in STIM1-ΔK, the MSD versus ∆t curve is not as strongly confined (Figure 4C, blue vs. green). However, in the presence of Orai1, STIM1-∆K becomes trapped, as shown by its MSD versus ∆t curve, which approaches a plateau like those of STIM1 ± Orai1 (Figure 4C, red, green, and black). The MSD versus ∆t plots for STIM1 only and STIM1-∆K + Orai1 plateau at slightly higher levels than STIM1 + Orai1, probably due to the larger average puncta sizes in these experiments (Figure 4D), which would result in larger areas of confinement. Consistent with previous studies (Park et al., 2009; Walsh et al., 2010), our data indicate that either PM binding or Orai1 binding is sufficient to trap STIM1 within the ER–PM junction.
Trapping of Orai1 requires binding to STIM1
Previous studies have suggested that Orai1 becomes trapped at ER–PM junctions through binding of the CAD domain of STIM1 to the C-terminus of Orai1 (Kawasaki et al., 2009; Park et al., 2009; Yuan et al., 2009). To test this model at the single-particle level, we examined the mobility of Orai1-L273D-GFP, an Orai1 mutant that does not bind STIM1 (Li et al., 2011). When expressed at very low levels, Orai1-L273D-GFP is expected to form multimers (presumably hexamers; Hou et al., 2012) with endogenous Orai1, consistent with the low fluorescence intensity and primarily one- to two-step photobleaching of Orai1-L273D-GFP particles (unpublished data). Thus these channels would be expected to have a reduced capacity to bind STIM1 compared with the wild-type (wt)-Orai1 channel. In depleted cells, diffusion of Orai1-L273D-GFP within junctions was nearly as slow as wt-Orai1-GFP (Figure 5, A and B). The slowed diffusion of Orai1-L273D in puncta could be due to STIM1 binding to endogenous wt-Orai1 subunits assembled with Orai1-L273D as well as to molecular crowding within the junction. The MSD versus ∆t graphs clearly show that in depleted cells the Orai1-L273D curves, while displaying anomalous diffusion, do not approach an asymptote as wt-Orai1 does (Figure 5C), even though L273D- and wt-Orai1 molecules start in puncta of equivalent size (Figure 5D). These data indicate that diffusion of Orai1-L273D is significantly less confined than diffusion of wt-Orai1, which is consistent with Orai1-L273D–containing channels having reduced binding to STIM1. Accordingly, we observed more frequent examples of escape of Orai1-L273D than wt-Orai1 from puncta (Figure 3D and Video S7).
FIGURE 5:
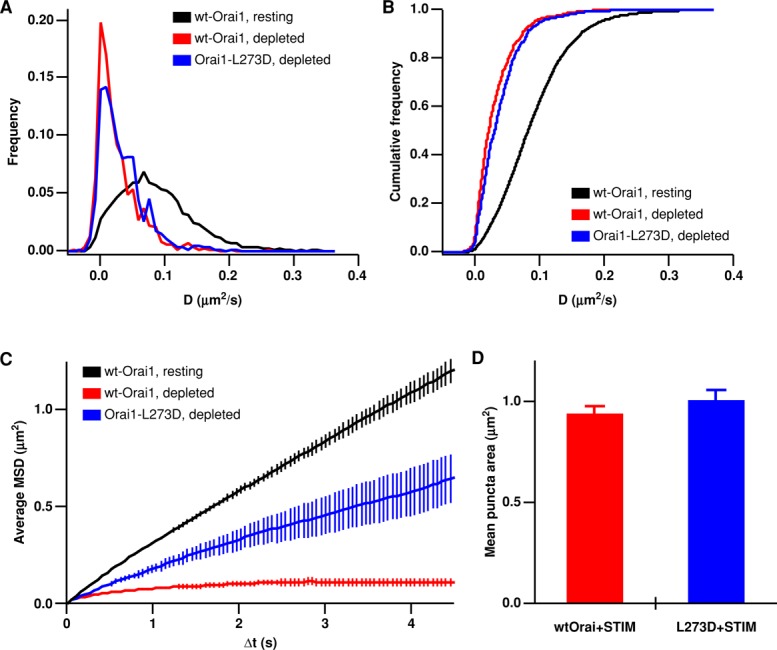
Diminished STIM1 binding affinity allows Orai1-L273D to escape more readily from ER–PM junctions. ER Ca2+ stores in HEK cells were depleted with 1 μM TG in 0 Ca2+ Ringer's, and mCh-STIM1 was expressed at moderate levels in all experiments. (A) Histograms of diffusion coefficients for Orai1 (from Figures 1C and 2C) and Orai1-L273D (blue, 588 tracks, 4 cells) in store-depleted cells. (B) Cumulative histograms of the data shown in (A). (C) Average MSD vs. ∆t for Orai1 and Orai1-L273D (431 tracks, 4 cells) for particle trajectories that started in puncta (for depleted cells). Orai1 MSD plots from resting and depleted cells (black and red) are reproduced from Figure 2. (D) Average area of all puncta containing a tracked Orai particle, measured from thresholded mCh-STIM1 images (for Orai1, n = 439 puncta; for Orai1-L273D, n = 431 puncta).
Dynamic equilibrium of STIM1 and Orai1 across ER–PM junctional boundaries
Our SPT analyses of STIM1 and Orai1 movement show directly that STIM1 and Orai1 accumulate at ER–PM junctions as the result of confinement by a diffusion trap. An important question is to what extent these proteins are able to escape from the trap when Ca2+ stores are depleted. GFP-labeled STIM1 and Orai1 usually bleached within 5 s, making it difficult to capture escape events by SPT. Therefore we estimated the strength of the diffusion trap in two ways: 1) using simulations to estimate the maximal escape probability from the MSD versus ∆t plots and 2) using photoactivation of an ensemble of STIM1 or Orai1 proteins labeled with PAGFP to measure their exit from ER–PM junctions over long time periods (>200 s).
We estimated the escape rate of single particles from junctions using Monte Carlo simulations (see Figure S8). To simulate STIM1 escape from puncta, we placed 1225 particles (the number of particles tracked by SPT) randomly within a circular punctum of diameter 0.68–0.78 μm (dimensions estimated from MSD vs. ∆t graphs from depleted cells, Figure 2E). Particles were assigned diffusion coefficients and track lengths corresponding to values measured experimentally. We ran several simulations using these parameters and varied the escape probability, which specified the fraction of time a particle colliding with the junction boundary was allowed to escape rather than be reflected back into the junction. Even with an escape rate as low as 1%, the simulated MSD versus ∆t curve lacked a clear plateau (Figure S8), unlike our experimental STIM1 and Orai1 data. These results indicate that under our experimental conditions, fewer than 1% of STIM1 and Orai1 particles escape junctions.
Such a tight trap appears to be consistent with observations that STIM1 and Orai1 fluorescence in puncta is constant over minutes in cells with depleted stores (Figure 6). However, constant fluorescence can also result from particles entering and leaving the puncta at the same rate. Furthermore, escape of STIM1 and Orai1 particles may have been limited in the SPT experiments, because each particle was surrounded by a great excess of its binding partner (to facilitate identification of the junctions), whereas levels of endogenous STIM1 and Orai1 are probably more evenly matched. To examine the rate of escape under more physiological conditions, we examined puncta dynamics in cells expressing roughly equal amounts of PAGFP-STIM1 and mCh-Orai1 or PAGFP-Orai1 and mCh-STIM1. In its resting state, PAGFP is minimally fluorescent when excited at 488 nm, but its fluorescence increases ∼100-fold after activation by 405-nm light (Patterson and Lippincott-Schwartz, 2002). Transfected cells were treated with TG, and a 405-nm laser was focused on a mCh-labeled punctum (Figure 7A). A 5- to 20-ms flash activated PAGFP-STIM1 or PAGFP-Orai1 within the punctum, causing the fluorescence to reach a peak and subsequently decay over several minutes, revealing the loss of STIM1 or Orai1 from the punctum by diffusion (Figure 7, B and C). Because the membrane area into which the fluorescent protein diffuses is essentially infinite, the decay of fluorescence from photoactivation approximates the effective leaving rate for STIM1 and Orai1 from puncta. For both proteins, fluorescence decayed with a double-exponential time course with time constants of 22–25 and 357–376 s. The STIM1 exit rate was somewhat slower than that of Orai1 (t1/2 of fluorescence decay was ∼100 s for PAGFP-STIM1 and ∼50 s for PAGFP-myc-Orai1; Figure 7C). Given that the fluorescence intensities of mCh-labeled STIM1 and Orai1 puncta are constant over time (Figure 6), these results imply that when expressed at comparable levels, STIM1 and Orai1 in puncta are in diffusional equilibrium with their surrounding pools.
FIGURE 6:
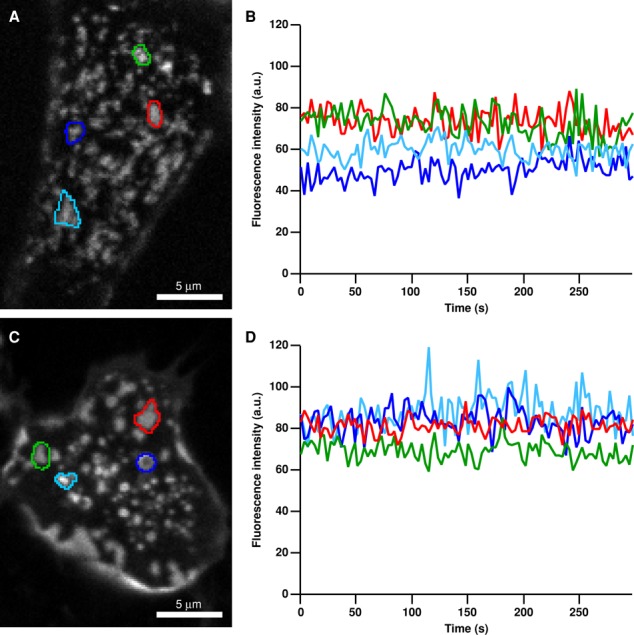
STIM1 and Orai1 puncta fluorescence is stable over time. HEK cells expressing moderate levels of mCh-STIM1 (A and B) or mCh-myc-Orai1(C and D) together with PAGFP-labeled partner protein were store depleted with 1 μM TG in 0 Ca2+ Ringer's and imaged by confocal microscopy over time. Regions of interest marking selected puncta are superimposed on average images of 100-frame movies for mCh-STIM1 (A) and mCh-myc-Orai1 (C). Mean fluorescence vs. time (corrected for photobleaching) of the selected mCh-STIM1 (B) and mCh-myc-Orai1 (D) puncta shows stable puncta fluorescence over >5 min.
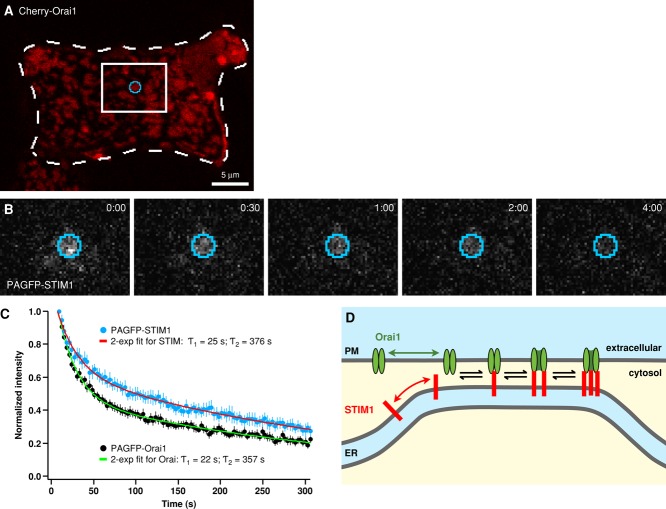
FIGURE 7:
Photoactivation reveals continuous escape of STIM1 and Orai1 across ER–PM junctional boundaries. (A) Cells expressing roughly equal amounts of either mCh-myc-Orai1+PAGFP-STIM1 (shown in A and B) or mCh-STIM1+PAGFP-Orai1 were store depleted, and a punctum (identified by the corresponding mCh marker) was illuminated briefly with a focused 405-nm laser beam to activate PAGFP. Dashed lines outline the cell border; white box indicates the zoomed-in area shown in B. (B) Fluorescence of activated PAGFP-STIM1 within the activation region shown in A (blue) was monitored over time by excitation at 488 nm. Times shown in min:s. (C) Mean fluorescence decay (mean ± SEM) from 15 PAGFP-STIM1 puncta (blue) and 17 PAGFP-Orai1 puncta (black) is shown after correction for photobleaching (∼15% over 300 s) and normalization to the initial fluorescence following photoactivation. Biexponential curves (red and green) were fit by least-squares to each data set; time constants are indicated. Half-times for fluorescence decay were ∼50 s for Orai1 and ∼100 s for STIM1. (D) A simple model to explain dynamic exchange of STIM1 and Orai1 across ER–PM junctional boundaries. Formation of STIM1-Orai1 complexes traps both proteins within the junction as they become constrained by their connections across the junctional gap. Only free STIM1 and Orai1 can escape; free proteins are generated at a slow rate that is controlled by the reversible binding reactions between STIM1 and Orai1 subunits (see Discussion).
DISCUSSION
A diffusion trap mechanism has been widely hypothesized to account for STIM1-Orai1 accumulation at ER–PM junctions but never directly tested. Our analysis of STIM1 and Orai1 movement at the single-particle level, together with photoactivation experiments and Monte Carlo simulations, provides clear evidence for free diffusion of STIM1 and subdiffusion of Orai1 in resting cells and trapping of STIM1 and Orai1 at ER–PM junctions in store-depleted cells. Furthermore, our studies reveal previously unknown behaviors of STIM1 and Orai1, including mobility within junctions and bidirectional exchange across junctional boundaries even when stores are fully depleted.
Diffusion of STIM1 and subdiffusion of Orai1 in resting cells
SPT measurements yielded average D values for STIM1 and Orai1 in resting cells similar to those measured previously by FRAP, validating the single-particle analysis approach. However, SPT also revealed a range of particle speeds and their specific mode of motion, which were not previously detectable. In resting cells, STIM1 moved by Brownian diffusion, as the MSD versus ∆t curve for STIM1 fell within the range of variation expected for freely diffusing particles (Figure S4A). Although STIM1 can associate with the microtubule plus end–tracking protein EB1, and several groups have reported “comets” that move with microtubules in cells overexpressing labeled STIM1 (Baba et al., 2006; Wu et al., 2007; Grigoriev et al., 2008; Smyth et al., 2008), we found no evidence for directed movement of individual STIM1 molecules, even for the fastest-moving STIMs (Figure S5). Thus our results indicate that STIM1 moves primarily by Brownian diffusion in resting cells.
Our SPT data provide the first evidence that Orai1 diffusion in resting cells is subdiffusive and not purely Brownian as had been reported previously (Madl et al., 2010). This discrepancy may reflect the short maximal time interval of ∼250 ms used by Madl et al. (2010); in our experiments, Orai1 diffusion does appear Brownian at 250 ms, but longer trajectories (∼1–2 s) reveal clear subdiffusion (Figure 1E). The diffusion of many if not all PM proteins is thought to be anomalous (Alenghat and Golan, 2013), and some well-studied sources of protein subdiffusion in the PM include association with cytoskeletal networks (Andrews et al., 2008; Jaqaman et al., 2011; Weigel et al., 2011) and transit in and out of microdomains formed either by lipid rafts (Dietrich et al., 2002) or by proteins (Douglass and Vale, 2005; Weigel et al., 2013). Several Orai1-interacting proteins, including calmodulin, CRACR2A, adenylyl cyclase 8, and caveolin (Mullins et al., 2009; Srikanth et al., 2010; Yu et al., 2010; Willoughby et al., 2012), could in principle contribute to anomalous diffusion of Orai1. A recent study found that knockdown of septin proteins alters PM Orai1 distribution in resting cells and disrupts both PIP2 and Orai1 arrangement in the PM of depleted cells (Sharma et al., 2013), raising the possibility that septins may contribute to Orai1's anomalous diffusion. In yeast, septins are required for the formation of the PM diffusion barrier between mother and bud, and there is suggestive evidence that septins are involved in creating diffusion barriers in mammalian cells as well (Caudron and Barral, 2009). More experiments will be needed to assess the effects of septins on Orai1 diffusion in resting and store-depleted cells.
SPT revealed a broad range of D values for both Orai1 and STIM1 in resting cells (from 0 to >0.4 μm2/s), well beyond what would be expected from statistical uncertainty in estimating D (Figure S2). The D distribution we measured for Orai1 in resting cells is similar to the broad ranges of diffusion coefficients measured by SPT for other PM-localized proteins (Douglass and Vale, 2005; Ehlers et al., 2007; Andrews et al., 2008). Together with its subdiffusive behavior, Orai1's range of mobilities implies that the PM contains a heterogeneous mix of Orai1 molecules moving at different speeds due to diverse environmental interactions. STIM1 on average diffuses more slowly than most single-transmembrane ER proteins (Lippincott-Schwartz et al., 2001); this has been attributed to its large cytoplasmic domain interacting with local structures (Liou et al., 2007; Covington et al., 2010). Collisions with sparsely distributed obstacles slow diffusion without creating subdiffusive behavior (Saxton, 1987) and therefore would be consistent with the slow but Brownian nature of STIM1 diffusion. While it is possible that a fraction of the STIM1 particles we measured in resting cells were within ER–PM junctions, it is unlikely that interactions of their polybasic domain with the PM created a population of slower particles, because STIM1-ΔK particles showed a similar range of diffusion coefficients (Figure 4A). Other potential contributors to the wide range of Ds include the effects of heterogeneous protein density in the ER (Anderluh et al., 2014) or differences in STIM1 oligomerization state. While STIM1 is widely considered to be dimeric in resting cells, a subset of monomers has not been ruled out experimentally. Monomers may have higher mobility, although the effect of transmembrane radius on membrane protein diffusion is still debated (Saffman and Delbrück, 1975; Gambin et al., 2006; Weiß et al., 2013).
Characteristics of the STIM1-Orai1 diffusion trap at ER–PM junctions
In addition to showing the junctional trapping of STIM1 and Orai1 directly, SPT analysis reveals important features of the diffusion trap. Slowing of diffusion within ER–PM junctions alone could cause proteins to accumulate and appear to be trapped. However, the high degree of STIM1 and Orai1 accumulation in puncta, threefold more than would be expected due to slowed diffusion alone (Figure S6), and the clearly defined plateau of the MSD versus ∆t curves for STIM1 and Orai1 (Figure 2E) show that accumulation occurs because of a physical confinement of STIM1 and Orai1 within ER–PM junctions. This confinement and the boundaries of the diffusion trap are created by STIM-Orai complex formation and the geometry of the ER–PM junction. STIM-Orai binding is only possible where the ER and PM are close enough (Wu et al., 2006; Orci et al., 2009) to allow the two proteins to physically interact. Thus formation of the complex restricts movement to the junction, and only upon dissociation into free STIM1 or Orai1 can the proteins escape (Figure 7D; see A model for the STIM-Orai diffusion trap below).
Trapped STIM1 and Orai1 were not completely immobilized, but generally dynamic within junctions, with >60% of D values between 0.005 and 0.05 μm2/s. Because the encounter frequency of two proteins is proportional to the sum of their diffusion coefficients (Bell, 1978), mobility within junctions is expected to promote interactions between STIM1 and Orai1 and therefore facilitate STIM-Orai complex formation and dynamics. Several potential factors governing the speed of STIM1 and Orai1 within the junctions are considered below.
Interactions with cytosolic components. STIM1 diffusion in junctions may be slowed not only by interaction of the polybasic domain with the PM but also by interactions with cytosolic components. Our SPT experiments revealed that, even though STIM1-ΔK oligomerizes upon store depletion, it does not slow down (Figures S7 and 4, A and B). In contrast, wt-STIM1 diffusion slows by twofold after store depletion, even when measured in bulk ER, where interaction with the PM would not be expected (Covington et al., 2010). This discrepancy raises the possibility that STIM1's polybasic domain interacts with other proteins after ER Ca2+ depletion to slow its diffusion.
Collisions with junctional boundaries. In SPT experiments, if the sampling interval (∆t) is long enough that a particle collides with the corral boundary (i.e., when 4D∆t > corral area; Ritchie et al., 2005), the particle's position becomes averaged toward the center of the corral, resulting in an underestimate of D. Random walk simulations of STIM1 particles with experimentally measured D values from resting cells placed in a 0.76-μm diameter circular corral showed that collisions with the boundary slow the apparent diffusion rate, but not to the degree that we observed in depleted cells (Figure S9). In particular, boundary collisions cannot account for the large increase in immobile particles we observe upon store depletion.
Molecular crowding. Slowed and anomalous diffusion due to immobile or mobile obstacles is well recognized (Saxton, 1987; Calvert et al., 2001; Frick et al., 2007) and could contribute to the behavior of STIM1 and Orai1 in puncta. In a study of overexpressed STIM1 and Orai1 in HEK cells, the estimated distance between Orai1 channels in puncta was estimated to be ∼40 nm (Ji et al., 2008); at this density, molecular crowding would be likely to slow diffusion. Further experiments will be required to determine how much the junctional protein density slows diffusion and whether crowding is significant at physiological concentrations of STIM1 and Orai1.
-
STIM-Orai complex formation. Several lines of evidence suggest that the mobility of STIM1 and Orai1 are significantly reduced by formation of the STIM-Orai complex. In the absence of Orai1, STIM1 diffusion in depleted cells is slowed, but not as much as when Orai1 is coexpressed (Figure 4, A and B). When both proteins are present in store-depleted cells, STIM1 and Orai1 D histograms are indistinguishable, consistent with the idea that complexes are mostly mobile and that complex assembly drastically slows diffusion of both proteins (Figure 2, C and D). In fact, STIM1-ΔK in the presence of Orai1 diffuses with the same range of speeds as wt-STIM1 + Orai1 (Figure 4A). Because STIM1-ΔK cannot bind the PM, its mobility is primarily determined by forming complexes with Orai1. Finally, the slowing of STIM1 and Orai1 by store depletion is quantitatively consistent with the increased drag caused by binding of 3 STIM1 dimers to a hexameric Orai1 channel (based on the dOrai structure of Hou et al. [2012] and the proposed binding stoichiometry of Stathopulos et al. [2013]). In this case,
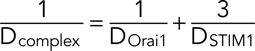
where Dcomplex is the predicted D of the STIM-Orai complex, and DOrai1 and DSTIM1 are the respective mean diffusion coefficients measured in resting cells (0.09 and 0.116 μm2/s). Dcomplex = 0.027 μm2/s, close to the measured mean D of 0.03 μm2/s. Given that we expressed an excess of mCh-STIM1 to label puncta with low amounts of Orai1-GFP for SPT, most channels were likely maximally bound to 2–3 dimers of STIM1. The minority of more mobile Orai1 particles may therefore represent channels with 0 or 1 STIM1 dimer bound. This notion may also explain why faster particles were more likely to escape the puncta (e.g., Figure 3A).
A model for the STIM-Orai diffusion trap
Our SPT and PAGFP data are consistent with a diffusion trap created by STIM-Orai complex formation at ER–PM junctions (Figure 7D). This working model has several major tenets: 1) STIM-Orai complexes can diffuse only within areas where the ER–PM intermembrane distance is compatible with STIM-Orai binding; 2) STIM and Orai are in a state of dynamic equilibrium in which STIM1 continually binds and unbinds from Orai1; and 3) only free STIM1 and Orai1 can escape from the junction. This model makes several key predictions about the escape of proteins from puncta that are consistent with our experimental results:
At high STIM:Orai ratios, the STIM-Orai equilibrium will be biased heavily toward complex formation; there will be little free Orai1 in junctions, so it will rarely escape the trap. Correspondingly, in SPT experiments in which Orai1 was expressed at very low levels compared with mCh-STIM1, escape events were quite rare (<1% escape probability; Figure S8).
Reducing the number or affinity of STIM1 binding sites on Orai1 channels should increase the proportion of free Orai1, thus increasing the frequency of Orai1 escape. This prediction was confirmed in the Orai1-L273D SPT experiments. Under low expression conditions, Orai1-L273D would be expected to combine with endogenous wt-Orai1 to form heteromeric channels with partially impaired STIM1 binding. In agreement with the model, particles containing Orai1-L273D showed significantly less confinement than wt-Orai1 (Figure 5C).
At roughly equal expression of STIM1 and Orai1, a shift toward free STIM1 and Orai1 in junctions will occur, enabling the exchange of STIM1 and Orai1 across the junctional boundary. This prediction was borne out in our photoactivation experiments, in which STIM1 and Orai1 were expressed at approximately equal amounts, and both proteins left the puncta at moderate rates (Figure 7C). The biexponential decay of PAGFP-Orai1 in the puncta can be explained by a kinetic scheme in which the fast exponential component largely reflects the escape of the Orai1 pool that was free at the time of photoactivation, while the slow phase reflects the kinetics of Orai1 reequilibration among the various bound states to generate new free molecules that can then escape (Figure 7D).
STIM1 is retained at junctions not only by interactions with Orai1 but also by binding to PIP2. Considering that STIM1 alone is tightly confined to junctions through binding to PIP2 (Figure 4C), it is surprising that STIM1 (in the presence of Orai1) escapes the junctions in our photoactivation experiments. A possible resolution to this discrepancy is suggested by recent findings that in cells overexpressing STIM1 and Orai1, PIP2 was reduced in puncta where STIM1 and Orai1 colocalized, and this effect was dependent on septins (Sharma et al., 2013). Thus a reduction of PIP2 in the junctions may account for the increased escape frequency of STIM1 in the photoactivation experiments, and the somewhat slower time course of STIM1 escape relative to Orai1 may reflect slowing of STIM1 by reversible binding to residual PIP2.
Given the high efficiency of STIM-Orai accumulation in puncta, and the constancy of STIM/Orai puncta fluorescence over time, it is surprising that the diffusion trap for STIM1 and Orai1 is so dynamic. Our SPT and photoactivation studies suggest that complexes form with varying stoichiometries and move randomly inside junctional boundaries while stochastically binding and unbinding from their partners. Because free STIM1 and free Orai1 are at diffusional equilibrium across junctional boundaries, when a free protein is generated in the junction, it has a chance to escape if it reaches the edge before it rebinds to a partner. These behaviors suggest that STIM1 and Orai1 interact with relatively low affinity, perhaps allowing for modulation of SOCE, even under conditions of maintained Ca2+ store depletion. Further analysis of STIM1 and Orai1 movement within and across junctional boundaries (e.g., in the presence of calcium influx or the absence of accessory proteins) may offer new insights into the dynamic control of STIM-Orai binding and Orai channel activation.
MATERIALS AND METHODS
Plasmids, cells, and solutions
GFP-myc-Orai1, mCh-STIM1, GFP-STIM1, and mCh-STIM1-∆K have been described previously (Luik et al., 2006; Wu et al., 2006; Park et al., 2009). Human STIM1 was from Origene (Rockville, MD). Cyan fluorescent protein (CFP)-STIM1 and yellow fluorescent protein (YFP)-STIM1 were gifts from T. Meyer (Stanford University); Orai1-GFP was a gift from T. Xu (Chinese Academy of Sciences, Beijing, China), and mCh-myc-Orai1 was from C. Y. Park and R. Dolmetsch (Stanford University). STIM1-∆K, CFP-STIM1-∆K, GFP-STIM1-∆K, and YFP-STIM1-∆K were made by site-directed mutagenesis (QuikchangeII XL; Agilent Technologies, Santa Clara, CA) of STIM1, CFP-STIM1, GFP-STIM1, or YFP-STIM1 (respectively) to introduce a premature stop codon after aa 670 with primer 5′-GACTCCAGCCCAGGCTGAAAGAAGTTTCCCCTC-3′. Orai1-L273D-GFP was made by mutagenesis of Orai1-GFP using primer 5′-ACCGACAGTTCCAGGAGGACAACGAGCTGGCGGAG-3′. PAGFP-myc-Orai1 and PAGFP-STIM1 were made from GFP-myc-Orai1 or GFP-STIM1, respectively. Three rounds of mutagenesis were made to introduce four point mutations (L64F, T65S, V163A, and T203H) into GFP to convey photoactivatability (Patterson and Lippincott-Schwartz, 2002) using the following primers:
for L64F and T65S (QuikChangeII XL):
5′-CCCTCGTGACCACCTTCAGCTACGGCGTGCAGTG-3′
for V163A (QuikChangeII XL):
5′-GAAGAACGGCATCAAGGCCAACTTCAAGATCCGCC-3′
for T203H (QuikChange Multi):
5′-AACCACTACCTGAGCCACCAGTCCGCCCTGAG-3′
PAGFP-STIM1 was PCR-amplified and cloned into pcDNA5/FRT/TO vector to construct an inducible PAGFP-STIM1–stable HEK cell line using the Flp-In T-REx system (Life Technologies, Carlsbad, CA). HEK 293 cells (American Type Culture Collection, Manassas, VA) and HEK 293 cell lines stably expressing inducible mCh-STIM1 (Hoover and Lewis, 2011) or inducible PAGFP-STIM1 were cultured as previously described (Park et al., 2009; Hoover and Lewis, 2011). For stable cell lines, protein expression (of mCh-STIM1 or PAGFP-STIM1) was induced with 1 μg/ml tetracycline for 18–25 h before imaging.
Cells were imaged in 2 Ca Ringer's solution containing (in mM): 155 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 10 d-glucose, and 5 Na-HEPES (pH 7.4). For depletion of Ca2+ stores, cells were perfused with 0 Ca Ringer's solution prepared with 1 μM TG or ionomycin (EMD, Darmstadt, Germany) and (in mM): 155 NaCl, 4.5 KCl, 3 MgCl2, 1 EGTA, 10 d-glucose, and 5 Na-HEPES (pH 7.4).
Cell transfections
For Förster resonance energy transfer (FRET) and photoactivation experiments, 6-cm dishes of HEK cells were transfected with 0.66–1.5 μg total DNA (e.g., 0.33 μg CFP-STIM1 + 0.33 μg YFP-STIM1) with Lipofectamine 2000 (Life Technologies) for 5–6 h. Cells were plated on coverslips coated with poly-d-lysine (Sigma-Aldrich, St. Louis, MO), and experiments were performed 24- to 48-h posttransfection.
For SPT experiments, 0.08–0.20 μg of plasmid DNA encoding GFP-labeled protein (e.g., GFP-STIM1, Orai1-GFP) was transiently cotransfected with 0.20–0.40 μg of DNA encoding mCh-labeled proteins (e.g., mCh-myc-Orai1, mCh-STIM1, used to mark location of ER–PM junctions in depleted cells) into HEK cells with Lipofectamine 2000 for 3–5 h. In some experiments, unlabeled STIM1 was cotransfected to ensure proper puncta formation. For example, in the GFP-STIM1 SPT experiments, in which GFP-STIM1 expression was low (0.08–0.15 μg), unlabeled STIM1 (0.3–0.5 μg) and mCh-myc-Orai1 (∼0.2 μg) were both cotransfected. GFP-STIM1–only SPT experiments were performed using an inducible mCh-STIM1–stable HEK cell line (induced with 1 μg/ml tetracycline for 18–25 h; Hoover and Lewis, 2011) to mark puncta. For GFP-STIM1-∆K SPT experiments, unlabeled STIM1-∆K (0.10–0.34 μg) and/or mCh-STIM1-∆K (0.25 μg) were cotransfected to discourage GFP-STIM1-∆K from oligomerizing with endogenous wt-STIM1.
Cells showed a range of expression levels; higher expressers clearly showed GFP-STIM1 correctly localized in the ER and Orai1-GFP and GFP-myc-Orai1 in the PM. For single-molecule imaging, transfected cells were plated 2–20 h before imaging on coverslips (high performance from Zeiss, Jena, Germany) that were sonicated in 2% RBS detergent (Pierce, Rockford, IL), rinsed several times in nanopure water, aspirated dry, coated with filtered poly-d-lysine for 30 min at 37°C, and rinsed well with phosphate-buffered saline. We imaged cells either <24 h or >72 h posttransfection, when cells expressed low enough levels of GFP suitable for single-molecule imaging (Videos S1–S3).
Photoactivation
HEK cells stably expressing mCh-STIM1 or PAGFP-STIM1 were transiently transfected with 1–1.5 μg PAGFP-myc-Orai1 or mCh-myc-Orai1, respectively, and imaged 24 h posttransfection on a Leica (Wetzlar, Germany) SP2 AOBS confocal microscope with HCS PL Apo Cs 100× oil-immersion objective (1.4 NA). Cells were bathed in 0 Ca2+ Ringer's solution + 1 μM TG to deplete stores. mCh-STIM1 or mCh-myc-Orai1 fluorescence (excited with a 594-nm line of a 2 mW HeNe laser, PMT collection from 610 to 840 nm) was used to locate puncta for photoactivation of PAGFP-myc-Orai1 or PAGFP-STIM1, respectively. The selected punctum was illuminated for 5–25 ms with a 405-nm diode laser at 100% power (pinhole set to 1 Airy unit) to photoactivate PAGFP. Laser-spot size (∼1.5–1.8 μm2) was measured by photobleaching diffuse GFP-myc-Orai1 in resting cells fixed with 4% paraformaldehyde. PAGFP fluorescence was monitored with 488-nm excitation at low power (5–25% of 20 mW Ar laser, PMT collection set to 500–570 nm for GFP). Fluorescence decay within a 10-pixel (∼1.5-μm diameter) circle surrounding the punctum was analyzed using ImageJ (Schneider et al., 2012). Before analysis, all fluorescence decay data were background subtracted and corrected for photobleaching (∼15% over 300 s) using fixed cells expressing the same PAGFP construct and imaged with identical laser settings. Experiments were performed at 22–25°C.
FRET
HEK 293 cells expressing CFP- and YFP-labeled STIM1 or STIM1-∆K were imaged at 22–25°C 24–48 h posttransfection using the three-cube FRET (E-FRET) method as described by Covington et al. (2010), except the YFP excitation filter was 490 ± 5 nm (Chroma, Bellows Falls, VT). FRET efficiency (FRET-E) was calculated as described by Zal and Gascoigne (2004), using the following measured bleedthrough factors: a = 0.150 ± 0.002 (n = 30 cells), b = 0.021 ± 0.005 (n = 30), c = 0, d = 0.442 ± 0.004 (n = 43), and G parameter = 1.83 ± 0.06 (n = 21). FRET-E values are expressed as mean ± SEM.
TIRF and single-particle imaging
GFP and mCh were imaged at 22–25°C with through-the-objective TIRF microscopy using a microscope (Axiovert S100TV; Zeiss) with a Fluar 100×, 1.45 NA oil-immersion objective (Zeiss). The 488-nm and/or 561-nm laser light (Coherent, Santa Clara, CA) was filtered, combined, expanded (10× beam expander; Edmund Optics, Barrington, NJ), and focused at the back focal plane of the objective. For simultaneous excitation of GFP and mCh, we used a Di01-R405/488/561/533 dichroic (Semrock, Rochester, NY). For simultaneous collection of GFP and mCh images, fluorescence emission was split using an Optosplit-II (Cairn Research, Faversham, UK) containing dichroic (FF580-FDi01; Semrock) and emission filters for GFP (FF02-525/50; Semrock) and mCh (E600LP; Chroma). In some experiments, a neutral density filter was placed in the mCh emission path of the Optosplit to prevent mCh image saturation. In some experiments 488-nm excitation alone was sufficient to excite both GFP and mCh. Images were acquired with an Andor (Belfast, UK) iXon DU897E electron multiplying charge-coupled device camera, with EM gain set to 1000 (maximum). Laser shutters and image acquisition were controlled by Micro-Manager (Edelstein et al., 2010). All fluorescence images were acquired with 1 × 1 pixel binning.
To image single molecules of STIM1 or Orai1, we tested various 488-nm TIRF illumination settings and frame exposures from 10 to 100 ms and found that a 488-nm laser power density (at the coverslip) of 30 W/cm2 and ∼50 ms camera exposure yielded the best balance between signal-to-noise ratio and GFP lifetime before bleaching. Continuous bursts of 1000 or 2000 images (at 51.74 ms/frame) were collected to disk. To minimize the overlap among particle trajectories in cells expressing a higher than ideal density of GFP molecules, we applied 488 laser illumination before burst movie acquisition to photobleach and reduce the number of fluorescent particles.
Single-particle tracking
Simultaneously acquired GFP and mCh burst movies were aligned to each other using fluorescent bead images acquired on the same day (100-nm TetraSpeck Microspheres; Life Technologies) and the MultiStackReg plugin (Micheva et al., 2010) for Fiji (Schindelin et al., 2012). u-track software (Jaqaman et al., 2008) was applied to GFP burst movies to detect single particles at subpixel resolution and to link them across frames into trajectories (“tracks”). Gaps occurred due to intermittent transitions to and from a nonfluorescent dark state (“blinking”). Tracks were linked across gaps using parameters that optimized the accuracy of track identification. Tracks generally terminated due to irreversible GFP bleaching, and lengths were approximately exponentially distributed with means from 1.8 s (GFP-STIM1 in resting cells) to 4.1 s (Orai1-GFP in depleted cells). Our localization precision was ∼30 nm in both x and y dimensions based on the precision of Gaussian fitting to particle profiles in u-track (Jaqaman et al., 2008).
SPT analysis
Analysis of SPT data obtained with u-track was performed using custom procedures written in Igor Pro (Wavemetrics, Portland, OR). Each particle's MSD versus n∆t was calculated by averaging the square displacements over all pairs of points in the track for increasing time intervals, where ∆t was the sampling interval in our experiments, 51.74 ms, and n the number of time intervals. For each tracked particle, the average microscale diffusion coefficient, D, was estimated from the initial slope of the MSD versus ∆t curve (D = MSD/4∆t), between points 2 and 5 (∼50–200 ms). To collectively analyze SPT data acquired under the same experimental conditions, we combined individual MSD versus ∆t curves into an ensemble average MSD versus ∆t. Because the MSDs for short time intervals (which will be the mean of many points) are more reliable than the MSDs for longer time intervals (means of fewer points), all average MSD versus ∆t plots are displayed as a weighted ensemble average MSD (± weighted SD), where each individual MSD was weighted by that particle's track length. Only particles with tracks longer than 20 points (1.03 s) were included in the ensemble average MSD versus ∆t calculations.
SPT analysis based on particle location relative to ER–PM junctions
For all analyses of STIM1 or Orai1 tracks in resting cells, we computed MSD versus ∆t and diffusion coefficients from all tracks (e.g., Figure 1, C–E). For SPT analysis in store-depleted cells, STIM1 and Orai1 tracks were analyzed based on their location relative to ER–PM junctions. We defined ER–PM junction boundaries by summing all mCh-STIM1 or mCh-myc-Orai1 frames in a burst movie and thresholding the summed mCh image using the Bernsen local contrast method (radius = 5) in Fiji. The Bernsen method produced thresholded images that very closely approximated the location of puncta of mCh-STIM1 or mCh-myc-Orai1 (Figure S10). Any time the x-y position of a STIM1 or Orai1 particle was in a white pixel (Figure S10B), that particle was considered to be in a punctum or ER–PM junction. For all SPT analyses in store-depleted cells, we analyzed either junctional sojourns or tracks starting in puncta (see explanations below), with the exception of STIM1-∆K (without Orai) in depleted cells. Because STIM1-∆K does not form puncta in the absence of Orai, we calculated MSD versus ∆t graphs and D values from all tracks for STIM1-∆K (–Orai).
When estimating the D values of STIM1 and Orai1 within junctions in depleted cells, we analyzed junctional sojourns, which we defined as only the segments of tracks within puncta (delineated by Bernsen thresholding). Tracks may not have started or ended in puncta, but only the time intervals spent within puncta were included in the MSD versus ∆t curves used to compute D values in store-depleted cells (Figures 2, C and D, 4, A and B, and 5, A and B). This procedure provided an estimate of the mean D for each particle only during the time it was within a junction.
For determining whether STIM1 or Orai1 in depleted cells experienced restricted diffusion, only tracks starting in puncta were included in the MSD versus ∆t graphs (e.g., Figures 2E, 4C, and 5C). MSD versus ∆t was calculated from the entire particle's track, even if the particle left the punctum, as long as the track started in a punctum. If we limit the MSD versus ∆t graph to only junctional sojourns, the curve approaches an asymptote that is artificially imposed by the Bernsen-thresholded image, because the MSD versus ∆t plot is limited to track segments that never leave puncta boundaries (Figure S10, D and E, blue curves). If we assume the edges of the corral are determined by the edges of ER–PM junctions, then comparing the MSD versus ∆t graphs of Orai1 tracks starting in puncta versus junctional sojourns tells us how well the Bernsen threshold method approximates the size of the ER–PM junctions. Figure S10, D and E, shows that the Orai1 and STIM1 MSD versus ∆t curves for junctional sojourns reach asymptotes slightly faster than the curves for tracks starting in puncta. The lower plateaus of the MSD versus ∆t curves for junctional sojourns would estimate smaller confinement radii than the MSD plateaus for tracks starting in puncta, indicating that the Bernsen threshold slightly underestimates the size of ER–PM junctions.
Diffusion simulations
Monte Carlo simulations of diffusion were performed using custom procedures written in Igor Pro (Wavemetrics). For each time interval ∆t (set to 51.74 ms as in our experiments), new particle positions were calculated based on a step size of (4D∆t)1/2 and a randomly chosen trajectory angle (0–360o). All particles were either assigned a common D and track length, or, in some simulations, each particle was given a unique D and track length from an experimentally analyzed STIM1 or Orai1 particle. For simulations of escape from puncta, when particles crossed the simulated boundary they were reflected back into the punctum with a frequency of 1 − Pesc, where Pesc is the escape probability.
Supplementary Material
Acknowledgments
We thank Paul Hoover and Marija Vrljic for initiating our foray into single-particle imaging, Ludmila Lokteva for making the STIM-∆K and Orai1-L273D constructs, Khuloud Jaqaman for help with u-track, and members of the Lewis Lab for helpful discussions and comments on the manuscript. Photoactivation experiments were performed in the Cell Sciences Imaging Facility at Stanford. This work was supported by National Institutes of Health grant R37GM45374 and the Mathers Charitable Foundation.
Abbreviations used:
- CAD
CRAC activation domain
- CFP, cyan fluorescent protein; CRAC
calcium release–activated calcium
- D
diffusion coefficient
- ER
endoplasmic reticulum
- FRAP
fluorescence recovery after photobleaching
- FRET, Förster resonance energy transfer; GFP
green fluorescent protein
- mCh
mCherry
- MSD
mean square displacement
- PAGFP
photoactivatable GFP
- PIP2, phosphatidylinositol 4,5-bisphosphate; PM
plasma membrane
- SOCE
store-operated calcium entry
- SPT
single-particle tracking
- STIM1
stromal interaction protein 1
- TG
thapsigargin
- TIRF
total internal reflection fluorescence; YFP, yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1107) on July 23, 2014.
REFERENCES
- Alenghat FJ, Golan DE. Membrane protein dynamics and functional implications in mammalian cells. Curr Top Membr. 2013;72:89–120. doi: 10.1016/B978-0-12-417027-8.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderluh A, Klotzsch E, Ries J, Reismann AW, Weber S, Fölser M, Koban F, Freissmuth M, Sitte HH, Schütz GJ. Tracking single serotonin transporter molecules at the endoplasmic reticulum and plasma membrane. Biophys J. 2014;106:L33–L35. doi: 10.1016/j.bpj.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NL, Lidke KA, Pfeiffer JR, Burns AR, Wilson BS, Oliver JM, Lidke DS. Actin restricts FcεRI diffusion and facilitates antigen-induced receptor immobilization. Nat Cell Biol. 2008;10:955–963. doi: 10.1038/ncb1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Hayashi K, Fujii Y, Mizushima A, Watarai H, Wakamori M, Numaga T, Mori Y, Iino M, et al. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Calvert PD, Govardovskii VI, Krasnoperova N, Anderson RE, Lem J, Makino CL. Membrane protein diffusion sets the speed of rod phototransduction. Nature. 2001;411:90–94. doi: 10.1038/35075083. [DOI] [PubMed] [Google Scholar]
- Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Chvanov M, Walsh CM, Haynes LP, Voronina SG, Lur G, Gerasimenko OV, Barraclough R, Rudland PS, Petersen OH, Burgoyne RD, et al. ATP depletion induces translocation of STIM1 to puncta and formation of STIM1-ORAI1 clusters: translocation and re-translocation of STIM1 does not require ATP. Pflugers Arch. 2008;457:505–517. doi: 10.1007/s00424-008-0529-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington ED, Wu MM, Lewis RS. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol Biol Cell. 2010;21:1897–1907. doi: 10.1091/mbc.E10-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C, Yang B, Fujiwara T, Kusumi A, Jacobson K. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys J. 2002;82:274–284. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010;Chap 14 doi: 10.1002/0471142727.mb1420s92. Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD, Heine M, Groc L, Lee MC, Choquet D. Diffusional trapping of GluR1 AMPA receptors by input-specific synaptic activity. Neuron. 2007;54:447–460. doi: 10.1016/j.neuron.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan E, Momburg F, Engel U, Temmerman K, Nickel W, Seedorf M. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic. 2009;10:1802–1818. doi: 10.1111/j.1600-0854.2009.00995.x. [DOI] [PubMed] [Google Scholar]
- Frick M, Schmidt K, Nichols BJ. Modulation of lateral diffusion in the plasma membrane by protein density. Curr Biol. 2007;17:462–467. doi: 10.1016/j.cub.2007.01.069. [DOI] [PubMed] [Google Scholar]
- Gambin Y, Lopez-Esparza R, Reffay M, Sierecki E, Gov NS, Genest M, Hodges RS, Urbach W. Lateral mobility of proteins in liquid membranes revisited. Proc Natl Acad Sci USA. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JW, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover PJ, Lewis RS. Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci USA. 2011;108:13299–13304. doi: 10.1073/pnas.1101664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, Kuwata H, Touret N, Collins R, Trimble WS, Danuser G, Grinstein S. Cytoskeletal control of CD36 diffusion promotes its receptor and signaling function. Cell. 2011;146:593–606. doi: 10.1016/j.cell.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Xu P, Li Z, Lu J, Liu L, Zhan Y, Chen Y, Hille B, Xu T, Chen L. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc Natl Acad Sci USA. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun. 2009;385:49–54. doi: 10.1016/j.bbrc.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski MK, Manjarrés IM, Varnai P, Balla T. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci Signal. 2010;3:ra82. doi: 10.1126/scisignal.2001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Sako Y, Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RS. Store-operated calcium channels: new perspectives on mechanism and function. Cold Spring Harb Perspect Biol. 2011;3:a003970. doi: 10.1101/cshperspect.a003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu L, Deng Y, Ji W, Du W, Xu P, Chen L, Xu T. Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits. Cell Res. 2011;21:305–315. doi: 10.1038/cr.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Luik R, Wang B, Prakriya M, Wu M, Lewis R. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik R, Wu M, Buchanan J, Lewis R. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl J, Weghuber J, Fritsch R, Derler I, Fahrner M, Frischauf I, Lackner B, Romanin C, Schütz GJ. Resting-state Orai1 diffuses as homotetramer in the plasma membrane of live mammalian cells. J Biol Chem. 2010;285:41135–41142. doi: 10.1074/jbc.M110.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva D, O'Rourke N, Busse B, Smith J. Array tomography: semiautomated image alignment. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5527. 11pdb.prot5527. [DOI] [PubMed] [Google Scholar]
- Muik M, Fahrner M, Derler I, Schindl R, Bergsmann J, Frischauf I, Groschner K, Romanin C. A cytosolic homomerization and a modulatory domain within STIM1 C terminus determine coupling to ORAI1 channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins F, Park C, Dolmetsch R, Lewis R. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc Natl Acad Sci USA. 2009;106:15495–15500. doi: 10.1073/pnas.0906781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Rock RS, Spudich JA. A myosin II mutation uncouples ATPase activity from motility and shortens step size. Nat Cell Biol. 2001;3:311–315. doi: 10.1038/35060110. [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Le Coadic M, Shen W-W, Demaurex N, Cosson P. STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:19358–19362. doi: 10.1073/pnas.0911280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh AB, Putney JW. Store-operated calcium channels. Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Ritchie K, Shan XY, Kondo J, Iwasawa K, Fujiwara T, Kusumi A. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys J. 2005;88:2266–2277. doi: 10.1529/biophysj.104.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman PG, Delbrück M. Brownian motion in biological membranes. Proc Natl Acad Sci USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Lateral diffusion in an archipelago. The effect of mobile obstacles. Biophys J. 1987;52:989–997. doi: 10.1016/S0006-3495(87)83291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Quintana A, Findlay GM, Mettlen M, Baust B, Jain M, Nilsson R, Rao A, Hogan PG. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Feske S. Regulation of lymphocyte function by ORAI and STIM proteins in infection and autoimmunity. J Physiol. 2012;590:4157–4167. doi: 10.1113/jphysiol.2012.233221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth J, Dehaven W, Bird G, Putney J., Jr Ca2+-store-dependent and -independent reversal of Stim1 localization and function. J Cell Sci. 2008;121:762–772. doi: 10.1242/jcs.023903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S, Jung H-J, Kim K-D, Souda P, Whitelegge J, Gwack Y. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat Cell Biol. 2010;12:436–446. doi: 10.1038/ncb2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos P, Li G, Plevin M, Ames J, Ikura M. Stored Ca2+ depletion-induced oligomerization of STIM1 via the EF-SAM region: an initiation mechanism for capacitive Ca2+ entry. J Biol Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- Stathopulos PB, Schindl R, Fahrner M, Zheng L, Gasmi-Seabrook GM, Muik M, Romanin C, Ikura M. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat Commun. 2013;4:2963. doi: 10.1038/ncomms3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P, Tóth B, Tóth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- Walsh CM, Chvanov M, Haynes LP, Petersen OH, Tepikin AV, Burgoyne RD. Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry. Biochem J. 2010;425:159–168. doi: 10.1042/BJ20090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel AV, Simon B, Tamkun MM, Krapf D. Ergodic and nonergodic processes coexist in the plasma membrane as observed by single-molecule tracking. Proc Natl Acad Sci USA. 2011;108:6438–6443. doi: 10.1073/pnas.1016325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel AV, Tamkun MM, Krapf D. Quantifying the dynamic interactions between a clathrin-coated pit and cargo molecules. Proc Natl Acad Sci USA. 2013;110:E4591–E4600. doi: 10.1073/pnas.1315202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiß K, Neef A, Van Q, Kramer S, Gregor I, Enderlein J. Quantifying the diffusion of membrane proteins and peptides in black lipid membranes with 2-focus fluorescence correlation spectroscopy. Biophys J. 2013;105:455–462. doi: 10.1016/j.bpj.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby D, Everett L, Halls L, Pacheco J, Skroblin P, Vaca L, Klussmann E, Cooper DMF. Direct binding between Orai1 and AC8 mediates dynamic interplay between Ca2+ and cAMP signaling. Sci Signal. 2012;5:ra29. doi: 10.1126/scisignal.2002299. [DOI] [PubMed] [Google Scholar]
- Wu M, Buchanan J, Luik R, Lewis R. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Luik R, Lewis R. Some assembly required: constructing the elementary units of store-operated Ca2+ entry. Cell Calcium. 2007;42:163–172. doi: 10.1016/j.ceca.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Lu J, Li Z, Yu X, Chen L, Xu T. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- Yu F, Sun L, Machaca K. Constitutive recycling of the store-operated Ca2+ channel Orai1 and its internalization during meiosis. J Cell Biol. 2010;191:523–535. doi: 10.1083/jcb.201006022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zeng W, Dorwart M, Choi Y, Worley P, Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T, Gascoigne NRJ. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys J. 2004;86:3923–3939. doi: 10.1529/biophysj.103.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yu Y, Roos J, Kozak J, Deerinck T, Ellisman M, Stauderman K, Cahalan M. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Meraner P, Kwon HT, Machnes D, Masatsugu O-H, Zimmer J, Huang Y, Stura A, Rao A, Hogan PG. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Srinivasan P, Razavi S, Seymour S, Meraner P, Gudlur A, Stathopulos PB, Ikura M, Rao A, Hogan PG. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat Struct Mol Biol. 2013;20:973–981. doi: 10.1038/nsmb.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.