Abstract
Background
Findings related to the influence of the −160C → A promoter polymorphism and haplotypes of the E-cadherin (CDH1) gene have not been consistent in previous studies regarding the risk for sporadic gastric cancer. Investigators in most previous studies detected those genotypes using restriction fragment length polymorphism analysis. Therefore, we conducted a case–control study to investigate the association of the CDH1 − 160C → A promoter polymorphism and haplotypes for cancer risk related to sporadic diffuse and intestinal gastric cancer by direct sequencing analysis.
Methods
We included 107 diffuse gastric cancer cases, 60 intestinal gastric cancer cases and 134 controls. The genotypic polymorphisms in the −160 promoter region, exons and intron–exon boundaries of CDH1 were detected by direct sequencing analysis. Genotype frequencies were compared. The CDH1 − 160C → A promoter polymorphism and four polymorphisms (48 + 6 T → C, 2076C → T, 2253C → T and 1937–13 T → C) were included in the haplotype analyses, which were estimated using the expectation–maximization algorithm.
Results
Compared to controls, the frequency of the −160A allele was significantly higher in diffuse gastric cancer cases (P = 0.005), but it was not significantly different in intestinal gastric cancer cases (P = 0.119). Two sets of three-marker haplotypes (−160C → A, 48 + 6 T → C, 2076C → T and −160C → A, 1937–13 T → C, 2253C → T) were associated with the risk of diffuse gastric cancer (P = 0.011 and P = 0.042, respectively).
Conclusion
Based on direct sequencing analysis, our findings suggest that the CDH1 − 160C → A promoter polymorphism and haplotypes play significant roles in cancer risk for sporadic diffuse gastric cancer, but not for intestinal gastric cancer, in a Taiwanese population.
Keywords: CDH1, Direct sequencing, Haplotypes, Single-nucleotide polymorphisms, Sporadic diffuse and intestinal gastric cancer
Background
E-cadherin (encoded by the CDHI gene [dbSNP:NC_000016.9]) is a member of a family of transmembrane glycoproteins expressed in epithelial cells and is responsible for calcium-dependent cell-to-cell adhesion [1-3]. Loss of cell adhesion may contribute to loss of growth contact inhibition, which is an early step in the neoplastic process [4,5]. Furthermore, loss of cadherin activity may result in cancer cell detachment and metastasis [6]. Hence CDHI is considered to be a tumor suppressor gene.
A C/A single-nucleotide polymorphism (SNP) at the −160 position of the CDH1 promoter region has been reported to result in downregulation of the transcription of this gene in a prostate cancer cell line, DU145, such that the A allele at this site decreased transcriptional efficiency by 68% compared with the C allele [7]. Therefore, the −160A allele variant has been considered to be a potential genetic marker for susceptibility to cancer. However, findings related to the influence of the −160C → A promoter polymorphism and haplotypes of the E-cadherin (CDH1) gene have not been consistent in previous studies regarding the risk for sporadic gastric cancer [8-16]. Some studies have reported that the CDH1 − 160C → A promoter polymorphism was not found to be associated with the risk of gastric cancer [8-11], and others have reported that the −160A allele increased the risk for diffuse gastric cancer [12,13]. Conversely, two studies in Asian populations reported that the −160A allele decreased the risk of gastric cancer [15,16].
The genotypes of the −160C → A promoter polymorphism in most of the previous studies were detected by restriction fragment length polymorphism (RFLP) analysis [8-13,15,16]. RFLP analysis is an indirect method applied extensively in the past [17]. However, it involves a manual process that is based on gel-based processing techniques and the results it produces are subjectively evaluated by direct observation. In contrast, direct sequencing is a standardized procedure, and the results are processed by a computer [18].
By literature review, we compared the CDH1 − 160A allele frequency in controls in studies in which RFLP analysis was used, and the −160A allele frequency in healthy samples reported by HapMap based on direct sequencing derived from the National Center for Biotechnology Information dbSNP Short Genetic Variations online database [19]. The frequency of the −160A allele was calculated using the formula CA% × ½ + AA%, where CA is the C and A alleles and AA is the double A alleles. Humar et al. reported that the frequency of the −160A allele in controls in an Italian population was 23.6% [12], and Corso et al. reported a frequency of 32.05% in a different Italian population [11]. In a study by Pharoah et al., the frequency of the −160A allele was found to be 30% for Canadian, German and Portuguese populations [8]. Park et al. reported that the frequency of the −160A allele in controls in a Korean population was 23% [9]. Lu et al. reported that the frequency of the −160A allele was 24.3% for Chinese individuals in Jiangsu Province, China [10]. Medina-Franco et al. reported that the frequency of the −160A allele was 24.35% in a Mexican population [13]. Wu et al. reported that the frequency of the -160A allele was 33.67% in a Taiwanese population [15]. Kuraoka et al. reported that the frequency of the −160A allele was 35% in a Japanese population [16]. The HapMap database, however, gives information based on direct sequencing that the frequency of the −160A allele in healthy samples is 26.0% for Europeans in Italy, 25.2% for Han Chinese in Beijing, 19.9% for Japanese in Tokyo and 15.0% for Africans in Nigeria [19]. In aforementioned studies in which RFLP analysis was used, the reported frequencies of the −160A allele in controls were diverse within the same ethnic groups, and some were much higher than the values in the HapMap database. Therefore, genotype detection by RFLP analysis in some of previous studies might be inaccurate, which might contribute to the inconsistency of results in previous studies. Tanahashi et al. and Davis et al. also reported that RFLP analysis was less accurate than direct sequencing in their studies [20,21]. However, researchers in many studies have suggested that environmental factors, lifestyles and ethnic differences might account for opposite directions in associations of the CDH1 − 160C → A polymorphism with gastric cancer among some Asian and Caucasian studies [8-14,22,23]. Gene-based haplotypes, which are collections of SNPs located throughout the functional regions of candidate genes, may have greater power than any individual polymorphism in influencing a clinical response [24].
In one study of haplotype analysis using RFLP methods, the investigators suggested that the CDH1 − 160C → A polymorphism might be in linkage disequilibrium with other distinct CDH1 polymorphisms in sporadic diffuse gastric cancer [12] Researchers in another study using direct sequencing reported a statistically nonsignificant risk of −160C → A polymorphism containing haplotypes associated with gastric cancer without differentiation of histopathologic subgroups [14]. Furthermore, diffuse and intestinal types of gastric cancer are different in terms of their epidemiology, etiology, pathogenesis and behavior [25]. Therefore, we investigated the influence of the CDH1 − 160C → A promoter polymorphism and haplotypes on risk for diffuse and intestinal gastric cancer separately by direct sequencing.
Methods
Patients and samples
Eligible cases comprised 167 sporadic gastric cancer patients who had undergone surgical treatment between 2001 and 2005 in the Division of General Surgery, Department of Surgery, Tri-Service General Hospital, Taipei, Taiwan. All of the Taiwanese patients were Han Chinese, but no aborigines were included, as they belong to a different ethnic group that account for only 0.02% of the population in Taiwan. None of the patients had familial gastric cancer. All patient DNA obtained from the noncancerous gastric epithelia of resected specimens were stored in liquid nitrogen. Control DNA was obtained from the peripheral blood of 134 healthy individuals who attended general health checkup including upper gastrointestinal tract endoscopy in this hospital. The study participants provided written informed consent prior to participation. The study was approved by the Institutional Review Board of the Tri-Service General Hospital. All surgical samples were classified into diffuse and intestinal types according to the Laurén criteria [25].
Immunohistochemical staining and evaluation of E-cadherin expression
Specimens from paraffin blocks were cut into 5-μm sections and stained with hematoxylin and eosin for histological diagnosis. E-cadherin expression was evaluated using a monoclonal antibody (Cappel, Aurora, OH, USA) followed by a secondary antibody [26]. The signal was detected using an avidin-biotin complex and a 3,3′-diaminobenzidine (DAB) kit (Vector Laboratories, Burlingame, CA, USA). DAB produced a yellowish brown stain if a sample was positive. If more than 90% of the tumor cells showed intense membranous staining similar to that of normal cells, the result was considered positive (++). If the staining intensity was demonstrably reduced relative to that of normal cells and/or if the staining pattern was heterogeneous (10% to 90% positive), the result was recorded as weakly positive (+). If immunohistochemical expression was completely absent or positive in less than 10% of the cells, the result was defined as negative (−).
Genotyping
DNA samples from all cases were extracted from noncancerous gastric epithelia. Control DNA samples were isolated from peripheral blood lymphocytes by proteinase K digestion and phenol-chloroform extraction. All CDH1 exons and the corresponding intron–exon boundaries of case DNA and control DNA were amplified using primers developed by Berx et al. [27]. The primers used to amplify the −160 promoter region were described by Li et al. [7]. PCR was carried out in volumes of 25 μl containing 20 ng of genomic DNA as a template, 2 mM MgSO4, 0.4 μM sense and antisense primers, 0.25 mM deoxyribonucleotide triphosphate, and buffer containing Taq DNA polymerase. The amplification program was as follows: 40 cycles with a denaturing temperature of 95°C for 30 seconds, annealing for 30 seconds and extension at 72°C for 30 seconds in a DNA thermal cycler. All amplified products were purified, and direct sequencing was performed using an ABI PRISM 377 automated sequencer and an ABI PRISM Dye Terminator Cycle Sequencing Kit (PerkinElmer, Greenville, SC, USA).
Statistical analysis
Stata 8 software (StataCorp, College Station, TX, USA) was used to manage data and for statistical analyses. The observed genotype frequencies were compared between cases and controls using a χ2 test. Fisher’s exact test was used to assess the genotype and allele distributions in diffuse and intestinal type cases. The allelic distribution was in Hardy-Weinberg equilibrium in the case and control groups (P > 0.82). Genotype-specific risks were estimated as the odds ratio (OR) with associated 95% confidence interval (CI) by unconditional logistic regression. ORs were adjusted according to age and gender. A P-value less than 0.05 was considered statistically significant. The study power was 0.7758 to 0.9509 with an α of 0.05 [28]. The haplotype frequencies for various marker combinations were estimated separately for the two type cases and the controls by using an expectation-maximization algorithm.
Results
Characteristics of diffuse and intestinal type cases
The 134 controls were composed of 86 males and 48 females. Their mean age was 51.06 ± 13.04 years (range = 19 to 89 years). The 167 gastric cancer cases comprised 114 males and 53 females. Their mean age was 69.11 ± 13.04 years (range = 27 to 90 years). According to Laurén’s classification system, 107 cases were of the diffuse type and 60 cases were of the intestinal type. The mean age of the diffuse type cases was 66.87 ± 13.81 years (range = 27 to 90 years) and that of the intestinal type cases was 73.82 ± 9.76 years (range = 48 to 88 years). The mean age of the diffuse type cases was approximately 7 years younger than that of the intestinal type cases. No differences were observed with respect to gender or TNM stages I to III in these two types. There were significantly more diffuse gastric cancer cases classified as stage IV. Reduced E-cadherin expression (Figure 1) was more frequent in the diffuse type cases than in the intestinal type cases. Comparison of the characteristics between these two case types is summarized in Table 1.
Figure 1.
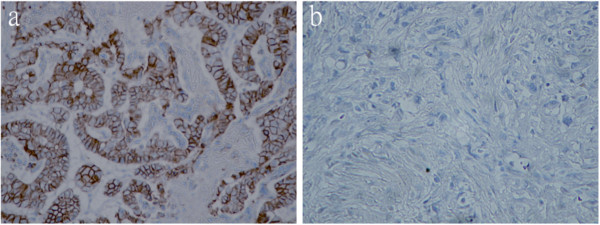
Immunohistochemical staining for E-cadherin. (a) E-cadherin-positive (++) in intestinal type cancer. (b) E-cadherin-negative (−) in diffuse type cancer.
Table 1.
Comparison of clinicopathologic characteristics between intestinal and diffuse type tumors
| Characteristics | Intestinal type, n (%) (N = 60) | Diffuse type, n (%) (N = 107) | P- value |
|---|---|---|---|
| Gender | |||
| Male |
46 (76.7%) |
68 (63.6%) |
0.081a |
| Female |
14 (23.3%) |
39 (36.5%) |
|
| Age (yr) | |||
| Mean ± SD |
73.82 ± 9.76 |
66.87 ± 13.81 |
0.0001b |
| TNM staging | |||
| Stage I |
16 (26.7%) |
16 (15.0%) |
0.065c |
| Stage II |
8 (13.3%) |
24 (22.4%) |
0.152c |
| Stage III |
28 (46.7%) |
35 (32.7%) |
0.074c |
| Stage IV |
8 (13.3%) |
32 (30.0%) |
0.016c |
| Reduced E-cadherin expression | 32 (53.3%) | 93 (86.9%) | <0.001c |
aχ2 test. bt-test. cProportion test.
Genotype detected by direct sequencing
The genotypes of the CDH1 − 160C → A (dbSNP:rs16260), 48 + 6 T → C (dbSNP:rs3743674), 2076C → T (dbSNP:rs1801552), 2253C → T (dbSNP:rs33964119) and 1937–13 T → C (dbSNP:rs2276330) polymorphisms detected by direct sequencing in the cases and controls are shown in Figure 2.
Figure 2.
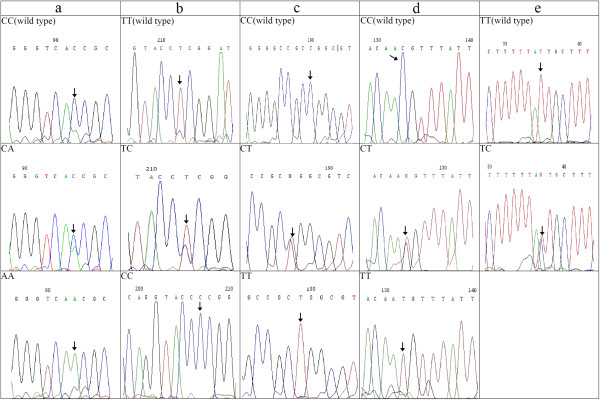
Genotypes of CDH1 polymorphisms detected by direct sequencing. (a) Promoter polymorphism −160C → A. (b) Intron 1 polymorphism 48 + 6 T → C. (c) Exon 13 polymorphism 2076C → T. (d) Exon 14 polymorphism 2253C → T. (e) Intron 12 polymorphism 1937–13 T → C.
Single-locus analysis in diffuse type cases
For promoter polymorphism −160C → A, the frequency of the −160A allele was significantly higher in diffuse type cases compared to the controls (P = 0.005). The OR associated with the A allele was 1.750 (95% CI = 1.014 to 3.022) for CA heterozygotes and 4.375 (95% CI = 1.467 to 14.565) for AA homozygotes. For other four polymorphisms (48 + 6 T → C, 2076C → T, 1937–13 T → C and 2253C → T), the genotype frequencies were not significantly different between the diffuse type cases and the controls (P = 0.441, 0.649, 0.147 and 0.982, respectively) (Table 2).
Table 2.
Genotype frequencies of CDH1 polymorphisms in diffuse gastric cancer cases and controls
| Variant | Cases vs. controls | n | (%) | n | (%) | n | (%) | n | (%) |
Statistical tests |
Fisher’s exact test | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| χ 2 test | P -value | |||||||||||
| Promoter |
|
CC |
|
CA |
|
AA |
|
Total |
|
10.7858 |
0.005 |
0.005 |
| −160C → A |
Diffuse |
48 |
44.86% |
44 |
41.12% |
15 |
15.00% |
107 |
100.00% |
|
|
|
| |
Controls |
84 |
62.69% |
44 |
32.84% |
6 |
4.48% |
134 |
100.00% |
|
|
|
| Odds ratioa |
|
1 |
|
1.750 (1.014 to 3.022) |
4.375 (1.467 to 14.565) |
|
|
|
|
|
||
| Intron 1 |
|
TT |
|
TC |
|
CC |
|
|
|
1.6397 |
0.441 |
0.489 |
| 48 + 6 T → C |
Diffuse |
71 |
66.36% |
32 |
29.91% |
4 |
3.74% |
107 |
100.00% |
|
|
|
| |
Controls |
92 |
68.66% |
33 |
24.63% |
9 |
6.72% |
134 |
100.00% |
|
|
|
| Odds Ratioa |
|
1 |
|
1.257 (0.677 to 2.328) |
0.576 (0.125 to 2.170) |
|
|
|
|
|
||
| Exon 13 |
|
CC |
|
CT |
|
TT |
|
|
|
0.8651 |
0.649 |
0.641 |
| 2076C → T |
Diffuse |
45 |
42.06% |
48 |
44.83% |
14 |
13.08% |
107 |
100.00% |
|
|
|
| |
Controls |
55 |
41.05% |
66 |
49.25% |
13 |
9.70% |
134 |
100.00% |
|
|
|
| Odds ratioa |
|
1 |
|
0.889 (0.499 to 1.584) |
1.136 (0.514 to 3.381) |
|
|
|
|
|
||
| Intron 12 |
|
TT |
|
TC |
|
CC |
|
|
|
2.100 |
0.147 |
0.182 |
| 1937-13 T → C |
Diffuse |
89 |
83.18% |
18 |
16.82% |
0 |
0.00% |
107 |
100.00% |
|
|
|
| |
Controls |
120 |
89.55% |
14 |
10.45% |
0 |
0.00% |
134 |
100.00% |
|
|
|
| Odds ratioa |
|
1 |
|
1.734 (0.766 to 3.976) |
to |
|
|
|
|
|
|
|
| Exon 14 |
|
CC |
|
CT |
|
TT |
|
|
|
0.037 |
0.982 |
1.000 |
| 2253C → T |
Diffuse |
89 |
83.18% |
17 |
15.89% |
1 |
0.93% |
107 |
100.00% |
|
|
|
| |
Controls |
111 |
82.84% |
22 |
16.42% |
1 |
0.75% |
134 |
100.00% |
|
|
|
| Odds ratioa | 1 | 0.964 (0.451 to 1.842) | 1.247 (0.016 to 98.766) | |||||||||
aAdjusted for age and sex.
Single-locus analysis in intestinal type cases
In the intestinal type cases, no significant association with disease was found for any of the five polymorphisms (−160C → A, 48 + 6 T → C, 2076C → T, 1937–13 T → C and 2253C → T; P = 0.119, 0.329, 0.185, 0.889 and 0.375, respectively).
Haplotype analysis in diffuse type cases
A three-marker haplotype (−160C → A, 48 + 6 T → C, 2076C → T) showed a significant association with disease and the ATC, ACC haplotypes were found to be associated with increased risk for diffuse type cancer (Table 3). A different three-marker haplotype (−160C → A, 1937–13 T → C, 2253C → T) also exhibited a significant association with disease and the ATC haplotypes was associated with increased risk for diffuse gastric cancer. The two other sets of three-marker haplotypes (−160C → A, 2076C → T, 2253C → T and −160C → A, 48 + 6 T → C, 1937–13 T → C) exhibited no significant association with disease (Table 3).
Table 3.
CDH1 three-marker haplotype analysis in diffuse gastric cancer cases
| Three polymorphisms | Haplotype | Cases (%) | Controls (%) | OR | Lower 95% CI | Upper 95% CI | P -value |
|---|---|---|---|---|---|---|---|
| −160C → A |
CTT |
24.70 |
25 |
1.132 |
0.860 |
1.489 |
0.011 |
| 48 + 6 T → C |
ATT |
5.93 |
4.48 |
1.511 |
0.926 |
2.466 |
|
| 2076C → T |
CCC |
12.07 |
13.27 |
1.030 |
0.731 |
1.452 |
|
| |
CTC |
31.78 |
36.38 |
1 |
|
|
|
| |
CCT |
3.80 |
4.48 |
0.954 |
0.552 |
1.648 |
|
| |
ATC |
18.81 |
15.11 |
1.431 |
1.051 |
1.949 |
|
| |
ACC |
2.52 |
0.93 |
3.272 |
1.322 |
8.096 |
|
| |
ACT |
0.38 |
0.37 |
0.954 |
0.158 |
5.761 |
|
| −160C → A |
CTT |
7.72 |
8.02 |
1.079 |
0.729 |
1.597 |
0.042 |
| 1937-13 T → C |
ATT |
1.41 |
0.75 |
2.073 |
0.732 |
5.872 |
|
| 2253C → T |
CCC |
3.20 |
3.36 |
1.075 |
0.599 |
1.929 |
|
| |
CTC |
61.22 |
67.53 |
1 |
|
|
|
| |
CCT |
0.21 |
0.19 |
0.691 |
0.062 |
7.648 |
|
| |
ATC |
23.34 |
18.47 |
1.4 |
1.078 |
1.819 |
|
| |
ACC |
2.86 |
16.79 |
0.184 |
0.111 |
0.306 |
|
| |
ACT |
0.04 |
0 |
– |
– |
– |
|
| −160C → A |
CCT |
1.62 |
14.93 |
0.108 |
0.055 |
0.212 |
0.096 |
| 2076C → T |
ACT |
0.09 |
0.19 |
0.691 |
0.058 |
7.227 |
|
| 2253C → T |
CTC |
37.54 |
42.91 |
0.909 |
0.703 |
1.176 |
|
| |
CCC |
26.88 |
27.99 |
1 |
|
|
|
| |
CTT |
6.31 |
6.72 |
0.987 |
0.628 |
1.551 |
|
| |
ACC |
6.23 |
4.66 |
1.406 |
0.862 |
2.293 |
|
| |
ATC |
19.97 |
15.49 |
1.332 |
0.971 |
1.827 |
|
| |
ATT |
1.37 |
0.56 |
2.341 |
0.771 |
7.111 |
|
| −160C → A |
CTC |
53.84 |
58.79 |
0.904 |
0.470 |
1.739 |
0.103 |
| 48 + 6 T → C |
ATC |
22.01 |
17.91 |
1.21 |
0.613 |
2.389 |
|
| 1937-13 T → C |
CCT |
0.77 |
0.93 |
0.882 |
0.237 |
3.281 |
|
| |
CTT |
2.65 |
2.61 |
1 |
|
|
|
| |
CCC |
15.10 |
16.79 |
0.888 |
0.445 |
1.772 |
|
| |
ATT |
2.73 |
1.68 |
1.588 |
0.616 |
4.094 |
|
| |
ACT |
0.17 |
0 |
– |
– |
– |
|
| ACC | 2.73 | 1.31 | 2.021 | 0.754 | 5.416 |
Haplotype analysis in intestinal type cases
In the intestinal type cases, no significant association with disease was found for any of the four sets of three-marker haplotypes (−160C → A, 48 + 6 T → C, 2076C → T; −160C → A, 1937–13 T → C, 2253C → T; −160C → A, 2076C → T, 2253C → T and −160C → A, 48 + 6 T → C, 1937–13 T → C; P = 0.164, 0.319, 0.408 and 0.607, respectively).
Discussion
To detect SNP variations, both direct sequencing and RFLP are suitable. RFLP is subject to experimental problems, however, such as incomplete digestion and relatively poor resolution, resulting in misclassification of genotypic status. Nevertheless, many previous studies have depended on RFLP because it is cheaper and easy to perform. However, the cost of direct sequencing has decreased significantly, and regular automatic sequencing platforms are able to provide reliable results easily. The use of sequencing in the present study can be considered a major strength.
In this study, specimens were obtained from 167 Taiwanese patients with sporadic gastric cancer who underwent surgical treatment. The patients with intestinal gastric cancer are generally older, have more comorbidities and tend not to undergo surgical treatment, which accounts for more diffuse gastric cancer cases than intestinal gastric cancer cases. Although we used an acceptable number of cases with statistical power, the sample size was still relatively small. Further studies including more cases are needed.
The frequency of the A allele in the CDH1 − 160C → A promoter polymorphism in this study was significantly greater in diffuse type cases than in the controls. Humar et al. [12] and Medina-Franco et al. [13] reported similar results.
It has been suggested that CDH1 may contribute to gastric cancer risk in a complex manner due to multiple polymorphic variants [12,14]. Humar et al. confirmed that the three-marker haplotype (−160C → A, 48 + 6 T → C, 2076C → T) was associated with diffuse gastric cancer and suggested that haplotype ATT (−160A, 48 + 6 T, 2076 T) was a marker for diffuse gastric cancer susceptibility, whereas haplotype CTT had a protective effect [12].
We also found that the three-marker haplotype (−160C → A, 48 + 6 T → C, 2076C → T) was associated with diffuse gastric cancer and the ATC and ACC haplotypes were associated with increased risk. However, the haplotype CTT had no protective effect on diffuse gastric cancer which differs from that reported by Humar et al. [12]. Furthermore, we found another three-marker haplotype (−160C → A, 1937–13 T → C, 2253C → T) that was associated with diffuse gastric cancer, and the ATC haplotypes showed increased risk.
In the three-marker haplotype in this study (−160C → A, 48 + 6 T → C, 2076C → T), the haplotype ATC, with only one polymorphic variant (−160A) showed increased diffuse gastric cancer risk. Therefore, with respect to the CDH1 − 160C → A polymorphism, the A allele might result in a defect in gene transcription and increased risk for diffuse type cancer. The ACC haplotype, with two polymorphic variants (−160A and 48 + 6C), exhibited a higher OR. This finding indicates that the presence of two polymorphic variants in this haplotype produces synergic effects related to cancer risk.
Conclusions
We conclude that, based on direct sequencing analysis, the −160C → A promoter polymorphism and the investigated haplotypes play significant roles in the risk for sporadic diffuse gastric cancer, but not for intestinal gastric cancer, in a Taiwanese population. In the future, additional polymorphisms in other regions of the same gene, such as the promoter region and splicing sites, should be evaluated, and haplotype analysis should include additional polymorphisms for the purpose of increasing knowledge about how combinations of polymorphisms can influence cancer risk.
Abbreviations
RFLP: Restriction fragment length polymorphism; SNP: Single-nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LYC and SCY designed the research. LYC, CCJ, CDC, CCM, WHS, CTM, YJC, HHJ, YCP and YMH performed the research and analyzed the data. CCM performed the statistical analysis. LYC, CCJ and CDC wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Chi-Ming Chu, Email: chuchiming@web.de.
Cheng-Jueng Chen, Email: doc20227@ndmctsgh.edu.tw.
De-Chuan Chan, Email: chrischan1168@yahoo.com.tw.
Hurng-Sheng Wu, Email: hswu1297@yahoo.com.tw.
Yao-Chi Liu, Email: dlyaochi@gmail.com.
Chen-Yang Shen, Email: bmcys@ibms.sinica.edu.tw.
Tzu-Ming Chang, Email: tzumchang@hotmail.com.
Jyh-cherng Yu, Email: doc20106@ndmctsgh.edu.tw.
Horng-Jyh Harn, Email: dukeharn@cmuh.org.tw.
Cheng-Ping Yu, Email: cpyupath@yahoo.com.tw.
Ming-Hsin Yang, Email: yangming@mail.ndmctsgh.edu.tw.
Acknowledgements
This study was supported by the Clinical Research Fund of the Tri-Service General Hospital, Taipei, Taiwan (TSGH-C97-15). The authors are grateful to Ye-Chien Mau for her wonderful technical assistance.
References
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: a molecular family important in selective cell–cell adhesion. Annu Rev Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Grunwald GB. The structural and functional analysis of cadherin calcium-dependent cell adhesion molecules. Curr Opin Cell Biol. 1993;5:797–805. doi: 10.1016/0955-0674(93)90028-O. [DOI] [PubMed] [Google Scholar]
- Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli M, Vessey CJ. Adhesion molecules: novel molecular tools in tumor pathology. Hum Pathol. 1994;25:849–856. doi: 10.1016/0046-8177(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Bussemakers MJ, van Moorselaar RJ, Giroldi LA, Ichikawa T, Isaacs JT, Takeichi M, Debruyne FM, Schalken JA. Decreased expression of E-cadherin in the progression of rat prostatic cancer. Cancer Res. 1992;52:2916–2922. [PubMed] [Google Scholar]
- Li LC, Chui RM, Sasaki M, Nakajima K, Perinchery G, Au HC, Nojima D, Carroll P, Dahiya R. A single nucleotide polymorphism in the E-cadherin gene promoter alters transcriptional activities. Cancer Res. 2000;60:873–876. [PubMed] [Google Scholar]
- Pharoah PDP, Oliveira C, Machado JC, Keller G, Volgelsang H, Laux H, Becker KF, Hahn H, Paproski SM, Brown LA, Caldas C, Huntsman D. CDH1 c-160a promoter polymorphism is not associated with risk of stomach cancer. Int J Cancer. 2002;101:196–197. doi: 10.1002/ijc.10590. [DOI] [PubMed] [Google Scholar]
- Park WS, Cho YG, Park JY, Kim CJ, Lee JH, Kim HS, Lee JW, Song YH, Park CH, Park YK, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY. A single nucleotide polymorphism in the E-cadherin gene promoter-160 is not associated with risk of Korean gastric cancer. J Korean Med Sci. 2003;18:501–504. doi: 10.3346/jkms.2003.18.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xu YC, Shen J, Yu RB, Niu JY, Guo JT, Hu X, Shen HB. E-cadherin gene C-160A promoter polymorphism and risk of non-cardia gastric cancer in a Chinese population. World J Gastroenterol. 2005;11:56–60. doi: 10.3748/wjg.v11.i1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corso G, Berardi A, Marrelli D, Pedrazzani C, Garosi L, Pinto E, Roviello F. CDH1 C-160A promoter polymorphism and gastric cancer risk. Eur J Cancer Prev. 2009;18:46–49. doi: 10.1097/CEJ.0b013e32830c8d70. [DOI] [PubMed] [Google Scholar]
- Humar B, Graziano F, Cascinu S, Catalano V, Ruzzo AM, Magnani M, Toro T, Burchill T, Futschik ME, Merriman T, Guilford P. Association of CDH1 haplotypes with susceptibility to sporadic diffuse gastric cancer. Oncogene. 2002;21:8192–8195. doi: 10.1038/sj.onc.1205921. [DOI] [PubMed] [Google Scholar]
- Medina-Franco H, Ramos-De la Medina A, Vizcaino G, Medina-Franco JL. Single nucleotide polymorphisms in the promoter region of the E-cadherin gene in gastric cancer: case–control study in a young Mexican population. Ann Surg Oncol. 2007;14:2246–2249. doi: 10.1245/s10434-006-9054-4. [DOI] [PubMed] [Google Scholar]
- Al-Moundhri MS, Al-Khanbashi M, Al-Kindi M, Al-Nabhani M, Burney IA, Al-Farsi A, Al-Bahrani B. Association of E-cadherin (CDH1) gene polymorphisms and gastric cancer risk. World J Gastroenterol. 2010;16:3432–3436. doi: 10.3748/wjg.v16.i27.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MS, Huang SP, Chang YT, Lin MT, Shun CT, Chang MC, Wang HP, Chen CJ, Lin JT. Association of the −160C → A promoter polymorphism of E-cadherin gene with gastric carcinoma risk. Cancer. 2002;94:1443–1448. doi: 10.1002/cncr.10371. [DOI] [PubMed] [Google Scholar]
- Kuraoka K, Oue N, Yokozaki H, Kitadai Y, Ito R, Nakayama H, Yasui W. Correlation of a single nucleotide polymorphism in the E-cadherin gene promoter with tumorigenesis and progression of gastric carcinoma in Japan. Int J Oncol. 2003;23:421–427. [PubMed] [Google Scholar]
- de Martinville B, Wyman AR, White R, Francke U. Assignment of first random restriction fragment length polymorphism (RFLP) locus (D14S1) to a region of human chromosome 14. Am J Hum Genet. 1982;34:216–226. [PMC free article] [PubMed] [Google Scholar]
- Guo LH, Yang RC, Wu R. An improved strategy for rapid direct sequencing of both strands of long DNA molecules cloned in a plasmid. Nucleic Acids Res. 1983;11:5521–5540. doi: 10.1093/nar/11.16.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information: dbSNP Short Genetic Variations (Online Database) Available at http://hapmap.ncbi.nlm.nih.gov/cgi-perl/snp_details_phase3?name=rs16260&source=hapmap28_B36&tmpl=snp_details_phase3 (accessed 03 March 2014)
- Tanahashi T, Kita M, Kodama T, Sawai N, Yamaoka Y, Mitsufuji S, Katoh F, Imanishi J. Comparison of PCR-restriction fragment length polymorphism analysis and PCR-direct sequencing methods for differentiating Helicobacter pylori ureB gene variants. J Clin Microbiol. 2000;38:165–169. doi: 10.1128/jcm.38.1.165-169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AH, Wang J, Tsang TC, Harris DT. Direct sequencing is more accurate and feasible in detecting single nucleotide polymorphisms than RFLP: using human vascular endothelial growth factor gene as a model. Biol Res Nurs. 2007;9:170–178. doi: 10.1177/1099800407308083. [DOI] [PubMed] [Google Scholar]
- Gao L, Nieters A, Brenner H. Meta-analysis: tumour invasion-related genetic polymorphisms and gastric cancer susceptibility. Aliment Pharmacol Ther. 2008;28:565–573. doi: 10.1111/j.1365-2036.2008.03760.x. [DOI] [PubMed] [Google Scholar]
- Wang GY, Lu CQ, Zhang RM, Hu XH, Luo ZW. The E-cadherin gene polymorphism −160C→A and cancer risk: a HuGE review and meta-analysis of 26 case–control studies. Am J Epidemiol. 2008;167:7–14. doi: 10.1093/aje/kwm264. [DOI] [PubMed] [Google Scholar]
- Judson R, Stephens JC, Windemuth A. The predictive power of haplotypes in clinical response. Pharmacogenomics. 2000;1:15–26. doi: 10.1517/14622416.1.1.15. [DOI] [PubMed] [Google Scholar]
- Laurén P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type of carcinoma. An attempt at histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- Bànkfalvi A, Terpe HJ, Breukelmann D, Bier B, Rempe D, Pschadka G, Krech R, Lellè RJ, Boecker W. Immunophenotypic and prognostic analysis of E-cadherin and β-catenin expression during breast carcinogenesis and tumour progression: a comparative study with CD44. Histopathology. 1999;34:25–34. doi: 10.1046/j.1365-2559.1999.00540.x. [DOI] [PubMed] [Google Scholar]
- Berx G, Cleton-Jansen AM, Nollet F, de Leeuw WJ, van de Vijver M, Cornelisse C, van Roy F. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995;14:6107–6115. doi: 10.1002/j.1460-2075.1995.tb00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–191. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]


