Abstract
The PhysioNet/CinC 2013 Challenge aimed to stimulate rapid development and improvement of software for estimating fetal heart rate (FHR), fetal interbeat intervals (FRR), and fetal QT intervals (FQT), from multichannel recordings made using electrodes placed on the mother’s abdomen. For the challenge, five data collections from a variety of sources were used to compile a large standardized database, which was divided into training, open test, and hidden test subsets. Gold-standard fetal QRS and QT interval annotations were developed using a novel crowd-sourcing framework.
The challenge organizers used the hidden test subset to evaluate 91 open-source software entries submitted by 53 international teams of participants in three challenge events, estimating FHR, FRR, and FQT using the hidden test subset, which was not available for study by participants. Two additional events required only user-submitted QRS annotations to evaluate FHR and FRR estimation accuracy using the open test subset available to participants.
The challenge yielded a total of 91 open-source software entries. The best of these achieved average estimation errors of 187bpm2 for FHR, 20.9 ms for FRR, and 152.7 ms for FQT. The open data sets, scoring software, and open-source entries are available at PhysioNet for researchers interested on working on these problems.
1. Introduction
Since the late 19th century, decelerations of fetal heart rate (FHR) have been known to be associated with fetal distress. The 14th annual PhysioNet/Computing in Cardiology Challenge was aimed at accelerating the development of accurate algorithms for locating fetal QRS (FQRS) complexes and estimating fetal QT intervals in noninvasive fetal electrocardiograms (FECGs), acquired using electrodes placed on the mother’s abdomen. Unlike direct FECGs obtained using a fetal scalp electrode, noninvasive FECGs can be observed throughout the second half of pregnancy with negligible risk, but it is often difficult to detect fetal QRS complexes in non-invasive FECGs, since maternal QRSs are usually of greater amplitude.
Beyond FHR, features such as FHR variability and fetal QT interval may be useful independent indicators of fetal status. There are no accepted techniques for assessing such features from non-invasive FECGs, however. Multiple challenge events were designed to test basic FHR estimation accuracy, as well as accuracy in measurement of inter-beat (RR) and QT intervals needed as a basis for derivation of other FECG features. The PhysioNet/CinC 2013 Challenge was therefore conducted as five different events:
Events 1 and 2: Open-Source FHR and FRR Estimation
Each open-source entry produced a set of estimated R-peak locations that was used to construct an FHR (Event 1) and an FRR (Event 2) time series for each recording in the hidden test set.
Event 3: Open-Source FQT Estimation
Each open-source entry produced an estimate of the median QT interval for each recording in the hidden test set.
Events 4 and 5: FHR and FRR Estimation
Participants submitted a set of estimated R-peak locations that was used to construct an FHR (Event 4) and an FRR (Event 5) time series for each recording in the open test set.
2. Challenge data
Data for the challenge consist of a collection of one-minute, four-channel non-invasive FECGs sampled at 1 kHz. The data were obtained from multiple sources using a variety of instrumentation with differing frequency response, resolution, and configuration. The 447 records used in the challenge were drawn from five data collections (Table 1).
Table 1.
FECG database reference.
The 447 records were partitioned into three subsets. Training set A contains 75 records; its reference annotations were provided to the participants. Open test set B contains 100 records; its signals, but not its reference annotations, were available for study by the participants. Hidden test set C contains the remaining 272 records; it was withheld from the competitors for the purpose of testing the open-source entries (Events 1, 2, and 3). Sets A and B, and the reference annotations for set A, remain freely available [4]. Figure 1 shows a short excerpt of a record from set A.
Figure 1.
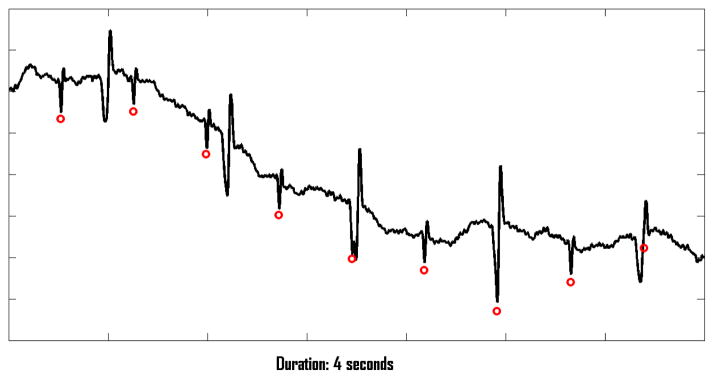
A four-second excerpt of a one-minute record from the challenge training set; the red circles mark the locations of the fetal QRS complexes, coinciding in three cases with the larger-amplitude maternal QRS complexes.
2.1. Reference annotations
The reference annotations used for the challenge were revised during the competition. During the first phase of the challenge, preliminary scores were based on reference QRS annotations derived by the authors of the respective data sets. During the later stages of the challenge, the original annotations were replaced by a set of crowd-sourced reference annotations, derived by applying a novel probabilistic voting algorithm [5, 6] to multiple sets of manual annotations and those made by the open-source entries in Phase 1. For Event 3, reference QT durations were also derived by applying the probabilistic voting algorithm to manual annotations done by seven observers.
3. Scoring criteria
Entries were scored using tach, mxm, and ann2rr (components of the WFDB software package [7]) and custom software developed for the challenge. The software processed the reference annotations and the test annotations (i.e., those generated by the participant’s entry) to obtain a score for each test record, then calculated an average score for all records in the test set.
3.1. FHR time series estimation (Events 1 and 4)
Reference and test FHR time series were derived from the reference and test FQRS annotations by tach, which used the IPFM method [8] to obtain 12 uniformly sampled and smoothed instantaneous FHR estimates for each record. The score for each record was the mean squared error (in bpm2) between the fetal heart rate signals estimated from the reference and test annotations. (For records with no test annotations, the score was 8000.) Any test annotations that preceded the first reference QRS, and any that followed the last reference QRS, were ignored. The final aggregate score for the event was the average score for all records.
3.2. FRR time series estimation (Events 2 and 5)
Reference and test FRR time series were obtained from the reference and test FQRS annotations using ann2rr. Scoring for Events 2 and 5 was performed by comparing these time series using mxm, which derived the root mean square difference (in milliseconds) between corresponding RR intervals. (For records with no test FRR intervals, the score was 200.) Any test RR intervals that preceded the first reference RR, and any that followed the last reference RR, were ignored. The final aggregate score for the event was the average score for all records.
3.3. FQT interval estimation (Event 3)
Scoring for Event 3 was the root mean square difference between the reference and test FQT intervals for all records, in milliseconds.
4. Results
Participants submitted a total of 208 sets of FQRS annotations for Events 4 and 5, and 91 open-source entries for Events 1, 2, and 3 during the challenge period. Results for events 1, 2, 4 and 5 are shown in Fig. 2. Table 2 shows the top scores and the teams who achieved them for all five events.
Figure 2.
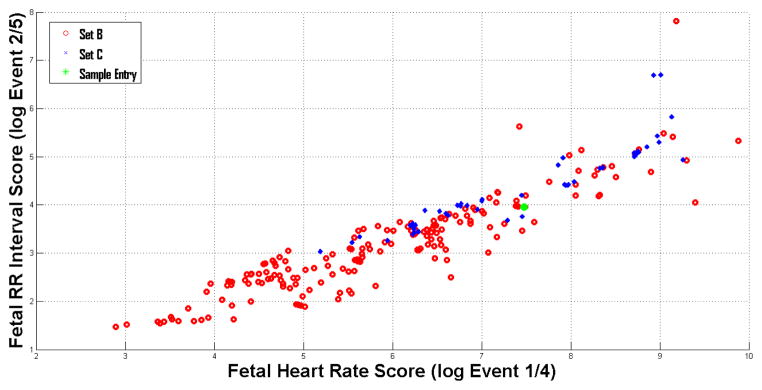
Scores for events 1,2, 4 and 5.
Table 2.
Top scores for Challenge events. Results in parentheses are from unofficial participants.
5. Discussion
The PhysioNet/CinC 2013 challenge attracted a total of 53 teams attempting non-invasive extraction of fetal ECG information from maternal abdominal leads. Most teams employed on a two-step approach, where the first step typically consisted of the removal of the maternal QRS, followed by a second step with the aim of the extracting the fetal QRS. The removal of the maternal component was achieved using techniques that included subspace decomposition or reconstruction [13–20], adaptive filtering/averaging [12, 21–25], de-noising via wavelets [26–30], and a fusion of several approaches [10]. The final step of fetal QRS detection used a variety of approaches including matched filtering [14,15,31], Christov’s beat detection [32], entropy [14, 33], RS slope [11], expectation weighting [34], echo state recurrent neural network [35], or fusion of multiple methods [36].
Interestingly, most of the top-performing entries in the challenge made use of strategies that differentiated them from their competitors. For instance, Behar et al [10] instead of relying on a single technique for maternal ECG extraction, decided to implement a fusion of several different extraction methods. Another successful approach was the use adaptive QRS templates for the either the mother, fetus, or both; thus allowing for realistic non-stationary conditions [12].
Acknowledgments
A significant part of the fetal ECG records for this challenge was contributed by Prof. Vyacheslav Shulgin and Dr. Anton Tokarev, from the Biomedical Signal Processing Laboratory of National Aerospace University, Kharkov, Ukraine. The authors are thankful to Qiao Li and Marisol Martinez Alanis for their help with the QT annotations. This work was funded in part by the National Institute of Biomedical Imaging and Bio-engineering and by the National Institute of General Medical Sciences, under NIH cooperative agreement U01-EB-008577 and NIH grant R01-EB-001659. JB is supported by the UK Engineering and Physical Sciences and Research Council, the Balliol French Anderson Scholarship Fund and MindChild Medical Inc.
References
- 1.Matonia A, Jezewski J, Kupka T, Horoba K, Wrobel J, Gacek A. The influence of coincidence of fetal and maternal QRS complexes on fetal heart rate reliability. Medical Biological Engineering Computing. 2006;44(5):393–403. doi: 10.1007/s11517-006-0054-0. [DOI] [PubMed] [Google Scholar]
- 2.Sameni R, Clifford GD, Jutten C, Shamsollahi M. Multi-channel ECG and noise modeling: Application to maternal and fetal ECG signals. EURASIP Journal on Advances in Signal Processing. 2007:043407. [Google Scholar]
- 3.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng CK, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101(23):E215–E220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 4.Noninvasive fetal ECG: the PhysioNet/Computing in Cardiology Challenge. 2013 http://physionet.org/challenge/2013/ [PMC free article] [PubMed]
- 5.Raykar VC, Yu S, Zhao LH, Valadez GH, Florin C, Bogoni L, Moy L. Learning from crowds. Journal of Machine Learning Research. 2010;11:1297–1322. [Google Scholar]
- 6.Zhu T, Johnson AE, Behar J, Clifford GD. Bayesian voting of multiple annotators for improved QT interval estimation. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 7.Moody GB. the WFDB software package. http://physionet.org/physiotools/wfdb.shtml.
- 8.Berger RD, Akselrod S, Gordon D, Cohen RJ. An efficient algorithm for spectral analysis of heart rate variability. IEEE TBME. 1986;9:900–904. doi: 10.1109/TBME.1986.325789. [DOI] [PubMed] [Google Scholar]
- 9.Varanini M, Tartarisco G, Billeci L, Macerata A, Pioggia G, Balocchi R. A multi-step approach for non-invasive fetal ECG analysis. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 10.Behar J, Oster J, Clifford GD. Non invasive FECG extraction from a set of abdominal sensors. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 11.Podziemski P, Gieraltowski J. Fetal heart rate discovery: algorithm for detection of fetal heart rate from noisy, non-invasive fetal ECG recordings. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 12.Andreotti F, Riedl M, Himmelsbach T, Wedekind D, Zaunseder S, Wessel N, Malberg H. Maternal signal estimation by Kalman filtering and template adaptation for fetal heart rate extraction. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 13.Fatemi M, Niknazar M, Sameni R. A robust framework for noninvasive extraction of fetal electrocardiogram signals. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 14.Lipponen JA, Tarvainen MP. Advanced maternal ECG removal and noise reduction for application of fetal QRS detection. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 15.Maier C, Dickhaus H. Fetal QRS detection and RR interval measurement in noninvasively registered adbominal ECGs. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 16.Niknasar M, Rivet B, Jutten C. Fetal QRS complex detection based on three-way tensor decomposition. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 17.Akhbari M, Niknazar M, Jutten C, Shamsollahi MB, Rivet B. Fetal electrocardiogram R-peak detection using robust tensor decomposition and extended Kalman filtering. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 18.Llamedo M, Martín-Yebra A, Laguna P, Martínez JP. Fetal ECG estimation based on linear transformations. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 19.Petrolis R, Krisciukaitis A. Multi stage principal component analysis based method for detection of fetal heart beats in abdominal ECG. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 20.Starc V. Noninvasive fetal ECG detection from maternal abdominal recording: the PhysioNet/CinC Challenge 2013. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 21.Rodrigues R. Fetal ECG detection in abdominal recordings: a method for QRS location. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 22.Perlman O, Katz A, Zigel Y. Noninvasive fetal QRS detection using linear combination of abdomen ECG signals. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 23.Haghpanahi M, Borkholder DA. Fetal ECG extraction from abdominal recordings using array signal processing. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 24.Liu C, Li P. Systematic methods for fetal electrocardiographic analysis: Determining the fetal heart rate, RR interval and QT interval. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 25.Kropf M, Schreier G, Modre-Osprian R, Hayn D. A robust algorithm for fetal QRS detection using the noninvasive maternal abdomen ECG. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 26.Razavipour F, Haghpanahi M, Sameni R. Fetal QRS detection using semi-blind source separation framework. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 27.Di Maria C, Duan W, Bojarnejad M, Pan F, King S, Zheng D, Murray A, Langley P. An algorithm for the analysis of foetal ECG from 4-channel non-invasive abdominal recordings. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 28.Almeida R, Gonçalves H, Rocha AP, Bernardes J. A wavelet-based method for assessing fetal cardiac rhythms from abdominal ECGs. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 29.Ali G, Atyabi SA, Mollakazemi MJ, Niknazar M, Niknami M, Soleimani A. PhysioNet/CinC Challenge 2013: A novel noninvasive technique to recognize the fetal QRS complexes from noninvasive fetal electrocardiogram signals. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 30.Dessi A, Pani D, Raffo L. Identification of fetal QRS complexes in low density non-invasive biopotential recordings. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 31.Plešinger F, Halámek J, Jurák P. Extracting R-wave position from an FECG record using recognition of its multi-channel shapes. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 32.Christov I, Simova I, Abächerli R. Cancellation of the maternal and extraction of the fetal ECG in noninvasive recordings. Computing in Cardiology. 2013:40. doi: 10.1088/0967-3334/35/8/1713. (this volume) [DOI] [PubMed] [Google Scholar]
- 33.Kuzilek J, Lhotska L. Advanced signal processing techniques for fetal ECG analysis. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 34.Di Marco LY, Marzo A, Frangi A. Multichannel foetal heartbeat detection by combining source cancellation with expectation-weighted estimation of fiducial points. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]
- 35.Lukoševičius M, Marozas V. Noninvasive fetal QRS detection using echo state network. Computing in Cardiology. 2013:40. doi: 10.1088/0967-3334/35/7/1685. (this volume) [DOI] [PubMed] [Google Scholar]
- 36.Xu-Wilson M, Carlson E, Cheng L, Vairavan S. Spatial filtering and adaptive rule based fetal heart rate extraction from abdominal fetal ECG recordings. Computing in Cardiology. 2013:40. (this volume) [Google Scholar]


