Abstract
Background
Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have been successfully used to knock out endogenous genes in stem cell research. However, the deficiencies of current gene-based delivery systems may hamper the clinical application of these nucleases. A new delivery method that can improve the utility of these nucleases is needed.
Results
In this study, we utilized a cell-penetrating peptide-based system for ZFN and TALEN delivery. Functional TAT-ZFN and TAT-TALEN proteins were generated by fusing the cell-penetrating TAT peptide to ZFN and TALEN, respectively. However, TAT-ZFN was difficult to purify in quantities sufficient for analysis in cell culture. Purified TAT-TALEN was able to penetrate cells and disrupt the gene encoding endogenous human chemokine (C-C motif) receptor 5 (CCR5, a co-receptor for HIV-1 entry into cells). Hypothermic treatment greatly enhanced the TAT-TALEN-mediated gene disruption efficiency. A 5% modification rate was observed in human induced pluripotent stem cells (hiPSCs) treated with TAT-TALEN as measured by the Surveyor assay.
Conclusions
TAT-TALEN protein-mediated gene disruption was applicable in hiPSCs and represents a promising technique for gene knockout in stem cells. This new technique may advance the clinical application of TALEN technology.
Electronic supplementary material
The online version of this article (doi:10.1186/2045-9769-2-5) contains supplementary material, which is available to authorized users.
Keywords: CCR5, HIV-1, Cell-penetrating peptide, TALEN, TAT, Protein delivery, Induced pluripotent stem cells
Background
Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are two types of artificial site-specific nucleases that are generated by fusing the DNA cleavage domain of the FokI restriction endonuclease with a custom-designed DNA biding domain. In ZFNs and TALENs, these DNA-binding domains are the C2H2 zinc-finger motif [1] and the transcription activator-like effector (TALE) domain respectively [2]. Because the FokI nuclease domain functions as a dimer, ZFNs and TALENs are typically used in pairs. These chimeric nucleases induce DNA double-strand breaks at defined sites in living cells that can be repaired via homology-directed repair (HDR) pathway, which utilizes a homologous donor DNA, or imprecise nonhomologous end joining (NHEJ) pathway [3]. These engineered nucleases have been widely used to modify endogenous genes in human stem cells [4–7]. However, gene-based delivery systems are problematic. In particular, the use of viral vectors entails the risk of insertional mutagenesis [8], whereas plasmid DNA has cell-type restrictions and cytotoxicity.
Cell-penetrating peptides (CPPs), also known as protein transduction domain (PTD) peptides, have been used to deliver a variety of cargoes (including drug molecules, nucleic acids, liposome nanoparticles and large proteins) into cells both in vivo and in vitro and have therefore shown great promise as therapeutic delivery mechanisms for the treatment of several diseases [9]. The CPP delivery system has various advantages including applicability to all cell types, high transduction efficiency and controlled administration. Among the CPPs, TAT (YGRKKRRQRRR) is the most widely investigated and widely used peptide. TAT is a short peptide consisting of 11 amino acids from the human immunodeficiency virus-1 (HIV-1) TAT protein and is rich in arginine and lysine; therefore, TAT is highly positively charged and hydrophilic [10]. Since the first use of TAT as a delivery vehicle for introducing molecules into cells in the late 1990s, numerous studies have reported the use of TAT for the delivery of various biomolecules (especially proteins) into cells in the form of TAT fusion proteins or TAT-protein conjugates [11]. Use of TAT as a delivery vehicle has been considered as one of the most promising gene-free strategies.
In this study, we designed TAT-ZFN and TAT-TALEN fusion proteins by in-frame cloning. We succeeded in purifying functional TAT-TALEN proteins, and demonstrated their cell penetrating properties. When incubated with living cells, TAT-TALENs efficiently disrupted the endogenous CCR5 gene under hypothermic conditions in a dose-dependent manner and we observed a disruption efficiency of up to 5% in human induced pluripotent stem cells (hiPSCs).
Results
Purification and activity testing of TAT-ZFNs and TAT-TALENs
The structure of the TAT-ZFN construct is shown in Figure 1A. Soluble proteins were purified as described in Methods. TAT-ZFN purification was greatly limited by its low protein expression level and low binding affinity; thus, only a small amount of TAT-ZFN could be obtained (Figure 1B). Because the purification tag could be detected by anti-His antibodies (Figure 1C), we speculated that the His tag was likely to be hidden by the tertiary structure of the native protein. Nevertheless, TAT-ZFNs showed specific nuclease activity in vitro (Figure 1D).
Figure 1.
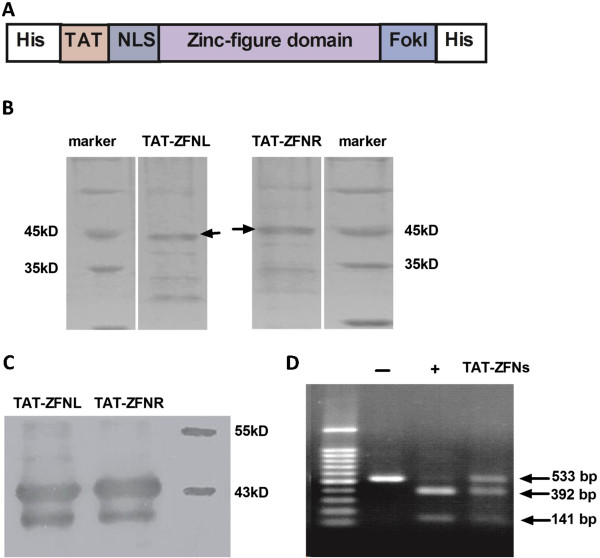
Purification of TAT-ZFNs. (A) Schematic diagram of the TAT-ZFN construct. (B) SDS-PAGE analysis of TAT-ZFNs purified under native conditions. (C) Western blot analysis of expressed TAT-ZFNs using an anti-His antibody. (D) In vitro activity testing of TAT-ZFNs purified under native conditions. The negative control is indicated by “-”. The recognition sequence of Hpy188III is located within the ZFN recognition sequence; therefore, Hpy188III was used as the positive control (+).
No problems were encountered with TAT-TALEN purification. Standard TALEN was also constructed and purified for use as a control in subsequent experiments. The structure of standard TALEN and TAT-TALEN is illustrated in Figure 2A. The purified proteins were visualized using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie Brilliant Blue staining (Figure 2B). The in vitro DNA cleavage assay demonstrated that both purified standard TALEN and TAT-TALEN were functional (Figure 3). Notably, a high TALEN concentration resulted in non-specific DNA cleavage.
Figure 2.
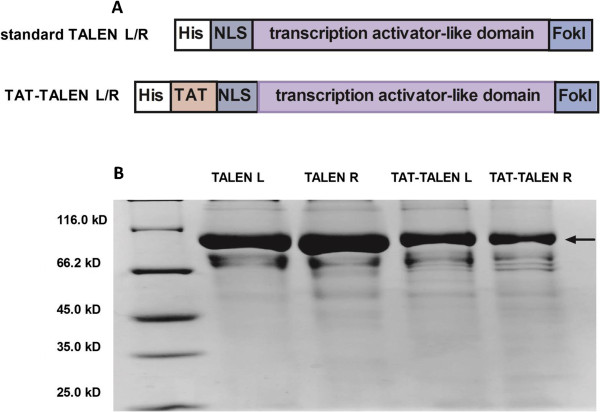
Purification of standard TALENs and TAT-TALENs. (A) Schematic diagram of standard TALENs and TAT-TALENs. (B) SDS-PAGE analysis of four TALEN proteins stained with Coomassie blue after purification.
Figure 3.
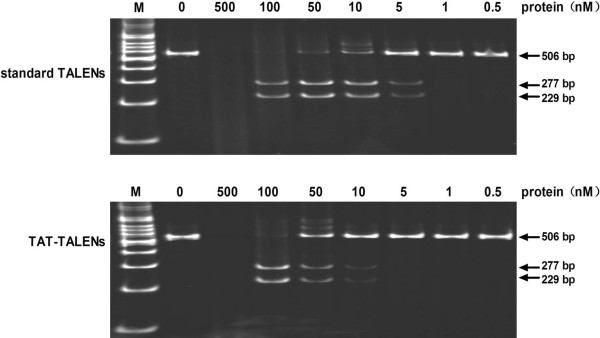
Activity testing of purified standard TALENs and TAT-TALENs. Specific activity was determined based on the presence of two cleavage products with lengths of 229 bp and 277 bp.
Transduction of proteins into living cells
To evaluate the ability of the purified proteins to penetrate cells, HeLa cells were treated with 1.5 μM TALEN proteins for 1 hour at 37°C and subsequently washed with 1 mg/ml heparin to remove proteins bound to the cell surface. Immunoblotting of the treated HeLa lysate showed that TALEN itself could not penetrate the cells; however, the 11-amino acid TAT was able to deliver the 110 kD TALEN into the cells (Figure 4).
Figure 4.
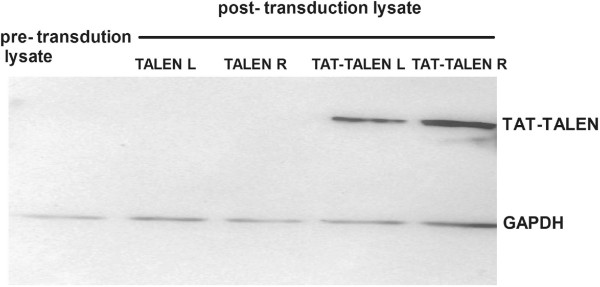
Western blot analysis of lysate from the treated HeLa cells using an anti-FLAG antibody. HeLa cells were incubated with TALEN for 1 hour at 37°C and subsequently washed three times with 1 mg/ml heparin-PBS to remove proteins bound to the cell surface before lysis.
CCR5 disruption in HeLa cells and hiPSCs
HeLa cells were subjected to treatment with TALENs at 37°C for 1 hour and then continuously cultured at 37°C for 24 hours before the next treatment. After three treatment cycles, a Surveyor nuclease assay was performed to evaluate gene disruption. TAT-TALENs disrupted CCR5 at 37°C in a dose-dependent manner, whereas standard TALENs exerted no effects on CCR5 disruption. As shown in Figure 5A, HeLa cells subjected to three consecutive treatments with 3 μM TAT-TALEN proteins showed a modification frequency of 3% as measured by the Surveyor assay. This modification frequency increased to 16% when the cells treated with TAT-TALEN were incubated at 30°C. Standard TALENs still did not disrupt CCR5 gene under hypothermic conditions. To verify the reliability of the Surveyor assay, TAT-TALEN mediated CCR5 disruption was confirmed by sequence analysis of cloned CCR5 alleles amplified from treated HeLa cells (Figure 5B).
Figure 5.
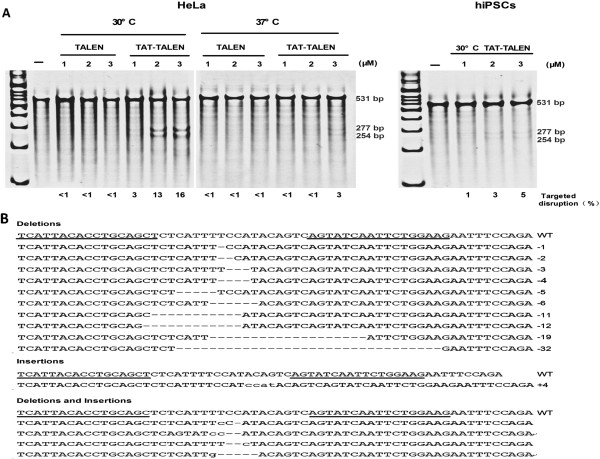
Disruption of endogenous genes by direct delivery of TALEN proteins. (A) Frequency of endogenous CCR5 gene disruption in HeLa cells and hiPSCs subjected to three consecutive treatments with TALEN proteins as determined by the Surveyor assay. Surveyor nuclease cleavage at mismatches produces products of 254 bp and 277 bp. (B) Representative sequence analysis of the CCR5 locus in HeLa cells after three consecutive treatments with 3 μM TAT-TALEN proteins under hypothermic conditions. Multiple deletions (dashed) and insertions (lowercase) are aligned with the cleavage site (wild type [WT]). Underlines highlight binding sites for TALEN L and TALEN R.
To explore the potential of this method for practical application, we evaluated the effects of CCR5 gene disruption via TAT-TALEN proteins in hiPSCs. Under hypothermic conditions, hiPSCs subjected to three consecutive treatments with 3 μM TAT-TALEN proteins showed gene disruption frequencies of up to 5% (Figure 5A). It is unknown whether cell membrane composition or endocytosis capabilities could explain the observed differences between HeLa cells and hiPSCs with regard to CCR5 gene disruption frequency.
TAT-TALEN cytotoxicity analysis
To examine the cytotoxicity of TAT-TALENs, the proliferation of HeLa cells and hiPSCs treated with TAT-TALENs was measured. We observed a decrease in the proliferation of each cell type after three consecutive treatments with TAT-TALENs (Figure 6). Cytotoxicity was also dose-dependent, suggesting that consecutive low doses might be required to minimize potential toxic effects.
Figure 6.
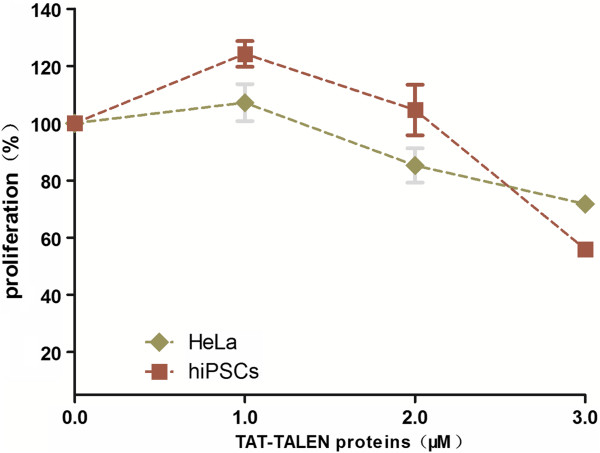
Proliferation of HeLa cells and iPSCs subjected to three consecutive treatments with various concentrations of TAT-TALEN proteins. Values are normalized to mock treated cells. Error bars, s.d. (n = 3).
Discussion
Targeted genomic engineering in stem cells at desired endogenous gene loci by ZFNs or TALENs has significant therapeutic implications [4]. The correction of disease-causing mutations in patient-derived stem cells is a goal of curative regenerative medicine. The disruption of a normal gene in stem cells can also be beneficial for the treatment of certain diseases. CCR5 is a major co-receptor for HIV-1 that is present on the surface of target cells [12]. Currently, autologous transplantation of CCR5-deficient human hematopoietic stem cells (hHSCs) has been a promising strategy for acquired immunodeficiency syndrome (AIDS) treatment [13–17]. Notably, HIV-1 can establish latent viral reservoirs in hHSCs [18] prior to genome engineering; thus, caution must be exercised when using these cells. Our lab has successfully disrupted the CCR5 locus of hiPSCs using ZFNs and demonstrated that these CCR5-disrupted hiPSCs can differentiate into CD34+ cells with multipotency in vitro[19]. This work contributes to the advancement of patient-specific therapies with genetically modified hiPSCs produced from the healthy tissues of HIV-infected patients. However, some problems such as safety concerns regarding the gene-based delivery method of ZFNs and TALENs need to be addressed [8, 20]. In addition to mRNA delivery system, our TAT-TALEN protein provides an alternative to eliminate the risk of insertional mutagenesis and expands the range of cell types that the nucleases can be applied to modify.
The concept of CPP-ZFN proteins has been proposed previously [21, 22]. However, we failed to utilize TAT-ZFN to modify the endogenous gene because of a purification problem. Recently, a study by Gaj and colleagues demonstrated the intrinsic cell-penetrating capabilities of standard ZFN proteins and their ability to successfully modify CCR5 in living cells [23]. Compared with ZFNs, TALENs are more mutagenic [24] and more specific [25]; additionally, the design and assemble of TALENs is very simple and straightforward [26, 27]. In all these aspects, TALENs are superior to ZFNs.
We utilized TAT-TALEN to successfully disrupt endogenous CCR5 in HeLa cells and hiPSCs. It has been reported that hypothermia treatment improves ZFN-driven [28] and TALEN-driven [29] gene disruption in human cells transfected with eukaryotic expression vectors. We found that hypothermic treatment was also effective in cells transduced with TAT-TALENs. A high TALEN concentration may lead to an increase in off-target cleavage events in cells; cells treated with excessive amounts of TAT-TALENs are likely to die as a result of non-specific genome cleavage. This effect may account for the moderate cytotoxicity observed in this experiment. In our study, TAT was fused to the N-terminus of TALEN. It is unknown whether shifting the TAT peptide to the C- terminus of TALEN or using other CPPs such as polyarginine could further enhance protein penetration efficiency; however, similar adjustments need to be considered in future studies.
In summary, we generated functional TAT-TALEN proteins that were capable of penetrating cells. These constructs were used to efficiently disrupt CCR5 in HeLa cells and hiPSCs. Combined with our previous work, this gene-free delivery system has great potential for the clinical application of CCR5-disrupted hiPSCs. In addition, this new TALEN delivery method may be applied to the gene therapy for several monogenic diseases as well, a circumstance in which a DNA donor template is needed.
Methods
Construction of prokaryotic expression plasmids
ZFN mammalian expression vectors were previously constructed in our lab [19]. TALEN mammalian expression vectors were constructed as described [26, 29]. The corresponding prokaryotic expression plasmids were constructed as follows: encoding sequences of ZFN and TALEN were cloned from their mammalian expression vectors. TAT-encoding sequences were introduced by primer design. All primer sequences are provided in Additional file 1: Table S1. The PCR products were digested with NdeI and BamHI for (ZFN) or NdeI and HindIII (for TALEN), and the digested products were inserted into pET28a (Novagen) to yield the following six plasmids: pET.TAT-ZFNL, pET.TAT-ZFNR, pET.TAT-TALENL, pET.TAT-TALENR, pET.TALENL, pET.TALENR. L and R stand for left and right, respectively. Left ZFN (or TALEN) and right ZFN (or TALEN) constitute a functional ZFN (or TALEN) pair. Proper construction of each expression cassette was verified by sequence analysis. The amino acid sequences of each protein are provided in Additional file 1: Table S2.
Expression and purification of fusion proteins
Recombinant expression plasmids were transformed into competent Escherichia-coli BL21 (DE3) cells. LB medium (40 ml) supplemented with 100 μg/ml kanamycin was inoculated with a single colony and incubated overnight at 37°C with shaking. On the following day, the overnight culture was diluted 50-fold into fresh LB medium supplemented with 100 μg/ml kanamycin and incubated at 37°C with shaking until the optical density at 600 nm (OD600) reached 0.5. The culture was then incubated at 16°C for 1 hour. Protein synthesis was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.7 mM, and the culture was grown overnight at 16°C with shaking. The cells were harvested and lysed five times using a French press (JN-3000 PLUS, JNBIO) at 10,000 psi in lysis buffer (25 mM Tris–HCl, pH 8.0, 300 mM NaCl, 5% V/V glycerol) at 4°C. After the cell debris was removed by centrifugation at 20,000 g for 30 minutes, TALEN proteins in the soluble fraction were purified using Ni-NTA agarose resin (QIAGEN) and eluted with elution buffer (lysis buffer with 500 mM imidazole, pH 8.0). For TAT-ZFN expression and purification, 100 μM ZnCl2 was added to all solutions.
Each protein was desalted using PD-10 columns (GE Healthcare) into PBS (pH 7.4) with 20% glycerol and sterilized by filtration through a 0.22 μM filter. The protein was then aliquoted and stored at −80°C. Protein purity was assessed by SDS-PAGE, and the total protein concentration was determined using a BCA protein assay kit (Beyotime, China). The total protein concentrations of standard TALEN and TAT-TALEN were between 0.9 and 1.2 mg/ml and between 0.3 and 0.5 mg/ml, respectively.
In vitro activity of purified proteins
Because the recognition sites of ZFN and TALEN are distinct from each other, CCR5 loci were amplified separately from the human genomic DNA with different primer pairs (Primer Pair 1 for ZFNs and Primer Pair 2 for TALENs). The substrate DNA was incubated with ZFN or TALEN in NEB buffer 4 for 1 hour at 37°C [30]. Cleavage products were separated by agarose gel electrophoresis (1.5%) or polyacrylamide gel electrophoresis (8%) in 1 × TBE buffer and stained with ethidium bromide solution.
Cell culture
HeLa cells were seeded onto 48-well plates at a density of 5 × 104 cells per well and cultured in a humidified atmosphere containing 5% CO2 at 37°C. Cells were maintained in high-glucose DMEM (HyClone) containing 10% fetal bovine serum (FBS) and 1% (v/v) penicillin-streptomycin.
hiPSCs were induced from the urine cells of a healthy individual using episomal vectors. Initial populations were seeded onto 48-well plates at the proper density and grown until cell confluence reached 60-70%. These cells were maintained in a feeder-free culture system using mTesR1 (Stem Cell Technologies, Vancouver, BC, Canada) and Matrigel (BD Bioscience, Bedford, MA) with 1% (v/v) penicillin-streptomycin.
Protein treatments
TALEN proteins, including standard TALENs and TAT-TALENs were prepared for treatment as follows: TALEN proteins were diluted into serum-free high-glucose DMEM. The cells were washed with serum-free medium and incubated with TALEN proteins for 1 hour at 37°C. After the initial treatment, the cells were washed and maintained at 37°C or shifted to 30°C for 24 hours before the next treatment. This process was repeated three times over four consecutive days.
Western blot analysis to determine protein internalization
HeLa cells were treated with either 1.5 μM TALEN or 1.5 μM TAT-TALEN for 1 hour at 37°C and then washed three times with 1 mg/ml heparin-PBS to remove proteins bound to the cell surface. After trypsinization, the cells were collected and lysed in RIPA lysis buffer (Beyotime) and subsequently boiled in the presence of loading buffer (50 mM Tris–HCl, pH 6.8, 100 mM dithiothreitol, 2% sodium dodecyl sulfate (SDS), 20% glycerol and 0.2 mg/mL bromophenol blue) for 10 minutes and subjected to electrophoresis using 10% SDS-PAGE. The gel was transferred to a polyvinylidene difluoride membranes using a Trans-blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad Laboratories, Hercules, CA). TALENs were verified using monoclonal anti-FLAG M2 antibodies (Sigma, 1:1000 dilution) and secondary HRP-labeled goat anti-mouse antibodies (Sigma, 1:5000 dilution). The internal loading control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was detected with peroxidase-conjugated anti-GAPDH antibody. The western blots were developed with DAB substrate according to the manufacturer’s instructions.
Surveyor nuclease assay
Genomic DNA was extracted from TALEN-treated cells with a DNeasy Blood and Tissue Kit (QIAGEN). The CCR5 locus was amplified by nested PCR, and the sequences of the outer and inner primers are provided in Additional file 1: Table S1. After amplification of the CCR5 locus using Platinum® Taq DNA Polymerase (Invitrogen), a Surveyor nuclease assay was performed following the instructions included with the SURVEYOR Mutation Detection Kit (Transgenomic). Analysis of the gene disruption frequency was performed as described [31]. The gel image was processed with the ImageJ software.
Cell proliferation assay
HeLa cells and hiPSCs were seeded onto 48-well plates at the optimal densities. The cells were then treated with TAT-TALEN proteins as described above. Cytotoxicity assays were conducted using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.) according to the manufacturer’s instructions.
Sequence analysis
Genomic DNA from TAT-TALEN-treated HeLa cells was isolated with the DNeasy Blood and Tissue Kit (QIAGEN). The CCR5 locus was amplified by PCR with the primers BamHI-5’hCCR5 and HindIII-3’hCCR5. Subsequently, the PCR products were cloned into PUC19 plasmids using the BamHI and HindIII restriction sites. CCR5 disruption was confirmed by sequence analysis of individual cloned transformants.
Ethical approval
This experiments was approved by the Human Subject Research Ethics Committee (IRB), Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences with the reference number: GIBH-IRB02-2009002.
Electronic supplementary material
Additional file 1: Table S1.: The sequences of primers used in this study. Table S2. The amino acid sequences of the ZFN and TALEN proteins used in this study. (PDF 118 KB)
Acknowledgments
We are grateful to Miguel A. Esteban and his group for supplying the hiPSCs. This work was financially supported by the National Science and Technology Major Project (2013ZX10001-004-002-004) and the National Natural Science Foundation of China (No. 81200398).
Footnotes
Competing interests
The authors confirm that there are no competing interests associated with this publication and there has been no significant financial support that could have influenced the outcome of this work.
Authors’ contributions
RR and YY conceived and designed the experiment, collected and/or assembled the data, performed data analysis and interpretation, and wrote the manuscript. SY, WX and BY performed cell culture and molecular cloning. SZ and LQ assisted with manuscript revisions. XC supervised the work, revised the manuscript and approved the final version of the manuscript prior to submission for publication. All Authors read and approved the final manuscript
Contributor Information
Renli Ru, Email: ru_renli@gibh.ac.cn.
Yongchao Yao, Email: yao_yongchao@gibh.ac.cn.
Songlin Yu, Email: yu_songlin@gibh.ac.cn.
Benpeng Yin, Email: yin_benpeng@gibh.ac.cn.
Wanwan Xu, Email: xu_wanwan@gibh.ac.cn.
Siting Zhao, Email: zhao_siting@gibh.ac.cn.
Li Qin, Email: qin_li@gibh.ac.cn.
Xiaoping Chen, Email: chen_xiaoping@gibh.ac.cn.
References
- 1.Mandell JG, BarbasIII CF. Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li T, Yang B. TAL Effector Nuclease (TALEN) Engineering. Methods Mol Biol. 2013;978:63–72. doi: 10.1007/978-1-62703-293-3_5. [DOI] [PubMed] [Google Scholar]
- 3.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collin J, Lako M. Concise review: putting a finger on stem cell biology: zinc finger nuclease-driven targeted genetic editing in human pluripotent stem cells. Stem Cells. 2011;29:1021–1033. doi: 10.1002/stem.658. [DOI] [PubMed] [Google Scholar]
- 5.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hockemeyer D, Wang HY, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng XD, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, Trevisan M, Gupta RM, Moisan A, Banks E, Friesen M, Schinzel RT, Xia F, Tang A, Xia Y, Figueroa E, Wann A, Ahfeldt T, Daheron L, Zhang F, Rubin LL, Peng LF, Chung RT, Musunuru K, Cowan CA. A TALEN Genome-Editing System for Generating Human Stem Cell-Based Disease Models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 9.Morris MC, Deshayes S, Heitz F, Divita G. Cell-penetrating peptides: from molecular mechanisms to therapeutics. Biol Cell. 2008;100:201–217. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]
- 10.Vyas PM, Payne RM. TAT opens the door. Mol Ther. 2008;16:647–648. doi: 10.1038/mt.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brasseur R, Divita G. Happy birthday cell penetrating peptides: already 20 years. Biochim Biophys Acta. 2010;1798:2177–2181. doi: 10.1016/j.bbamem.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Lopalco L. CCR5: From Natural Resistance to a New Anti-HIV Strategy. Viruses. 2010;2:574–600. doi: 10.3390/v2020574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 14.Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, Schneider T. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 15.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Symonds GP, Johnstone HA, Millington ML, Boyd MP, Burke BP, Breton LR. The use of cell-delivered gene therapy for the treatment of HIV/AIDS. Immunol Res. 2010;48:84–98. doi: 10.1007/s12026-010-8169-7. [DOI] [PubMed] [Google Scholar]
- 18.Huddleston JE. HIV: HIV hides in haematopoietic stem cells. Nat Rev Microbiol. 2011;9:311. doi: 10.1038/nrmicro2564. [DOI] [PubMed] [Google Scholar]
- 19.Yao Y, Nashun B, Zhou T, Qin L, Zhao S, Xu J, Esteban MA, Chen X. Generation of CD34+ Cells from CCR5-disrupted Human Embryonic and Induced Pluripotent Stem Cells. Hum Gene Ther. 2011;23:238–242. doi: 10.1089/hum.2011.126. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Troilo PJ, Wang X, Griffiths TG, Pacchione SJ, Barnum AB, Harper LB, Pauley CJ, Niu Z, Denisova L, Follmer TT, Rizzuto G, Ciliberto G, Fattori E, Monica NL, Manam S, Ledwith BJ. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11:711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- 21.Planel S, Salomon A, Jalinot P, Feige JJ, Cherradi N. A novel concept in antiangiogenic and antitumoral therapy: multitarget destabilization of short-lived mRNAs by the zinc finger protein ZFP36L1. Oncogene. 2010;29:5989–6003. doi: 10.1038/onc.2010.341. [DOI] [PubMed] [Google Scholar]
- 22.Nain V, Sahi S, Verma A. CPP-ZFN: a potential DNA-targeting anti-malarial drug. Malar J. 2010;9:258. doi: 10.1186/1475-2875-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas III CF. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Oikonomou G, Chiu CN, Niles BJ, Liu J, Lee DA, Antoshechkin I, Prober DA. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41:2769–2778. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, Deng WM, Jiao R. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics. 2012;39:209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 29.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 30.Tovkach A, Zeevi V, Tzfira T. A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J. 2009;57:747–757. doi: 10.1111/j.1365-313X.2008.03718.x. [DOI] [PubMed] [Google Scholar]
- 31.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1.: The sequences of primers used in this study. Table S2. The amino acid sequences of the ZFN and TALEN proteins used in this study. (PDF 118 KB)


