Abstract
Cells sense biochemical, electrical, and mechanical cues in their environment that affect their differentiation and behavior. Unlike biochemical and electrical signals, mechanical signals can propagate without the diffusion of proteins or ions; instead, forces are transmitted through mechanically stiff structures, flowing, for example, through cytoskeletal elements such as microtubules or filamentous actin. The molecular details underlying how cells respond to force are only beginning to be understood. Here we review tools for probing force-sensitive proteins and highlight several examples in which forces are transmitted, routed, and sensed by proteins in cells. We suggest that local unfolding and tension-dependent removal of autoinhibitory domains are common features in force-sensitive proteins and that force-sensitive proteins may be commonplace wherever forces are transmitted between and within cells. Because mechanical forces are inherent in the cellular environment, force is a signal that cells must take advantage of to maintain homeostasis and carry out their functions.
INTRODUCTION
Physical forces act on us every day when we push open a door or accelerate while riding in a car. These experiences provide us with an intuitive understanding of force at the scale of our bodies. Similarly, cells interact with their local environment by sensing and generating forces, but the magnitudes of these forces and the mechanisms by which cells and proteins respond to force are not as intuitive. This Perspective aims to provide an intuition for the magnitudes of forces involved in cellular processes and a framework for thinking about how molecules transmit, route, and sense mechanical signals.
Cells must respond to mechanical force to carry out vital processes. B cells selectively develop high-affinity antibodies by mechanically testing the strength of antigens on the surface of antigen-presenting cells (Natkanski et al., 2013); dividing cells rely on mechanical cues to control progression through several phases of mitosis (Rajagopalan et al., 2004; Pinsky and Biggins, 2005; Lafaurie-Janvore et al., 2013; Hotz and Barral, 2014); and cells sense mechanical properties of their surroundings to direct cell differentiation, wound healing, and tumor progression (Moore et al., 2010). Our senses of touch and hearing are also examples of cellular force transduction involving mechanosensitive ion channels that convert mechanical signals into electrical (ionic) currents. We will not focus on this class of mechanosensitive proteins, which are the subject of several reviews (Sackin, 1995; Hamill and Martinac, 2001; Kung, 2005). Instead, we focus on proteins that route forces through the cytoskeleton and on proteins that transduce these mechanical signals into biochemical signals.
By definition, a signal can be transmitted, routed, and transduced, and each of these steps can be a point for regulation. The study of cellular signaling has traditionally rested on biochemical concepts, in which chemical signals are transmitted via diffusion, routed by specific binding interactions, and transduced via activation of key effectors (e.g., ion channels, enzymes, or transcription machinery). Propagation of biochemical signals often occurs through allosteric changes in a protein's conformation or through phosphoregulation. Changes in phosphorylation state can directly control the activity of a binding site or active site of an enzyme, and they can induce conformational changes similar to allosteric mechanisms. Because mechanical loads on a protein can also change the conformation of a protein, it should be no surprise that evolution has produced molecules that use force-dependent conformational changes to route and transduce mechanical signals (Ingber, 1997).
In cells, mechanical signals are transmitted by cytoskeletal filaments such as actin and microtubules, routed by rearrangements of the cytoskeletal network, and transduced into biochemical signals by force-sensitive proteins. How cytoskeletal filaments transmit mechanical signals is straightforward; however, what molecular features allow certain proteins to modulate when and where mechanical signals are routed or transduced is an active area of research. Dembo et al. (1988) provided an early hypothesis about how proteins might exhibit sensitivity to forces when they introduced the concept of binding interactions, called catch bonds, that become stronger under a tensile force (Evans and Leung, 1984). Marshall et al. (2003) confirmed the existence of catch bonds by applying precise mechanical loads to individual L-selectin and P-selectin glycloprotein ligand-1 (PSLG-1) interactions and observing longer lifetimes under a range of tensile loads compared with interactions that were not under tension. Because catch bonds are preferentially stable once engaged, they can be likened to switches or routers of mechanical signals: when a catch bond is engaged, force is preferentially propagated through the adjoining proteins and cytoskeletal filaments. Other mechanosensitive proteins exhibit tension-dependent modulation of enzyme activity, tension-dependent accessibility of substrate sites, and tension-dependent availability of cryptic binding sites. These types of force-sensitive proteins can be likened to transducers that convert mechanical signals into biochemical signals.
FORCES SUSTAINED BY INDIVIDUAL PROTEINS
To estimate the forces that individual proteins sustain, consider that molecular interactions, such as enzyme-catalyzed reactions, require fluctuations in thermal energy to overcome transition states. Thus the energies involved must be similar in magnitude to thermal energy, which is given by Boltzmann's constant, kB, multiplied by temperature, T. At room temperature, kBT = 4.1 pN nm (4.1 × 10−21 J), and since proteins are nanometer sized, forces acting on individual proteins must be in the piconewton range. One piconewton is 10−12 N, about the weight of one red blood cell, approximately the force exerted by a standard laser pointer on a screen, and about the drag force on a 30-nm vesicle moving at 500 nm/s through cytosol (Courty et al., 2006; Howard, 2001). One can also consider the molecular interactions that enable proteins to bind other proteins. Electrostatic interactions between the charged surfaces of two proteins generate forces on the order of 1–100 pN that decay with d−2, where d represents the distance between the charges. For instance, according to Coulomb's law, two point charges separated by 0.5 nm in water exert a force on one another of ∼10 pN. van der Waals forces and hydrogen bonds, which are due to permanent or transient dipole moments in polar molecules, are on the order of 10 pN and decay with d−3 (Howard, 2001). Finally, hydrophobic interfaces associate with strengths similar to those of hydrogen bonds (Chandler, 2005) due to the hydrophobic effect, which is an entropy-driven process that reduces the amount of ordered water around nonpolar molecules. Hence essentially all types of forces relevant to biological protein–protein interactions and biological enzyme catalysis are on the piconewton scale (see Table 1 for examples).
TABLE 1:
Common cellular events in which forces are critical to biochemical function.
| Event | Speed or lifetime | Relevant force (pN) | Note | References |
|---|---|---|---|---|
| Measurements in reconstituted systems | ||||
| Kinesin movement on a microtubule | 800 nm/s | 5–7.5 | Maximum load on motor before it stalls | Svoboda and Block (1994), Kojima et al. (1997), Visscher et al. (1999) |
| Dynein movement on a microtubule | 85 nm/s | 7–10 | Maximum load on motor before it stalls | Reck-Peterson et al. (2006, 2012) |
| Myosin movement on an actin filamenta | 0.03 s at 6 pN | 10 and 80 | Rupture forces at ramp rates ∼5 and 1000 pN/s | Guo and Guilford (2006) |
| Activation of titin kinase by removal of inhibitory peptide | — | 30 | Equivalent to the activity of ∼5 or 6 myosin units | Puchner et al. (2008) |
| VWF tethering platelets to endothelial cellsa | 0.2 s at 20 pN | 5–80 | Force required to reveal protease cleavage site | Zhang et al. (2009), Wu et al. (2010) |
| One kinetochore complex binding to one microtubulea | 50 min at 5 pN | 9 | Rupture force at a ramp rate of 0.25 pN/s | Akiyoshi et al. (2010) |
| FimH-mannose bonda | — | ∼150 | Rupture force at a ramp rate of 250 pN/s | Yakovenko et al. (2008) |
| Single integrin in vitroa | 10 s at 30 pN | 13–50 | Rupture force at ramp rates of 50–100 pN/s | Thoumine et al. (2000), Li et al. (2003), Kong et al. (2009) |
| Unfolding of talin to reveal vinculin binding sites | 5 | Force at a ramp rate of 5 pN/s | Yao et al. (2014) | |
| Measurements and estimates in live cells | ||||
| Single kinesin transporting a 30-nm quantum dot | 570 ± 20 nm/s | 0.6 | Estimated drag force on quantum dot during transport | Courty et al. (2006) |
| Chromosome segregation in anaphase | 100 nm/s | 0.1–10 | Estimated force to move chromosome in vivo during anaphase | Nicklas (1988), Alexander and Rieder (1991), Marshall et al. (2001), Fisher et al. (2009), Civelekoglu-Scholey and Scholey (2010) |
| Force to stop chromosome movement during anaphase | — | 700; 50 | Force per chromosome; force per kinetochore microtubule | Nicklas (1988) |
| Single integrin in cells to RGD on surface | — | 1–5 | FRET sensor in ECM | Ivaska (2012), Morimatsu et al. (2013) |
| Single vinculin connecting talin to F‑actin in cells | Minutes | 2.5 ± 1.0 up to 10 | FRET sensor in cells | Grashoff et al. (2010) |
| Activation of Notch during cell adhesion | 5–15 min | <12 | Based on tension-gauge-tether sensor | Wang and Ha (2013) |
| Contractile forces through focal adhesion complexes | Minutes to hours | 100–165 | Estimate for a complex of 3–5 integrins | Moore et al. (2010) |
aEvent involving protein with catch-bond behavior.
TOOLS FOR DETERMINING HOW PROTEINS RESPOND TO FORCE
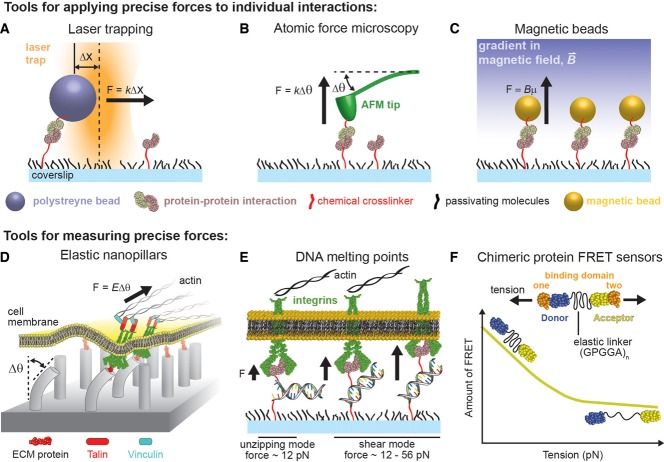
Table 1 shows that tools for direct observation of individual protein interactions must operate with nanometer precision and on the piconewton force scale. Biophysical tools capable of this precision can be grouped into two categories: those that actively control position and therefore apply force (Figure 1, A–C) and those that passively measure force (similar to a strain gauge; Figure 1, D–F). To apply force actively to a protein interaction of interest, the interaction is typically recapitulated in vitro with one protein tethered to a glass coverslip and its binding partner tethered to a polystyrene bead for laser trapping (Ashkin and Dziedzic, 1987; Neuman and Block, 2004; Matthews, 2009), to a magnetic bead for magnetic tweezers (Smith et al., 1992; Strick et al., 1996; Guttenberg et al., 2000), or to the tip of a submicrometer cantilever for atomic force microscopy (AFM; Binnig et al., 1986). Another method, not illustrated in Figure 1, called the biomembrane force probe, can also be used to apply forces to protein–protein interactions; however, most investigators have moved toward the laser trapping, AFM, or magnetic beads platforms (Evans et al, 1995; Gourier et al., 2008; Neuman and Nagy, 2008). Proteins are typically tethered using well-established conjugation chemistries (Hermanson, 2013; Kim and Herr, 2013), and in general tethering is not a major limitation. The physical principles underlying these instruments are outside the scope of this Perspective (see review by Neuman and Nagy, 2008). However, all of them share the ability to manipulate the position of a tethered protein or complex with nanometer precision while observing the deflection of a sensor element (i.e., bead or AFM tip) from its resting position. This deflection is usually proportional to the force applied on the protein–protein interaction with sub-piconewton sensitivity.
FIGURE 1:
Methods for applying and measuring precise forces to single molecules and molecular complexes. (A–C) Instruments often used to apply precise forces to individual macromolecules or complexes. (A) In laser trapping, a focused laser beam behaves roughly like a Hookean spring, pulling a submicrometer bead toward its center with a force proportional to the stiffness of the laser trap, k, multiplied with the displacement of the bead from the trap center, Δx; beads are often decorated with a protein or receptor of interest and can be controlled by manipulating the position of the laser beam relative to the microscope slide. (B) Atomic force microscopes employ a micrometer-width cantilever, at the tip of which is a nanometer-sized pointer that can be decorated with proteins or receptors; once these proteins bind their receptors on the surface of a glass slide, the cantilever is retracted causing it to deflect. (C) Magnetic tweezers employ magnetic beads with a magnetic moment, μ; when subjected to a magnetic field, the force on the beads is proportional to the magnetic field strength multiplied by μ. Up to several hundred magnetic beads can be pulled at the same time. (D–F) Techniques for measuring forces precisely between and within molecules. (D) Pillars with diameters and lengths on the nanometer to submicrometer scale can be formed from elastic polymers and decorated with extracellular matrix proteins, such that cultured cells adhere and form focal adhesions; the deflection of each nanopillar from its resting position reveals the contractile forces exerted at the corresponding focal adhesion. (E) Hybridized dsDNA molecules for which one strand is tethered to a surface and the complementary strand is tethered to a protein or receptor can act as a “tension-gauge-tether” by which the number of base pairs within the dsDNA that support the load dictates a well-defined force at which the dsDNA will unzip or melt; unzipping of the dsDNA can be observed using fluorescent tags on the DNA molecules or by cell phenotypes, allowing estimation of the range of forces to which a protein–ligand interaction might be subjected during a cellular event such as early stages of cell adhesion. (F) An intramolecular strain sensor based on FRET can be used to determine the forces exerted through a protein by engineering the probe into the protein structure and monitoring the level of FRET.
Experiments using these instruments can be performed in several ways. To quantify the strength of a binding interaction, the force on an interaction may be steadily increased (“ramped”) until the interaction dissociates (i.e., until it mechanically fails). The force at which failure occurs is called the rupture force, and it is a function of the intrinsic binding strength, as well as of the rate at which the force was increased (Evans, 2001). To test for force-dependent stabilization of an interaction, as occurs in catch bonds, the average lifetime of an interaction can be measured as a function of a constant applied force (i.e., with a “force clamp”); any increase in the lifetime with increasing force indicates catch-bond behavior (Dembo et al., 1988). More complex force profiles, such as sinusoids or rapid changes in force, can also be applied and may reveal time dependence that would otherwise be difficult or impossible to observe.
Techniques for passively measuring piconewton-scale forces use cleverly engineered molecules or nanoscale materials. For instance, arrays of pillars formed in elastic substrates can be decorated with adhesion molecules, and when cells adhere, their contractile forces are applied to the pillars via focal adhesion complexes. Subsequent bending of the pillars indicates the forces at individual focal adhesions (Figure 1D; Tan et al., 2003). The unzipping behavior of individual double-stranded DNA (dsDNA) molecules can be used as a molecular force gauge, since dsDNA unzips at well-defined and tunable levels of force, depending on how many base pairs support the load (Figure 1E). Another type of molecular force gauge relies on insertion of a stress sensor into a structural protein host (Figure 1F). The stress sensor consists of elastic α-helical peptides (Meng et al., 2008) or elastic derivatives of the spider silk flagelliform protein (Grashoff et al., 2010), which connect a pair of fluorophores or fluorescent proteins capable of Förster resonance energy transfer (FRET). When this sensor is inserted into the flexible region of a protein believed to transmit force, the magnitude of the FRET signal indicates the amount of stretching in the sensor and, when calibrated, the force being transmitted through the protein in vivo (Grashoff et al., 2010).
MOLECULAR REARRANGEMENTS TRANSDUCE FORCES INTO BIOCHEMICAL SIGNALS
The foregoing single-molecule approaches, in combination with atomic structures and mutational analyses, are beginning to reveal how cells route and transduce mechanical signals (Thomas, 2009; Forties and Wang, 2014). In many cases, mechanosensitivity derives from tension-dependent allosteric regulation or local unfolding of a protein. In this section we provide brief details for several well-studied examples.
P-selectin proteins exhibit catch-bond behavior when bound to PSGL-1. Together these two proteins play a crucial role after injuries as P‑selectin, which is exposed on the surface of epithelial cells after exposure to inflammatory cytokines, captures PSGL-1–presenting platelet cells to hold them at the location of injury. AFM experiments revealed that tensile loads stabilize selectin–PSGL-1 interactions (Marshall et al., 2003; Sarangapani et al., 2004). This stabilization stems from the allosteric regulation between selectin's two domains, which are connected through a hinge region that can adopt one of two conformations. Tension across selectin straightens the hinge region, concomitantly positioning residues in the binding pocket to make high-quality molecular interactions with PSGL-1 and improving the strength of the interaction (Lou et al., 2006; Phan et al., 2006; Klopocki et al., 2008; Waldron and Springer, 2009). For an illustrated example see Rakshit and Sivasankar (2014). A similar mechanism appears in intracellular adhesion molecule-1 (ICAM-1), which exhibits catch-bond behavior when binding to lymphocyte function-associated antigen-1 (LFA-1; Chen et al., 2010). The bacterial adhesive protein FimH also has two domains and exhibits catch-bond behavior through a slightly different mechanism. Its lectin domain, which binds mannosylated proteins, connects through a flexible peptide to its pilin domain, which anchors the protein to fimbriae. Binding of the pilin to the lectin domain reduces lectin's affinity for mannose (Aprikian et al., 2007; Yakovenko et al., 2007). AFM experiments revealed that individual lectin–mannose interactions in vitro typically fall into a low- or a high-strength population (Yakovenko et al., 2008), but applying an intermediate tension before increasing the force shifts most interactions into the high-strength population (Yakovenko et al., 2008). A crystal structure revealed that when the pilin and lectin domains are associated, a β-sheet sandwich in lectin is twisted such that the mannose binding pocket is in a low-affinity state (Le Trong et al., 2010). In contrast, tension-dependent dissociation of the pilin from the lectin domain allows the β-sheet sandwich to untwist and tighten the ligand pocket. Similar sandwich domains are found in many adhesive, matrix, and extracellular proteins believed to be under tensile loads (Le Trong et al., 2010; Puchner and Gaub, 2012) and thus are likely well suited to propagate mechanical signals. In general, the common mechanism underlying these adhesion proteins is the tension-dependent removal of an autoinihibitory domain, which concomitantly induces a conformational change in the binding pocket of protein–protein interactions that are under tension. Because these types of force-stabilized bonds influence where and when forces are transmitted through the cytoskeleton, it is not surprising that catch-bond behavior is commonly observed in cell-adhesion proteins.
Clusters of cell-adhesion proteins occur at focal adhesions and contain multiple mechanosensitive proteins involved in coupling transmembrane α/β-integrins to the actin network. The extracellular portion of α/β-integrins connects to fibronectin in the extracellular matrix (ECM) and exhibits classic catch-bond behavior, likely through an allosteric pathway (Kong et al., 2009). Contractile forces generated in the actomyosin network are transmitted through talin to the integrins and the ECM. Talin contains a C-terminal rod-like structure consisting of 13 α-helical bundles. Under ∼5 pN of tension, several α-helical bundles unfold, revealing binding sites for vinculin (Figure 2; del Rio et al., 2009; Yao et al., 2014). Vinculin binds to these cryptic sites with high affinity (nanomolar range), preventing the α-helical bundles from refolding and recruiting additional actin filaments (Ciobanasu et al., 2014; Yao et al., 2014). The onset of vinculin binding to talin correlates with an increase in the strength of the focal adhesion (Ciobanasu et al., 2014). An intracellular FRET sensor (Figure 1D) inserted into vinculin indicated that each vinculin in a stable focal adhesion supports an average of 2.5 pN, that recruitment of vinculin and force transmission are independently controlled processes, and that the ability of vinculin to transmit force determines whether a focal adhesion will assemble or disassemble under tension (Grashoff et al., 2010).
FIGURE 2:
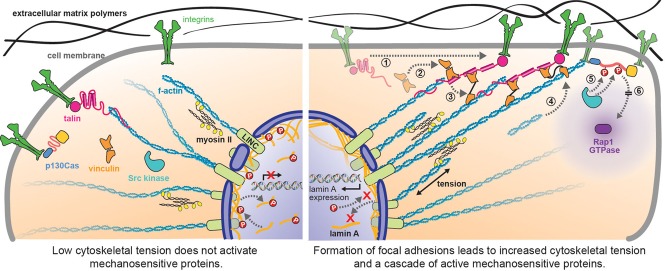
Forces in cells are routed, transmitted, and transduced by mechanically sensitive proteins and have cell-wide implications, influencing biochemical signaling in the cytoplasm and gene expression in the nucleus. Forces generated in the actin cytoskeleton by myosin II cross-bridges are transmitted several micrometers between adhesion proteins in cell membrane and LINC complexes in the nuclear cortex. Tension-dependent unfolding of talin (step 1) reveals substrates for vinculin binding (step 2), which in turn recruits additional actin filaments (steps 3 and 4) as part of focal adhesion development (del Rio et al., 2009; Ciobanasu et al., 2014; Yao et al., 2014). Tension-dependent unfolding of p130Cas reveals phosphorylation sites for Src kinase as part of integrin signaling and ultimately generates the active form of a diffusible GTPase Rap1(steps 5 and 6; Sawada et al., 2006). Concomitantly, tension in the actin cytoskeleton is transmitted through LINC complexes to the nuclear cortex. Lamin A, an intermediate filament of the nuclear cortex, mechanically couples the nuclear cortex to LINC complexes and therefore the cytoplasmic cytoskeleton; it affects DNA transcription of the gene for lamin A and the transcription of stress fiber genes (Swift et al., 2013). Increased cytoskeletal tension on LINC complexes correlates with decreasing phosphorylation of lamin A, decreasing turnover of lamin A in the nuclear cortex, increasing stiffness of the nuclear cortex, and ultimately, through the retnonic acid pathway, increasinglevels of lamin A. Note that many intermediate proteins are not shown, for simplicity.
Whereas catch bonds influence when and where forces are transmitted, other mechanosensitive proteins transduce forces into biochemical signals with downstream effectors. For instance, local unfolding of a protein under tension can reveal substrates for enzymatic modification (e.g., protease cleavage sites, phosphorylation sites, or glutathionylation sites). For example, the blood protein von Willebrand Factor (VWF) is partially unfolded by forces on the order of 10–20 pN, the same range of forces it is expected to experience in blood vessels due to shear flow, revealing an otherwise hidden protease site (Interlandi and Thomas, 2010). Cleavage at this site reduces the size of VWF aggregates and inhibits blood coagulation. VWF also exhibits catch-bond-like behavior in which force increases its interaction with platelet receptor protein Iβ/α. The p130Cas protein, which associates with integrins as part of integrin signaling, also unfolds under tension to reveal phosphorylation sites for the Src family of kinases (Sawada et al., 2006). Phosphorylation of these sites leads to the activation of a diffusible secondary messenger called Rap1, which is a GTPase involved in integrin signaling (Hattori and Minato, 2003; Tamada et al., 2004).
Local unfolding can also reveal the active site of a kinase and is the mechanism underlying the tension-dependent activity of the muscle protein titin (Puchner et al., 2008). Titin connects myosin bundles in the A band to the Z-disk in sarcomeres, where it is optimally positioned to sense muscle tension and thereby control the size of sarcomere units (Agarkova et al., 2003; Agarkova and Perriard, 2005). The kinase domain of titin is closely related to the myosin light-chain kinase (MLCK) class of enzymes, which are activated by repositioning of an autoinhibitory tail upon Ca2+ binding. Unlike MLCKs, the active site of titin is revealed only upon forceful repositioning of the autoinhibitory tail under tension (Puchner et al., 2008; Gautel, 2011). Once the active site is revealed, titin phosphorylates downstream signaling proteins that affect the expression of muscle genes and the turnover of proteins (Lange et al., 2005). Another domain of titin, the immunoglobulin G (IgG) domain, unfolds under ∼100 pN of force in vitro, revealing cysteines for S-glutathionylation. Glutathionylation of these cysteines prevents the IgG domain from completely refolding, thereby affecting the intrinsic length of titin and the elasticity of cardiomyocytes (Alegre-Cebollada et al., 2014). Thus titin exhibits features of tension-dependent kinase activity and tension-dependent unveiling of cryptic substrates, which ultimately enable titin to participate in regulating muscle elasticity.
The foregoing examples illustrate that local unfolding is a shared molecular mechanism in many mechanosensitive proteins.
MECHANOREGULATION AT A DISTANCE
Mechanical signaling has the potential to transmit information very quickly. Mechanical forces are transmitted at the speed of sound, orders of magnitude faster than diffusion for any distances greater than tens of nanometers, arriving in microseconds, compared with seconds or minutes for diffusion. There is precedence in biology for rapid transmission of signals across cells: in muscle cells, electrical signals, which also travel much faster than diffusive signals, penetrate deep into the cell along t-tubules, where they arrive very close to sarcomeres; at this point, the electrical signal is transduced into a biochemical signal: release of calcium from the sarcoplasmic reticulum.
Could analogous mechanisms exist in mechanical signaling? A recent report suggests that mechanoregulation occurring at integrins in the cell membrane may route forces through the cytoplasmic cytoskeleton directly to the nuclear cortex and ultimately to transcription factors in the nucleus (Figure 2; Swift et al., 2013). LINC complexes are the nuclear envelope equivalent of integrins in the cell membrane. They span the nuclear envelope, with their cytosolic side binding actin and their nuclear side binding the intermediate filaments, lamin A and lamin C, an ideal arrangement for transmitting forces within cytosolic actin into the intranuclear cortex. Like integrins, LINC complexes may form catch bonds or exhibit mechanical-to-biochemical transducing behavior. Consistent with this expectation, high cytoskeletal tension correlates with low turnover and dephosphorylation of lamin A, perhaps because phosphorylation sites on lamin A are sequestered by tension in a mechanism similar to the tension-dependent sequestration of protease sites in collagen (Camp et al., 2011; Swift et al., 2013). Moreover, actin filaments emanating from a subset of focal adhesions appear to connect directly to LINC complexes in the nuclear membrane, and these actin-cap-associated focal adhesions respond more readily to changes in the stiffness of the extracellular matrix than non–actin-cap-associated focal adhesions (Kim et al., 2012). Thus it certainly seems plausible that mechanical signals can be routed through catch-bond behavior at the cell membrane, transmitted long distances through cytoskeletal elements, and transduced into a biochemical signals that affect gene expression directly in the nucleus.
FORCE SENSING DURING CELL DIVISION
The key steps of cell division are regulated by checkpoints such as the spindle assembly checkpoint, which delays anaphase until all chromosomes are properly attached to microtubules emanating from each spindle pole. Whether force-sensing proteins participate in mitotic checkpoints is an active area of research and is likely understudied due to the difficulties of measuring precise forces in vivo and reconstituting these complex systems in vitro. Given the presence of force-sensitive proteins in many aspects of cell behavior, however, it seems likely that they participate in routing mechanical signals through spindle microtubules and transducing mechanical signals into biochemical signals by which the ends of microtubules are anchored to their substrates.
On chromosomes, the multiprotein structure that binds the plus ends of spindle microtubules is the kinetochore. Even before kinetochores and the roles of microtubules were understood, tension-dependent processes were implicated in orienting chromosomes for proper segregation when Dietz in the 1950s noticed that bioriented chromosomes remained stably in that configuration, whereas maloriented chromosomes were unstable and constantly reorienting. Dietz proposed that the defining feature of properly oriented chromosomes was tension, and that all chromosomes are biased toward the bioriented state through tension-dependent stabilization of their attachments to the spindle (Dietz, 1958). In support of this hypothesis, Nicklas later showed that maloriented chromosomes could remain stable for hours if tension was artificially applied using a microneedle, whereas relaxed, maloriented bivalents reoriented within minutes (Nicklas and Koch, 1969). We now know that kinetochores contain at least 80 different proteins in various copy numbers (Cheeseman and Desai, 2008; Biggins, 2013), but determining the strength of kinetochore coupling to microtubules in vivo has remained difficult, in part due the unknown and size-dependent viscosity of the nucleoplasm, the complex architecture of the spindle, and likely differences in the kinetochore–microtubule coupling strength in different organisms. Despite these challenges, theoretical and experimental efforts to determine the strength of kinetochore attachments to microtubules in vivo indicate that kinetochores couple to individual microtubules during metaphase with strengths on the order of 1–100 pN (Nicklas, 1988; Marshall et al., 2001; Chacón et al., 2014). The number of microtubules attached to kinetochores varies among organisms, ranging from one microtubule/kinetochore in yeast to 15–20 microtubules/kinetochore in humans (Cheeseman and Desai, 2008), and thus the net strength of kinetochore attachments to the spindle could approach nanonewton forces in some organisms. Chromosomes in grasshopper spermatocytes, which have ∼15 microtubules/kinetochore, resisted opposing forces up to 700 pN (Nicklas, 1988). It remains an open question as to why chromosomes can couple to the spindle with such high and widely varying strengths, since the forces to segregate chromosomes in vivo during anaphase are estimated to be between 0.1 and 10 pN (Marshall et al., 2001; Fisher et al., 2009; Ferraro-Gideon et al., 2013). In vitro tests using the laser trapping method (Figure 1) revealed that isolated kinetochore complexes can bind dynamically to microtubules and harness the energy of disassembling microtubule tips to move loads with forces up to an average of 9 pN (Table 1). Moreover, these kinetochore particles exhibited catch-bond-like behavior, in which the lifetime of kinetochore–microtubule interactions increased with tension over the range of 1–5 pN (Akiyoshi et al., 2010). The physiologically relevant consequence of this catch-bond behavior is that kinetochores on bioriented sister chromatids, which are under tension, form more stable attachments to microtubules than those attachments to kinetochores on chromatids that are not bioriented and therefore not under tension, in agreement with the hypothesis of Dietz. Adding to the complexity, these laser trapping experiments also revealed that tension applied across the kinetochore-–microtubule interface affected microtubule dynamics. Ongoing efforts to reconstitute the load-bearing components of the kinetochore piece by piece may reveal the force-sensitive elements and the effects of microtubule dynamics (Asbury et al., 2006; Franck et al., 2007, 2010; Gestaut et al., 2008, 2010; Powers et al., 2009; Tien et al., 2010, 2013; Umbreit et al., 2012; Volkov et al., 2013). There is strong evidence that the activity of several enzymes also facilitates biorientation of sister chromatids. For example, Aurora B kinase is known to phosphorylate various kinetochore proteins and thereby weaken kinetochore–microtubule attachments. Aurora B activity declines with the onset of tension across the microtubule–kinetochore interface (Tanaka et al., 2002; Dewar et al., 2004; Welburn et al., 2010; DeLuca et al., 2011), but how the activity of Aurora B responds to tension is unclear. Several components of the kinetochore have been proposed as tension sensors that could affect the activity of Aurora B (Biggins, 2013). One candidate is the Sli15-Bir1 protein complex, which localizes to kinetochores and contains an Aurora B–activating domain. Tension across Sli15-Bir1 might reveal a binding site to sequester the Aurora B–activating domain, thus preventing Aurora B–mediated phosphorylation of kinetochore proteins when chromosomes are properly bioriented (Sandall et al., 2006). Alternatively, tension within kinetochore proteins might induce a conformational change that 1) sequesters Aurora B substrates, 2) physically separates Aurora B from its substrates, or 3) induces a conformational change in the catalytic site of kinetochore-associated Aurora B, thereby inhibiting its activity (Lampson and Cheeseman, 2011; Biggins, 2013; Sarangapani and Asbury, 2014).
Mechanical signals in the spindle might also regulate processes at centrosomes. Centrosomes, called spindle pole bodies in yeast, are mechanical hubs balancing forces from astral, kinetochore, and interpolar microtubules. In yeast, they serve as signaling platforms for the mitotic exit network, sporulation, and possibly the spindle position checkpoint (Jaspersen and Winey, 2004); therefore mechanosensitive proteins may reside at centrosomes to transduce mechanical signals into biochemical signals during regulation of these processes. One study implicates kendrin proteins, called Pcp1 in fission yeast and Spc110 in budding yeast, as possible tension sensors (Rajagopalan et al., 2004) to control progress through the spindle position checkpoint. So far, however, the lack of suitable techniques for mechanically probing protein interactions with centrosomes has precluded a clear understanding of the extent to which centrosomes participate in mechanical signaling pathways.
Mechanical signaling in other aspects of cell division are also being actively investigated. For instance, the final step of cell division is abscission of the intercellular bridge, which takes anywhere from 1 to 3 h, even in the same cell lines (Steigemann et al., 2009). Despite this large time window, abscission occurs promptly if tension in the intercellular bridge is suddenly removed by cutting the bridge with a laser (Lafaurie-Janvore et al., 2013), suggesting that elements of the abscission machinery may be inhibited by tension. Other indications for mechanical signaling in mitotic processes stem from observations that confined cells undergoing mitosis that are prohibited from rounding up appear to have difficulty capturing chromosomes, maintaining defined spindle poles, and stably positioning their spindles (Fink et al., 2011; Lancaster et al., 2013; Cadart et al., 2014). These defects could stem from disruptions in the normal routing of mechanical signals important for functionality of the checkpoints that govern kinetochore–microtubule attachment and spindle positioning. The precise role of mechanical signaling in all aspects of cell division warrants thorough investigation and will likely benefit from molecular force probes that can be integrated into in vivo structures.
CONNECTING FORCES TO SIGNALS
This review has highlighted some well-understood mechanisms for transmitting, routing, and sensing mechanical signals, and it has discussed emerging areas of mechanobiology in which mechanical signaling is likely to be important. In many instances, combinations of protein structures, molecular dynamic simulations, and single-molecule force spectroscopy were required to uncover the molecular basis for mechanosensation. Single-molecule techniques have been particularly powerful since they allow careful control of the force, force history, and loading rates on individual molecules. These tools have enabled researchers to uncover how numerous mechanically active protein systems work, including how myosin, kinesin, and dynein convert chemical energy into work and how simple catch-bond systems become stronger under load (FimH, selectins). Various examples of mechanically regulated enzymes have also been demonstrated directly (Src kinase, titin kinase, collagenases). Thus it seems certain that these tools will be crucial for understanding at the molecular level all forms of mechanoregulation. A major challenge will be applying these tools to even more complex systems, such as the eukaryotic replisome, the ribosome, the spliceosome, the kinetochore, and centrosomes. In these systems, force may affect multiple proteins simultaneously, or the complex as a whole, in ways that will be difficult to discern without studying the whole macromolecular complex for emergent behaviors and without understanding the entire structure. Fortunately, it is becoming easier to study large complexes, for two reasons. First, advances in traditional biochemical techniques are making it possible to isolate larger protein assemblies in vitro. Second, next-generation single-molecule tools are more capable, multimodal (e.g., combined laser trap and total internal reflection fluorescence microscopes), reliable, and accessible than ever. There is a synergy between these two advances because the single-molecule techniques allow one to work with much smaller amounts of material than bulk techniques and thus make it possible for useful information to be gained even for large assemblies for which large amounts of concentrated material can be difficult or impossible to obtain. As the molecular components sensitive to force become better characterized, it may become possible to complete the picture of mechanoregulatory cascades that involve transmitting, routing, and transducing mechanical signals.
Acknowledgments
We thank Trisha Davis for critical reading of the manuscript and valuable comments. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award F32GM109493 to E.C.Y. and Grant RO1GM079373 to C.L.A. and also by Packard Fellowship 2006-30521 to C.L.A.
Abbreviations used:
- AFM
atomic force microscopy
- FRET
Förster resonance energy transfer
- ICAM-1
intracellular adhesion molecule-1
- LFA-1
lymphocyte function-associated antigen-1
- PSLG-1
P-selectin glycloprotein ligand-1
- VWF
von Willebrand Factor
Footnotes
REFERENCES
- Agarkova I, Ehler E, Lange S, Schoenauer R, Perriard JC. M-band: a safeguard for sarcomere stability. J Muscle Res Cell Motil. 2003;24:191–203. doi: 10.1023/a:1026094924677. [DOI] [PubMed] [Google Scholar]
- Agarkova I, Perriard JC. The m-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre-Cebollada J, Kosuri P, Giganti D, Eckels E, Rivas-Pardo, Jaime A, Hamdani N, Warren, Chad M, Solaro RJ, et al. S-glutathionylation of cryptic cysteines enhances titin elasticity by blocking protein folding. Cell. 2014;156:1235–1246. doi: 10.1016/j.cell.2014.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: Initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprikian P, Tchesnokova V, Kidd B, Yakovenko O, Yarov-Yarovoy V, Trinchina E, Vogel V, Thomas W, Sokurenko E. Interdomain interaction in the fimh adhesin of escherichia coli regulates the affinity to mannose. J Biol Chem. 2007;282:23437–23446. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic J. Optical trapping and manipulation of viruses and bacteria. Science. 1987;235:1517–1520. doi: 10.1126/science.3547653. [DOI] [PubMed] [Google Scholar]
- Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 2013;194:817–846. doi: 10.1534/genetics.112.145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Cadart C, Zlotek-Zlotkiewicz E, Le Berre M, Piel M, Matthews HK. Exploring the function of cell shape and size during mitosis. Dev Cell. 2014;29:159–169. doi: 10.1016/j.devcel.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Camp RJ, Liles M, Beale J, Saeidi N, Flynn BP, Moore E, Murthy SK, Ruberti JW. Molecular mechanochemistry: low force switch slows enzymatic cleavage of human type i collagen monomer. J Am Chem Soc. 2011;133:4073–4078. doi: 10.1021/ja110098b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón JM, Mukherjee S, Schuster BM, Clarke DJ, Gardner MK. Pericentromere tension is self-regulated by spindle structure in metaphase. J Cell Biol. 2014;205:313–324. doi: 10.1083/jcb.201312024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the αa domain generates lfa-1/icam-1 catch bonds. J Biol Chem. 2010;285:35967–35978. doi: 10.1074/jbc.M110.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanasu C, Faivre B, Le Clainche C. Actomyosin-dependent formation of the mechanosensitive talin–vinculin complex reinforces actin anchoring. Nat Commun. 2014;5:3095. doi: 10.1038/ncomms4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Scholey J. Mitotic force generators and chromosome segregation. Cell Mol Life Sci. 2010;67:2231–2250. doi: 10.1007/s00018-010-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Tracking individual kinesin motors in living cells using single quantum-dot imaging. Nano Lett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu RC, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca KF, Lens SMA, DeLuca JG. Temporal changes in hec1 phosphorylation control kinetochore–microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment; Proc R Soc B Biol Sci; 1988. pp. 55–83. [DOI] [PubMed] [Google Scholar]
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Dietz R. Multiple geschlechchromosomen bei den cypriden ostracoden, ihre evolution und ihr teilungsverhalten. Chromosoma. 1958;8:183. [PubMed] [Google Scholar]
- Evans E. Probing the relation between force-lifetime-and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- Evans E, Leung A. Adhesivity and rigidity of erythrocyte-membrane in relation to wheat-germ-agglutinin binding. J Cell Biol. 1984;98:1201–1208. doi: 10.1083/jcb.98.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Ritchie K, Merkel R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys J. 1995;68:2580–2587. doi: 10.1016/S0006-3495(95)80441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro-Gideon J, Sheykhani R, Zhu Q, Duquette ML, Berns MW, Forer A. Measurements of forces produced by the mitotic spindle using optical tweezers. Mol Biol Cell. 2013;24:1375–1386. doi: 10.1091/mbc.E12-12-0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J, Carpi N, Betz T, Betard A, Chebah M, Azioune A, Bornens M, Sykes C, Fetler L, Cuvelier D, et al. External forces control mitotic spindle positioning. Nat Cell Biol. 2011;13:771–778. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- Fisher JK, Ballenger M, O’Brien ET, Haase J, Superfine R, Bloom K. DNA relaxation dynamics as a probe for the intracellular environment. Proc Natl Acad Sci USA. 2009;106:9250–9255. doi: 10.1073/pnas.0812723106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forties RA, Wang MD. Discovering the power of single molecules. Cell. 2014;157:4–7. doi: 10.1016/j.cell.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Davis TN, Asbury CL. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 2010;51:242–250. doi: 10.1016/j.ymeth.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–U171. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M. Cytoskeletal protein kinases: titin and its relations in mechanosensing. Pflugers Arch. 2011;462:119–134. doi: 10.1007/s00424-011-0946-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut DR, Cooper J, Asbury CL, Davis TN, Wordeman L. Reconstitution and functional analysis of kinetochore subcomplexes. Methods Cell Biol. 2010;95:641–656. doi: 10.1016/S0091-679X(10)95032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–U470. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourier C, Jegou A, Husson J, Pincet F. A nanospring named erythrocyte. The biomembrane force probe. Cell Mol Bioeng. 2008;1:263–275. [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci USA. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenberg Z, Bausch AR, Hu B, Bruinsma R, Moroder L, Sackmann E. Measuring ligand−receptor unbinding forces with magnetic beads: molecular leverage. Langmuir. 2000;16:8984–8993. [Google Scholar]
- Hamill OP, Martinac B. Molecular basis of mechanotransduction in living cells. Physiol Rev. 2001;81:685–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- Hattori M, Minato N. Rap1 gtpase: functions, regulation, and malignancy. J Biochem. 2003;134:479–484. doi: 10.1093/jb/mvg180. [DOI] [PubMed] [Google Scholar]
- Hermanson GT. Bioconjugate Techniques. 3rd ed. London: Elsevier; 2013. [Google Scholar]
- Hotz M, Barral Y. The mitotic exit network: new turns on old pathways. Trends Cell Biol. 2014;24:145–152. doi: 10.1016/j.tcb.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- Ingber DE. Tensegrity: the architectural basis of cellular mechanotransduction. Annu Rev Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- Interlandi G, Thomas W. The catch bond mechanism between von willebrand factor and platelet surface receptors investigated by molecular dynamics simulations. Proteins. 2010;78:2506–2522. doi: 10.1002/prot.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaska J. Unanchoring integrins in focal adhesions. Nat Cell Biol. 2012;14:981–983. doi: 10.1038/ncb2592. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kim D, Herr AE. Protein immobilization techniques for microfluidic assays. Biomicrofluidics. 2013;7:041501. doi: 10.1063/1.4816934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Khatau SB, Feng Y, Walcott S, Sun SX, Longmore GD, Wirtz D. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki AG, Yago T, Mehta P, Yang J, Wu T, Leppanen A, Bovin NV, Cummings RD, Zhu C, McEever RP. Replacing a lectin domain residue in l-selectin enhances binding to p-selectin glycoprotein ligand-1 but not to 6-sulfo-sialyl lewis x. J Biol Chem. 2008;283:11493–11500. doi: 10.1074/jbc.M709785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Muto E, Higuchi H, Yanagida T. Mechanics of single kinesin molecules measured by optical trapping nanometry. Biophys J. 1997;73:2012–2022. doi: 10.1016/S0006-3495(97)78231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C. A possible unifying principle for mechanosensation. Nature. 2005;436:647–654. doi: 10.1038/nature03896. [DOI] [PubMed] [Google Scholar]
- Lafaurie-Janvore J, Maiuri P, Wang I, Pinot M, Manneville J-B, Betz T, Balland M, Piel M. Escrt-iii assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science. 2013;339:1625–1629. doi: 10.1126/science.1233866. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora b and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, Oscar M, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell. 2013;25:270–283. doi: 10.1016/j.devcel.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, Rodriguez V, Interlandi G, Klevit R, Vogel V, et al. Structural basis for mechanical force regulation of the adhesin fimh via finger trap-like β sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FY, Redick SD, Erickson HP, Moy VT. Force measurements of the alpha(5)beta(1) integrin-fibronectin interaction. Biophys J. 2003;84:1252–1262. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou JZ, Yago T, Klopocki AG, Mehta P, Chen W, Zarnitsyna VI, Bovin NV, Zhu C, McEver RP. Flow-enhanced adhesion regulated by a selectin interdomain hinge. J Cell Biol. 2006;174:1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Marko JF, Agard DA, Sedat JW. Chromosome elasticity and mitotic polar ejection force measured in living drosophila embryos by four-dimensional microscopy-based motion analysis. Curr Biol. 2001;11:569–578. doi: 10.1016/s0960-9822(01)00180-4. [DOI] [PubMed] [Google Scholar]
- Matthews JNA. Commercial optical traps emerge from biophysics labs. Phys Today. 2009;62:26–28. [Google Scholar]
- Meng F, Suchyna TM, Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 2008;275:3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natkanski E, Lee W-Y, Mistry B, Casal A, Molloy JE, Tolar P. B cells use mechanical energy to discriminate antigen affinities. Science. 2013;340:1587–1590. doi: 10.1126/science.1237572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Block SM. Optical trapping. Rev Sci Instruments. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA. Chromosome micromanipulation iii. Spindle fiber tension and reorientation of mal-oriented chromosomes. J Cell Biol. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan UT, T Waldron T, Springer TA. Remodeling of the lectin-egf-like domain interface in p- and l-selectin increases adhesiveness and shear resistance under hydrodynamic force. Nat Immunol. 2006;7:883–889. doi: 10.1038/ni1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schäfer LV, Brandmeier B, Gräter F, Grubmüller H, Gaub HE, Gautel M. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchner EM, Gaub HE. Single-molecule mechanoenzymatics. Annu Rev Biophys. 2012;41:497–518. doi: 10.1146/annurev-biophys-050511-102301. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Bimbo A, Balasubramanian MK, Oliferenko S. A potential tension-sensing mechanism that ensures timely anaphase onset upon metaphase spindle orientation. Curr Biol. 2004;14:69–74. doi: 10.1016/j.cub.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Rakshit S, Sivasankar S. Biomechanics of cell adhesion: how force regulates the lifetime of adhesive bonds at the single molecule level. Phys Chem Chem Phys. 2014;16:2211–2223. doi: 10.1039/c3cp53963f. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Vale RD, Gennerich A. Motile properties of cytoplasmic dynein. In: Hirose K, Amos LA, editors. Handbook of Dynein. Singapore: Pan Stanford Publishing; 2012. pp. 145–172. [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackin H. Mechanosensitive channels. Annu Rev Phys. 1995;57:333–353. doi: 10.1146/annurev.ph.57.030195.002001. [DOI] [PubMed] [Google Scholar]
- Sandall S, Severin F, McLeod IX, Yates JR, III, Oegema K, Hyman A, Desai A. A bir1-sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Asbury CL. Catch and release: how do kinetochores hook the right microtubules during mitosis. Trends Genet. 2014;30:150–159. doi: 10.1016/j.tig.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Yago T, Klopocki AG, Lawrence MB, Fieger CB, Rosen SD, McEver RP, Zhu C. Low force decelerates l-selectin dissociation from p-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279:2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the src family kinase substrate p130cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- Steigemann P, Wurzenberger C, Schmitz MHA, Held M, Guizetti J, Maar S, Gerlich DW. Aurora b-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Strick TR, Allemand J-F, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal P, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev Cell. 2004;7:709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K. Evidence that the ipl1-sli15 (aurora kinase-incenp) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Thomas WE. Mechanochemistry of receptor–ligand bonds. Curr Opin Struct Biol. 2009;19:50–55. doi: 10.1016/j.sbi.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Thoumine O, Kocian P, Kottelat A, Meister JJ. Short-term binding of fibroblasts to fibronectin: optical tweezers experiments and probabilistic analysis. Eur Biophys J Biophys Lett. 2000;29:398–408. doi: 10.1007/s002490000087. [DOI] [PubMed] [Google Scholar]
- Tien JF, Fong KK, Umbreit NT, Payen C, Zelter A, Asbury CL, Dunham MJ, Davis TN. Coupling unbiased mutagenesis to high-throughput DNA sequencing uncovers functional domains in the ndc80 kinetochore protein of saccharomyces cerevisiae. Genetics. 2013;195:159–170. doi: 10.1534/genetics.113.152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the dam1 and ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora b. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL, Davis TN. The ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci USA. 2012;109:16113–16118. doi: 10.1073/pnas.1209615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- Volkov VA, Zaytsev AV, Gudimchuk N, Grissom PM, Gintsburg AL, Ataullakhanov FI, McIntosh R, Grishchuk EL. Long tethers provide high-force coupling of the dam1 ring to shortening microtubules. Proc Natl Acad Sci USA. 2013;110:7708–7713. doi: 10.1073/pnas.1305821110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron TT, Springer TA. Transmission of allostery through the lectin domain in selectin-mediated cell adhesion. Proc Natl Acad Sci USA. 2009;106:85–90. doi: 10.1073/pnas.0810620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JPI, Vleugel M, Liu D, Yates JR 3rd, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Lin J, Cruz MA, Dong J-F, Zhu C. Force-induced cleavage of single vwf a1a2a3-tridomains by adamts-13. Blood. 2010;115:370–378. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas WE. Fimh forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovenko O, Tchesnokova V, Aprikian P, Forero M, Vogel V, Sokurenko E, Thomas W. Mechanical force activates an adhesion protein through allosteric regulation. Biophys J. 2007:348A–349A. [Google Scholar]
- Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]