Cbx2 is immobilized at mitotic chromosomes, and the immobilization is independent of PRC1 or PRC2. Cbx2 plays important roles in recruiting PRC1 complex to mitotic chromosomes. This study provides novel insights into the PcG epigenetic memory passing down through cell divisions.
Abstract
Polycomb group (PcG) proteins are epigenetic transcriptional factors that repress key developmental regulators and maintain cellular identity through mitosis via a poorly understood mechanism. Using quantitative live-cell imaging in mouse ES cells and tumor cells, we demonstrate that, although Polycomb repressive complex (PRC) 1 proteins (Cbx-family proteins, Ring1b, Mel18, and Phc1) exhibit variable capacities of association with mitotic chromosomes, Cbx2 overwhelmingly binds to mitotic chromosomes. The recruitment of Cbx2 to mitotic chromosomes is independent of PRC1 or PRC2, and Cbx2 is needed to recruit PRC1 complex to mitotic chromosomes. Quantitative fluorescence recovery after photobleaching analysis indicates that PRC1 proteins rapidly exchange at interphasic chromatin. On entry into mitosis, Cbx2, Ring1b, Mel18, and Phc1 proteins become immobilized at mitotic chromosomes, whereas other Cbx-family proteins dynamically bind to mitotic chromosomes. Depletion of PRC1 or PRC2 protein has no effect on the immobilization of Cbx2 on mitotic chromosomes. We find that the N-terminus of Cbx2 is needed for its recruitment to mitotic chromosomes, whereas the C-terminus is required for its immobilization. Thus these results provide fundamental insights into the molecular mechanisms of epigenetic inheritance.
INTRODUCTION
The transcriptional state of genes is programmed during development and differentiation. Once established, the activated or repressed state of genes tends in most cases to be maintained for the rest of development, although gene activity is flexible and can be switched. Polycomb group (PcG) proteins are epigenetic transcriptional regulators that repress hundreds of developmental regulators that control key developmental processes by mediating trimethylation on lysine 27 of histone H3 (H3K27me3; Kerppola, 2009; Di Croce and Helin, 2013; Sharif et al., 2013; Simon and Kingston, 2013). The PcG-mediated repressed state is established during development and in most cases is maintained from mother to daughter cells via mitosis. During mitosis, chromatin structure changes extraordinarily. Most transcription factors and structural chromatin proteins are excluded from mitotic chromosomes, yet mitotic chromosomes are accessible (Chen et al., 2005; Hemmerich et al., 2011). Therefore one of the major questions is how the PcG-mediated repressed state of genes persists during mitosis.
PcG proteins comprise two repressive complexes, Polycomb repressive complex (PRC) 1 and PRC2, both of which possess enzymatic activities for modifying histones and repressing target genes (Kerppola, 2009; Di Croce and Helin, 2013; Sharif et al., 2013; Simon and Kingston, 2013). In mammals, PRC1 complexes are composed of a multiplicity of variants and vary in biochemical composition (Kerppola, 2009; Di Croce and Helin, 2013; Sharif et al., 2013; Simon and Kingston, 2013). Ring1b is the assemblage of PRC1 complexes, and in some cases, Ring1a can substitute for Ring1b. Ring1b forms a heterodimer with alternative Pcgf-family proteins (Pcgf1/2/3/4/5/6) to render the activity of ubiquitin ligase for lysine 119 on H2A (H2AK119ub1; de Napoles et al., 2004; Wang et al., 2004a). The Ring1b-Pcgf2 (Mel18) or Ring1b-Pcgf4 (Bmi1) dimers can further form complexes with one of five alternative Cbx protein homologues (Cbx2/4/6/7/8) and one of three alternative Phc protein homologues (Phc1/2/3). The Cbx-containing PRC1 complexes have been defined as the canonical PRC1 complexes (Gao et al., 2012; Tavares et al., 2012; Morey et al., 2013). The other Ring1b-Pcgf dimers can associate with different proteins to form the noncanonical PRC1 complexes. The biological functions of individual PRC1 complexes remain largely unknown.
The recruitment of canonical PRC1 complexes to interphasic chromatin has been extensively investigated (Cao et al., 2002; Ogawa et al., 2002; Wang et al., 2004b; Gearhart et al., 2006; Sanchez et al., 2007; Boukarabila et al., 2009; Ren and Kerppola, 2011; Vandamme et al., 2011; Gao et al., 2012; Tavares et al., 2012; Morey et al., 2013; Cheng et al., 2014), yet it remains relatively unknown whether 1) PRC1 complexes remain at mitotic chromosomes during mitosis, and 2) if so, what mechanisms target PRC1 complexes to mitotic chromosomes. Early studies of vertebrate PRC1 proteins provided varying views on how PRC1 proteins interact with mitotic chromosomes (Wang et al., 1997; Saurin et al., 1998; Koga et al., 1999; Voncken et al., 1999; Akasaka et al., 2002; Suzuki et al., 2002; Miyagishima et al., 2003; Aoto et al., 2008; Vincenz and Kerppola, 2008). More recently, studies of Drosophila PRC1 proteins by live-cell imaging and genome-wide sequencing provided evidence that most of PRC1 proteins dissociate from mitotic chromosomes, but a quantitative subpopulation of PRC1 proteins remain associated with mitotic chromosomes (Follmer et al., 2012; Fonseca et al., 2012; Steffen et al., 2013). Quantitative analysis of binding dynamics of PRC1 proteins with chromatin revealed remarkable differences between interphase and mitosis (Fonseca et al., 2012). The residence time of a subpopulation of PRC1 proteins at mitotic chromosomes is up to 300-fold longer than that at interphasic chromatin. If the retention and immobilization of PRC1 at mitotic chromosomes is central for epigenetic inheritance, then the mechanism should be conserved across organisms that contain PRC1.
Here we show, using quantitative live-cell imaging, that Cbx2 is the primary Cbx-family protein that remains associated with mitotic chromosomes. The association of Cbx2 protein with mitotic chromosomes is independent of PRC2 or PRC1. We demonstrate that the Cbx2 protein is the major factor for the recruitment of PRC1 proteins to mitotic chromosomes. By using quantitative fluorescence recovery after photobleaching (FRAP), we reveal that PRC1 proteins rapidly exchange at interphasic chromatin, as reported previously (Ren et al., 2008), and identify that a special Cbx2-PRC1 complex is selectively immobilized at mitotic chromosomes. The immobilization of Cbx2 protein at mitotic chromosomes is independent of PRC2 or PRC1. Finally, we reveal that the Cbx2 recruitment and immobilization are mechanistically uncoupled.
RESULTS
PRC1 proteins vary in association with mitotic chromosomes
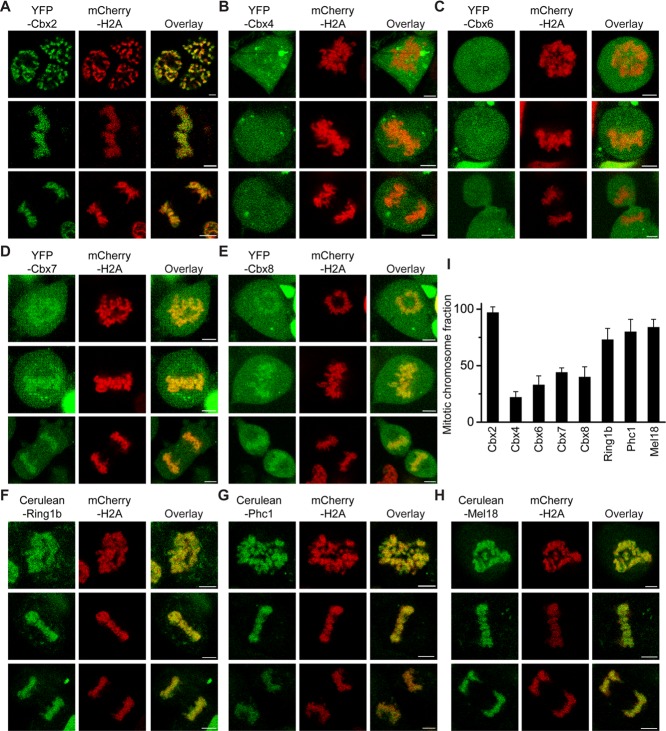
Early studies of the association of mammalian PRC1 proteins with mitotic chromosomes reached divergent opinions (Wang et al., 1997; Saurin et al., 1998; Koga et al., 1999; Voncken et al., 1999; Akasaka et al., 2002; Suzuki et al., 2002; Miyagishima et al., 2003; Aoto et al., 2008; Vincenz and Kerppola, 2008). To appreciate fully whether the Cbx-containing PRC1 complexes associate with mitotic chromosomes, we established ES cell lines that inducibly express PRC1 protein fused with either Cerulean or yellow fluorescent protein (YFP) in a doxycycline-controlled manner. To facilitate live-cell imaging and mark mitotic chromosomes, we coexpressed histone H2A fused with Cerulean or mCherry with PRC1 fusion protein. We performed multicolor Z-scan imaging of live cells at 37°C with a confocal laser-scanning microscope. The PRC1 fusion proteins (Cbx family proteins [Cbx2, Cbx4, Cbx6, Cbx7, Cbx8], Ring1b, Phc1, and Mel18) exhibited variable capacities of association with mitotic chromosomes. YFP-Cbx2 fusion was the primary protein accumulated at mitotic chromosomes (Figure 1, A–H). These PRC1 fusion proteins were granularly distributed at mitotic chromosomes. Quantitative analysis of Z-stack images showed that (96 ± 5)% of YFP-Cbx2 protein associates with mitotic chromosomes, whereas YFP-Cbx4, (22 ± 5)%; YFP-Cbx6, (26 ± 7)%; YFP-Cbx7, (44 ± 4)%; Cbx8, (40 ± 9)%; Cerulean-Ring1b, (63 ± 10)%; Cerulean-Phc1, (84 ± 10)%; and Cerulean-Mel18, (84 ± 7)%, all showed reduced association with mitotic chromosomes (Figure 1I). These results are consistent with previous bimolecular fluorescence complementation analysis of Cbx2 and Cbx6 association with mitotic chromosomes (Vincenz and Kerppola, 2008), but the present studies provide quantitative insights into the entire set of Cbx-family proteins and other PRC1 proteins. Thus these data indicate that the Cbx-containing PRC1 complexes associate with mitotic chromosomes, and Cbx2 is the primary protein enriched at mitotic chromosomes.
FIGURE 1:
Mitotic chromosomal association of PRC1 fusion proteins. (A–H) Confocal fluorescence images of PRC1 fusion proteins tagged with either YFP or Cerulean expressed in PGK12.1 ES cells in various phases of mitosis. The mCherry-H2A fusion protein was used to mark mitotic chromosomes. Prometaphase (top), metaphase (middle), and anaphase/telophase (bottom). Representative images from Z-scan stacks are presented in the figures. Scale bar, 5 μm. (I) Quantitative analysis of mitotic binding fraction of PRC1 fusion proteins at mitotic chromosomes. The mitotic binding fraction is the average of individual slices of Z-scan stacks. The data represent at least 10 cells analyzed. Error bars, SD of the mean.
Because Cbx2 is the primary protein associated with mitotic chromosomes, we sought to determine whether the mitotic chromosomal association of Cbx2 protein is cell-type specific. Therefore we established HEK293 and HeLa cell lines that stably and inducibly express YFP-Cbx2 and mCherry-H2A fusion proteins. Z-scan imaging of live HEK293 and HeLa cells showed that YFP-Cbx2 fusion protein profoundly accumulated at mitotic chromosomes in both cell lines (Supplemental Figure S1). Consistent with the YFP-Cbx2 fusion protein localization at mitotic chromosomes in ES cells, quantitative analysis of Z-stack images showed that (96 ± 4)% of YFP-Cbx2 is associated with mitotic chromosomes in both HEK293 and HeLa cells, indicating the mitotic chromosomal association of Cbx2 protein is not restricted to ES cells.
To ask whether the expression level of Cbx2 fusion protein affects the mitotic chromosomal association, we administrated a wide range of concentrations of doxycycline, from 0.1 to 1.0 μM, into HEK293 cells stably and inducibly expressing YFP-Cbx2 and Cerulean-H2A. Epifluorescence imaging of live HEK293 cells showed that the fraction of mitotic chromosomal association of Cbx2 fusion protein was similar to one another under various concentrations of doxycycline (Supplemental Figure S2). Unless otherwise indicated, we used doxycycline at a concentration of 0.2–0.5 μM throughout subsequent studies. At 0.5 μM doxycycline, the expression levels of the PRC1 proteins tested were either similar to or twofold to threefold higher than those of their endogenous counterparts (Supplemental Figure S3).
To provide independent evidence of PRC1 protein association with mitotic chromosomes, we performed biochemical fractionation followed by Western blot analysis. ES cells were synchronized at M phase by sequential treatment with thymidine and nocodazole based on a published report (Ballabeni et al., 2011). At least 85% of ES cells were mitotic, as revealed by cell morphology and immunostaining of H3S10p (Supplemental Figure S4, A and B). The nonsynchronized (control) and synchronized (mitotic) cells were fractionated according to the scheme showed in Supplemental Figure S4C based on previous reports (Mendez and Stillman, 2000; Follmer et al., 2012). In control and mitotic cells, the histone H3 was primarily found in the chromatin fraction (P3), whereas α-tubulin was found primarily in the cytosolic fraction (S2; Supplemental Figure S4D). We tested the chromatin association of four PRC1 proteins (Ring1b, Phc1, Cbx2, and Cbx7) in both control and mitotic cells. In mitotic cells, we found that Cbx2 protein is primarily in the fraction P3, whereas Cbx7 is primarily in the cytosolic fraction S2 (Supplemental Figure S4D). Thus these data are consistent with live-cell imaging data that show that Cbx2 is the primary protein enriched at mitotic chromosomes.
The mitotic chromosomal association of Cbx2 is independent of PcG proteins
The Cbx-containing PRC1 complexes are recruited to interphasic chromatin via Cbx-family protein interactions with H3K27me3 mediated by PRC2 (Cao et al., 2002; Wang et al., 2004b; Bernstein et al., 2006; Tavares et al., 2012; Morey et al., 2013). Therefore we asked whether PRC2 is required for the accumulation of Cbx2 protein at mitotic chromosomes. To test the hypothesis, we stably and inducibly expressed both YFP-Cbx2 and Cerulean-H2A fusion proteins in Eed knockout (KO) ES cell lines, which lack the H3K27me3 modification. Z-scan of confocal images showed that YFP-Cbx2 fusion protein was granularly distributed at mitotic chromosomes, consistent with its distribution in wild-type ES cells. Quantitative analysis of Z-stack images showed that (97 ± 5)% of Cbx2 protein was enriched at mitotic chromosomes in Eed KO ES cell lines (Figure 2, B and D), consistent with YFP-Cbx2 associating with mitotic chromosomes in wild-type ES cells (Figure 2, A and D), indicating that a core component of the PRC2 complex, Eed, is not required for accumulation of Cbx2 protein at mitotic chromosomes.
FIGURE 2:
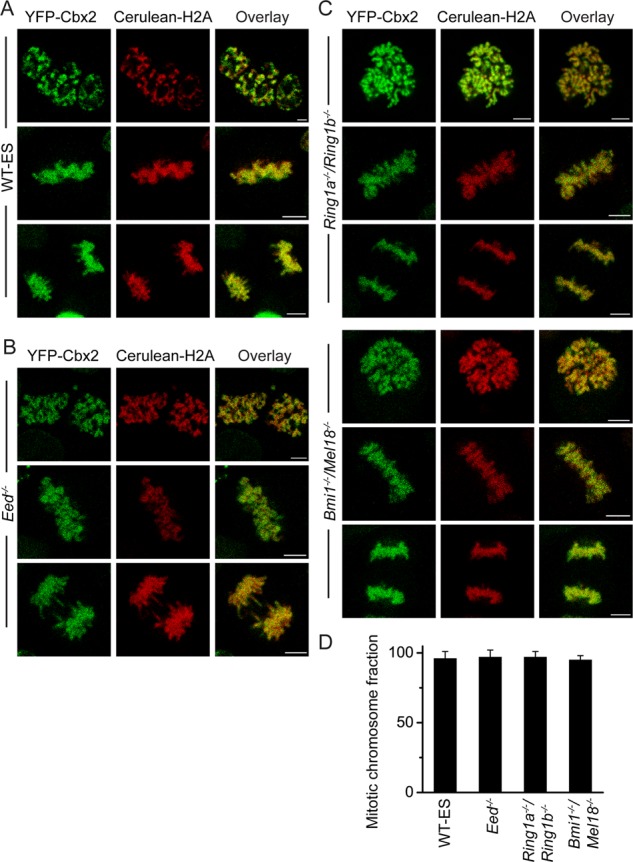
The association of Cbx2 fusion protein with mitotic chromosomes in PRC1 and PRC2 gene-knockout ES cells. (A) Confocal fluorescence images of YFP-Cbx2 fusion protein expressed in PGK12.1 ES cells (WT-ES). The different version of YFP-Cbx2 of Figure 1A is presented here in order to compare with B and C. Scale bar, 5 μm. (B) Confocal fluorescence images of YFP-Cbx2 fusion protein expressed in Eed−/− ES cells in various phases of mitosis. Prometaphase (top), metaphase (middle), and anaphase/telophase (bottom). Scale bar, 5 μm. (C) Confocal fluorescence images of YFP-Cbx2 fusion protein expressed in Ring1a−/−/Ring1bfl/fl and Bmi1−/−/Mel18−/− ES cell lines in various phases of mitosis. Prometaphase (top), metaphase (middle), and anaphase/telophase (bottom). The Ring1a−/−/Ring1bfl/fl was treated with OHT to induce depletion of Ring1b for 3 d before imaging. The protein level of Ring1b in Ring1a−/−/Ring1bfl/fl cells with or without OHT is presented in Supplemental Figure S5. Scale bar, 5 μm. (D) Quantitative analysis of mitotic binding fractions of YFP-Cbx2 fusion protein at mitotic chromosomes. The data represent at least 10 cells analyzed. Error bars, SD of the mean.
To determine whether the formation of integral canonical PRC1 complexes is required for accumulation of Cbx2 protein at mitotic chromosomes, we stably and inducibly expressed YFP-Cbx2 and Cerulean-H2A fusion proteins in Ring1a constitutively KO and Ring1b conditionally KO ES cell lines (Ring1a−/−/Ring1bfl/fl). We administered 4-hydroxytamoxifen (OHT) and doxycycline for 3 d. The addition of OHT induced depletion of endogenous Ring1b protein (Supplemental Figure S5). We performed Z-scan of live-cell imaging of these cells using a confocal microscope. Quantitative analysis of Z-stack images showed that (97 ± 4)% of Cbx2 fusion protein is accumulated at mitotic chromosomes (Figure 2, C and D), consistent with localization of the YFP-Cbx2 fusion protein in wild-type and Eed KO ES cells. To investigate further whether other canonical PRC1 subunits affect the mitotic chromosomal accumulation of Cbx2, we stably and inducibly expressed YFP-Cbx2 and Cerulean-H2A fusion proteins in Bmi1 and Mel18 (Bmi1/Mel18) double-KO ES cell lines. Quantitative analysis of Z-stack images indicated that (95 ± 3)% of Cbx2 fusion protein accumulated at mitotic chromosomes, consistent with the result in wild-type ES cells (Figure 2, C and D). Thus these data show that the mitotic chromosomal accumulation of Cbx2 protein is independent of PRC1 proteins Ring1a/Ring1b and Bmi1/Mel18.
Cbx2 is required for recruiting the canonical PRC1 proteins to mitotic chromosomes
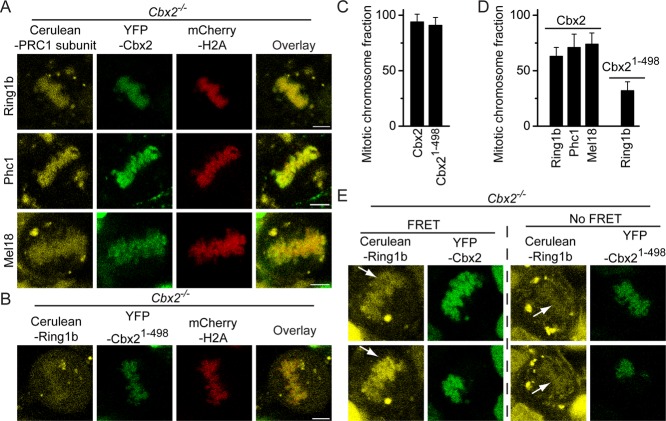
Because 1) Cbx2 is the primary Cbx-family protein accumulated at mitotic chromosomes, and 2) mitotic chromosomal accumulation of Cbx2 protein is independent of PRC1 or PRC2 complex proteins, we reasoned that Cbx2 protein is required for recruiting canonical PRC1 proteins to mitotic chromosomes. To this end, we inducibly expressed Cerulean-PRC1 protein (Cerulean-Ring1b, Cerulean-Phc1, or Cerulean-Mel18) and mCherry-H2A fusion protein in Cbx2−/− ES cell lines. Z-scan imaging of live cells by using confocal microscopy revealed that fluorescence intensities of Cerulean-Ring1b, Cerulean-Phc1, and Cerulean-Mel18 fusion proteins at mitotic chromosomes in Cbx2 KO ES cells were greatly reduced in comparison to that seen in wild-type ES cells (compare Figures 1, F–H, and 3A). Quantitative analysis of Z-stack images revealed that (29 ± 8)% of Cerulean-Ring1b protein associated with mitotic chromosomes, (17 ± 7)% of Cerulean-Phc1 protein did, and (27 ± 5)% of Cerulean-Mel18 protein did (Figure 3B). To test whether Cbx2 gene knockout affects the level of Phc1, Mel18, and Ring1b proteins, we performed immunoblotting, using extracts from WT and Cbx2−/− ES cells. Western blots showed that the level of Phc1, Mel18, and Ring1b proteins in Cbx2−/− ES cells was similar to that seen in WT cells (Figure 3C). Thus these data suggest that Cbx2 protein is required for the accumulation of canonical PRC1 proteins Ring1b, Phc1, and Mel18 at mitotic chromosomes.
FIGURE 3:
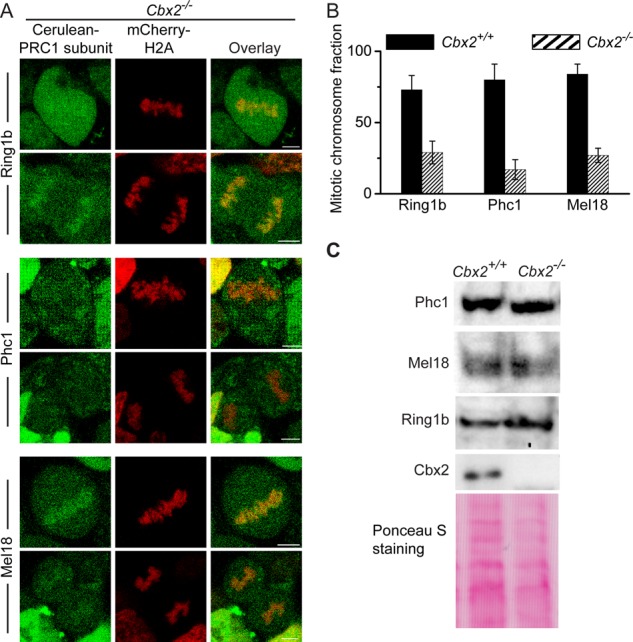
The mitotic chromosomal association of PRC1 fusion proteins Ring1b, Phc1, and Mel18 in Cbx2−/− ES cells. (A) Confocal fluorescence images of Cerulea-Ring1b, Cerulean-Phc1, and Cerulean-Mel18 fusion proteins expressed in Cbx2−/− ES cells in metaphase (top) and anaphase (bottom). Scale bar, 5 μm. (B) Quantitative comparison of mitotic chromosomal association of Cerulea-Ring1b, Cerulean-Phc1, and Cerulean-Mel18 fusion proteins in Cbx2+/+ and Cbx2−/− ES cells. The data represent an average of at least 10 cells analyzed. Error bars, SD of the mean. (C) Western blots of cell extracts from Cbx2+/+ and Cbx2−/− ES cells. Ponceau S staining indicates the loading control.
To investigate further whether Cbx2 is required for recruiting Phc1 and Ring1b to mitotic chromosomes, we performed immunostaining in WT and Cbx2−/− ES cells by using antibodies that detect endogenous Phc1 and Ring1b (Supplemental Figure S6). Immunostaining showed that endogenous Phc1 and Ring1b associate with mitotic chromosomes in wild-type ES cells. However, endogenous Phc1 and Ring1b were excluded from mitotic chromosomes in Cbx2-knockout ES cells. These data suggest that Cbx2 is required to recruit Phc1 and Ring1b to mitotic chromosomes.
Cbx2 directly recruits the canonical PRC1 proteins to mitotic chromosomes
Because Cbx2 protein affects the accumulation of canonical PRC1 proteins at mitotic chromosomes, we reasoned that Cbx2 directly recruits canonical PRC1 proteins to mitotic chromosomes. To this end, we coexpressed the three fusion proteins in Cbx2 KO ES cell lines: YFP-Cbx2, Cerulean-PRC1 subunit (Cerulean-Ring1b, or Cerulean-Phc1, or Cerulean-Mel18), and mCherry-H2A. We expected that the introduction of an YFP-Cbx2 fusion protein to the Cbx2-null background ES cells would restore the mitotic chromosomal association of the three PRC1 proteins (Cerulean-Ring1b, Cerulean-Phc1, and Cerulean-Mel18). We performed three-color Z-scan imaging of live cells by using confocal microscope. Quantitative analysis of Z-stack images from three ES cell lines expressing Ring1b, Phc1, and Mel18 fusion proteins showed an average of (95 ± 7)% of YFP-Cbx2 associated with mitotic chromosomes, consistent with YFP-Cbx2 localization in wild-type ES cells (Figure 4, A and C), indicating that mitotic chromosomal association of YFP-Cbx2 fusion protein is independent of endogenous Cbx2 protein. Of note, quantitative image analysis revealed that (63 ± 8)% of Cerulean-Ring1b, (71 ± 12)% of Cerulean-Phc1, and (74 ± 10)% of Cerulean-Mel18 also associated with mitotic chromosomes (Figure 4, A and D). The fraction of retention of the three fusion proteins at mitotic chromosomes in Cbx2 KO ES cell lines complemented with YFP-Cbx2 was similar to that seen in wild-type ES cells, indicating that YFP-Cbx2 fusion protein recruits the three canonical PRC1 fusion proteins to mitotic chromosomes.
FIGURE 4:
Directly recruiting PRC1 fusion proteins to mitotic chromosomes by YFP-Cbx2 but not by YFP-Cbx21-498. (A) Confocal fluorescence images of Cerulean-Ring1b, Cerulean-Phc1, and Cerulean-Mel18 fusion proteins coexpressed with YFP-Cbx2 and mCherry-H2A fusion proteins in Cbx2−/− ES cells in metaphase. Scale bar, 5 μm. (B) Confocal fluorescence images of Cerulean-Ring1b fusion protein coexpressed with YFP-Cbx21-498 and mCherry-H2A fusion protein in Cbx2−/− ES cells in metaphase. Scale bar, 5 μm. (C) Quantification of mitotic chromosomal association of YFP-Cbx2 and YFP-Cbx21-498 fusion proteins in Cbx2−/− ES cells. The data represent an average of at least 10 cells analyzed. Error bars, SD of the mean. (D) Quantitative analysis of mitotic chromosomal association of Cerulea-Ring1b, Cerulean-Phc1, and Cerulean-Mel18 fusion proteins coexpressed with YFP-Cbx2 and of Cerulea-Ring1b coexpressed with YFP-Cbx21-498 in Cbx2−/− ES cells in metaphase. The data represent an average of at least 10 cells analyzed. Error bars, SD of the mean. (E) Photobleaching FRET images of Cerulean-Ring1b interaction with YFP-Cbx2 and YFP-Cbx21-498 at mitotic chromosomes. The YFP-Cbx2, YFP-Cbx21-498, and Cerulean-Ring1b fusion proteins as indicated above images expressed in Cbx2−/− ES cells. Half-area of fluorescence of YFP-Cbx2 or YFP-Cbx21-498 fusion proteins at mitotic chromosomes was photobleached. Z-scan imaging of live cells by confocal laser microscope was performed before (top) and after (bottom) photobleaching. The arrowheads indicate the bleaching areas.
To test whether the Cbx2 interaction with Ring1b is required for the recruitment of Ring1b protein to mitotic chromosomes, the three fusion proteins mCherry-H2A, Cerulean-Ring1b, and YFP-Cbx21-498 were expressed in Cbx2 KO ES cells. The YFP-Cbx21-498 fusion protein lacks the chromobox (Cbox) domain required for interaction with Ring1b (Satijn et al., 1997; Schoorlemmer et al., 1997; Bardos et al., 2000). We expected that the Cbx2 mutant fusion protein would not be able to recruit Cerulean-Ring1b to mitotic chromosomes. Quantitative image analysis showed that (92 ± 7)% of YFP-Cbx21-498 fusion protein accumulated at mitotic chromosomes, indicating the deletion of the Cbox domain of Cbx2 protein does not affect its mitotic chromosomal association (Figure 4, B and C; also see later discussion of Figure 7B). However, only (32 ± 8)% of Cerulean-Ring1b fusion protein associated with mitotic chromosomes (Figure 4, B and D). The fraction of mitotic retention of Ringb1b fusion protein in Cbx2 KO ES cell lines complemented with Cbx21-498 was similar to that observed in Cbx2 KO ES cells. These data indicate that the Cbx2 interaction with Ring1b is required for the recruitment of Cerulean-Ring1b fusion protein to mitotic chromosomes.
FIGURE 7:
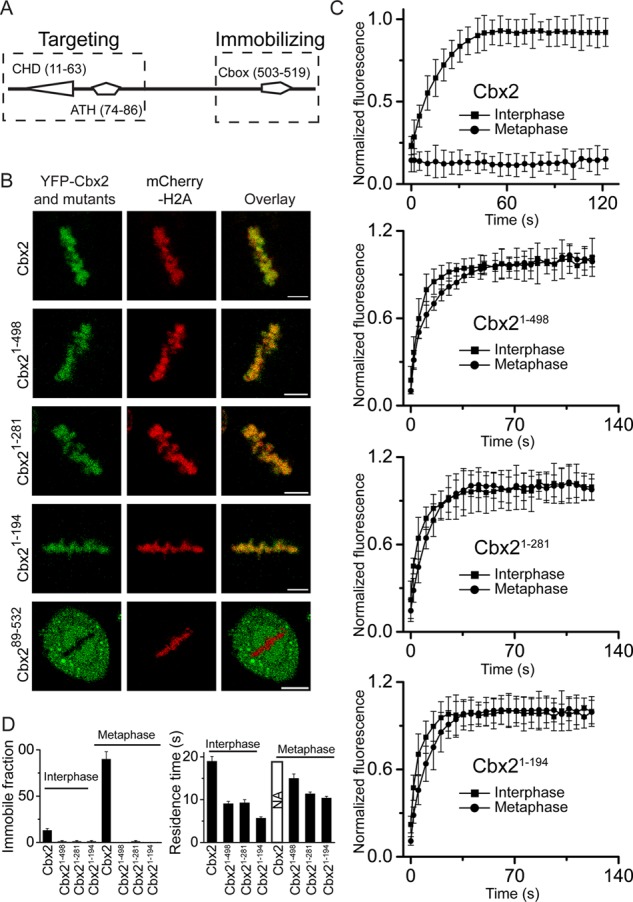
Analysis of structural elements of YFP-Cbx2 fusion protein required for its targeting and immobilizing. (A) Diagram of structural domains of Cbx2. The dashed rectangles indicate the region required for targeting Cbx2 to mitotic chromosomes (left) and the region required for immobilizing Cbx2 at mitotic chromosomes (right). ATH, AT-hook domain; Cbox, chromobox domain; CHD, chromodomain domain. The numbers in parentheses indicate the starting and ending of amino acid sequence. (B) Confocal images of YFP-Cbx2 and its variant fusion proteins in metaphase of HeLa cells. YFP-Cbx2 mutant and mCherry-H2A fusion proteins were stably expressed in HeLa cells. The mCherry-H2A was used to mark mitotic chromosomes. Scale bar, 5 μm. (C) FRAP curves of interphases and metaphases of YFP-Cbx2 and its variant fusion proteins expressed in HeLa cells. FRAP analysis is described in Figure 5. (D) Residence time and immobile fraction of YFP-Cbx2 variant fusion proteins at interphasic and mitotic chromatins. The residence time and immobile fraction were calculated from FRAP curves by fitting a first-order kinetic model.
The direct recruitment of the canonical PRC1 proteins to mitotic chromosomes by Cbx2 implies that there is direct interaction between Cbx2 and PRC1 subunits at mitotic chromosomes. To test this hypothesis, we performed photobleaching fluorescence resonance energy transfer (FRET) analysis between YFP-Cbx2 and Cerulean-Ring1b at mitotic chromosomes (Figure 4E). Fluorescence of YFP-Cbx2 fusion protein at half of mitotic chromosomes was photobleached. The ratio of fluorescence intensity of Cerulean-Ring1b in photobleached versus nonphotobleached areas was calculated and compared before and after photobleaching. Quantitative image analysis indicated that the fluorescence intensity of Cerulean-Ring1b fusion protein was increased (1.5 ± 0.1)-fold by photobleaching YFP-Cbx2 fusion protein, indicating that there is energy transfer between YFP-Cbx2 and Cerulean-Ring1b. As a control, we photobleached the YFP-Cbx21-498 fusion protein and quantified the fluorescence change of Cerulean-Ring1b fusion protein. Image analysis revealed that the fluorescence intensity of Cerulean-Ring1b fusion protein after photobleaching YFP-Cbx21-498 fusion protein was (1.1 ± 0.2)-fold of that before photobleaching, indicating that there is no energy transfer between YFP-Cbx21-498 and Cerulean-Ring1b. Thus these data demonstrate that YFP-Cbx2 fusion protein interacts with Cerulean-Ring1b fusion protein at mitotic chromosomes.
The Cbx2-PRC1 complex is immobilized at mitotic chromosomes, but other Cbx family proteins rapidly exchange at mitotic chromosomes
Several studies demonstrated that mammalian PRC1 proteins are highly dynamic during interphase in cells ( Hernandez-Munoz et al., 2005; Ren et al., 2008; Isono et al., 2013; Vandenbunder et al., 2014). Recent studies of Drosophila Pc and Ph proteins showed that a subpopulation of the two proteins bind to mitotic chromosomes with up to 300-fold-longer residence time than during interphase (Fonseca et al., 2012; Steffen et al., 2013). To determine the dynamic properties of mammalian PRC1 proteins binding to chromatin in both interphase and mitosis of ES cells, we performed quantitative FRAP on the Cbx family of proteins (Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8), as well as on the three core components of the canonical PRC1 complex (Ring1b, Phc1, and Mel18; Figure 5, A–I). The mCherry-H2A fusion protein served as a guide for placing bleach spots at mitotic chromosomes. Comparison of recovery kinetics of the Cbx-family fusion proteins binding to mitotic chromosomes revealed striking differences among Cbx proteins. Greater than 90% of YFP-Cbx2 fusion protein was immobilized at mitotic chromosomes without exchange over a time period of 120 s. Conversely, >85% of the YFP-Cbx4, YFP-Cbx6, YFP-Cbx7, and YFP-Cbx8 fusion proteins rapidly exchanged at mitotic chromosomes, with a residence time of 10–15 s (Figure 5, A– F, J, and K). During interphase, >90% of the Cbx-family fusion proteins showed fluorescence recovery, with a residence time of ∼10–20 s (Figure 5, A– F, J, and K), which is consistent with previous studies (Ren et al., 2008). Thus these data reveal that the YFP-Cbx2 fusion protein stably binds to mitotic chromosomes but rapidly exchanges at interphasic chromatin, whereas other Cbx-family proteins dynamically exchange on both interphasic and mitotic chromatin.
FIGURE 5:
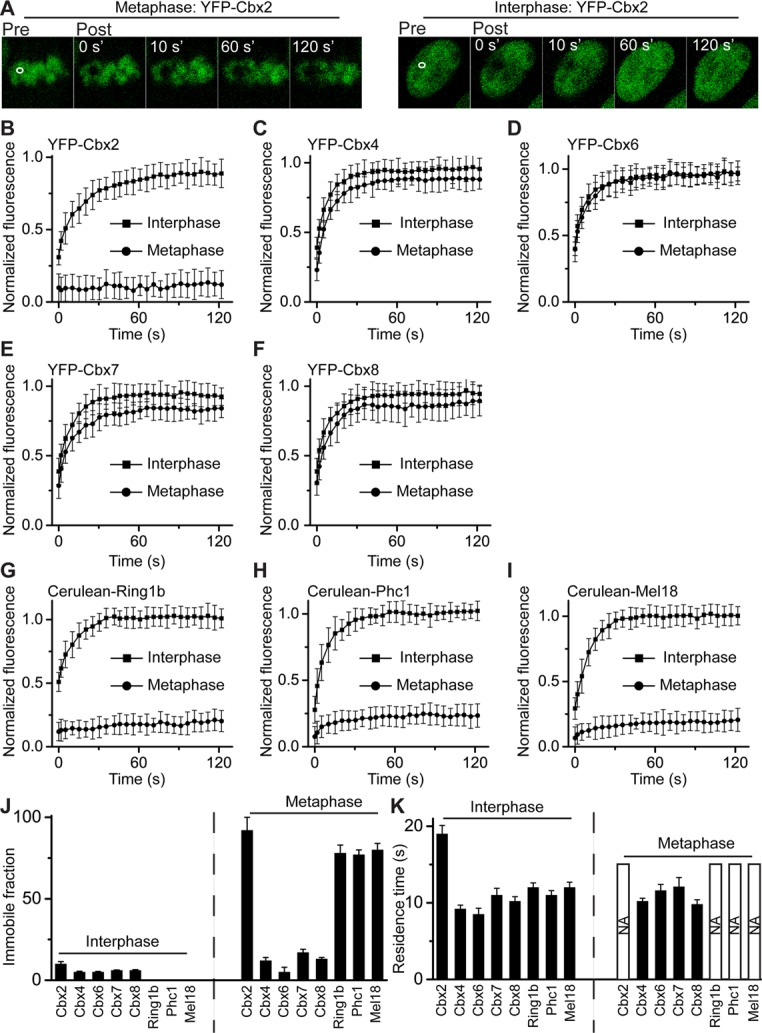
FRAP analysis of PRC1 fusion proteins binding to interphasic and mitotic chromatins. (A) Representative FRAP images of YFP-Cbx2 fusion protein at metaphasic and interphasic chromatins of ES cells. The images were taken before (pre) and after (post) photobleaching. The bleaching area is indicated and outlined in white. (B–I) FRAP curves of PRC1 fusion proteins at interphases and metaphases of PGK12.1 ES cells. The FRAP curves are the normalized fluorescence intensities of the bleached areas as a function of time after photobleaching and are an average of at least eight cells. Error bars, SDs of means. (J) Immobile faction of PRC1 fusion protein at interphasic and metaphasic chromatins. The immobile fraction was calculated from FRAP curves by fitting a first-order kinetic model. The dashed line indicates the contrast between interphase and metaphase. The data are an average of at least eight cells. (K) Residence time of PRC1 fusion protein at interphasic and metaphasic chromatins. The residence time was calculated from FRAP curves by fitting a first-order kinetic model. The bar with NA indicates that the fusion protein is immobilized at mitotic chromatin, and thus the residence time is not measurable within the time scale of the experiments. The dashed line indicates the contrast between interphase and metaphase.
To ask whether the binding kinetics of the Cbx2 fusion protein is cell-type specific, we performed FRAP analysis of YFP-Cbx2 fusion protein in both HeLa and HEK293 cells (Supplemental Figure S7; also see later discussion of Figure 7C). Analysis of FRAP curves revealed that interphasic and mitotic dynamic properties of YFP-Cbx2 fusion protein in both cell lines were similar to those in ES cells. Thus these data demonstrate that the YFP-Cbx2 fusion protein possesses inherently different properties of interaction with interphasic versus mitotic chromatin.
The various dynamic binding properties of Cbx family members to interphasic and mitotic chromatin prompted us to explore other core components of the canonic PRC1 complex. We performed FRAP analysis of Cerulean-Ring1b, Cerulean-Phc1, and Cerlean-Mel18 fusion proteins bound to chromatin in both interphase and mitosis in ES cells. Analysis of FRAP curves of mitotic chromosomal binding of Cerulean-Ring1b, Cerulean-Phc1, and Cerlean-Mel18 fusion proteins revealed that nearly 80% of these fusion proteins stably bind to mitotic chromosomes without exchange (Figure 5, G–K), which is similar to YFP-Cbx2 but differs from other Cbx-family fusion proteins. Calculation of the recovery kinetics and measured parameters of interphasic FRAP curves of the Cerulean-Ring1b, Cerulean-Phc1, and Cerlean-Mel18 proteins revealed that these three fusion proteins showed complete recovery, with residence time of 10–15 s, and had no immobile fraction at interphasic chromatin. Thus these data indicate that the four PRC1 fusion proteins—YFP-Cbx2, Cerulean-Ring1b, Cerulean-Phc1, and Cerulean-Mel18—bind to mitotic chromosomes with similar kinetic characteristics yet differ from other Cbx-family proteins.
The immobilization of Cbx2 at mitotic chromosomes is independent of PcG proteins
Because Cbx2 protein is recruited to mitotic chromosomes by a PRC2-independent mechanism, we reasoned that depletion of PRC2 complex gene Eed would not affect the immobilization of Cbx2 on mitotic chromosomes. To test the hypothesis, we performed FRAP analysis of YFP-Cbx2 protein binding to both mitotic and interphasic chromosomes in Eed KO ES cells. Analysis of mitotic FRAP curves revealed that >85% of YFP-Cbx2 fusion protein showed no recovery of fluorescence within 120 s (Figure 6, A and D). Calculation of interphasic FRAP curves of YFP-Cbx2 fusion protein revealed that the residence time for the mobile fraction is 25 s, slightly higher than that observed in wild-type ES cells, whereas the immobile fraction is the same as seen in wild-type ES cells. Thus these data indicate that the dynamics of YFP-Cbx2 fusion protein binding to interphase and mitotic chromatin is independent of the PRC2 gene Eed.
FIGURE 6:
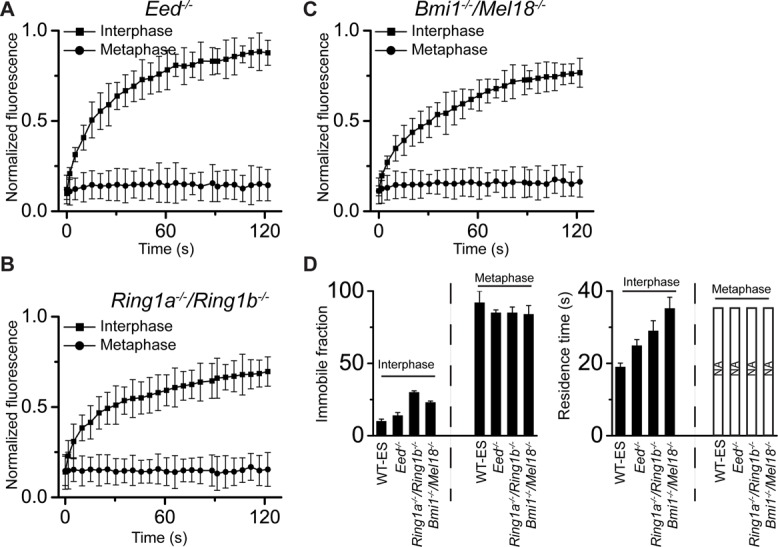
FRAP analysis of YFP-Cbx2 fusion protein binding to chromatins in Eed−/−, Ring1a−/−/Ring1b−/−, and Bmi1−/−/Mel18−/− ES cells. (A–C) FRAP curves of interphases and metaphases of YFP-Cbx2 fusion protein in Eed−/−, Ring1a−/−/Ring1b−/−, and Bmi1−/−/Mel18−/− ES cells. The FRAP curves were normalized and plotted as described in Figure 5. More than eight cells were analyzed. Error bars, SDs of means. (D) Residence time and immobile fraction of YFP-Cbx2 on chromatins. The residence time and immobile fraction were calculated from FRAP curves by fitting a first-order kinetic model. Bars with NA indicate that the residence time is not available due immobilization of YFP-Cbx2 on mitotic chromosomes. The dashed lines indicate the contrast between interphase and metaphase. WT-ES is denoted as PGK12.1 ES cells. The data are an average of at least eight cells.
To interrogate whether the interphasic and mitotic kinetics of Cbx2 fusion protein binding to chromatin are affected by PRC1 proteins, we performed FRAP analysis of YFP-Cbx2 fusion protein in Ring1a/Ring1b double-KO and Bmil1/Mel18 double-KO ES cells. Analysis of mitotic FRAP curves in both double-KO ES cell lines revealed that >85% of YFP-Cbx2 showed no recovery of fluorescence, indicating that the YFP-Cbx2 fusion protein binds to mitotic chromosomes without exchange (Figure 6, B– D). Next we analyzed the interphasic FRAP curves of YFP-Cbx2 fusion protein in the double-KO ES cell lines. Calculation of residence time and immobile fraction of YFP-Cbx2 fusion protein in interphase of the two double-KO ES cell lines revealed striking differences in comparison to wild-type ES cells (Figure 6, B– D). The YFP-Cbx2 fusion protein was much less dynamic in the double-KO ES cells than in wild-type ES cells, with residence time 30–35 s. A further difference is that the immobile fraction of YFP-Cbx2 fusion protein in interphase of the double-KO ES cell lines was 23–30% of total protein, which is >2.0-fold of that seen in wild-type ES cells. Thus these data suggest that the immobilization of YFP-Cbx2 fusion protein at mitotic chromosomes is independent of the PRC1 proteins Ring1a/Ring1b and Mel18/Bmi1, whereas the dynamic behavior of YFP-Cbx2 during interphase of the cell cycle can be affected by the PRC1 proteins.
The recruitment and immobilization of Cbx2 to mitotic chromosomes requires its distinct regions
To dissect the domains (regions) of Cbx2 required for targeting mitotic chromosomes, we generated a variety of Cbx2 mutants tagged with YFP and introduced them into HeLa cells. The mCherry-H2A protein was used to mark mitotic chromosomes. Imaging of live cells by using a confocal fluorescence microscope showed that the deletion of the C-terminus of Cbx2 protein (Cbx21-498, Cbx21-281, and Cbx21-194) does not affect targeting of the Cbx2 fusion mutants to mitotic chromosomes (Figure 7, A and 7B). On the other hand, deletion of the N-terminus of Cbx2 (Cbx289-532) resulted in complete loss of the Cbx2 fusion variant from mitotic chromosomes. These data suggest that the N-terminus of Cbx2 protein is required for targeting the Cbx2 fusion protein to mitotic chromosomes.
To explore the molecular basis for the immobilization of Cbx2 at mitotic chromosomes, we performed FRAP assays on the YFP-Cbx2 fusion variants (Cbx21-498, Cbx21-281, Cbx21-194) during interphase and mitosis of the cell cycle (Figure 7, C and D). Kinetic analysis of interphasic FRAP curves of YFP-Cbx2 fusion variants revealed that the residence time of three YFP-Cbx2 variants was 6–9 s, which is half of that for full-length Cbx2 fusion protein. In contrast to the existence of immobile fraction of Cbx2 fusion protein in interphase cells, there was no immobile fraction for the three YFP-Cbx2 fusion variants. Analysis of mitotic FRAP curves of the three YFP-Cbx2 variants showed striking kinetic differences in comparison to the YFP-Cbx2 fusion protein. The three YFP-Cbx2 variants rapidly exchanged at mitotic chromosomes, with residence time of 8–11 s. In contrast to full-length YFP-Cbx2, YFP-Cbx2 variants were fully recovered at mitotic chromosomes. Thus these data indicate that the immobilization of Cbx2 fusion proteins at mitotic chromosomes requires its extreme C-terminus.
DISCUSSION
We used quantitative live-cell imaging analysis to investigate the mitotic chromosomal association of the canonical PRC1 proteins and interrogate the dynamics of these proteins binding to chromatin in both interphase and mitosis. Our results revealed several striking findings, summarized as follows: 1) The canonical PRC1 subunits tested vary at the level of association with mitotic chromosomes, and Cbx2 is the primary protein accumulated at mitotic chromosomes; 2) the mitotic chromosomal association of Cbx2 protein is independent of PRC1 or PRC2 complex proteins; 3) the Cbx2 protein directly targets the canonical PRC1 proteins to mitotic chromosomes; 4) the Cbx2-containing PRC1 complex is immobilized at mitotic chromosomes, whereas other Cbx-family proteins dynamically exchange at mitotic chromosomes; 5) the immobilization of Cbx2 protein at mitotic chromosomes is independent of PRC1 or PRC2 proteins; and 6) the recruitment of Cbx2 protein to mitotic chromosomes requires its N-terminus, whereas the immobilization of Cbx2 protein at mitotic chromosomes requires its C-terminus. Thus these data provide insights into the mechanisms underlying how canonical PRC1 proteins interact with interphasic and mitotic chromatin and also have implications for understanding PRC1-mediated epigenetic inheritance.
Early studies of mammalian PRC1 proteins by immunofluorescence in fixed cells provided divergent opinions as to whether PRC1 proteins are substantially retained at mitotic chromosomes (Wang et al., 1997; Saurin et al., 1998; Koga et al., 1999; Voncken et al., 1999; Akasaka et al., 2002; Suzuki et al., 2002; Miyagishima et al., 2003; Aoto et al., 2008; Vincenz and Kerppola, 2008). These variations could be due to inaccessibility of the protein to the antibody or may be due to damage or loss of epitopes during the experimental procedures or to differences in how cells were prepared and imaged, as well as in the cell types that were used. Consistent with these notions, we noticed that there were remarkable differences in the mitotic chromosomal association of PRC1 proteins if subtle experimental variations were applied. For instance, upon adding Hoechst to cells before fixing with formaldehyde, we observed that Cbx-family proteins are completely excluded from mitotic chromosomes. By fixing cells with formaldehyde before adding Hoechst, we observed that Cbx2 protein then showed a punctate pattern at mitotic chromosomes (unpublished data). Thus we performed quantitative live-cell imaging to interrogate mitotic chromosomal association and chromatin binding of PRC1 proteins. The quantitative live-cell imaging requires that PRC1 proteins are fused with fluorescence proteins. Many PRC1 fusion proteins were reported to function normally in cells or animals. The Cbx-family proteins fused with Venus are able to form PRC1 complex and bind to PcG target genes (Ren et al., 2008; Ren and Kerppola, 2011). The knock-in mice expressing Mel18 and Ring1b proteins fused with enhanced green fluorescent protein or YFP function normally like their endogenous counterparts (Isono et al., 2013). Drosophila Ph and Pc proteins fused with green fluorescent protein can fulfill the functions of the endogenous proteins (Fonseca et al., 2012). These data suggest that the PRC1 proteins can tolerate the addition of fluorescence protein tag. The inducible gene delivery vector used in the present studies allows us to control the level of fusion protein expression. The protein expression level of the test fusion protein under the doxycycline concentration used is similar to or slightly higher than in the endogenous counterparts. The immunostaining of endogenous Ring1b and Phc1 in wild-type and Cbx2-knockout ES cells also supports that PRC1 proteins associate with mitotic chromosomes and that Cbx2 affects the mitotic chromosomal association of PRC1 proteins.
Although Cbx-family proteins share conserved domains (Cbox and CHD; Simon and Kingston, 2009), accumulating evidence suggests that they have both overlapping and nonoverlapping functions (Core et al., 1997; Katoh-Fukui et al., 1998; Vincenz and Kerppola, 2008; Forzati et al., 2012; Gao et al., 2012; Morey et al., 2012; Klauke et al., 2013). Cbx2 accumulates at mitotic chromosomes, yet other Cbx-family proteins show greatly reduced association with mitotic chromosomes. Deletion of the CHD domain causes dissociation of Cbx2 variants from mitotic chromosomes, suggesting that the CHD domain plays a role in recruiting Cbx2 to mitotic chromosomes. Other Cbx-family proteins also contain a CHD domain but display a much-reduced association with mitotic chromosomes, indicating that other, unknown factors must also contribute to the unique binding properties of Cbx2. These factors may include posttranslational modifications of Cbx2, which could lead to a switch in binding platform. A previous report showed that phosphorylation of Cbx2 changes its binding specificity for methylated histone H3 (Hatano et al., 2010). Another report also indicated that methylation of Cbx4 switches its binding partners (Yang et al., 2011). Another possibility is that Cbx2 protein on its own has unique physical properties—for example, intrinsic charge properties (Grau et al., 2011). It is also possible that the accumulation of Cbx2 proteins at mitotic chromosomes is due to changes of recruiting or competing molecules. Finally, Cbx2 may form unique protein complexes at mitotic chromosomes. Further studies will help in understanding mechanisms by which mitotic Cbx-family proteins are selectively displaced and retained.
We provided several lines of evidence to demonstrate that Cbx2 protein is essential for the recruitment of the canonical PRC1 proteins to mitotic chromosomes. First, we observed that the mitotic fraction of the three PRC1 proteins Ring1b, Phc1, and Mel18 in Cbx2 KO ES cells are reduced at least twofold in comparison to that observed in wild-type ES cells, suggesting that Cbx2 plays a major role in recruiting PRC1 proteins to mitotic chromosomes. Second, the mitotic chromosomal association of the three PRC1 proteins Ring1b, Phc1, and Mel18 in Cbx2 KO ES cells can be restored by supplementing YFP-Cbx2 fusion protein but not YFP-Cbx21-498 fusion protein, which is unable to interact with Ring1b (Satijn et al., 1997; Schoorlemmer et al., 1997; Bardos et al., 2000). Finally, we observed by FRET imaging that the YFP-Cbx2 fusion protein interacts with the Cerulean-Ring1b fusion protein at mitotic chromosomes. Taken together, these data reveal that Cbx2 directly recruits canonical PRC1 proteins to mitotic chromosomes.
It is interesting to note that the Cbx2-containing PRC1 complex (Cbx2, Ring1b, Phc1, and Mel18) is immobilized at mitotic chromosomes without exchange, whereas other Cbx-family proteins (Cbx4, Cbx6, Cbx7, and Cbx8) dynamically bind to mitotic chromosomes with kinetics similar to their binding to interphasic chromatin. It is not clear which factors dictate the transition between a dynamic and a stable Cbx2-PRC1 complex during different phases of the cell cycle. Because the C-terminus of Cbx2 is required for the immobilization of Cbx2 proteins at mitotic chromosomes, the C-terminus may dictate the dynamic switching between interphase and mitosis. The C-terminus contains the Cbox domain, which interacts with Ring1b (Satijn et al., 1997; Schoorlemmer et al., 1997; Bardos et al., 2000), but depletion of Ring1a/Ring1b proteins did not alter the immobilization of Cbx2 to mitotic chromosomes, suggesting that other factors play roles in immobilizing the Cbx2-PRC1 complex at mitotic chromosomes. Previous studies of transcription factors, epigenetic regulators, and chromosomal structural proteins showed that most of them either rapidly exchange at or stably bind to chromatin (Phair et al., 2004; Cherukuri et al., 2008; Ueda et al., 2008; Souza et al., 2009; Hemmerich et al., 2011), and yet a subset of these factors switch binding dynamics upon signaling stimuli or cell cycle transition (Angus et al., 2003; Schmiedeberg et al., 2004; Chen et al., 2005; Mekhail et al., 2005; Gerlich et al., 2006; Meshorer et al., 2006; Yao et al., 2006; Ren et al., 2008; Giglia-Mari et al., 2009; Hellwig et al., 2011; Hemmerich et al., 2011). We hypothesize that the dynamic switching of the Cbx2-PRC1 complex between interphase and mitosis may be regulated through covalent modifications or additional interacting partners (Figure 8).
FIGURE 8:
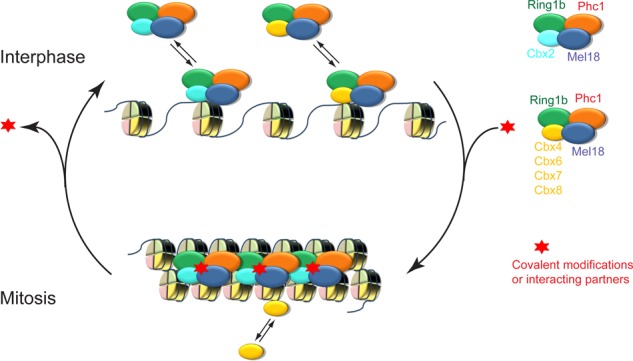
A hypothetic model for the interaction of canonical PRC1 complex with interphasic and mitotic chromatin. In interphases of cells, Cbx2-PRC1 and Cbx4/6/7/8-PRC1 complexes dynamically bind to chromatin. During mitosis, the Cbx2-PRC1 complex is immobilized at mitotic chromosomes, whereas other Cbx family members (Cbx4, Cbx6, Cbx7, and Cbx8) rapidly exchange at mitotic chromosomes. Red star implies that factors such as covalent modification and protein interactor stabilize the Cbx2-PRC1 complex binding to mitotic chromosomes.
The Ringrose laboratory (Fonseca et al., 2012) and the Francis laboratory (Follmer et al., 2012) identified a fraction of Drosophila PRC1 proteins association with mitotic chromosomes, and the Ringrose laboratory (Fonseca et al., 2012) also revealed that 0.2–2% of PRC1 proteins (PC and PH) remain stably bound to mitotic chromatin, with up to 300-fold-longer residence times than in interphase, which supports our findings of mitotic chromosomal association and stable binding to mitotic chromosomes of mammalian PRC1 proteins. All mammalian PRC1 proteins tested in our research have the capacity of stably binding to mitotic chromosomes; however, the fraction of their stable binding to mitotic chromosomes varies greatly. We revealed that >85% of the Cbx2-PRC1 complex is selectively and specifically immobilized at mitotic chromosomes, whereas >85% of other Cbx-family proteins dynamically exchange at mitotic chromosomes. Because mammalian PRC1 complexes comprise a multiplicity of variants and are far more biochemically diverse than their Drosophila counterparts, the selective immobilization of the Cbx2-PRC1 complex at mitotic chromosomes implies that the PRC1 complexes became functionally divergent during evolution.
MATERIALS AND METHODS
Cell lines
The Cbx2−/− (Katoh-Fukui et al., 1998), Ring1a−/−/Ring1bfl/fl; Rosa26::CreERT2 (Ring1a knockout, Ring1b conditional knockout; Endoh et al., 2008), Bmi1−/−/Mel18−/− (Bmi1 and Mel18 double knockout; Elderkin et al., 2007), Eed−/− (Endoh et al., 2008), and PGK12.1 mouse ES cell (Penny et al., 1996) lines were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 15% fetal bovine serum (FBS; BioExpress, Kaysville, UT), 2 mM glutamine (Life Technologies, Carlsbad, CA), 100 U/ml penicillin G sodium (Life Technologies), 0.1 mg/ml streptomycin sulfate (Life Technologies), 0.1 mM β-mercaptoethanol (Life Technologies), 103 units/ml leukemia inhibitor factor, and 0.1 mM nonessential amino acids (Life Technologies) at 37°C in 5% CO2. To deplete Ring1b alleles in Ring1a−/−/Ring1bfl/fl; Rosa26::CreERT2 ES cells, we administrated OHT (Sigma-Aldrich) for 3 d under the concentration of 1.0 μM. HeLa, HEK293, and HEK293T cells were maintained in DMEM supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin G sodium, 0.1 mg/ml streptomycin sulfate at 37°C in 5% CO2.
Plasmids
The pTRIPZ shRNAmir Lentivirus vector (Open Biosystems, Waltham, MA) was engineered to remove both the turboRFP and the regulatory sequences of shRNAmir to produce pTRIPZ(M). The sequences coding Cerulean (Addgene, Cambridge, MA), YFP (Ren et al., 2008), and mCherry (Addgene) fluorescence proteins were amplified by PCR and inserted into the pTRIPZ(M) to produce vectors pTRIPZ(M)-Cerulean, pTRIPZ(M)-YFP, and pTRIPZ(M)-mCherry. The sequences coding Ring1b (Ren et al., 2008), Phc1 (Addgene), Mel18 (Addgene), H2A (Addgene), Cbx2 (Ren et al., 2008), Cbx4 (Ren et al., 2008), Cbx6 (Ren et al., 2008), Cbx7 (Ren et al., 2008), and Cbx8 (Ren et al., 2008) were amplified by PCR and inserted downstream of the coding sequence of fluorescence protein in pTRIPZ(M) vector. The same strategy was used to construct Cbx2 variants tagged with YFP. The Cbx2 variants were as follows: 1) Cbx21-498, deletion of amino acids 499–532; 2) Cbx21-281, deletion of amino acids 282–532; 3) Cbx21-194, deletion of amino acids 195–532; and 4) Cbx289-532, deletion of amino acids 1–88. The sequences encoding fusion proteins have been verified by DNA sequencing.
Generation of stable cell lines by lentivirus infection
Approximately 24 h before transfection, HEK293T cells were seeded in 10-cm dishes at density (3.5–4.0) × 106 to reach 90% confluence at the time of transfection. Cells were cotransfected with 21 μg of pTRIPZ(M) containing the gene of interest, 21 μg of psPAX2, and 10.5 μg of pMD2.G using calcium phosphate precipitation. After 12 h of transfection, the medium was replaced with fresh medium containing 10% FBS, 2 mM glutamine, 100 U/ml penicillin G sodium, and 0.1 mg/ml streptomycin sulfate. At 48 h after changing medium, the medium was collected and centrifuged at 1600 × g for 10 min at room temperature. The supernatants were collected and used for infection of mouse ES, HeLa, and HEK293 cells. If coexpression of multiple proteins was needed, lentiviruses were produced separately and mixed at the time of infection. Polybrene (Sigma-Aldrich) was added at the final concentration of 8 μg/ml, and the cells were seeded at ∼15% confluence on gelatin-coated plates or mitotically inactivated MEF cells. At 16 h after infection, the medium was replaced with fresh medium. At 48 h after changing medium, 1.0–2.0 μg/ml puromycin (Life Technologies) was added to select for infected cells. The expression of transgenes was induced with doxycycline (Sigma-Aldrich) at a concentration of 0.1–1.0 μg/ml.
Confocal microscope imaging of live cells and quantification of mitotic fraction
Zeiss LSM 700 observer Z1 equipped with a 10× oil objective (numerical aperture, 1.4) and an electron-multiplying charge-coupled density (EMCCD) camera was used for Z-scan imaging. For Cerulean fluorescence, 435-nm excitation and 476-nm emission filters were used. For YFP fluorescence, 514-nm excitation and 527-nm emission filters were used. For mCherry fluorescence, 587-nm excitation and 610-nm emission filters were used. The section size was 1 μm for the three fluorescence proteins Cerulean, YFP, and mCherry. The frame size was 512 × 512 pixels. Scan time was 1.56 s. Average number of images was four. For live-cell imaging, cells were incubated with doxycycline (0.1–1.0 μM) to induce protein expression. At 48 h after induction of protein expression, cells were seeded to gelatin-coated cover glass dishes (MatTek, Ashland, MA) in the presence of doxycycline. One day after seeding, medium was replaced with either Ring buffer (155 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 2 mM NaH2PO4, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 10 mM glucose, pH 7.2) or phenol-free DMEM supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin G sodium, and 0.1 mg/ml streptomycin sulfate. Cells were maintained at 37°C using heater controller (TC-324; Warner Instrument, Hamden, CT) during imaging. The grayscale images were converted into pseudocolor, merged, and cropped using Photoshop (Adobe, San Jose, CA).
The Z-stack movies were exported as individual images using Zeiss Zen software. The intensity of each imaging section was quantified using ImageJ software (National Institutes of Health, Bethesda, MD). The mean fluorescence intensities of a region of interest corresponding to the mitotic chromosomes of the metaphase plate marked with H2A were measured. The mean fluorescence intensities of cell nucleus were also measured. The fluorescence intensities of a region without cells were measured as background. The fluorescence values were sums of intensities of individual sections of three-dimensional stacks. The mitotic fraction RM was calculated as RM = (Imitosis − Ib)/(Inucleus − Ib), where Imitosis is the fluorescence intensity of PRC1 fusion protein at mitotic chromosomes; Inucleus is the fluorescence intensity of PRC1 fusion protein in the cell nucleus; and Ib is the background fluorescence intensity corresponding to regions without cells.
FRAP imaging and quantification
FRAP imaging was performed using a Zeiss LSM 700 observer. Cells were maintained as described for confocal laser-scanning imaging of live cells. The expression of fusion proteins was induced by 0.1–0.2 μM doxycycline for 2 d. The pinhole was fully open for FRAP imaging. The scan speed was 1.56 s. Images were taken without average. Before photobleaching, four images were taken. Immediately after photobleaching, 30 images were taken with 5-s intervals. The images were analyzed and fluorescence intensities were quantified using ImageJ software. To correct for movement in the xy plane, the images were aligned using TurboReg. The fluorescence intensities were corrected for fluctuations in background and total signal and normalized to the signal before bleaching to obtain the fluorescence recovery (IR) as described previously (Ren et al., 2008). IR was plotted as a function of time (t) after bleaching. FRAP curves were fitted by one-binding-state kinetic model, IR = 1 − me−kt, where m is the mobile fraction. The immobile fraction was calculated as 1 − m. Residence time was calculated as 1/k.
FRET imaging
FRET measurements were performed using a Zeiss LSM 700 observer. Two images were acquired in the same field of view in the Cerulean-Ring1b (donor) and YFP-Cbx2 (donor) channels. Half-area of YFP-Cbx2 at mitotic chromosomes was bleached with a 514-nm laser, and a second set of images of Cerulean-Ring1b and YFP-Cbx2 was acquired. The FRET ratio was calculated as
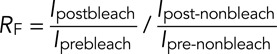 |
where Ipostbleach and Iprebleach are the mean fluorescence intensities of Cerulean-Ring1b after and before bleaching at the bleached half-area, respectively; and Ipost-nonbleach and Ipre-nonbleach are the mean fluorescence intensities of Cerulean-Ring1b after and before bleaching at the nonphotobleached half-area, respectively.
Epifluorescence imaging of live cells
The images were acquired using an Axio Observer D1 Microscope (Zeiss) equipped with a 100× oil objective (numerical aperture, 1.4) and an EMCCD camera. For Cerulean fluorescence, 438/24-nm excitation and 483/32-nm emission filters were used. For YFP fluorescence, 500/24-nm excitation and 542/27-nm emission filters were used. For mCherry fluorescence, 560/10-nm excitation and 610/35-nm emission filters were used. For Hoechst fluorescence, 387/11-nm excitation and 447/60-nm emission filters were used. For live-cell imaging, cells were maintained as described. Images were presented as described.
Cell synchronization and fractionation
ES cell synchronization was carried out as described previously (Ballabeni et al., 2011). Briefly, ES cells were cultured in the presence of 1.25 mM thymidine (T1895-1G; Sigma-Aldrich) for 14 h. After removal of medium and washing with phosphate-buffered saline (PBS), we added fresh ES cell medium with 200 ng/ml nocodazole to the plate (M1404-2MG; Sigma-Aldrich) and cultured cells for 7 h. Cells were harvested and washed with PBS, followed by chromatin isolation or immunofluorescence.
Chromatin fractionation was performed as previously (Mendez and Stillman, 2000; Follmer et al., 2012) with minor modifications. To prepare total cell extracts, synchronized (mitotic) and nonsynchronized (control) cells were directly resuspended in Laemmli buffer, followed by sonication. To isolate chromatin, control and mitotic cells were resuspended (5 × 107 cells/ml) in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, protein inhibitors [P8340; Sigma-Aldrich], 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 1.0 mM dithiothreitol [DTT]) and incubated on ice for 5 min. Nuclei (P1) were collected by centrifugation (1300 × g, 4 min, 4°C). The supernatant (S1) was centrifuged (13,000 × g, 15 min, 4°C) to give supernatant (S2) and pellet (P2). P1 was washed once with buffer A and incubated with buffer B (3 mM EDTA, 0.2 mM ethylene glycol tetraacetic acid, 0.2 mM PMSF, 1.0 mM DTT, and protein inhibitors [P8340; Sigma-Aldrich]). The sample was centrifuged (1700 × g, 4 min, 4°C) to give supernatant (S2) and pellet (P3). P3 was resuspended in Laemmli buffer, followed by sonication. The fractions were run on 4–12% SDS–PAGE gels and immunoblotted.
Immunoblotting
To quantify protein level, cells were collected and lysed with buffer (20 mM Tris-HCl, pH 7.4, 2.0% NP-40, 1.0% Triton X-100, 500 mM NaCl, 0.25 mM EDTA, 0.1 mM Na3VO4, 0.1 mM PMSF, and protease inhibitors [P8340; Sigma–Aldrich]). After centrifugation, the protein concentration was quantified and normalized to the same concentration. Proteins were separated using SDS–PAGE, transferred to 0.45-μm Immobilon-FL polyvinylidene fluoride membrane (Millipore, Darmstadt, Germany), and probed with anti-Cbx2 (ab80044; Abcam, Cambridge, MA), anti-Phc1 (6-1-3; Active Motif, Carlsbad, CA), anti-Ring1b (D139-3; MBL, Woburn, MA), and anti-Mel18 (sc-10744; Santa Cruz Biotechnology, Dallas, TX). Proteins were detected using ECL Plus detection reagents (GE Healthcare, Pittsburgh, PA). Membranes were imaged using a ChemiDoc XRS system (Bio-Rad, Hercules, CA).
Immunofluorescence
Wild-type and Cbx2-knockout ES cells were plated on coverslips and cultured for 24 h. Cells were extracted with detergent buffer (10 mM 1,4-piperazinediethanesulfonic acid, pH 6.8), 300 mM sucrose, 3 mM MgCl2, 0.2% Triton X-100, protein inhibitors [P8340; Sigma-Aldrich], and 100 mM NaCl), and fixed using 1.0% paraformaldehyde for 10 min. Cells were washed with PBS three times for 5 min and incubated with 0.2% Triton X-100 for 10 min. After washing with basic blocking buffer (10 mM PBS, pH 7.2, 0.1% Triton X-100, 0.05% Tween 20) three times for 5 min, cells were incubated with blocking buffer (basic blocking buffer plus 3% goat serum and 3% bovine serum albumin) for 1 h. Primary antibody diluted in blocking buffer was incubated with cells for 2 h at room temperature. After washing with basic blocking buffer three times for 5 min, the secondary antibody diluted in blocking buffer was incubated for 1 h. Cells were rinsed with PBS three times and washed with basic blocking buffer twice for 5 min. After incubation with 0.1 μg/ml Hoechst, cells were washed and mounted on slides with ProLong Antifade reagents (Life Technologies). The primary antibodies were anti-Phc1 and anti-Ring1b. The primary antibodies were detected using fluorescein isothiocyanate (FITC)–labeled goat anti–mouse antibodies (Sigma-Aldrich).
To immunostain synchronized cells, after trypsinization, mitotic cells were collected by centrifugation and washed with PBS. Cells were spun onto glass slides at 1000 rpm for 10 min in a Shandon Cytospin 2. Cells were fixed with 2.0% formaldehyde at room temperature for 10 min and immunostained as described. The primary antibody is anti–histone H3 (phospho S10) antibody (ab5176; Abcam) and was detected using FITC-labeled goat anti–rabbit antibodies (Sigma-Aldrich).
Supplementary Material
Acknowledgments
We thank Tom Kerppola for continuous support and scientific advice and critical comments on the manuscript. We also thank Haruhiko Koseki for providing the Cbx2−/−, Ring1a−/−/Ring1bfl/fl; Rosa26::CreERT2, Bmi1−/−/Mel18−/−, and Eed−/− ES cell lines. This work was supported by the University of Colorado Denver and by American Cancer Society Grant IRG #57-001-53 to X.R.
Abbreviations used:
- Cerulean
cerulean fluorescent protein
- DTT
dithiothreitol
- FRAP
fluorescence recovery after photobleaching
- FRET
fluorescence resonance energy transfer
- H3K27me3
H3 trimethyl K27
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- KO
knockout
- OHT
4-hydroxytamoxifen
- PBS
phosphate-buffered saline
- PcG
polycomb group
- PMSF
phenylmethylsulfonyl fluoride
- PRC
polycomb repressive complex
- YFP
yellow fluorescent protein
Footnotes
*These authors contributed equally to this work.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1109) on September 17, 2014.
REFERENCES
- Akasaka T, Takahashi N, Suzuki M, Koseki H, Bodmer R, Koga H. MBLR, a new RING finger protein resembling mammalian Polycomb gene products, is regulated by cell cycle-dependent phosphorylation. Genes Cells. 2002;7:835–850. doi: 10.1046/j.1365-2443.2002.00565.x. [DOI] [PubMed] [Google Scholar]
- Angus SP, Solomon DA, Kuschel L, Hennigan RF, Knudsen ES. Retinoblastoma tumor suppressor: analyses of dynamic behavior in living cells reveal multiple modes of regulation. Mol Cell Biol. 2003;23:8172–8188. doi: 10.1128/MCB.23.22.8172-8188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto T, Saitoh N, Sakamoto Y, Watanabe S, Nakao M. Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression. J Biol Chem. 2008;283:18905–18915. doi: 10.1074/jbc.M709322200. [DOI] [PubMed] [Google Scholar]
- Ballabeni A, Park IH, Zhao R, Wang W, Lerou PH, Daley GQ, Kirschner MW. Cell cycle adaptations of embryonic stem cells. Proc Natl Acad Sci USA. 2011;108:19252–19257. doi: 10.1073/pnas.1116794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardos JI, Saurin AJ, Tissot C, Duprez E, Freemont PS. HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J Biol Chem. 2000;275:28785–28792. doi: 10.1074/jbc.M001835200. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukarabila H, Saurin AJ, Batsche E, Mossadegh N, van Lohuizen M, Otte AP, Pradel J, Muchardt C, Sieweke M, Duprez E. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23:1195–1206. doi: 10.1101/gad.512009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Ren X, Kerppola TK. KAP1 represses differentiation-inducible genes in embryonic stem cells through cooperative binding with PRC1 and derepresses pluripotency-associated genes. Mol Cell Biol. 2014;34:2075–2091. doi: 10.1128/MCB.01729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherukuri S, Hock R, Ueda T, Catez F, Rochman M, Bustin M. Cell cycle-dependent binding of HMGN proteins to chromatin. Mol Biol Cell. 2008;19:1816–1824. doi: 10.1091/mbc.E07-10-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core N, Bel S, Gaunt SJ, Aurrand-Lions M, Pearce J, Fisher A, Djabali M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Elderkin S, Maertens GN, Endoh M, Mallery DL, Morrice N, Koseki H, Peters G, Brockdorff N, Hiom K. A phosphorylated form of mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol Cell. 2007;28:107–120. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura YI, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- Follmer NE, Wani AH, Francis NJ. A polycomb group protein is retained at specific sites on chromatin in mitosis. PLoS Genet. 2012;8:e1003135. doi: 10.1371/journal.pgen.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca JP, Steffen PA, Muller S, Lu J, Sawicka A, Seiser C, Ringrose L. In vivo Polycomb kinetics and mitotic chromatin binding distinguish stem cells from differentiated cells. Genes Dev. 2012;26:857–871. doi: 10.1101/gad.184648.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzati F, Federico A, Pallante P, Abbate A, Esposito F, Malapelle U, Sepe R, Palma G, Troncone G, Scarfo M, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122:612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Theil AF, Mari PO, Mourgues S, Nonnekens J, Andrieux LO, de Wit J, Miquel C, Wijgers N, Maas A, et al. Differentiation driven changes in the dynamic organization of Basal transcription initiation. PLoS Biol. 2009;7:e1000220. doi: 10.1371/journal.pbio.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano A, Matsumoto M, Higashinakagawa T, Nakayama KI. Phosphorylation of the chromodomain changes the binding specificity of Cbx2 for methylated histone H3. Biochem Biophys Res Commun. 2010;397:93–99. doi: 10.1016/j.bbrc.2010.05.074. [DOI] [PubMed] [Google Scholar]
- Hellwig D, Emmerth S, Ulbricht T, Doring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, et al. Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci. 2011;124:3871–3883. doi: 10.1242/jcs.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, Schmiedeberg L, Diekmann S. Dynamic as well as stable protein interactions contribute to genome function and maintenance. Chromosome Res. 2011;19:131–151. doi: 10.1007/s10577-010-9161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Endo TA, Ku M, Yamada D, Suzuki R, Sharif J, Ishikura T, Toyoda T, Bernstein BE, Koseki H. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Katoh-Fukui Y, Tsuchiya R, Shiroishi T, Nakahara Y, Hashimoto N, Noguchi K, Higashinakagawa T. Male-to-female sex reversal in M33 mutant mice. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- Kerppola TK. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke K, Radulovic V, Broekhuis M, Weersing E, Zwart E, Olthof S, Ritsema M, Bruggeman S, Wu X, Helin K, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol 15, 353–362. 2013 doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- Koga H, Matsui S, Hirota T, Takebayashi S, Okumura K, Saya H. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene. 1999;18:3799–3809. doi: 10.1038/sj.onc.1202732. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima H, Isono K, Fujimura Y, Iyo M, Takihara Y, Masumoto H, Vidal M, Koseki H. Dissociation of mammalian Polycomb-group proteins, Ring1B and Rae28/Ph1, from the chromatin correlates with configuration changes of the chromatin in mitotic and meiotic prophase. Histochem Cell Biol. 2003;120:111–119. doi: 10.1007/s00418-003-0551-2. [DOI] [PubMed] [Google Scholar]
- Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep. 2013;3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F-and Myc-responsive genes in G(0) cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Kerppola TK. REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol Cell Biol. 2011;31:2100–2110. doi: 10.1128/MCB.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Vincenz C, Kerppola TK. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol Cell Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JAA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Gunster MJ, van der Vlag J, Hamer KM, Schul W, Alkema MJ, Saurin AJ, Freemont PS, van Driel R, Otte AP. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J, MarcosGutierrez C, Were F, Martinez R, Garcia E, Satijn DPE, Otte AP, Vidal M. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 1997;16:5930–5942. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Endo TA, Ito S, Ohara O, Koseki H. Embracing change to remain the same: conservation of polycomb functions despite divergence of binding motifs among species. Curr Opin Cell Biol. 2013;25:305–313. doi: 10.1016/j.ceb.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza PP, Volkel P, Trinel D, Vandamme J, Rosnoblet C, Heliot L, Angrand PO. The histone methyltransferase SUV420H2 and heterochromatin proteins HP1 interact but show different dynamic behaviours. BMC Cell Biol. 2009;10:41. doi: 10.1186/1471-2121-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen PA, Fonseca JP, Ganger C, Dworschak E, Kockmann T, Beisel C, Ringrose L. Quantitative in vivo analysis of chromatin binding of Polycomb and Trithorax group proteins reveals retention of ASH1 on mitotic chromatin. Nucleic Acids Res. 2013;41:5235–5250. doi: 10.1093/nar/gkt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Mizutani-Koseki Y, Fujimura Y, Miyagishima H, Kaneko T, Takada Y, Akasaka T, Tanzawa H, Takihara Y, Nakano M, et al. Involvement of the Polycomb-group gene Ring1B in the specification of the anterior-posterior axis in mice. Development. 2002;129:4171–4183. doi: 10.1242/dev.129.18.4171. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678. 2012 doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Catez F, Gerlitz G, Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme J, Volkel P, Rosnoblet C, Le Faou P, Angrand PO. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol Cell Proteomics. 2011;10:M110.002642. doi: 10.1074/mcp.M110.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbunder B, Fourre N, Leray A, Mueller F, Volkel P, Angrand PO, Heliot L. PRC1 components exhibit different binding kinetics in Polycomb bodies. Biol Cell. 2014;106:111–125. doi: 10.1111/boc.201300077. [DOI] [PubMed] [Google Scholar]
- Vincenz C, Kerppola TK. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci USA. 2008;105:16572–16577. doi: 10.1073/pnas.0805317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, van Lohuizen M. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J Cell Sci. 1999;112:4627–4639. doi: 10.1242/jcs.112.24.4627. [DOI] [PubMed] [Google Scholar]
- Wang G, Horsley D, Ma A, Otte AP, Hutchings A, Butcher GW, Singh PB. M33, a mammalian homologue of Drosophila Polycomb localises to euchromatin within interphase nuclei but is enriched within the centromeric heterochromatin of metaphase chromosomes. Cytogenet Cell Genet. 1997;78:50–55. doi: 10.1159/000134626. [DOI] [PubMed] [Google Scholar]
- Wang HB, Wang LJ, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004a;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004b;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.