The latent properties of Stp1—the effector transcription factor of the SPS signaling pathway in yeast—depend on the RI motif in the N-terminal regulatory domain and three inner nuclear membrane proteins, Asi1, Asi2, and Asi3. RI functions in a modular and transferable manner as a cytoplasmic retention determinant and an Asi-dependent degron.
Abstract
The Ssy1-Ptr3-Ssy5 (SPS)–sensing pathway enables yeast to respond to extracellular amino acids. Stp1, the effector transcription factor, is synthesized as a latent cytoplasmic precursor with an N-terminal regulatory domain that restricts its nuclear accumulation. The negative regulatory mechanisms impinging on the N-terminal domain are poorly understood. However, Stp1 latency depends on three inner nuclear membrane proteins, Asi1, Asi2, and Asi3. We report that the N-terminal domain of Stp1 contains a small motif, designated RI, that fully accounts for latency. RI is modular, mediates interactions with the plasma membrane, and can retain histone Htb2 in the cytoplasm. A novel class of STP1 mutations affecting RI were isolated that are less efficiently retained in the cytoplasm but remain under tight negative control by the Asi proteins. Intriguingly, these mutant proteins exhibit enhanced stability in strains lacking ASI1. Our results indicate that RI mediates latency by two distinct activities: it functions as a cytoplasmic retention determinant and an Asi-dependent degron. These findings provide novel insights into the SPS-sensing pathway and demonstrate for the first time that the inner nuclear membrane Asi proteins function in a degradation pathway in the nucleus.
INTRODUCTION
The ability of cells to sense and respond to environmental cues is critical for survival. Accordingly, cells possess signal transduction pathways that generate appropriate responses to specific external signals. Often the cellular response effects adjustments in patterns of gene expression that enable cells to optimize growth. In such instances, the signaling pathways control the activity of specific transcription factors. Understanding the mechanisms that eukaryotic cells use to control signaling networks to achieve proper regulation of transcription in the nucleus remains a major challenge and an important task of cell biology.
The regulation of transcription factor activity can be achieved by different means. Multiple examples exist in which transcription factors are posttranslationally modified in a signal-dependent manner—for example, by phosphorylation. Such changes can affect protein stability, DNA-binding affinity, or the capacity to interact with other factors required for proper transcriptional control. In eukaryotic cells, signaling pathways can also control transcription factors by regulating their access to the nuclear compartment (Brivanlou and Darnell, 2002).
Regulated compartmentalization can be accomplished by several mechanisms. For example, membrane anchoring and signaling-induced release by proteolysis can serve as an efficient mechanism to control nuclear targeting of transcription factors. This type of regulation was first described for the sterol regulatory element–binding protein (SREBP). SREBP is an integral membrane protein of the endoplasmic reticulum (ER). The domain oriented toward the cytoplasm harbors transcription-inducing activity and is proteolytically liberated from the membrane. This mechanism, termed regulated intramembrane proteolysis, also regulates Notch signaling (Brown et al., 2000). Another documented proteolytic mechanism is termed regulated ubiquitin/proteasome-dependent processing (RUP). RUP relies on recruitment of the proteasome to the ER membrane, where, in an endoproteolytic processing event, it frees transcription factors Spt23 and Mga2 from C-terminal membrane anchors (Hoppe et al., 2000, 2001).
Alternatively, regulated compartmentalization of transcription factors can be achieved by interaction with cytosolic proteins. Well-studied examples include the nuclear factor-κB (NFκB), which is retained as an inactive cytoplasmic factor by interaction with the inhibitor of κB (IκB) family of proteins. The major isoform, IκB-β, sequesters NFκB in the cytoplasm by masking its nuclear localization signal, whereas IkB-α binding regulates NFκB localization via dynamic nucleocytoplasmic shuttling (Malek et al., 2001). A prominent example of nucleocytoplasmic shuttling in the yeast Saccharomyces cerevisiae is the transcription factor Yap1, which activates the transcription of antioxidant genes (Morano et al., 2012). On oxidative stress, Yap1 is activated by the oxidation of two cysteine residues, leading to the formation of a disulfide bond. This masks a nuclear export sequence (NES) and, consequently, leads to nuclear accumulation of the oxidized form of Yap1 (Delaunay et al., 2000). A second layer of regulation limits the duration of the oxidative stress response: the turnover of Yap1 is accelerated by its nuclear localization. Thus the enhanced rate of degradation sets temporal limits for the efficacy of Yap1-dependent transcription (Gulshan et al., 2012). Similarly, the degradative machinery in the nucleus governs the stability of Hac1, the transcription factor mediating the response to unfolded proteins in the lumen of the endoplasmic reticulum (Pal et al., 2007).
The Ssy1-Ptr3-Ssy5 (SPS) signaling pathway in S. cerevisiae enables cells to respond to the presence of extracellular amino acids and induce their uptake (for review see Ljungdahl and Daignan-Fornier, 2012). The most-downstream effectors of the SPS pathway, the homologous transcription factors Stp1 and Stp2 (Stp1/2), are synthesized as latent precursors with N-terminal regulatory domains that restrict their nuclear accumulation. The primary amino acid–induced signal, initiated by the plasma membrane–localized receptor Ssy1, leads via Ptr3 to the activation of the Ssy5 endoprotease. Active Ssy5 cleaves the regulatory domains of Stp1/2 (Abdel-Sater et al., 2011; Omnus et al., 2011; Omnus and Ljungdahl, 2013). As a consequence, the processed transcription factors lacking their N-termini accumulate in the nucleus, where they activate the transcription of amino acid permease genes, leading to an enhanced amino acid uptake capacity.
We reported that the first 125 amino acids of Stp1, comprising an N-terminal regulatory (REG) domain, contain all of the contextual information necessary for full SPS-sensing pathway control of Stp1 activity (Andréasson and Ljungdahl, 2004). The REG domain has two conserved sequence motifs, designated RI (residues 16–35) and RII (65–97). Results from rather crude mutational analyses suggest that RI is required to prevent the full-length nonprocessed form of Stp1 from inappropriately entering the nucleus and thus likely functions as a cytoplasmic retention determinant. RII contains the Ssy5 processing site (Andréasson et al., 2006). The REG domain conferred the entire spectrum of SPS-sensor pathway control when fused to the well-characterized synthetic transactivator lexA-AD (Andréasson and Ljungdahl, 2004).
Loss-of-function mutations in ASI1, ASI2, or ASI3 (amino acid sensor independent [Asi]) bypass the requirement of SPS-sensor processing and result in constitutive Stp1/2-dependent gene expression (Forsberg et al., 2001; Boban et al., 2006; Zargari et al., 2007). All three Asi proteins are integral membrane proteins that localize to the inner nuclear membrane (INM; Forsberg et al., 2001; Boban et al., 2006; Zargari et al., 2007). Asi1 and Asi3 are homologous proteins with five membrane-spanning segments. Asi1 and Asi3 copurify (Zargari et al., 2007), and both have conserved RING domains at their C-termini that are oriented to the nucleoplasm (Boban et al., 2006; Zargari et al., 2007). The conserved cysteine residues within the RING domains of Asi1 and Asi3 are required for function (Boban et al., 2006; Zargari et al., 2007). RING domains are found in a class of E3 ubiquitin ligases, where they play an active role in the transfer of ubiquitin from an E2-ubiquitin conjugate by destabilizing the thioester E2-ubiquitin bond. Consequently, the thioester bond is susceptible to nucleophilic attack by a lysine residue on a properly presented substrate protein (Budhidarmo et al., 2012; Berndsen and Wolberger, 2014). Despite extensive efforts, experimental evidence to corroborate the notion that Asi1 and Asi3 function as E3 ubiquitin ligases is lacking.
The fusion of amino acid residues 2–69 of Stp1 to lexA restricted binding to lexA operators in an Asi1-dependent manner (Boban et al., 2006). This finding confirmed that Asi1 exerts a negative regulatory function and contributes to Stp1 latency based on mechanisms impinging on its N-terminal domain (Boban et al., 2006). Of note, the deletion of ASI1 does not affect the intracellular distribution of the bulk Stp1; as in wild-type cells, Stp1 remained diffusely localized to the cytoplasm in asi1Δ cells (Boban et al., 2006). Consequently, the Asi proteins clearly function to prevent promoter accessibility, but it remains to be elucidated how these INM proteins exert their regulatory function in maintaining the off-state of SPS sensor–controlled gene expression.
In this study, we investigated the molecular basis of Stp1 latency. Our experiments indicate that the N-terminal RI motif comprised of 16 amino acids functions in a modular and transferable manner as a negative-acting determinant with two intrinsic activities: RI acts as a cytoplasmic retention determinant and as an Asi-dependent degron within the nucleus. The data show that both activities are required to prevent inappropriate transcription by Stp1. These results provide novel details regarding the SPS-sensing pathway and the first mechanistic insights into the function of the inner nuclear membrane proteins Asi1, Asi2, and Asi3.
RESULTS
The Stp1 N-terminal regulatory domain can control nuclear accumulation of histone Htb2
To test the efficacy of the Stp1 N-terminal REG domain, we examined whether it could control the localization of histone Htb2, a protein that normally efficiently targets to the nucleus. We posited that if the REG domain placed the chimeric protein under SPS-sensing pathway control, we could differentiate between the possibilities that the RI motif in the REG domain functions as a cytoplasmic retention determinant, or alternatively, functions as a nuclear export sequence (NES) that mediates efficient nuclear export. The N-terminal REG domain of Stp1 was fused to the N-terminus of histone Htb2 carrying a C-terminal green fluorescent protein (GFP)–hemagglutinin (HA) tag (Figure 1). A plasmid directing the expression of this chimera was introduced into a strain expressing chromosomal Htb2 fused with ymCherry. The Stp11-125Htb2-GFP construct was rapidly processed upon leucine induction (Figure 1, immunoblot), clearly demonstrating that the chimeric protein is subject to SPS-sensor control. The intracellular distribution of both the GFP- and Cherry-tagged Htb2 was assessed by fluorescence microscopy and followed over time after leucine induction. In contrast to the Htb2-ymCherry, which localized to distinct foci marking the nucleus, the unprocessed Stp11-125Htb2-GFP chimera displayed diffuse fluorescence in the absence of leucine induction (Figure 1, t = 0). On leucine induction, correlating precisely with SPS-sensor dependent processing, the GFP fluorescence associated with the shorter processed form of the chimera rapidly concentrated in foci that overlapped with the nuclear staining of the Htb2-ymCherry (Figure 1, merge). These results confirm and extend our previous finding that the N-terminal regulatory domain of Stp1 is modular and transferable (Andréasson and Ljungdahl, 2004). The fact that the RI-containing REG domain is able to prevent the nuclear localization of a histone protein with a high affinity for binding DNA diminishes the likelihood that RI functions as a NES mediating nucleocytoplasmic shuttling. Instead, the data suggest that the RI motif functions as a nuclear exclusion determinant that interacts and associates with cytoplasmic components.
FIGURE 1:
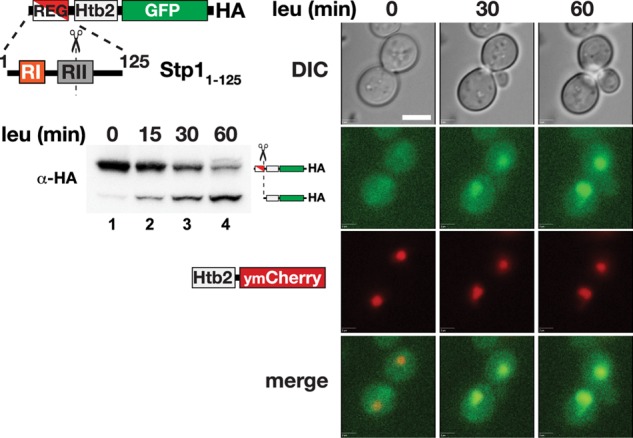
The N-terminal regulatory domain of Stp1 functions as modular cytoplasmic retention determinant. The Stp11-125-Htb2-GFP fusion protein is schematically presented. Immunoblot (left) of extracts prepared from and microscopic analysis (right) of strain CAY1259 (HTB2-ymCherry) carrying pDO39 (STP11-125-HTB2-GFP-HA) grown in SD media and at the indicated time points after leucine induction. For microscopic analysis, cells were immobilized on concanavalin A–coated (1 mg/ml) microscopy dishes and observed by fluorescence microscopy (Zeiss Axiovert 200M, 63× objective, differential interference contrast, GFP, and red fluorescent protein filters). Images were acquired using SlideBook software at 0, 30, and 60 min after cells were induced with leucine.
RI can mediate protein–protein interactions at the plasma membrane
To examine the notion that RI mediates an interaction with a cytoplasmic component, we used the SOS recruitment system (SRS). The SRS exploits the fact that the human Cdc25 homologue SOS (hSOS) can suppress the temperature-sensitive growth defect of a cdc25-2 strain when it is localized in close proximity to the plasma membrane, a property that is dependent on hSOS being fused to a plasma membrane–associated partner (Aronheim et al., 1997). We previously showed that the N-terminal REG domain of Stp1, amino acids 2–158 (hSOS-REG2-158), can direct hSOS to the plasma membrane (Andréasson and Ljungdahl, 2002). To examine specifically whether RI mediates the plasma membrane interaction of the hSOS-REG2-158 fusion protein, we constructed hSOS-REG2-158(Δ16-21) lacking the first six amino acid (aa) residues of RI (aa 16–21 of Stp1; Figure 2A). We introduced hSOS alone (ø), hSOS-REG2-158, and hSOS-REG2-158(Δ16-21) into a temperature-sensitive cdc25-2 reporter strain and assayed growth at permissive (25°C) and nonpermissive (37°C) temperatures. All strains grew equally well at 25°C; however, only the full-length hSOS-REG2-158 construct enabled growth at 37°C (Figure 2A; compare dilution 2 with 1 and 3). The hSOS-REG2-158(Δ16-21) mutant did not suppress the growth defect of the cdc25-2 strain, indicating that in the absence of RI, the REG domain is unable to associate with the plasma membrane.
FIGURE 2:
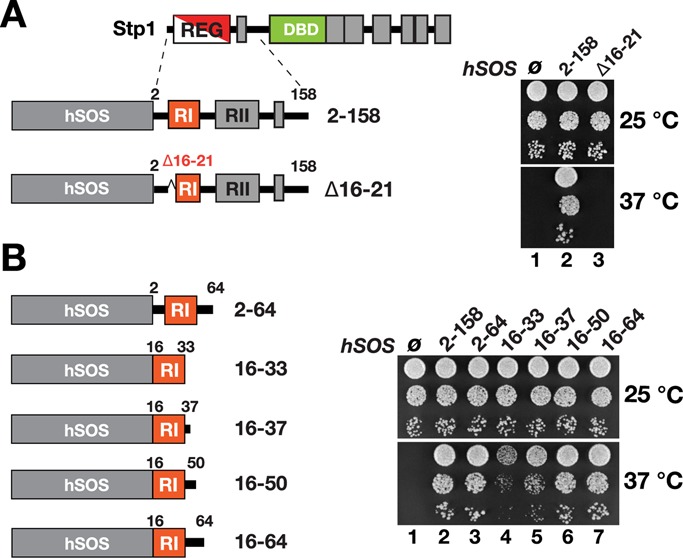
RI of Stp1 suffices to direct hSOS to the plasma membrane. Growth of (A) CAY187 (cdc25-2) carrying pCA094 (ø), pCA104 (hSOS-Stp12-158), or pDO196 (hSOS-Stp12-158(Δ16-21)) and of (B) CAY187 (cdc25-2) carrying pCA094 (ø), pCA104 (hSOS-Stp12-158), pDO203 (hSOS-Stp12-64), pDO205 (hSOS-Stp116-33), pDO230 (hSOS-Stp116-37), pDO231 (hSOS-Stp116-50), or pDO221 (hSOS-Stp116-64). Plates were incubated at 25 and 37°C as indicated. Right, hSOS-REG fusion constructs.
Next we asked whether the RI motif alone would suffice to mediate plasma membrane association. Indeed, we found that cdc25-2 cells expressing hSOS-REG2-64 grew equally well at 37°C as hSOS-REG2-158 (Figure 2B, compare dilution 3 with dilution 2). On the basis of this result, we sought to identify the minimal sequence required for plasma membrane association and constructed hSOS fusion constructs with Stp1 amino acid residues 16–33, 16–37, 16–50, and 16–64, respectively. When expressed in cdc25-2 cells, these constructs exhibited a graded effect on the ability to target the plasma membrane and suppress the temperature sensitivity (Figure 2B, compare dilutions 4–7). A sequence comprised of amino acids 16–50 conferred robust growth at 37°C, indistinguishable from that conferred by hSOS-REG2-64 and hSOS-REG2-158. The hSOS-REG16-37 construct noticeably suppressed the temperature sensitivity, indicating that amino acid residues 16–37, which closely match the boundaries of RI, suffice to localize hSOS to the plasma membrane. Together our results are consistent with RI functioning as a cytoplasmic retention determinant that is capable of directly or indirectly interacting with a cellular component associated with the plasma membrane.
RI is required for Stp1 latency
To directly test whether RI mediates Stp1 latency, we constructed a mutant allele in the context of a fully functional C-terminal HA epitope–tagged STP1, which encoded a protein lacking residues 16–35 (STP1ΔRI) (Figure 3A, schematic). On leucine induction, the Stp1ΔRI protein was processed as efficiently as wild type (Figure 3B, compare lanes 1a-b and 2a-b). The ability of Stp1ΔRI to induce transcription was assessed by monitoring growth on yeast extract/peptone/dextrose (YPD) medium containing 2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino]-sulfonyl}-benzoic acid (MM). MM is an inhibitor of branched-chain amino acid synthesis, and growth on YPD plus MM requires SPS sensor–induced, Stp1/Stp2-dependent expression of high-affinity permeases for leucine, isoleucine, and valine (Jørgensen et al., 1998). Cells expressing STP1ΔRI grew as well as cells expressing wild-type STP1 on YPD plus MM (Figure 3B, dilutions 1 and 2), indicating that Stp1ΔRI is capable of activating SPS sensor–regulated genes. Of importance, in cells lacking a functional SPS sensor (ssy5Δ), expression of wild-type STP1 did not activate amino acid permease expression. By contrast, ssy5Δ cells expressing STP1ΔRI exhibited robust growth (Figure 3B, dilutions 3 and 4). In conclusion, deletion of amino acid residues 16–35 bypasses the requirement of SPS sensor– dependent proteolytic processing, indicating that RI is essential for maintaining the latent properties of Stp1 in the absence of amino acid induction.
FIGURE 3:
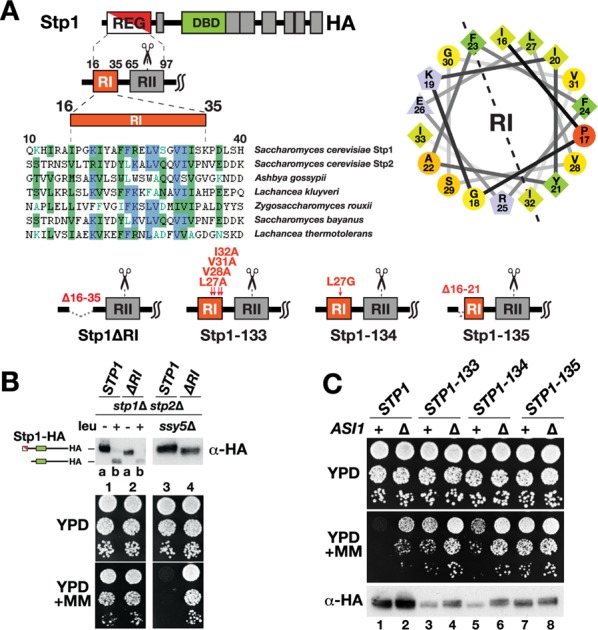
RI within the N-terminal domain of Stp1 is a conserved sequence motif required for latency. (A) Schematic representation of Stp1 and its N-terminal REG domain containing two conserved sequence motifs designated RI (aa 16–35) and RII (aa 65–97). Clustal X comparison of RI sequences of Stp1, corresponding to residues 10 and 40, and Stp2 of S. cerevisiae and the indicated fungal Stp1/Stp2 orthologues; the level of similarity is color coded as follows: conservative (blue) and similar (green) residues are highlighted; residues with weak (green text) or no similarity (black) are indicated. The RI motif is predicted to attain an amphipathic α-helix structure (Garnier et al., 1996); helical wheel projection of residues 16–33, created using Armstrong and Zidovetzki’s helical wheel script (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi). Schematic representation of the Stp1 mutant protein constructs Stp1ΔRI, Stp1-133, Stp1-134, and Stp1-135. (B) Immunoblot analysis of extracts prepared from CAY123 (stp1Δ stp2Δ) carrying plasmid pCA047 (STP1) or pDO248 (ΔR1) grown in SD medium and harvested 30 min after induction by leucine as indicated; immunoreactive forms of Stp1 species are indicated at their corresponding positions of migration (top left). Immunoblot analysis of extracts prepared from MBY93 (ssy5Δ stp1Δ stp2Δ) carrying plasmids pCA047 (STP1), pDO248 (ΔR1) grown in SD medium (top right). Growth of strains on YPD and YPD + MM; 10-fold dilutions of cultures grown in SD were spotted onto plates and incubated at 30°C (bottom). (C) Growth of MBY93 (ssy5Δ stp1Δ stp2Δ) and MBY102 (ssy5Δ stp1Δ stp2Δ asi1Δ) carrying plasmid pCA047 (STP1), pCA120 (STP1-133), pDO141 (STP1-134), or pDO142 (STP1-135) on YPD and YPD + MM; 10-fold dilutions of cultures grown in SD were spotted onto plates and incubated at 30°C (top). Immunoblot analysis of Stp1 protein levels in extracts prepared from cells grown in SD medium (bottom).
The constitutive gene expression conferred by RI mutations is enhanced in cells lacking ASI1
ASI1, ASI2, and ASI3 encode inner nuclear membrane proteins that exert a negative regulatory function required to prevent full-length Stp1 from binding promoters of SPS sensor–regulated genes. Deletion of any of the ASI genes leads to constitutive Stp1-dependent amino acid permease gene expression. Hence the Asi proteins collectively play an essential role in maintaining the “off state” of gene expression in the absence of amino acid induction (Forsberg et al., 2001; Boban et al., 2006; Zargari et al., 2007). Using a mutational analysis, we directly tested the importance of RI in mediating Asi-dependent control.
Secondary structure prediction algorithms suggest that RI folds into an amphipathic α-helix (Figure 3A, right). With this in mind, we exchanged the conserved amino acid residue leucine 27 with a potential helix-breaking glycine residue (L27G), creating the STP1-134 allele. In addition, we deleted the first 6 residues of RI, that is, residues 16–21 (IPGKIY), creating the STP1-135 allele. The latter construct, encoding a truncated RI motif, harbors the identical mutation that abolished the targeting of hSOS to the plasma membrane (Figure 2A). These new constructs and the dominant STP1-133 allele, carrying alanine substitutions at four conserved hydrophobic residues within RI (Figure 3A, sequence alignment) were introduced into ssy5Δ stp1Δ stp2Δ strains with (+) and without ASI1 (Δ), and growth was monitored in the presence of MM (Figure 3C). We previously showed that STP1-133 leads to constitutive expression of SPS sensor–regulated genes in the absence of processing (Andréasson and Ljungdahl, 2004) and bypasses Asi-dependent control without a detectable change in intracellular localization of Stp1 (Boban et al., 2006).
As expected, expression of wild-type STP1 induced permease expression in asi1Δ cells but not in ASI1 cells (Figure 3C, compare dilutions 1 and 2). In addition, expression of STP1-133 in ssy5Δ cells led to the same extend of resistance to MM as wild-type Stp1 in ssy5Δ asi1Δ cells (Figure 3C, compare dilutions 2 and 3). Of note, a higher degree of MM resistance was observed in ssy5Δ asi1Δ cells expressing STP1-133 (Figure 3C, compare dilutions 3 and 4). Expression of the STP1-134 allele conferred a substantial degree of resistance to MM compared with STP1 (Figure 3C, compare dilutions 5 and 1), indicating that Stp1-134 carrying the L27G mutation displays constitutive activity. Of interest, as observed for the constitutively active Stp1-133 mutant, we could also detect an even higher degree of resistance to MM in cells lacking ASI1 (Figure 3C, compare dilution 6 with dilution 5 and dilution 4 with dilution 3). The STP1-135 allele encoding a protein lacking RI residues 16–21 conferred robust resistance in both ASI1 and asi1Δ cells (Figure 3C, compare dilutions 7 and 8). Next we compared the levels of protein in the cells and found that, compared with wild-type Stp1, the steady-state levels of the mutant proteins were reduced in all cases (Figure 3C, bottom, compare lanes 3–8 with lanes 1 and 2). Strikingly, except for Stp1-135, the levels of the Stp1 proteins were significantly higher in cells lacking ASI1 (Figure 3C, compare lanes 1 and 2, 3 and 4, and 5 and 6).
Together the data indicate that mutations of RI, as in STP1-133 and STP1-134, lead to partial impairment of RI function, whereas the deletion of amino acid residues 16–21 (STP1-135) confers a complete loss of function of RI-mediated, Asi1-dependent latency. The observation that specific mutations of RI perturb but do not completely abolish its function, and deletion of ASI1 has additive negative effects on latency, is consistent with two independent mechanisms operating to ensure Stp1 latency. Of interest, the Asi-dependent mechanism appears to affect the level of Stp1 proteins.
Amino acids 17–33 define a minimal RI that suffices to mediate Stp1 latency
To ascertain the minimal sequence requirement of the RI motif, we created a series of mutant alleles encoding altered N-terminal domains of Stp1. Specifically, we deleted the sequence encoding amino acids 2–64 from STP1 and reinserted sequences encoding RI variants, that is, amino acids 15–35, 17–33, and 22–35 between minimal linker sequences, fused in-frame to the L65 codon of RII, respectively (Figure 4A). We expressed these RI constructs in ssy5Δ stp1Δ stp2Δ cells and analyzed Stp1 activity by assessing growth on YPD plus MM. Similar to wild-type Stp1, the RI15-35 and RI17-33 proteins were expressed as latent forms and did not facilitate growth (Figure 4B, compare dilutions 3 and 5 with dilution 1), indicating that amino acid residues RI comprising amino acids 15–35 or 17–33 suffice in isolation to confer latency to Stp1. However, in asi1Δ cells, the expression of RI15-35 and RI17-33 supported a more robust level of resistance to MM than that resulting from expression of wild-type Stp1 (Figure 4B, compare dilutions 4 and 6 with dilution 2). Consistent with our previous data, deletion of aa 17–21 creating the RI22-35 construct resulted in a constitutively active protein; robust growth on YPD plus MM was observed in ssy5Δ stp1Δ stp2Δ cells, and asi1Δ had no additional negative effect on latency (Figure 4B, dilutions 7 and 8).
FIGURE 4:
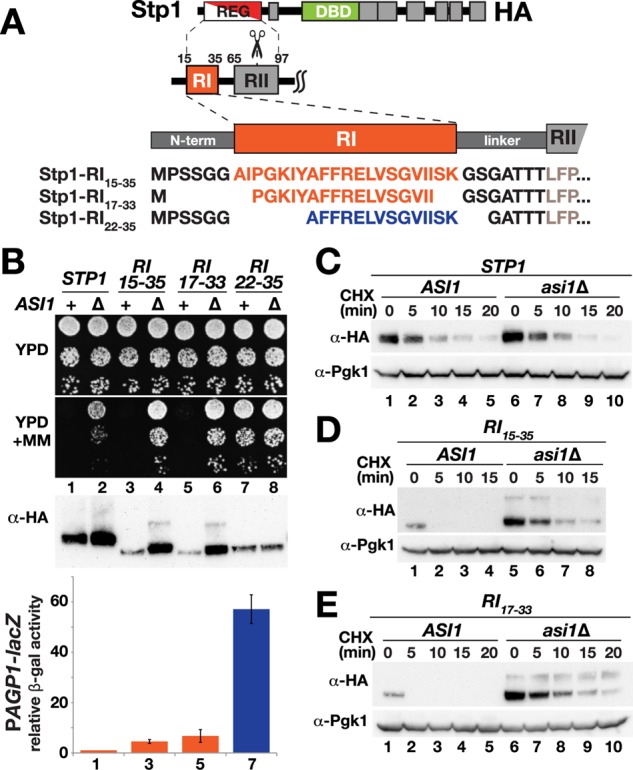
Amino acids 17–33 of RI suffice to mediate Stp1 latency. (A) Schematic representation and amino acid sequences of the modified Stp1 N-terminal domain constructs with altered forms of the RI motif. (B) Growth of MBY93 (ssy5Δ stp1Δ stp2Δ) and MBY102 (ssy5Δ stp1Δ stp2Δ asi1Δ) carrying pCA047 (STP1), pAB27 (RI15-35), pDO74 (RI17-33), or pDO73 (RI22-35) on YPD and YPD + MM (top). Immunoblot analysis of Stp1 levels in extracts from the strains (middle). β-Galactosidase activity in CAY265 (ssy5Δ PAGP1-lacZ) carrying pCA047 (STP1), pAB27 (RI15-35), pDO74 (RI17-33), or pDO73 (RI22-35) (bottom). Cells were grown in SD, and the β-galactosidase activities in three independent experiments were normalized to wild-type levels (STP1); error bars indicate standard deviation. (C) Immunoblot analysis of extracts from MBY93 (ssy5Δ stp1Δ stp2Δ) and MBY102 (ssy5Δ stp1Δ stp2Δ asi1Δ) carrying plasmid pCA047 (STP1) treated with CHX. Cells were pregrown in SD; at t = 0, the cultures received an aliquot of cycloheximide (final concentration, 100 µg/ml), and samples were taken at the indicated time points. (D) Immunoblot analysis of cell extracts from strains as in C carrying pAB27 (RI15-35) treated with CHX. (E) Immunoblot analysis of cell extracts from MBY89 (ssy5Δ) and MBY106 (ssy5Δ asi1Δ) carrying pDO74 (RI17-33) treated with CHX. Levels of Pgk1 serve as internal control for protein loading.
We examined the promoter-inducing activity of the minimal RI constructs in ssy5Δ cells by measuring activity of β-galactosidase driven by the Stp1/2-inducible AGP1 promoter. A gradual increase in promoter induction correlated with shorter RI sequences; RI15-35 induced the AGP1 promoter fivefold more than wild-type Stp1 (Figure 4B, compare bar 3 with bar 1); RI17-33, 1.5-fold more than RI15-35 (Figure 4B, compare bar 5 with bar 3); and RI22-35, 10-fold more than RI17-33 (Figure 4B, compare bar 7 with bar 5). We then analyzed protein levels by immunoblotting and found that the steady-state levels of RI15-35, RI17-33, and RI22-35 were lower than wild-type Stp1 levels in ssy5Δ stp1Δ stp2Δ cells (Figure 4B, middle, compare lanes 3, 5, and 7 with lane 1). In all cases, except for RI22-35, which lacks aa 16–21, the levels were significantly higher in asi1Δ cells than in ASI1 cells (Figure 4B, middle, compare lane 2 with lane 1, lane 4 with lane 3, lane 6 with lane 5, and lane 8 with lane 7).
A consistent observation that stems from our analysis is that mutations directly affecting RI express reduced steady-state levels of protein compared with wild-type Stp1. However, when expressed in asi1Δ cells, similar to Stp1-133 and Stp1-134, the RI15-35 and RI17-33 mutant proteins are present at higher levels than in ASI1 wild-type cells, correlating with more robust resistance to MM, which indicates enhanced promoter-inducing activity. Cells expressing RI22-35, a construct in which RI function is completely abolished, bypass Asi control and exhibit full promoter activity. The latter finding strongly suggests that the negative control exerted by the Asi proteins depends on the integrity of the RI motif.
Asi proteins affect the stability of Stp1
To test whether the increased steady-state protein levels of wild-type Stp1 and of the RI15-35 and RI17-33 proteins in asi1Δ cells were the consequence of altered rates of synthesis or degradation, we performed time-course experiments after addition of the translation inhibitor cycloheximide (CHX). As previously reported, wild-type Stp1 exhibited rapid turnover in ssy5Δ cells (Figure 4C, lanes 1–5), and no obvious difference in the rate of degradation was observed in ssy5Δ asi1Δ cells (Figure 4C, compare lanes 6–10 with lanes 1–5). For RI15-35 and RI17-33 proteins, however, a dramatic effect of asi1Δ on degradation was found; whereas in ASI1 wild-type cells, the levels of both proteins were below the detectable range at 5 min after addition of CHX (Figure 4, D, lane 2, and E, lane 2), the starting amounts of both proteins at time point 0 in asi1Δ cells were much higher (Figure 4, D, compare lane 5 with lane 1, and E, compare lane 6 with lane 1), and both RI15-35 and RI17-33 were readily detectable over the whole time course, that is, after 15 and 20 min, respectively (Figure 4, D, lanes 5–8, and E, lanes 6–10). Surprisingly, we observed a striking delay in degradation rates in cells lacking ASI1 (Figure 4, D, compare lanes 1 and 2 with lanes 7 and 8, and E, compare lanes 1 and 2 with lanes 9 and 10).
Next we asked whether the increased steady-state levels of RI15-35 and RI17-33 in asi1Δ cells—the consequence of enhanced stability in the absence of Asi1—could also be observed in cells lacking ASI2 or ASI3. Cells carrying deletions of ASI2 or ASI3 exhibit the same levels of Stp1/2-dependent promoter induction as asi1Δ mutants, and no additive effects are observed in cells carrying all possible double- and triple-null mutant combinations (Zargari et al., 2007). We analyzed the steady-state amount of proteins in asi2Δ and asi3Δ cells in comparison to those in asi1Δ and ASI wild-type cells. Indeed, RI15-35 and RI17-33 proteins accumulated to the same extent in cells lacking any of the ASI genes compared with the amount detected in ASI wild-type cells (Figure 5A, compare lanes 2–4 with lane 1 and lanes 6–8 with lane 5). In conclusion, deletion of ASI1, ASI2, or ASI3 results in an identical phenotype; consequently, Asi1, Asi2, and Asi3 appear to contribute equally to maintaining Stp1 latency and do so by affecting levels of Stp1 in a RI-dependent manner.
FIGURE 5:
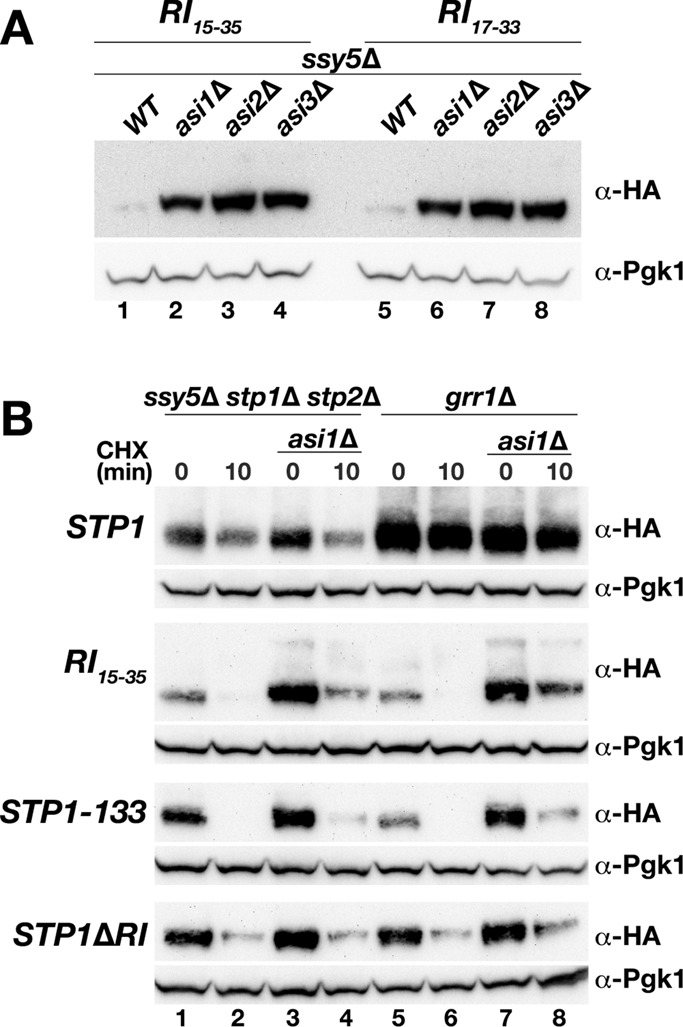
Turnover of nuclear-localized Stp1 is Asi dependent. (A) Immunoblot analysis of extracts from MBY89 (ssy5Δ; WT), MBY106 (ssy5Δ asi1Δ), MBY108 (ssy5Δ asi2Δ), and MBY110 (ssy5Δ asi3Δ) carrying pAB27 (RI15-35) or pDO74 (RI17-33) as indicated. (B) Immunoblot analysis of cell extracts from MBY93 (ssy5Δ stp1Δ stp2Δ), MBY102 (ssy5Δ stp1Δ stp2Δ asi1Δ), CAY86 (grr1Δ), and CAY109 (grr1Δ asi1Δ) carrying pCA047 (STP1), pAB27 (RI15-35), pCA120 (STP1-133), or pDO248 (STP1ΔR1) treated with CHX. Cells were pregrown in SD; samples were taken 0 and 10 min after cultures received an aliquot of CHX (final concentration, 100 µg/ml). Levels of Pgk1 serve as internal control for protein loading.
Asi-dependent Stp1 turnover occurs in the nucleus
Intrigued by the absence of a clear stabilization of wild-type Stp1 in asi1Δ cells and the fact that mutations diminishing the ability of RI to target hSOS to the plasma membrane correlate with clear Asi-dependent turnover, we proceeded to characterize the turnover of wild-type Stp1 and the RI mutants in more detail. Full-length Stp1 is strongly stabilized in cells lacking Grr1, the F-box component of the Skip-Cullin-F-box E3-ubiquitin ligase complex (SCFGrr1; Tumusiime et al., 2010; Omnus et al., 2011), whereas a truncated form of Stp1 lacking the N-terminal domain that constitutively localizes to the nucleus is only slightly stabilized in grr1Δ cells (Tumusiime et al., 2010). These results are consistent with the SCFGrr1 E3 ubiquitin ligase targeting cytoplasmic, but not nuclear, Stp1 for ubiquitylation. In full accord with these previous reports, we found that wild-type Stp1 is clearly stabilized in cells lacking Grr1. The steady-state levels were much higher, and almost no reduction was observed after 10 min of incubation with CHX (Figure 5B, top, compare lanes 5 and 6 with lanes 1 and 2).
Of interest, in contrast to wild-type Stp1, the degradation of RI15-35 was not dependent on Grr1; similar levels of protein were observed in grr1Δ and GRR1 cells (Figure 5B, third panel, compare lanes 5 and 6 with lanes 1 and 2). Strikingly, however, increased protein levels were clearly observed in cells lacking Asi1 (Figure 5B, third panel, compare lane 3 with lane 1 and lane 7 with lane 5). The same was true for Stp1-133, which also exhibited Grr1-independent and Asi1-dependent turnover (Figure 5B, fifth panel). Stp1ΔRI lacking aa 15–35 turned over with similar rates under all conditions tested, indicating that neither Grr1 nor Asi1 plays a role in its degradation (Figure 5B, seventh panel).
Together these results suggest that the different requirements for turnover reflect degradative processes located in different subcellular compartments. The Grr1-dependent and Asi-independent degradation of wild-type Stp1 occurs in the cytoplasm, whereas the Grr1-independent and Asi-dependent degradation of RI mutant Stp1 occurs within the nucleus. It is likely that the bulk of wild-type Stp1 with an intact RI motif is rapidly degraded in the cytoplasm and that the low levels that leak into the nucleus and are stabilized in asi1Δ cells are not efficiently visualized by immunoblotting.
Negative regulatory mechanisms are tuned; Stp1 overexpression constitutively activates gene expression
To analyze further whether turnover plays a role in Stp1 regulation, we sought to assess whether increased protein levels would lead to constitutive activity. To achieve higher steady-state levels of Stp1, we used two approaches. Initially, to effect a decrease in degradation, that is, to experimentally stabilize Stp1, we attempted to mutate potential degron sequences of Stp1. We examined the Stp1 amino acid sequence for putative conserved PEST motifs, that is, sequence motifs rich in proline, glutamic acid, serine, and threonine residues that are implicated in conferring protein instability (Rechsteiner and Rogers, 1996). We found five regions of sequence conservation that were rich in P, E, S, and T residues (Figure 6A). We performed a systematic deletion analysis targeting these conserved sequences. The stability of the mutant proteins was assessed in CHX chase assays, followed by immunoblotting; however, none of the conserved regions appeared to significantly affect Stp1 stability (Figure 6A, bottom). To elevate the protein pool by other means, we then expressed Stp1 from a 2μm plasmid, which led to increased steady-state levels of the protein (Figure 6B, compare lane 2 with lane 1). Of interest, we found that overexpression of Stp1 in ssy5Δ cells led to constitutive, processing-independent amino acid permease gene expression as judged by resistance to MM (Figure 6C, compare dilution 2 with dilution 1). This finding indicates that the mechanisms regulating RI-dependent Stp1 latency are saturable and thus sensitive to protein levels.
FIGURE 6:
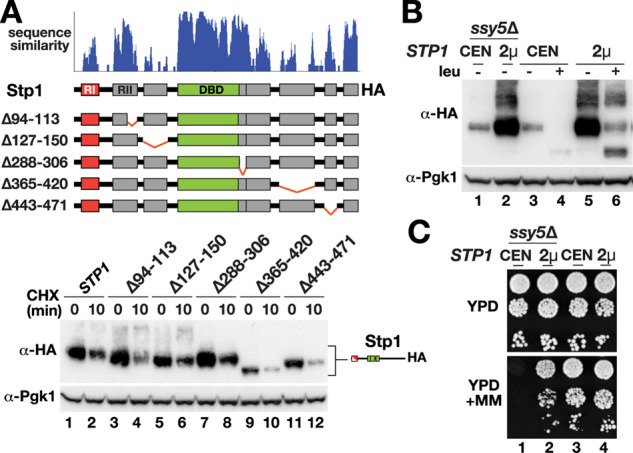
Overexpression of STP1 leads to constitutive promoter induction. (A) Amino acid sequences of Stp1 and Stp2 of S. cerevisiae and fungal Stp1 orthologues as in Figure 3 compared using Clustal X; the level of similarity of the aligned protein sequences is plotted. Schematic presentations of Stp1 and mutant proteins lacking discrete conserved domains. Immunoblot analysis of extracts from MBY93 (ssy5Δ stp1Δ stp2Δ) and carrying pCA047 (STP1), pDO243 (Δ94-113), pDO237 (Δ127-150), pDO244 (Δ288-306), pDO246 (Δ365-420), or pDO247 (Δ443-471) treated with CHX as in Figure 5. (B) Immunoblot analysis of extracts from MBY93 (ssy5Δ stp1Δ stp2Δ) and CAY123 (stp1Δ stp2Δ) carrying pCA072 (pRS316-FLAG-STP1-HA; CEN) or pCA078 (pRS202-FLAG-STP1-HA; 2μ). Cells were grown in SD medium and harvested 30 min after induction by leucine as indicated. Levels of Pgk1 serve as internal control for protein loading. (C) Growth of strains in B on YPD and YPD + MM.
RI functions as a modular promoter exclusion determinant and Asi-dependent degron
To ultimately test whether RI suffices to mediate cytoplasmic retention and constitutes a transferable Asi-dependent degron, we fused residues 16–50 of Stp1 containing RI to an artificial transcription factor composed of the bacterial DNA-binding protein lexA (lexA) and the strong viral VP16 transcription activation domain (AD). We used lexA operator–driven LYS2 and LEU2 genes as reporters for DNA binding of lexA-AD (Figure 7A). We then assessed cell growth in the absence of leucine and lysine as a functional readout for nuclear accumulation. When expressed in wild-type cells, the artificial transcription factor (lexA-AD) induced the reporter genes to facilitate growth in the absence of leucine and lysine (Figure 7B, dilution 1). In contrast, the fusion of lexA to Stp116-50 abolished promoter induction; no growth was observed (Figure 7B, compare dilution 2 with dilution 1). However, when expressed in cells lacking Asi1, robust growth was detected (Figure 7B, dilution 3). Of note, as judged by immunoblotting, the steady-state expression level of Stp116-50-lexA-AD was elevated in cells lacking Asi1 (Figure 7B, bottom, compare lane 3 with lane 2). Next we performed a CHX chase over a time course of 60 min to assess protein stability in wild-type and asi1Δ cells (Figure 7C). Strikingly, Stp116-50-lexA-AD was completely stable in asi1Δ cells (Figure 7C, lanes 6–10), whereas it efficiently turned over in wild-type cells (Figure 7C, lanes 1–5). Together these results show that the RI-containing domain of Stp1 comprising amino acids 16–50 contains the necessary information for Asi-dependent promoter exclusion, presumably by mediating efficient degradation.
FIGURE 7:
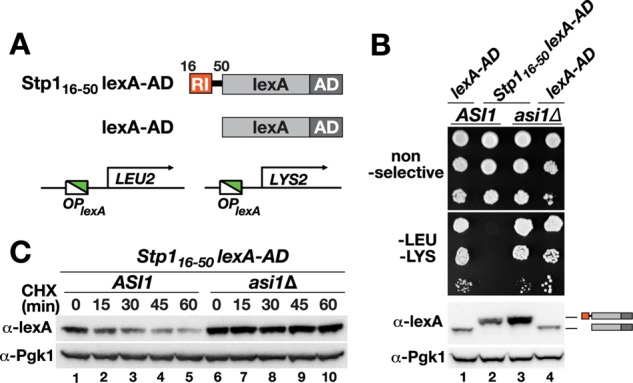
RI acts in a modular manner as a promoter exclusion determinant and an Asi-dependent degron. (A) Schematic representation of the artificial transcription factor composed of amino acids 16–50 of Stp1 fused to the bacterial DNA-binding protein lexA fused to the viral VP16 transcription AD. OPlexA-LEU2 and OPlexA-LYS2 reporter genes were used to assess binding to lexA operators. (B) Growth analysis of CAY235 (ASI1) and SHY016 (asi1Δ) carrying pDO211 (lexA-AD) or pDO260 (STP116-50-lexA-AD) on nonselective and selective media lacking leucine and lysine. Immunoblot of extracts prepared from strains. The immunoreactive forms of the lexA proteins present in cell extracts are schematically represented at their corresponding positions of migration. (C) Immunoblot analysis of extracts from strains CAY235 (ASI1) and SHY016 (asi1Δ) carrying pDO260 (STP116-50-lexA-AD) treated with CHX. Cells were pregrown in SD; at t = 0, the cultures received an aliquot of CHX (final concentration, 100 μg/ml), and samples were taken at the indicated time points. Levels of Pgk1 serve as internal control for protein loading.
DISCUSSION
This study addressed the mechanisms underlying the latent properties of Stp1 in the absence of amino acid induction. We defined the negatively acting RI motif in the Stp1 N-terminal regulatory domain and report that this motif, composed of amino acid residues 17–33, suffices to restrict promoter induction in an Asi protein–dependent manner. Specifically, the data indicate that RI harbors two distinct functions required for latency: it acts as a cytoplasmic retention determinant and as an Asi-dependent degron. The results demonstrate for the first time that the inner nuclear membrane Asi proteins function in a degradation pathway in the nucleus.
The RI-mediated mechanisms defined in this work and schematically presented in Figure 8 fully account for the latent properties of Stp1. Accordingly, RI retains the bulk of Stp1 in the cytoplasm, presumably by interacting with a cytoplasmically exposed protein determinant (Figure 8A; anchor). However, due to the noncovalent nature of protein–protein interactions, cytoplasmic anchoring is not absolute. Hence small amounts of full-length Stp1 possessing the RI motif constitutively escape anchoring and inappropriately enter the nucleus. In the nucleus, the integral inner nuclear membrane Asi proteins function in concert to effectively target the unprocessed Stp1 for degradation and thereby prevent Stp1 from binding SPS sensor–regulated promoters. Consequently, in cells lacking any of the Asi proteins, the small pool of Stp1 that enters the nucleus is not effectively degraded, leading to induced gene expression (Figure 8B).
FIGURE 8:
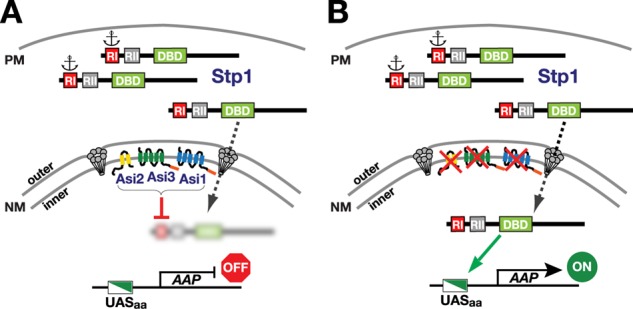
Model. Two functional activities intrinsic to the RI motif ensure Stp1 latency. (A) In wild-type cells growing in the absence of inducing amino acids, the RI motif functions as a cytoplasmic retention determinant due to its interaction with an unidentified cytoplasmic component (anchor) that associates with the plasma membrane. RI binding is not absolute; small amounts of Stp1 target the nucleus where Stp1 is degraded in an RI motif- and Asi-dependent manner. (B) In cells lacking any of the Asi proteins, the low levels of Stp1 that escape cytoplasmic anchoring can access and bind promoters of SPS sensor–regulated genes, leading to induction of amino acid permease (AAP) gene expression.
This model is supported by several lines of experimental evidence. First, the N-terminal domain of Stp1 containing RI functions efficiently in a modular manner to retain histone Htb2 in the cytoplasm (Figure 1). Second, amino acid residues 16–50 suffice to localize hSOS to the cytosolic face of the plasma membrane, and mutations specifically affecting RI abolish this activity (Figure 2). These findings suggest that RI serves as an interaction motif for a cytoplasmic component associated with the plasma membrane. However, we note that although the hSOS experiments demonstrate that RI can direct proteins to the PM, the diffuse cytoplasmic fluorescence of GFP fusion proteins carrying intact RI sequences (Figure 1) suggests that the interactions are not strictly restricted to the PM. Third, mutations affecting RI (Stp1-133, Stp1-134) or its surrounding sequences (RI15-35, RI17-33) that reduce cytoplasmic retention consequently lead to constitutive transcription and a concomitant decrease in the steady-state levels of Stp1 (Figures 3 and 4). Fourth, the increased nuclear pool of Stp1, resulting from reduced cytoplasmic retention, exhibits clear Asi-dependent stability. Taken together, the additive effects on promoter induction observed for mutations in RI- and asi-null mutations indicate that RI functions both as a cytoplasmic retention determinant and an Asi-dependent degron in the nucleus.
The role of RI within the nuclear compartment was unambiguously tested by fusing amino acid residues 16–50 of Stp1 to an artificial transcription factor (Figure 7). This experiment showed that this RI-containing sequence suffices to mediate promoter exclusion in an Asi-dependent manner. The enhanced stability of the RI-lexA-AD construct in asi1-null mutant cells revealed that the degron activity observed in the context of Stp1 is indeed intrinsic to the RI motif. These results demonstrate that the Asi proteins function in a posttranslational manner on RI-containing proteins by targeting them for degradation and thereby diminishing their nuclear abundance. The data fully account for the known negative regulatory activity of the Asi proteins (Boban et al., 2006; Boban and Ljungdahl, 2007; Zargari et al., 2007).
The ability of RI to restrict nuclear targeting is consistent with it mediating binding to a cytoplasmic and potentially PM associated component. The observation that mere overexpression of Stp1 induces SPS sensor–regulated genes is consistent with this model (Figure 6) and clearly suggests that the components responsible for conferring cytoplasmic retention are present in limiting and saturable amounts. Thus we consider it unlikely that RI directly interacts with an abundant nonprotein membrane component. Despite extensive efforts, including multiple genetic approaches aimed specifically at isolating genes required for Stp1 latency or screens aimed at identifying interacting proteins, the nature of the cytoplasmic components mediating latency or that physically interact with RI remains to be elucidated. The fact that these components have escaped identification may be explained if they catalyze essential functions or that cytoplasmic retention is the consequence of a redundant network of interacting partners. This latter possibility is also consistent with a promiscuous, more general retention mechanism involving multiple factors with low-affinity binding to RI.
The latent form of Stp1 is subject to degradation in two separate intracellular compartments. In the cytoplasm, full-length Stp1 is turned over in a SCFGrr1-dependent manner, and in the nucleus, Stp1 is turned over in an Asi-dependent manner (Figure 5). Of importance, the clear stabilization of the cytoplasmic pool of Stp1, as in grr1Δ cells, does not lead to promoter induction. Consequently, cytoplasmic retention and Asi-dependent turnover suffice to maintain nuclear levels of Stp1 below the critical threshold required to induce SPS sensor–regulated promoters. By contrast, in cells lacking Asi components, the decreased rate of nuclear localized Stp1 turnover leads to robust gene expression (Figures 3–5).
Significantly, our findings provide the first evidence that the Asi proteins participate within a degradative pathway in the nucleus. Consistently, Asi1 and Asi3 possess nucleoplasmically oriented RING motifs at their extreme C-termini. RING motifs are found in several E3-ubiquitin ligases; however, biochemical approaches failed to demonstrate auto- or trans-ubiquitylation activity (Zargari et al., 2007). The failure to observe E3-ubiquitn ligase activity may indeed be the consequence of the Asi proteins functioning together within a multiprotein complex; stable interactions with Stp1 and Stp2 may depend on contacts between more than one of the Asi proteins. Consistent with this notion, we found that combined presence of Asi1, Asi2, and Asi3 is required to efficiently recognize the RI degron (Figure 5).
Our data provide important and novel insights into the diverse function of INM proteins. The participation of INM proteins in regulating gene expression by the specific recruitment of chromosomal regions to discrete localizations at the nuclear periphery has been described (reviewed in Akhtar and Gasser, 2007; Heessen and Fornerod, 2007). However, only one set of studies reports the involvement of INM protein–dependent degradation to control transcription factor activity. Although Doa10 is a well-characterized integral membrane component of the ER that functions as an E3-ubiquitin ligase with an established role in the degradation of misfolded proteins, Doa10 has also been found to localize to the INM. In the INM, Doa10 controls the stability of the transcription factor Matα2 with a degron designated Deg1 (Chen et al., 1993; Deng and Hochstrasser, 2006).
The Asi proteins appear to control specifically the stability of the SPS sensor–regulated transcription factors that possess the RI degron, namely Stp1 and Stp2. No transcriptional affects other than the inappropriate activation of amino acid permease genes have been reported in strains lacking Asi function. It will be the focus of future studies to test whether the Asi proteins operate within a complex as a E3 ubiquitin ligase to directly recognize the RI degron sequence. In addition, sensitive genome-wide approaches addressing the question of whether additional transcription factors may be substrates of Asi-dependent regulation are needed. Our ongoing efforts will undoubtedly provide greater insight into the novel nuclear-localized degradative mechanisms regulating transcription. We expect that a detailed understanding of the cellular response to extracellular amino acids will provide the means to decipher similar regulatory events in signaling pathways in other organisms.
MATERIALS AND METHODS
Yeast strains and plasmids
S. cerevisiae strains and plasmids used in this work are listed in Tables 1 and 2, respectively. The yeast strains are isogenic descendants of the S288C-derived strain AA255/PLY115 (Antebi and Fink, 1992), with the exception of the BY4741-derived strain CAY1259 and EGY48-derived two-hybrid strains CAY235 and SHY016. The sequences of mutagenic oligonucleotides and PCR primers for homologous recombination are available upon request.
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Reference/source |
| CAY86 | MATa ura3-52 grr1Δ50::hphMX4 | Andréasson and Ljungdahl (2002) |
| CAY109 | MATa ura3-52 grr1Δ50::hphMX4 asi1Δ8::kanMX | Andréasson and Ljungdahl (2004) |
| CAY123 | MATa ura3-52 stp1Δ51::Agleu2 stp2Δ50::hphMX4 | Andréasson and Ljungdahl (2002) |
| CAY187 | MATa ura3-52 cdc25-2 | Andréasson and Ljungdahl (2002) |
| CAY235 | EGY48 derivative: MATα trp1 his3 leu2::6xOPlexA-LEU2 lys2::URA3-8xOPlexA-GAL1 GAL2 | Omnus et al. (2011) |
| CAY265 | MATa ura3-52 ssy5Δ2::hisG gap1Δ::PAGP1-LacZ | Andréasson et al. (2006) |
| CAY1259 | BY4741 derivative: MATa his3Δ1 leu2Δ0 ura3Δ0 HTB2::ymCherry-KanMX | Andréasson laboratory |
| MBY89 | MATα ura3-52 ssy5Δ77::natMX4 | Zargari et al. (2007) |
| MBY93 | MATa ura3-52 ssy5Δ1:: hisG stp1Δ51::Agleu2 stp2Δ50::hphMX4 | Boban and Ljungdahl (2007) |
| MBY102 | MATα ura3-52 ssy5Δ1:: hisG stp1Δ51::Agleu2 stp2Δ50::hphMX4 asi1Δ8::kanMX | Boban and Ljungdahl (2007) |
| MBY106 | MATα ura3-52 ssy5Δ77::natMX4 asi1Δ80::hphMX | Zargari et al. (2007) |
| MBY108 | MATα ura3-52 ssy5Δ77::natMX4 asi2Δ9::hisG | Zargari et al. (2007) |
| MBY110 | MATα ura3-52 ssy5Δ77::natMX4 asi3Δ5::kanMX | Zargari et al. (2007) |
| SHY016 | EGY48 derivative: MATα his3 trp1 ura3 leu2::6×OPlexA-LEU2 lys2::8×OPlexA-LYS2 asi1Δ8::loxP-kanMX-loxP GAL2 | Andréasson et al. (2006) |
TABLE 2:
Plasmids used in this study.
| Plasmid | Description | Reference/source |
|---|---|---|
| pAB1 | pRS313 carrying HIS3, MET15, and LEU2 | Omnus et al. (2011) |
| pAB27 | pRS316 (URA3) expressing Stp1-RI15-35 | This study |
| pCA047 | pRS316 (URA3) with STP1-3HA | Andréasson and Ljungdahl (2002) |
| pCA072 | pRS316 (URA3) with FLAG-STP1-HA | Andréasson and Ljungdahl (2002) |
| pCA078 | pRS202 (URA3) with FLAG-STP1-HA | Andréasson and Ljungdahl (2002) |
| pCA094 | URA3 derivative of pSOS | Andréasson and Ljungdahl (2002) |
| pCA104 | pCA094 (URA3) expressing hSOS-Stp12-158 | Andréasson and Ljungdahl (2002) |
| pCA120 | pRS316 (URA3) with STP1-133 | Andréasson and Ljungdahl (2002) |
| pDO39 | pRS316 (URA3) expressing Stp11-125-Htb2-GFP-HA | This study |
| pDO73 | pRS316 (URA3) expressing Stp1-RI22-35 | This study |
| pDO74 | pRS316 (URA3) expressing Stp1-RI17-33 | This study |
| pDO141 | pRS316 (URA3) with STP1-134 | This study |
| pDO142 | pRS316 (URA3) with STP1-135 | This study |
| pDO196 | pCA104 (URA3) expressing hSOS-Stp12-158(Δ16-21) | This study |
| pDO203 | pCA104 (URA3) expressing hSOS-Stp12-64 | This study |
| pDO205 | pCA104 (URA3) expressing hSOS-Stp116-33 | This study |
| pDO211 | pEG202 derivative (HIS3) KanR carrying PADH1-lexA-AD | This study |
| pDO221 | pCA104 (URA3) expressing hSOS-Stp116-64 | This study |
| pDO230 | pCA104 (URA3) expressing hSOS-Stp116-37 | This study |
| pDO231 | pCA104 (URA3) expressing hSOS-Stp116-50 | This study |
| pDO237 | pRS316 (URA3) with STP1Δ127-150-3HA | This study |
| pDO243 | pRS316 (URA3) with STP1Δ94-113-3HA | This study |
| pDO244 | pRS316 (URA3) with STP1Δ288-306-3HA | This study |
| pDO246 | pRS316 (URA3) with STP1Δ365-420-3HA | This study |
| pDO247 | pRS316 (URA3) with STP1Δ443-471-3HA | This study |
| pDO248 | pRS316 (URA3) with STP1ΔR1-3HA | This study |
| pDO260 | pEG202 derivative (HIS3) KanR carrying PADH1-STP116-50-lexA-AD | This study |
Media
Standard media, including YPD medium, ammonia-based synthetic minimal dextrose (SD) medium supplemented as required to enable growth of auxotrophic strains, and ammonia-based synthetic complete dextrose (SC), were prepared as described (Andréasson and Ljungdahl, 2002). Sensitivity to MM (100 μg/ml; E.I. du Pont de Nemours, Cernay, France) was monitored on YPD medium as described (Jørgensen et al., 1998; Andréasson and Ljungdahl, 2002). Briefly, 10-fold dilutions of cultures were spotted on YPD and YPD plus MM media. Plates were incubated at 30°C for 2–3 d and photographed.
Immunoblot analysis
Whole-cell extracts were prepared under denaturing conditions using NaOH and trichloroacetic acid as described previously (Silve et al., 1991). Cells were grown in SD medium, and when indicated (+), l-leucine (leu) was added at a concentration of 1.3 mM for 30 min to induce SPS-sensor signaling before extract preparation. Similarly, CHX (Sigma-Aldrich, St. Louis, MO) was added to the culture (CHX +) at a concentration of 100 μg/ml for the indicated time before extract preparation. Primary antibodies were diluted as follows: 3F10 anti–HA-horseradish peroxidase (Roche Applied Science, Basel, Switzerland), 1:2000; anti-Pgk1 (Molecular Probes), 1:1000; and anti-lexA (Abcam), 1:2000. Immunoreactive bands were visualized by chemiluminescence detection (SuperSignal West Dura Extended-Duration Substrate; Pierce, Rockford, IL) and quantified using the LAS1000 system (Fuji Photo Film, Tokyo, Japan).
β-Galactosidase activity assay
β-Galactosidase activity was determined with N-lauroyl-sarcosine–permeabilized cells (Kippert, 1995). Cells grown in SD were harvested by centrifugation, washed once, and resuspended in 200 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7), and the OD600 was measured. A 40-μl aliquot of cell suspension was mixed with 760 μl of 0.2% (wt/vol) Na N-lauroyl-sarcosine Z buffer and incubated at 30°C for 15 min. Then preheated 160 μl of Z buffer containing 4 mg/ml 2-nitrophenyl β-d-galactopyranoside was added. After a 10-min incubation at 30°C, the reaction was quenched by the addition of 400 μl of 1 M Na2CO3. Samples were centrifuged at 12,000 × g for 5 min, and the absorbance of the supernatant was measured at 420 nm.
Acknowledgments
We thank the members of the Andréasson, Ott, and Ljungdahl laboratories for constructive comments throughout the course of this work. We acknowledge Anna Schick and Albert Ferrer Herranz for constructing plasmids, Matthias Spiess for assistance with microscopy, and Claes Andréasson for providing strain CAY1259. This research was supported by funding from the Swedish Research Council (P.O.L.).
Abbreviations used:
- CHX
cycloheximide
- HA
hemagglutinin
- INM
inner nuclear membrane
- MM
2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino]-sulfonyl}-benzoic acid
- SCF
Skip-Cullin-F-box E3-ubiquitin ligase complex
- SD
synthetic minimal dextrose
- SPS
Ssy1-Ptr3-Ssy5
- YPD
yeast extract/peptone/dextrose.
Footnotes
*Present address: MRC Laboratory for Molecular Cell Biology, University College London, London WC1E 6BT, United Kingdom.
The authors declare no conflict of interest.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1140) on September 24, 2014.
REFERENCES
- Abdel-Sater F, Jean C, Merhi A, Vissers S, André B. Amino-acid signalling in yeast: activation of the Ssy5 protease is associated with its phosphorylation-induced ubiquitylation. J Biol Chem. 2011;286:12006–12015. doi: 10.1074/jbc.M110.200592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Andréasson C, Heessen S, Ljungdahl PO. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 2006;20:1563–1568. doi: 10.1101/gad.374206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Ljungdahl PO. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 2002;16:3158–3172. doi: 10.1101/gad.239202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Ljungdahl PO. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol Cell Biol. 2004;24:7503–7513. doi: 10.1128/MCB.24.17.7503-7513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Fink GR. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Biol Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- Boban M, Ljungdahl PO. Dal81 enhances Stp1- and Stp2-dependent transcription necessitating negative modulation by inner nuclear membrane protein Asi1 in Saccharomyces cerevisiae. Genetics. 2007;176:2087–2097. doi: 10.1534/genetics.107.075077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boban M, Zargari A, Andréasson C, Heessen S, Thyberg J, Ljungdahl PO. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J Cell Biol. 2006;173:695–707. doi: 10.1083/jcb.200601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biol Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Delaunay A, Isnard AD, Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Hammar M, Andréasson C, Moliner A, Ljungdahl PO. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics. 2001;158:973–988. doi: 10.1093/genetics/158.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- Gulshan K, Thommandru B, Moye-Rowley WS. Proteolytic degradation of the Yap1 transcription factor is regulated by subcellular localization and the E3 ubiquitin ligase Not4. J Biol Chem. 2012;287:26796–26805. doi: 10.1074/jbc.M112.384719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 2007;8:914–919. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol. 2001;13:344–348. doi: 10.1016/s0955-0674(00)00218-0. [DOI] [PubMed] [Google Scholar]
- Jørgensen MU, Bruun MB, Didion T, Kielland-Brandt MC. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast. 1998;14:103–114. doi: 10.1002/(SICI)1097-0061(19980130)14:2<103::AID-YEA203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Kippert F. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol Lett. 1995;128:201–206. doi: 10.1111/j.1574-6968.1995.tb07523.x. [DOI] [PubMed] [Google Scholar]
- Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics. 2012;190:885–929. doi: 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek S, Chen Y, Huxford T, Ghosh G. IkappaBbeta, but not IkappaBalpha, functions as a classical cytoplasmic inhibitor of NF-kappaB dimers by masking both NF-kappaB nuclear localization sequences in resting cells. J Biol Chem. 2001;276:45225–45235. doi: 10.1074/jbc.M105865200. [DOI] [PubMed] [Google Scholar]
- Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omnus DJ, Ljungdahl PO. Rts1-protein phosphatase 2A antagonizes Ptr3-mediated activation of the signaling protease Ssy5 by casein kinase I. Mol Biol Cell. 2013;24:1480–1492. doi: 10.1091/mbc.E13-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omnus DJ, Pfirrmann T, Andréasson C, Ljungdahl PO. A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol Biol Cell. 2011;22:2754–2765. doi: 10.1091/mbc.E11-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Chan NC, Helfenbaum L, Tan K, Tansey WP, Gething MJ. SCFCdc4-mediated degradation of the Hac1p transcription factor regulates the unfolded protein response in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:426–440. doi: 10.1091/mbc.E06-04-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Silve S, Volland C, Garnier C, Jund R, Chevallier MR, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumusiime S, Zhang C, Overstreet MS, Liu Z. Differential regulation of transcription factors Stp1 and Stp2 in the Ssy1-Ptr3-Ssy5 amino acid sensing pathway. J Biol Chem. 2010;286:4620–4631. doi: 10.1074/jbc.M110.195313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zargari A, Boban M, Heessen S, Andréasson C, Thyberg J, Ljungdahl PO. Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J Biol Chem. 2007;282:594–605. doi: 10.1074/jbc.M609201200. [DOI] [PubMed] [Google Scholar]


