Abstract
The early detection and biological investigation of esophageal cancer would benefit from the development of advanced imaging techniques to screen for the molecular changes that precede and accompany the onset of cancer. Surface-enhanced Raman scattering (SERS) nanoparticles (NPs) have the potential to improve cancer detection and the investigation of cancer progression through the sensitive and multiplexed phenotyping of cell-surface biomarkers. Here, a miniature endoscope featuring rotational scanning and axial pull back has been developed for 2D spectral imaging of SERS NPs topically applied on the lumenal surface of the rat esophagus. Raman signals from low-pM concentrations of SERS NP mixtures are demultiplexed in real time to accurately calculate the concentration and ratio of the NPs. Ex vivo and in vivo experiments demonstrate the feasibility of topical application and imaging of multiplexed SERS NPs along the entire length of the rat esophagus.
OCIS codes: (170.5660) Raman spectroscopy, (170.2150) Endoscopic imaging, (110.2350) Fiber optics imaging, (170.0170) Medical optics and biotechnology, (170.2680) Gastrointestinal, (160.4236) Nanomaterials
1. Introduction
Esophageal cancer is responsible for approximately one-sixth of all cancer-related mortalities worldwide [1, 2]. Although advanced endoscopy has improved the assessment accuracy and treatment outcome of esophageal cancer [3], a large portion of esophageal cancer patients still exhibit advanced metastatic disease at the time of diagnosis [4] resulting in poor outcomes with current treatment modalities [5, 6]. There is a need for improved imaging diagnostics to screen for the molecular changes that precede and accompany the onset of esophageal cancer, as well as to investigate the molecular mechanisms of tumor progression in the esophagus. In recent years, several imaging technologies has shown potential to improve cancer detection by providing “optical biopsy” such as confocal laser microendoscopy and fluorescence imaging with molecular contrast agents [7–10]. Since esophageal cancer originates from the epithelial cells at the lumenal surface of the esophagus, the imaging of cell-surface and tissue biomarkers at these surfaces could be used to monitor disease progression. However, due to the variability in protein biomarker expression patterns both between patients [11] and within a single patient over time, accurate disease diagnosis, patient stratification, and biological investigation would benefit from the ability to simultaneously image a large number of molecular targets.
In recent decades, a variety of exogenous contrast agents have been developed to enable the multiplexed molecular imaging of fresh ex vivo and in vivo tissues under time-constrained conditions that are relevant for point-of-care clinical applications and preclinical investigations [12–14]. Among these contrast agents, surface-enhanced Raman-scattering (SERS) nanoparticles (NPs), hereafter referred to as “SERS NPs” or “NPs”, have attracted interest due to their brightness, low toxicity and potential for sensitive and multiplexed biomarker detection [15]. The SERS NPs utilized in this study are available in multiple “flavors,” each of which emits a characteristic Raman fingerprint spectrum that allows for the identification and quantification of large multiplexed mixtures of different NP flavors when illuminated at a single wavelength [16–18]. Individual flavors of SERS NPs can be conjugated to unique antibodies or small-molecule ligands to target a diverse panel of cell-surface or tissue biomarkers. After orally introducing these biomarker-targeted NPs into the esophagus (e.g., by oral gavage or by having the subject ingest a cocktail of NPs), physicians and tumor biologists would be able to obtain a rich set of molecular information to detect cancer cells more accurately during endoscopic procedures and to study the molecular changes associated with tumor progression. Recently, a few studies have reported the multiplexed detection of large panels of nontargeted SERS NPs in live animals and human tissues [18–24]. Our lab and a few other groups have also developed biomarker-targeted SERS NPs and have demonstrated their specific binding abilities with cancer cells and tissues [25–30].
Most recently, we demonstrated a ratiometric method that quantifies the specific binding of a panel of biomarker-targeted NPs on fresh tissues and eliminates the ambiguities that often arise due to nonspecific sources of contrast [30]. Ratiometric quantification is important for molecular imaging due to the fact that contrast agents show varying levels of nonspecific accumulation, which is often misleading. For example, normal tissues can generate stronger signals than tumors due to higher nonspecific retention of contrast agents, i.e. inverse contrast [31, 32]. On the other hand, the opposite effect – enhanced penetration and retention (EPR) in tumors versus normal tissues – is also often observed [33–35]. Regardless of the functional mechanisms for the accumulation of contrast agents in tissues, there is increasing agreement that utilizing a dual-reporter ratiometric detection strategy, in which one negative reporter serves as a control for a simultaneously delivered positive probe, is valuable to accurately identify and quantify molecularly specific binding [23, 31, 32, 34]. SERS NPs are particularly suited for this ratiometric strategy, not only due to their excellent multiplexing capability but also due to the fact that they can be excited at a single illumination wavelength, ensuring that all NP flavors in a single measurement are interrogated identically in terms of illumination intensity, spot size and effective excitation depth.
In light of the diagnostic potential of SERS NPs, there has been interest in the development of instruments for the spectroscopic detection and imaging of SERS NPs in tissues [19, 21, 23]. For example, a hand-held spectroscopic pen device has been developed for intraoperative detection of SERS signals from malignant tumors in mice [21], and a small-animal Raman imaging system has been developed to detect multiplexed SERS signals in both superficial and deep tissue locations [20]. Recently, a fiber-optic Raman spectroscopy device was deployed through a clinical endoscope for use in human patients in vivo, and the detection and quantification of nontargeted SERS NPs on excised human colon tissues was demonstrated [19]. However, a spectral endoscope for comprehensive in vivo imaging of the esophagus has not been reported yet. Although recent studies have shown that SERS NPs applied topically in the rectum show no toxicity and result in negligible systemic uptake [36, 37], the systemic toxicity of these NPs when orally administered is unknown and SERS NPs are not currently approved for administration in the human esophagus. Therefore, we are developing a device to image the rat esophagus. In the future, this device may be used to investigate the molecular changes associated with tumor progression in rat models such as a chemically induced model of esophageal cancer [38, 39] as well as an orthotopic xenograft model [40].
In this article we describe the design of a rotational spectral-imaging endoscope for comprehensive imaging of SERS NPs topically applied in the rat esophagus with sub-millimeter resolution as well as validation studies demonstrating the sensitivity and accuracy of the device for the quantitative ratiometric imaging of multiplexed SERS NPs. While our previous studies have utilized three or more multiplexed NP flavors [30, 41], this study demonstrates the initial feasibility of our technologies with the simplest case of two NP flavors topically applied on cylindrical tissue phantoms and on fresh rat esophagus tissues (ex vivo and in vivo). Future studies using this device will include in vivo multiplexed molecular imaging of rat esophageal cancer models with targeted SERS NPs, such as the NPs recently described by us [30].
2. Methods
2.1 SERS nanoparticles
The SERS NPs used in this study were purchased from Cabot Security Materials Inc [42]. These NPs consist of a unique layer of Raman reporters adsorbed on a 60-nm gold core, which are encapsulated in silica, resulting in an overall diameter of ~120 nm. The silica encapsulation protects the Raman spectra in these NPs from environmental conditions and allows each flavor of NP to be accurately identified based on its stable and unique Raman fingerprint spectrum regardless of the buffer or medium in which it is suspended. Two “flavors” of NPs were used in this study, identified as S420 and S440. Additional details may be found in the literature [25].
2.2 Customized spectral-imaging endoscope
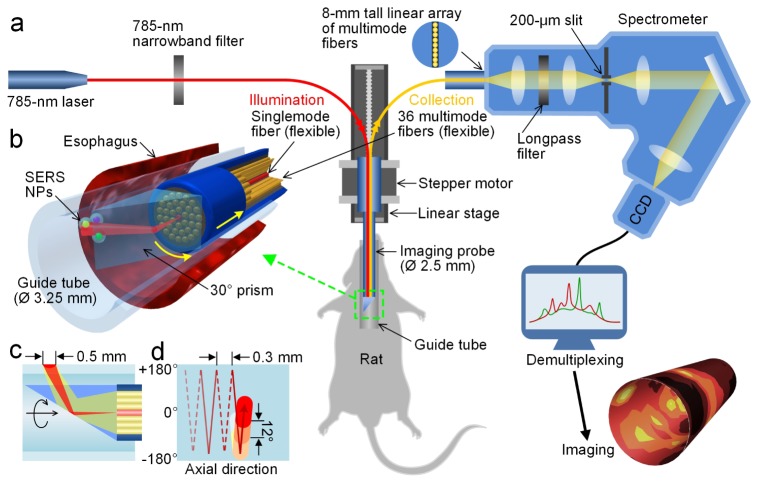
The endoscope developed in this study is comprised of a glass guide tube that is in contact with the rat esophagus and a fiber-optic probe inserted into the guide tube that is rotated and translated axially to image the lumen of the esophagus comprehensively (Figs. 1(a) and 1(b)). A miniature spectral-imaging probe with an outer diameter of 2.5 mm was developed to quantify and visualize SERS NPs applied on esophagus tissues. A singlemode fiber at the center of the fiber-bundle-based probe is used to illuminate the sample using a low-power 785-nm diode laser (~10 mW at the tissue) while 36 multimode fibers (200-μm core) surrounding the singlemode fiber are used to collect optical signals including Raman-scattered light (Fig. 1(b)). The fiber bundles are enclosed in a 0.4 mm-thick steel tube with an outer diameter of 2.5 mm (Fig. 1(b)). The light exiting the singlemode fiber diverges (1/e2 NA ~0.12) and illuminates a spot of roughly 0.5 mm in diameter (FWHM) at the tissue surface that is located ~3 mm from the distal end of the fiber bundle. A customized spectrometer (Bayspec Inc.) is used to disperse the light collected from the 36 multimode collection fibers onto a cooled deep-depletion spectroscopic CCD (Andor Newton, DU920P-BR-DD) with a spectral resolution of ~2 nm (~30 cm−1). Additional details about the detection system have been described previously [41].
Fig. 1.
Schematic of a customized spectral-imaging endoscope and SERS-endoscopy system. (a) Experimental platform to detect multiplexed NPs within the esophagus of a rat. (b) Zoom-in rendering of the prism and fiber-bundle imaging probe within a glass guide tube and rat esophagus (Media 1 (3.1MB, AVI) ). (c) Cross-sectional illustration of the illumination and collection beam paths. (d) Scanning trajectory for comprehensive imaging of the rat esophagus. To illustrate our scanning strategy, the guide tube is unfolded and the neutral position (0°) and turning points ( ± 180°) are marked. The probe rotates between ± 180° as it is slowly pulled out of the esophagus. The colored circles represent the spot size of the laser illumination and the spacing between consecutive spectral acquisitions (image pixels).
To accomplish rotational scanning, a customized 30° glass prism (Tower Optical Inc.) was adhered to the probe tip to deflect the illumination beam to the esophagus tissue as well as to collect Raman signal from the illuminated tissue area (Fig. 1). By angling the beam paths at 30° with respect to the direction normal to the tissue surface, the prism design reduces specular reflections and spectral background. The 2.5-mm diameter fiber-bundle probe is inserted within a 3.25-mm diameter borosilicate glass guide tube (2.6-mm inner diameter, Rayotek Scientific Inc). The glass guide tube is first inserted into the esophagus such that the lumen of the rat esophagus is gently stretched and flattened around the guide tube, thereby ensuring that the optical working distance is fixed and that the laser illuminates the tissue with a uniform spot size as it scans the lumenal surface of the tissue.
2.3 Rotational pull-back scanning of the spectral endoscope
For rotational scanning, a hollow-shaft stepper motor (11-mm inner diameter, Nanotec ST5918) positioned outside of the rat is used to hold and rotate the rigid imaging probe (Fig. 1(a)). The stepper motor is translated on a linear stage (Zaber Technologies, T-LSM200A) in the axial direction to pull the inner probe back at a programmed rate (1-1000 μm/s). The proximal end of the fiber bundle is flexible to allow for stable attachment of the collection fibers (linear array) into a spectrometer while the distal end is rotated (Figs. 1(c) and 1(d) depict the scanning trajectory of the endoscope). Images are reconstructed using custom LabVIEW software, with the stepper motor and translation stage controlled using a National Instruments data acquisition system. The spatial resolution of the endoscope is determined by the 0.5-mm diameter (FWHM) illumination spot on the esophagus tissue wrapped around the outer surface of the glass guide tube (Fig. 1(c)). In order to fully sample the esophagus, the imaging probe acquires 30 spectral acquisitions per rotation (12° per acquisition), and is translated 0.3 mm in the axial direction during each rotation with an adjustable frame rate of up to 10 spectra/s (up to 0.6 cm/min for esophagus imaging). To protect the fiber bundles from twisting, the imaging probe is alternately rotated in the positive and negative directions ( ± 180 deg) from a neutral position (0° in Fig. 1(d)).
2.4 Demultiplexing SERS spectra using direct classical least squares (DCLS)
To calculate the concentration and ratio of SERS NPs from a raw spectrum, this study employed a commonly used direct classical least squares (DCLS) demultiplexing method described previously [18, 41, 43]. Generally, other than sources of random noise, it is assumed that each measured spectrum consists of a linearly weighted sum of fixed nanoparticle spectra (Fn) and broadband background signals (Bi). Based on the assumption that the combination is linear, we employ a linear least-squares algorithm to compute the relative nanoparticle weights (wn). A third-order polynomial is included to account for broadband background signals that are not captured by the reference spectra (Bi).
| (1) |
where:
S = measured spectral data
wn = weight of SERS flavor n
Fn = known reference spectrum of SERS nanoparticle flavor n
ki = scaling factor for background signal magnitude
Bi = known reference spectra (principle components) of broadband background
am = weight of mth-order polynomial term
Pm = mth-order polynomial term (for baseline correction)
R = residual (minimized by least-squares algorithm)
In order to process raw spectral data, we first subtract the constant detector noise that is obtained with the laser illumination turned off. The spectra are cropped and only the range 700–2000 cm−1 (Raman shift) is used for demultiplexing analysis. Reference spectra are obtained for pure NP flavors, Fn, as well as for the broadband background, Bi. In order to obtain a NP reference spectrum, a 1-μL drop of stock NPs (800 pM concentration) is placed on the prism of the imaging probe, allowing the laser beam to pass through the center of the drop. A spectrum is recorded from the NP sample with a 1-s integration time. Next, another spectrum is recorded from a drop of the buffer in which the NPs were previously suspended within (water or PBS), under identical conditions (e.g., droplet volume, detection geometry, detector settings). These two spectra are subtracted from each other to obtain the pure reference spectra of the NPs. In order to obtain background reference spectra, Bi, the sample (e.g., tissue) is stained with an appropriate buffer solution (generally PBS) that is devoid of NPs. A spectrum of the buffer-stained sample is acquired under the exact same conditions (e.g., laser and detector settings, measurement geometry, etc.) used for measuring NP-stained samples. Multiple spectra are acquired to obtain an average background reference spectrum. In addition, up to 1 or 2 additional background components can be determined through principle component analysis (PCA) to form a sufficiently complete set of background references, Bi.
2.5 Imaging of small tissue phantoms
The NP flavors, S420 and S440, were mixed in either a 1:1 ratio or a 3:1 ratio, and diluted to high and low concentrations in Matrigel (100 pM and 10 pM for S440). A 0.5-μL drop of the low-concentration Matrigel phantom and a 1-μL drop of the high-concentration phantom were placed at adjacent sites on the exterior of a glass guide tube.
The imaging probe was used to scan a 9-mm section of the glass tube (scanning area 91.8 mm2), containing the high- and low-concentration NP phantoms, with an integration time of 1 s per spectrum. The 900 acquired spectra were demultiplexed with a LabVIEW program to calculate the weights of S420 and S440 at each time point. The concentrations and concentration ratio were calculated based on a calibration measurement with a stock mixture of S420 and S440 (see the linearity test in Section 3.1).
2.6 Animal imaging
Male Fischer 344 Inbred rats (7-9 weeks, Harlan Laboratories, Inc) were used for spectral imaging studies. All animal work was approved by the Institutional Animal Care and Use Committee (IACUC) at Stony Brook University.
For ex vivo experiments, rats were euthanized via inhalation of CO2, followed by the surgical removal of esophagus tissue (~8 cm in length). A 3.25-mm diameter guide tube (10 cm in length) was inserted into the rat’s esophagus and fixed by a holder. The imaging probe was then inserted within the guide tube to record the tissue background as well as to record SERS NP signals in later experiments.
For in vivo experiments, rats were anesthetized via intraperitoneal injection of ketamine and xylazine. A 3.25-mm diameter guide tube measuring 10 cm in length was inserted into the rat’s esophagus until gently opposed by the esophageal sphincter at the entrance to the stomach. The imaging probe was then inserted within the guide tube to record the tissue background as well as to record SERS NP signals in later experiments (similar to ex vivo experiments).
For the rotational scanning experiments, the measured weight of the SERS NPs (demultiplexing result) often exhibited a periodic fluctuation (0.5-2 pM level) due to optical and mechanical variations during rotation. While the ratiometric quantification of NP concentrations is relatively insensitive to many of these common-mode fluctuations in signal, these fluctuations can lead to increased errors in our measurements, especially at low NP concentrations. Therefore, we discarded any data points in which the demultiplexed NP weights were within the noise floor of our system (wi < 0.0015, corresponding to a NP concentration of less than 1 pM).
3. Results
3.1 Reproducibility and linearity of endoscopic spectral measurements
To assess detection uncertainty, a 1-μL drop from a mixture of NPs (S420 and S440, 200 pM for each) was placed on the guide tube, and 50 acquisitions were taken with the laser beam passing through the center of the drop. Different detector integration times were tested, including 0.1 s, 0.25 s, 0.5 s, 1 s and 2 s. Each set of 50 acquisitions was then demultiplexed to calculate the NP weights, which were used to evaluate the coefficient of variation (CV): CV = σ/μ, where σ is the standard deviation of the weight (or weight ratios) and µ is the average of the weight (or weight ratios). The CVs of the measured weights and ratios were below 1% for integration times of 0.5 s or greater, and below 3% at an integration time of 0.1 s (Fig. 2(a)).
Fig. 2.
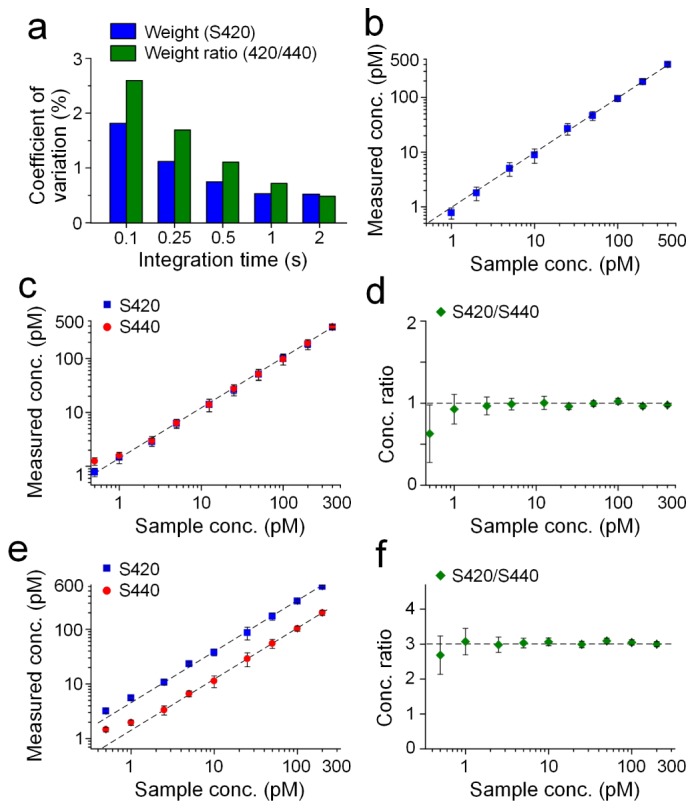
Reproducibility and linearity of endoscopic spectral measurements. (a) Measurement variability depends on integration time. (b) Single NP flavor and (c-f) a two-flavor linearity test. Error bars represent one standard deviation of the measured concentrations or concentration ratios. For the two-flavor experiments, the NP flavors S420 and S440 were mixed in either a 1:1 ratio (c, d) or a 3:1 ratio (e, f) to test the accuracy of the demultiplexing algorithm.
For linearity and limit-of-detection tests, both a single NP flavor and two-flavor NP mixtures were placed on a glass guide tube and detected with the spectral endoscope. Single-flavor samples included S440 solutions at 1, 2.5, 5, 10, 25, 50, 100, 200 and 400 pM. Two sets of two-flavor samples were prepared with different mixture ratios. One set was made by mixing S420 and S440 in an equimolar ratio (1:1) and diluted to different concentrations (0.5, 1, 2.5, 5, 12.5, 25, 50, 100, 200 and 400 pM). The other set was made by mixing S420 and S440 in a 3:1 volume ratio and diluted to different concentrations (for S440: 0.5, 1, 2.5, 5, 10, 25, 50, 100 and 200 pM). For these measurements, a 1-μL drop from each sample was placed on the guide tube, and 3 acquisitions were obtained with the laser beam directed through the center of the drop. The acquired spectra were then demultiplexed to calculate the NP weights. In order to calculate the concentration of NPs based on the measured weights, a calibration measurement was performed by recording the weights of known stock concentrations of S420 and S440. The concentrations and concentration ratios of S420/S440 are plotted in Figs. 2(b)-2(f). For both single-flavor and two-flavor experiments, the measured concentrations and concentration ratios exhibit good linearity in the range of 1 to 400 pM, with larger errors (>20%) appearing below 1 pM. Therefore, all data presented in this paper correspond to NP concentrations considerably greater than 1 pM.
3.2 Demonstration of endoscopic imaging with a tissue phantom
In order to test the endoscopy strategy, Matrigel (BD Biosciences, 354234) was mixed with two flavors of NPs and placed on a glass guide tube. In previous studies, a multiplexed panel of NP-mAb conjugates were allowed to preferentially bind to cell-surface biomarkers that were unique to different tissue types (tumor or normal) [30]. In these previous studies, which utilized targeted SERS NPs to stain tumor and normal tissues in mice, topical application of multiplexed NPs for a 5‒10 min period resulted in variable NP concentrations of 10‒100 pM on tissue surfaces, and NP concentration ratios ranging from 1 to 3. Therefore, to simulate these conditions, “tissue islands” were placed on the glass guide tube consisting of NP concentrations of 10‒100 pM and NP concentration ratios of 1:1 or 3:1 (for additional details, refer to Section 2.5).
Figures 3(a)-3(d) show the concentrations and concentration ratios of the two “tissue islands”. The NP flavors are clearly detectable even at low concentrations of 10 pM (Figs. 3(a) and 3(c)). Note that the maximum concentrations vary due to the uneven distribution of NPs as the Matrigel solidifies, as well as the variable thickness of the curved Matrigel droplet. However, in contrast to the NP concentrations, the measured concentration ratios show less fluctuation due to the fact that both NP flavors are distributed identically within the Matrigel and maintain a constant mixture ratio (Figs. 3(b) and 3(d)). For example, Figs. 3(e) and 3(g) are images of measured NP concentrations (S440), showing that the concentration gradually decreases from the center of the phantom to the edge. In comparison, the NP ratios (Figs. 3(f) and 3(h)) are more uniform within each phantom and also between the H and L phantoms.
Fig. 3.
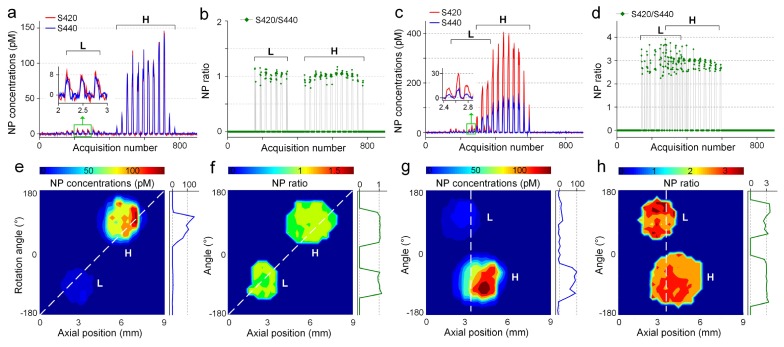
Tissue phantom imaging. Two flavors of NPs were mixed in either a 1:1 ratio (a, b, e, f) or a 3:1 ratio (c, d, g, h) within a Matrigel phantom at both high (H) and low (L) concentrations. (a, c) Measured concentrations of the two NP flavors; (b, d) ratio of measured NP concentrations; (e, g) image of the NP concentrations (S440); (f, h) image of the NP ratios.
3.3 Demonstration of endoscopic imaging in the rat esophagus
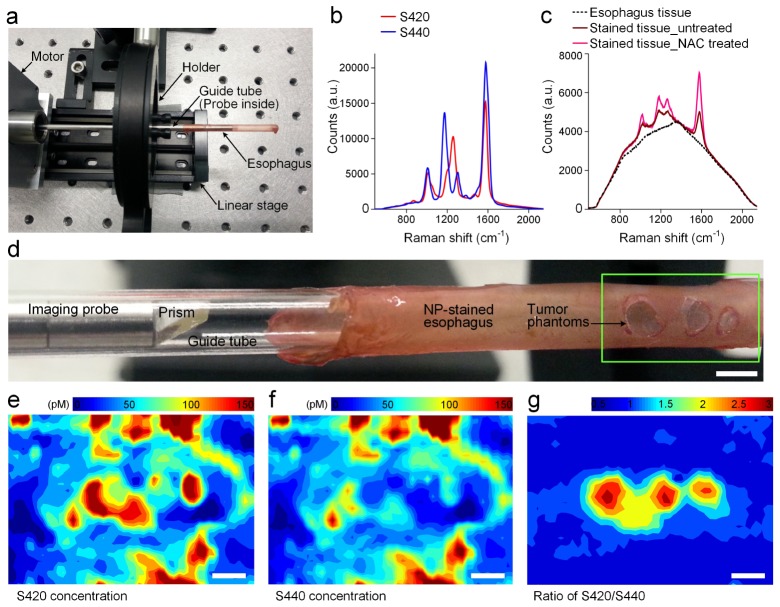
Ex vivo experiments were first performed to demonstrate the imaging ability of the spectral endoscope and to develop a NP-staining protocol (Fig. 4(a)). The excised esophagus was rinsed with PBS and then irrigated with a mucolytic agent (N-Acetyl-L-cysteine, or NAC for short, Sigma-Aldrich, part No. A7250) to allow the NPs to access and adhere to the tissue more effectively (0.5 mL at 0.1 g/mL). A glass guide tube was inserted within the esophagus lumen, along with the Raman probe, for acquiring tissue background measurements (300 acquisitions on a 1-mm section of the esophagus). The guide tube was then removed from the esophagus to allow for NP staining by pipetting in 50 μL of a two-flavor equimolar mixture (200 pM per flavor). After staining for 5 min, excess NPs were washed away by irrigating the esophagus with 1 mL of PBS. The guide tube and Raman probe were then re-inserted into the esophagus for spectral imaging of the NPs.
Fig. 4.
Ex vivo imaging of the rat esophagus. (a) Photograph of the Raman endoscope. (b) The reference spectra of the NP flavors S420 and S440; (c) Raw spectra from freshly excised esophagus and NP-stained esophagus (untreated or NAC-treated); (d) Zoom-in view showing the distal probe tip in a guide tube and tumor phantoms (green box) on a freshly excised rat esophagus. Images showing measured concentrations of S420 (e) and S440 (f), as well as the concentration ratio of S420/S440 (g). Scale bars = 2 mm.
The fresh esophagus was physically modified to test if the endoscope could detect tumor phantoms. In short, small holes were cut in the esophagus tissue as it was stretched around a glass guide tube. NP-matrigel phantoms were placed in those holes to simulate NP-labeled tumors. Based on our previous data with tumor-bearing mice [30] and human breast tumor specimens (unpublished data), the ratio of biomarker-targeted vs. nontargeted control NPs reaches 2–3 for tumors while for normal tissues the ratio remains at 1. More specifically, after staining the esophagus with a two-flavor equimolar mixture of NPs, three 1–2 mm holes were cut in the normal esophagus tissue, and matrigel phantoms containing a 3:1 ratio NP mixture (S440: 100 pM, S420: 300 pM) were placed in the holes as simulated tumors (1–2 μL depending on the hole size). The esophagus was imaged immediately after this modification by scanning a 1.5-cm section with a 0.25-s integration time per spectrum (1500 acquisitions). The image of the measured concentrations and ratios are shown in Figs. 4(e)-4(g). Note that for the concentration images (Fig. 4(e): S420, Fig. 4(f): S440), the tumor phantoms cannot be easily identified due to the high variability in NP concentrations across the tissue. In comparison, the ratio image (Fig. 4(g)) is highly insensitive to the variations in absolute NP concentrations and clearly reveals the location of the tumor phantoms. Only the tumor phantoms show an elevated ratio (1.5–3) while the normal tissues maintain a constant ratio near unity (0.8–1.2).
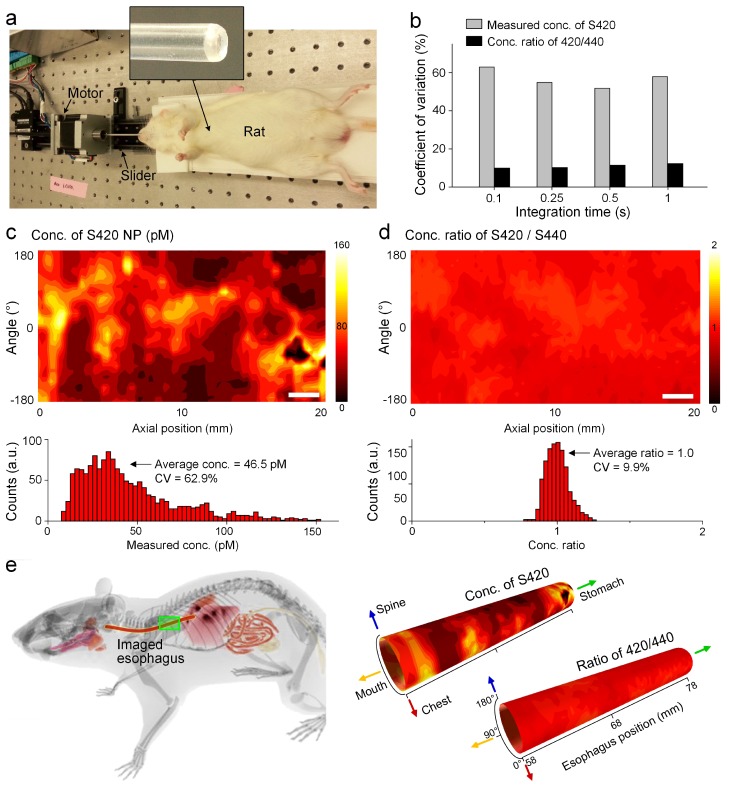
In vivo experiments were performed to explore the feasibility of topical application of NPs into the rat esophagus through oral gavage (n = 6), and to test the imaging speed and accuracy of the spectral endoscope (Fig. 5(a)). For in vivo experiments, one side of the guide tube was sealed to prevent liquids from entering the tube, and the distal tip of the guide tube was slightly tapered to ease the insertion of the tube into the esophagus (see top inset panel in Fig. 5(a)).
Fig. 5.
In vivo imaging performed after an oral-gavage protocol. (a) Photograph of the in vivo experimental system with an anesthetized rat. (b) CV of the imaged concentrations (S420) and concentration ratios (S420/S440) within the esophagus at different detector integration times showing that the NP ratios are relatively constant in spite of large fluctuations in absolute NP concentration. Images showing measured concentrations of S420 (c) and concentration ratios of S420/S440 (d). Scale bars = 2 mm. (e) Reconstructed esophagus images with physical coordinates. The lower teeth of the rat were used as a z = 0 reference point in the axial direction.
After anesthetizing the rat, the guide tube and imaging probe were first inserted to record tissue background spectra, and then gently pulled out. After the rat regained consciousness, the esophagus was irrigated with 0.5 mL of the NAC mucolytic agent (0.1 g/mL) through an oral gavage catheter placed within the esophagus, followed by irrigation with a two-flavor equimolar mixture of SERS NPs (400 pM/each, 0.2 mL). After 5 minutes, unbound NPs were rinsed away by irrigating the esophagus again with PBS (1 mL volume). The animal was anesthetized, and the guide tube and probe were inserted into the NP-stained esophagus to image the NPs. Results demonstrate that the oral gavage protocol is effective for the in vivo topical application of SERS NPs in the rat esophagus, with sufficient nonspecific accumulation of the NPs such that a short integration time of 0.1 s could be utilized for imaging of the NP concentrations and ratios (Figs. 4(c) and 5(b)). Figures 5(c) and 5(d) are images of the measured concentration (S420) and NP ratios (S420/S440). Note that although the esophagus is not evenly stained (NP concentrations vary from 10 pM to 160 pM as shown in the histogram below Fig. 5(c)), the ratio remains relatively constant (near unity) over the entire imaged area (Fig. 5(d)), thereby demonstrating a key advantage of the ratiometric strategy (see discussion section). The imaging coordinates of each acquisition were also recorded (Fig. 5(e)), which will be helpful for localizing lesions during detection, surgical guidance, and biological investigations.
4. Discussion
An emerging endoscopic strategy for detecting or investigating gastrointestinal cancers is to visualize the molecular phenotype of tissues through the use of molecularly targeted contrast agents [8, 19, 44]. Among various types of contrast agents, SERS NPs have attracted interest in recent years because of their high multiplexing capabilities and minimal systemic uptake and toxicity when applied to epithelial tissues [36, 45]. Here, we have described a miniature spectral endoscope custom developed to image SERS NPs topically applied within the rat esophagus through an oral gavage procedure. With a DCLS demultiplexing algorithm, the endoscope is able to measure the concentrations of multiplexed NPs with a limit of detection of 1 pM and an imaging speed of 0.6 cm/min (10 spectra/s). The disadvantage of rotational scanning is that it is designed to image the entire lumen of the esophagus and will therefore require more time than a localized imaging device. However, the advantage of the device developed in this study, versus a forward-looking device [19], is the ability to comprehensively image the entire organ to screen for molecular changes without prior knowledge about the location of potential lesions.
A calibrated ratiometric strategy has been demonstrated to accurately quantify the relative concentration of multiplexed NPs on esophagus tissue in spite of large variations in absolute NP concentrations (Figs. 3‒5). A significant motivation for introducing the ratiometric strategy is that contrast agents are not only sensitive to molecular targets but also behave differently in response to tissue morphology, porosity, nonspecific chemical targets, and topical-application parameters. For example, normal tissues can exhibit stronger signals than tumors due to higher retention of contrast agent, i.e. inverse contrast [31, 32], while in some other cases, tumors exhibit stronger signals than normal tissues due to EPR [33–35]. In addition, contrast agents are seldom distributed evenly when administered via systemic or topical application (Figs. 3(e), 3(g), 4(e), 4(f) and 5(c)). To eliminate these sources of image ambiguity, various groups are utilizing and witnessing the power of a dual-reporter ratiometric detection strategy, in which one negative reporter serves as a control to assess the behavior of a simultaneously delivered positive reporter for accurate identification and quantification of molecularly specific binding [23, 31, 32, 34]. SERS NPs are ideal reporters for such ratiometric strategies since they are excited at a single illumination wavelength (785 nm), ensuring that all NP reporters in a single measurement are interrogated identically in terms of illumination intensity, detection area, and effective excitation depth.
Through a series of imaging experiments with tissue phantoms and real tissues (Figs. 2‒5), we have demonstrated that our endoscope can accurately quantify the relative concentration ratios of NP flavors mixed in a 1:1 ratio or 3:1 ratio. These two conditions represent the range of ratios obtained when targeted and nontargeted SERS NPs were previously used to stain fresh tumor and normal tissues ex vivo and in vivo in animal models [30] and on human tissues (unpublished data).
Another important result of this study is that sufficient staining of the esophagus with multiplexed NPs (10‒160 pM in Fig. 5(c)) is achieved with a 5-min topical application (oral-gavage) procedure. In the future, our spectral endoscope will be utilized for multiplexed molecular imaging of rat esophageal cancer models with targeted SERS NPs. In addition, it will be interesting to test our oral application protocol and imaging strategy with larger animal models, such as swine, which more closely resemble the human esophagus in terms of size and geometry. Our previous studies have shown that the ratiometric imaging of SERS NP mixtures is relatively insensitive to variations in working distance and tissue geometry [30]. Therefore, in larger animal models and humans, a guide tube should not be necessary and a rotational Raman-imaging device could be deployed through the instrument channel of a standard endoscope. These future studies will leverage our recently developed protocols for the topical-delivery and rinsing of multiplexed SERS NPs (5‒10 min), along with a quantitative ratiometric detection strategy to unambiguously visualize cell-surface biomarkers on tissues. These tools have the potential to improve our ability to detect and investigate the molecular mechanisms of esophageal cancer progression.
Acknowledgments
The authors acknowledge support from the NIH / NIBIB – R21 EB015016 (Liu), the Department of Biomedical Engineering, and the Office of the Vice President of Research at Stony Brook University (SUNY).
References and links
- 1.Parkin D. M., Bray F. I., Devesa S. S., “Cancer burden in the year 2000. The global picture,” Eur. J. Cancer 37(Suppl 8), 4–66 (2001). 10.1016/S0959-8049(01)00267-2 [DOI] [PubMed] [Google Scholar]
- 2.Souza R. F., “Molecular and biologic basis of upper gastrointestinal malignancy--esophageal carcinoma,” Surg. Oncol. Clin. N. Am. 11(2), 257–272 (2002). 10.1016/S1055-3207(02)00003-0 [DOI] [PubMed] [Google Scholar]
- 3.Evans J. A., Early D. S., Chandraskhara V., Chathadi K. V., Fanelli R. D., Fisher D. A., Foley K. Q., Hwang J. H., Jue T. L., Pasha S. F., Sharaf R., Shergill A. K., Dominitz J. A., Cash B. D., ASGE Standards of Practice CommitteeAmerican Society for Gastrointestinal Endoscopy , “The role of endoscopy in the assessment and treatment of esophageal cancer,” Gastrointest. Endosc. 77(3), 328–334 (2013). 10.1016/j.gie.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 4.Layke J. C., Lopez P. P., “Esophageal cancer: a review and update,” Am. Fam. Physician 73(12), 2187–2194 (2006). [PubMed] [Google Scholar]
- 5.Polednak A. P., “Trends in survival for both histologic types of esophageal cancer in US surveillance, epidemiology and end results areas,” Int. J. Cancer 105(1), 98–100 (2003). 10.1002/ijc.11029 [DOI] [PubMed] [Google Scholar]
- 6.Younes M., Henson D. E., Ertan A., Miller C. C., “Incidence and survival trends of esophageal carcinoma in the United States: racial and gender differences by histological type,” Scand. J. Gastroenterol. 37(12), 1359–1365 (2002). 10.1080/003655202762671215 [DOI] [PubMed] [Google Scholar]
- 7.Sturm M. B., Joshi B. P., Lu S., Piraka C., Khondee S., Elmunzer B. J., Kwon R. S., Beer D. G., Appelman H. D., Turgeon D. K., Wang T. D., “Targeted imaging of esophageal neoplasia with a fluorescently labeled peptide: first-in-human results,” Sci. Transl. Med. 5(184), 184ra61 (2013). 10.1126/scitranslmed.3004733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiung P.-L., Hardy J., Friedland S., Soetikno R., Du C. B., Wu A. P., Sahbaie P., Crawford J. M., Lowe A. W., Contag C. H., Wang T. D., “Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy,” Nat. Med. 14(4), 454–458 (2008). 10.1038/nm1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiesslich R., Goetz M., Vieth M., Galle P. R., Neurath M. F., “Technology insight: confocal laser endoscopy for in vivo diagnosis of colorectal cancer,” Nat. Clin. Pract. Oncol. 4(8), 480–490 (2007). 10.1038/ncponc0881 [DOI] [PubMed] [Google Scholar]
- 10.Bird-Lieberman E. L., Neves A. A., Lao-Sirieix P., O’Donovan M., Novelli M., Lovat L. B., Eng W. S., Mahal L. K., Brindle K. M., Fitzgerald R. C., “Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett’s esophagus,” Nat. Med. 18(2), 315–321 (2012). 10.1038/nm.2616 [DOI] [PubMed] [Google Scholar]
- 11.Samoylova T. I., Morrison N. E., Globa L. P., Cox N. R., “Peptide phage display: opportunities for development of personalized anti-cancer strategies,” Anticancer. Agents Med. Chem. 6(1), 9–17 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Weissleder R., Ross B. D., Rehemtulla A., Gambhir S. S., Molecular Imaging: Principles and Practice (People's Medical Publishing House, 2010). [Google Scholar]
- 13.Hellebust A., Richards-Kortum R., “Advances in molecular imaging: targeted optical contrast agents for cancer diagnostics,” Nanomedicine (Lond) 7(3), 429–445 (2012). 10.2217/nnm.12.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen Q. T., Tsien R. Y., “Fluorescence-guided surgery with live molecular navigation--a new cutting edge,” Nat. Rev. Cancer 13(9), 653–662 (2013). 10.1038/nrc3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y. Q., Yan B., Chen L. X., “SERS Tags: Novel Optical Nanoprobes for Bioanalysis,” Chem. Rev. 113(3), 1391–1428 (2013). 10.1021/cr300120g [DOI] [PubMed] [Google Scholar]
- 16.Mulvaney S. P., Musick M. D., Keating C. D., Natan M. J., “Glass-Coated, Analyte-Tagged Nanoparticles: A New Tagging System Based on Detection with Surface-Enhanced Raman Scattering,” Langmuir 19(11), 4784–4790 (2003). 10.1021/la026706j [DOI] [Google Scholar]
- 17.Sha M. Y., Xu H., Natan M. J., Cromer R., “Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood,” J. Am. Chem. Soc. 130(51), 17214–17215 (2008). 10.1021/ja804494m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zavaleta C. L., Smith B. R., Walton I., Doering W., Davis G., Shojaei B., Natan M. J., Gambhir S. S., “Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy,” Proc. Natl. Acad. Sci. U.S.A. 106(32), 13511–13516 (2009). 10.1073/pnas.0813327106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zavaleta C. L., Garai E., Liu J. T. C., Sensarn S., Mandella M. J., Van de Sompel D., Friedland S., Van Dam J., Contag C. H., Gambhir S. S., “A Raman-based endoscopic strategy for multiplexed molecular imaging,” Proc. Natl. Acad. Sci. U.S.A. 110(25), E2288–E2297 (2013). 10.1073/pnas.1211309110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohndiek S. E., Wagadarikar A., Zavaleta C. L., Van de Sompel D., Garai E., Jokerst J. V., Yazdanfar S., Gambhir S. S., “A small animal Raman instrument for rapid, wide-area, spectroscopic imaging,” Proc. Natl. Acad. Sci. U.S.A. 110(30), 12408–12413 (2013). 10.1073/pnas.1301379110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohs A. M., Mancini M. C., Singhal S., Provenzale J. M., Leyland-Jones B., Wang M. D., Nie S., “Hand-held spectroscopic device for in vivo and intraoperative tumor detection: contrast enhancement, detection sensitivity, and tissue penetration,” Anal. Chem. 82(21), 9058–9065 (2010). 10.1021/ac102058k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McVeigh P. Z., Mallia R. J., Veilleux I., Wilson B. C., “Widefield quantitative multiplex surface enhanced Raman scattering imaging in vivo,” J. Biomed. Opt. 18(4), 046011 (2013). 10.1117/1.JBO.18.4.046011 [DOI] [PubMed] [Google Scholar]
- 23.Garai E., Sensarn S., Zavaleta C. L., Sompel D. V. d., Loewke N. O., Mandella M. J., Gambhir S. S., Contag C. H., “High-sensitivity, real-time, ratiometric imaging of surface-enhanced Raman scattering nanoparticles with a clinically translatable Raman endoscope device,” J. Biomed. Opt. 18, 096008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallia R. J., McVeigh P. Z., Veilleux I., Wilson B. C., “Filter-based method for background removal in high-sensitivity wide-field-surface-enhanced Raman scattering imaging in vivo,” J. Biomed. Opt. 17(7), 076017 (2012). 10.1117/1.JBO.17.7.076017 [DOI] [PubMed] [Google Scholar]
- 25.Jokerst J. V., Miao Z., Zavaleta C., Cheng Z., Gambhir S. S., “Affibody-Functionalized Gold-Silica Nanoparticles for Raman Molecular Imaging of the Epidermal Growth Factor Receptor,” Small 7(5), 625–633 (2011). 10.1002/smll.201002291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Qian X., Beitler J. J., Chen Z. G., Khuri F. R., Lewis M. M., Shin H. J. C., Nie S., Shin D. M., “Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles,” Cancer Res. 71(5), 1526–1532 (2011). 10.1158/0008-5472.CAN-10-3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S., Chon H., Yoon S.-Y., Lee E. K., Chang S.-I., Lim D. W., Choo J., “Fabrication of SERS-fluorescence dual modal nanoprobes and application to multiplex cancer cell imaging,” Nanoscale 4(1), 124–129 (2011). 10.1039/c1nr11243k [DOI] [PubMed] [Google Scholar]
- 28.Sun L., Sung K.-B., Dentinger C., Lutz B., Nguyen L., Zhang J., Qin H., Yamakawa M., Cao M., Lu Y., Chmura A. J., Zhu J., Su X., Berlin A. A., Chan S., Knudsen B., “Composite organic-inorganic nanoparticles as Raman labels for tissue analysis,” Nano Lett. 7(2), 351–356 (2007). 10.1021/nl062453t [DOI] [PubMed] [Google Scholar]
- 29.Su X., Zhang J., Sun L., Koo T. W., Chan S., Sundararajan N., Yamakawa M., Berlin A. A., “Composite organic-inorganic nanoparticles (COINs) with chemically encoded optical signatures,” Nano Lett. 5(1), 49–54 (2005). 10.1021/nl0484088 [DOI] [PubMed] [Google Scholar]
- 30.Wang Y. W., Khan A., Som M., Wang D., Chen Y., Leigh S. Y., Meza D., McVeigh P. Z., Wilson B. C., Liu J. T. C., “Rapid ratiometric biomarker detection with topically applied SERS nanoparticles,” Technology (Singap) 2(2), 118–132 (2014). 10.1142/S2339547814500125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tichauer K. M., Samkoe K. S., Sexton K. J., Gunn J. R., Hasan T., Pogue B. W., “Improved tumor contrast achieved by single time point dual-reporter fluorescence imaging,” J. Biomed. Opt. 17(6), 066001 (2012). 10.1117/1.JBO.17.6.066001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tichauer K. M., Samkoe K. S., Sexton K. J., Hextrum S. K., Yang H. H., Klubben W. S., Gunn J. R., Hasan T., Pogue B. W., “In vivo quantification of tumor receptor binding potential with dual-reporter molecular imaging,” Mol. Imaging Biol. 14(5), 584–592 (2012). 10.1007/s11307-011-0534-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kircher M. F., de la Zerda A., Jokerst J. V., Zavaleta C. L., Kempen P. J., Mittra E., Pitter K., Huang R., Campos C., Habte F., Sinclair R., Brennan C. W., Mellinghoff I. K., Holland E. C., Gambhir S. S., “A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle,” Nat. Med. 18(5), 829–834 (2012). 10.1038/nm.2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J. T. C., Helms M. W., Mandella M. J., Crawford J. M., Kino G. S., Contag C. H., “Quantifying Cell-Surface Biomarker Expression in Thick Tissues with Ratiometric Three-Dimensional Microscopy,” Biophys. J. 96(6), 2405–2414 (2009). 10.1016/j.bpj.2008.12.3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T. D., Friedland S., Sahbaie P., Soetikno R., Hsiung P. L., Liu J. T. C., Crawford J. M., Contag C. H., “Functional Imaging of Colonic Mucosa With a Fibered Confocal Microscope for Real-Time In Vivo Pathology,” Clin. Gastroenterol. Hepatol. 5(11), 1300–1305 (2007). 10.1016/j.cgh.2007.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakor A. S., Luong R., Paulmurugan R., Lin F. I., Kempen P., Zavaleta C., Chu P., Massoud T. F., Sinclair R., Gambhir S. S., “The fate and toxicity of Raman-active silica-gold nanoparticles in mice,” Sci. Transl. Med. 3(79), 79ra33 (2011). 10.1126/scitranslmed.3001963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zavaleta C. L., Hartman K. B., Miao Z., James M. L., Kempen P., Thakor A. S., Nielsen C. H., Sinclair R., Cheng Z., Gambhir S. S., “Preclinical evaluation of Raman nanoparticle biodistribution for their potential use in clinical endoscopy imaging,” Small 7(15), 2232–2240 (2011). 10.1002/smll.201002317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoner G. D., Gupta A., “Etiology and chemoprevention of esophageal squamous cell carcinoma,” Carcinogenesis 22(11), 1737–1746 (2001). 10.1093/carcin/22.11.1737 [DOI] [PubMed] [Google Scholar]
- 39.Stoner G. D., Wang L. S., “Chemoprevention of esophageal squamous cell carcinoma with berries,” Top. Curr. Chem. 329, 1–20 (2012). 10.1007/128_2012_343 [DOI] [PubMed] [Google Scholar]
- 40.Hori T., Yamashita Y., Ohira M., Matsumura Y., Muguruma K., Hirakawa K., “A novel orthotopic implantation model of human esophageal carcinoma in nude rats: CD44H mediates cancer cell invasion in vitro and in vivo,” Int. J. Cancer 92(4), 489–496 (2001). 10.1002/ijc.1234 [DOI] [PubMed] [Google Scholar]
- 41.Leigh S. Y., Som M., Liu J. T. C., “Method for Assessing the Reliability of Molecular Diagnostics Based on Multiplexed SERS-Coded Nanoparticles,” PLoS ONE 8(4), e62084 (2013). 10.1371/journal.pone.0062084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doering W. E., Piotti M. E., Natan M. J., Freeman R. G., “SERS as a Foundation for Nanoscale, Optically Detected Biological Labels,” Adv. Mater. 19(20), 3100–3108 (2007). 10.1002/adma.200701984 [DOI] [Google Scholar]
- 43.Lutz B. R., Dentinger C. E., Nguyen L. N., Sun L., Zhang J., Allen A. N., Chan S., Knudsen B. S., “Spectral analysis of multiplex Raman probe signatures,” ACS Nano 2(11), 2306–2314 (2008). 10.1021/nn800243g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bird-Lieberman E. L., Neves A. A., Lao-Sirieix P., O’Donovan M., Novelli M., Lovat L. B., Eng W. S., Mahal L. K., Brindle K. M., Fitzgerald R. C., “Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett’s esophagus,” Nat. Med. 18(2), 315–321 (2012). 10.1038/nm.2616 [DOI] [PubMed] [Google Scholar]
- 45.Chithrani B. D., Ghazani A. A., Chan W. C. W., “Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells,” Nano Lett. 6(4), 662–668 (2006). 10.1021/nl052396o [DOI] [PubMed] [Google Scholar]