Abstract
Type-III protein secretion systems are utilized by gram-negative pathogens to secrete building blocks of the bacterial flagellum, virulence effectors from the cytoplasm into host cells, and structural subunits of the needle complex. The flagellar type-III secretion apparatus utilizes both the energy of the proton motive force and ATP hydrolysis to energize substrate unfolding and translocation. We report formation of functional flagella in the absence of type-III ATPase activity by mutations that increased the proton motive force and flagellar substrate levels. We additionally show that increased proton motive force bypassed the requirement of the Salmonella pathogenicity island 1 virulence-associated type-III ATPase for secretion. Our data support a role for type-III ATPases in enhancing secretion efficiency under limited secretion substrate concentrations and reveal the dispensability of ATPase activity in the type-III protein export process.
Author Summary
The type-III protein secretion apparatus is a complex nanomachine responsible for secretion of building blocks and substrate proteins of the flagellum and the virulence-associated injectisome needle complex of many gram-negative pathogens. Type-III secretion systems utilize the energy of the proton motive force and ATP hydrolysis of a cytoplasmic ATPase to drive substrate export. The cytoplasmic components of the secretion system share strong homology to the FoF1 ATP synthase and it is thought that the flagellum was derived from a proto FoF1-ATP synthase where ATP hydrolysis energized the export process. Here, we report the dispensability of ATPase activity for the type-III protein export process in Salmonella. This finding has important implications for the evolution of the bacterial flagellum and type-III secretion systems, suggesting that a proto ATPase was added to a primordial proton-powered type-III export system with the evolutionary benefit of facilitating the export process.
Introduction
Many bacteria move by rotating a rigid, helical organelle, the flagellum [1]. The flagellum represents one of the smallest motor complexes known and enables bacteria to move through liquids (swimming) [2] and highly viscous environments or surfaces (swarming) [3]. In addition to the chemotactic behavior, flagellar motility contributes to bacterial pathogenesis by promoting bacteria-host interactions, adherence, biofilm formation and invasion of eukaryotic cells [4]. A schematic overview of flagellar biogenesis in Salmonella enterica is shown in Figure 1. Export of the building blocks of the flagellum is mediated by a flagellar-specific type-III secretion system (fT3SS), whose core cytoplasmic and inner-membrane export apparatus components (FliHIJ FliPQR FlhAB) are evolutionarily and functionally related to the virulence-associated type-III secretion systems (vT3SS) of pathogenic Gram-negative bacteria [5].
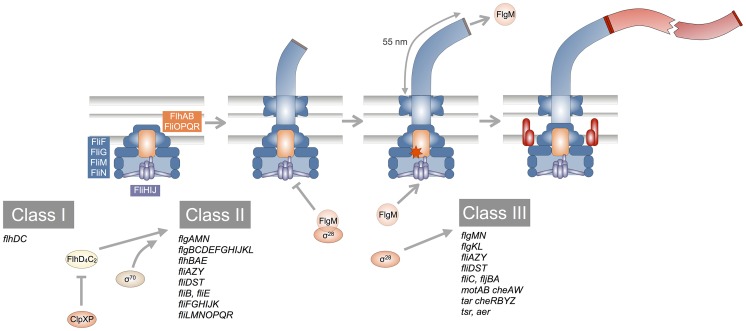
Figure 1. Schematic overview of the flagellar transcriptional hierarchy and biogenesis.
The flagellar transcriptional hierarchy of Salmonella enterica is composed of three classes of promoters. The Class I promoter transcribes a single operon encoding for the master regulator of the flagellar transcriptional hierarchy, the FlhD4C2 complex, which is negatively regulated by ClpXP protease. FlhD4C2, together with σ70, directs RNA polymerase to transcribe from Class II promoters. Genes transcribed from Class II promoters encode structural components of the hook-basal-body complex (shaded in blue), the flagellar type-III secretion apparatus (composed of the membrane proteins FlhA, FlhB, FliO, FliP, FliQ and FliR; and the soluble proteins FliH, FliI and FliJ), as well as regulatory proteins, in particular the flagellar-specific σ-factor, σ28 (encoded by fliA), and its cognate anti-σ factor, FlgM. The hook-basal-body is completed as soon as the hook reaches an approximate length of 55 nm, upon which the type-III secretion apparatus switches secretion specificity to its late-substrate secretion mode (indicated by the orange star). Subsequently, the late substrate FlgM is exported out of the cell, thereby freeing σ28 to turn on transcription from Class III promoters. Class III gene products include the filament subunits, motor-force generators and the chemotactic system (shaded in red).
The core export apparatus components of both fT3SS and vT3SS mediate the translocation of proteins across the inner membrane and exhibit stringent substrate recognition and high speed of protein translocation [6]. Core export apparatus assembly initiates in the cytoplasmic membrane with the stepwise addition of FliF, FliG, FliM and FliN to produce a structure resembling a cytoplasmic-facing cup [7]. The inner membrane export apparatus (export gate) of the fT3SS assembles within the cup-like structure and consists of the integral membrane proteins FlhB, FlhA, FliO, FliP, FliQ and FliR [8]. Functionally associated with the fT3SS is a cytoplasmic ATPase complex composed of the FliI, FliH and FliJ proteins [6]. The ATPase FliI forms a hexameric ring-shaped structure [9] and together with the negative regulator FliH [10] regulates the initial entry of substrates into the export gate by ensuring that FliI-dependent ATP hydrolysis occurs in concert with secretion. FliH interacts with the N-terminal region of FliI and prevents its ATPase activity until the export apparatus is competent to utilize ATP hydrolysis in the export process [10], [11]. Interestingly, FliI and FliJ show remarkable structural similarity to the α/β and γ-subunits of FOF1 ATPases, respectively [12], [13]. FliH is homologous to the stalk subunits b and δ of FOF1 ATPases [14]. Until recently, it was presumed that ATP hydrolysis was the energy source for type-III secretion. However, it was demonstrated that elimination of the proton motive force (PMF, consisting of the protein gradient ΔpH and the membrane potential Δψ) inhibited flagellar type-III secretion [15], [16]. ATP hydrolysis might energize chaperone release and secretion substrate unfolding [17]. Further functions of the FliHIJ proteins might be in localizing the ATPase/secretion substrate complexes to the vicinity of the export gate by interaction with the switch-complex protein FliN [18], [19] and the cytoplasmic domain of FlhA [12], [20]. An efficient interaction of FliJ with FlhA requires support of FliH and FliI upon which FliJ switches the conformation of the export gate from a low-efficient proton-protein antiporter to a highly efficient, Δψ-driven export system. FliJ is thus essential for the Δψ-driven type-III protein export and in the absence of FliH and FliI, the type-III export apparatus requires both Δψ and ΔpH of the PMF to couple the energy derived from the proton influx with substrate protein secretion [21]. Additional described functions of FliJ include chaperone-like activities by preventing premature aggregation of both early and late secretion substrates [22], as well as a function in substrate selectivity as a chaperone escort protein that specifically recruits unladen substrate-specific chaperones FlgN and FliT during assembly of the filament junction and cap substructures [23]. FliJ is also implicated in facilitating FliI hexamer ring formation, thereby stimulating the ATPase activity [12], [23].
Similar to the fT3SS, the vT3SS is an essential component of the virulence-associated needle complexes encoded on Salmonella Pathogenicity Islands 1 and 2 (Spi1 and Spi2) [5], [24]. The ATPase associated with the Spi1 vT3SS is encoded by invC [25]. InvC functions in effector substrate recognition and induces chaperone release and subsequent unfolding of the respective secretion substrate in an ATP-dependent manner [17].
Type-III protein secretion is a process of great complexity and the importance of ATP hydrolysis for the type-III secretion process was unclear. In the present work, we demonstrate that type-III protein secretion can efficiently occur in the absence of ATPase activity in both flagellar and virulence-associated T3SS.
Results
We demonstrate that flagellar ATPase activity is not essential for secretion by the isolation and characterization of spontaneous suppressor mutants that assemble flagella in a ΔfliHI genetic background. These motile revertants mapped to four locations on the Salmonella chromosome. One of these linkage groups included the flh flagellar region that likely included suppressors similar to the flhA and flhB mutations described before [16], [26] and were not characterized further. Based on the chromosomal location of the remaining three linkage groups, we identified candidates for genes that could account for the motile suppression phenotypes. One linkage group included the clpXP operon. Loss of ClpXP protease stabilizes the flagellar master regulatory complex, FlhDC [27], upregulating flagellar gene expression. It was previously shown that transposon insertions in the clpP gene increased flagellar secretion substrate levels sufficiently to overcome the requirement for the cytoplasmic C-ring for T3SS-dependent substrate translocation across the inner membrane [28]. The third group of motile revertants were linked to the ATP synthase structural genes (atp) (Fig. S1). ATP synthesis, flagellum assembly and flagellar rotation are all dependent on the PMF and it has been previously described that a null mutation in atp increases the PMF [29], [30]. Increased PMF could thus restore proper flagellar assembly and function if the fT3SS is primarily PMF-dependent. Consistently, Martinez-Argudo et al. [31] reported that mutations in atpB overcame the inhibitory effect of the unspecific type-III inhibitors salicylidene acylhydrazides on flagellar motility. The final linkage group was located to a set of flagellar genes that included a negative regulatory gene (flgM) of the flagellar-specific transcription factor σ28. Strains defective in genes required for flagellar hook-basal body assembly (including fliH, fliI and fliJ) accumulate the FlgM anti-σ28 factor in the cytoplasm, which prevents σ28-dependent flagellar class 3 promoter expression. Thus, loss of FlgM could account for suppression of the motility defect of the ΔfliHI allele.
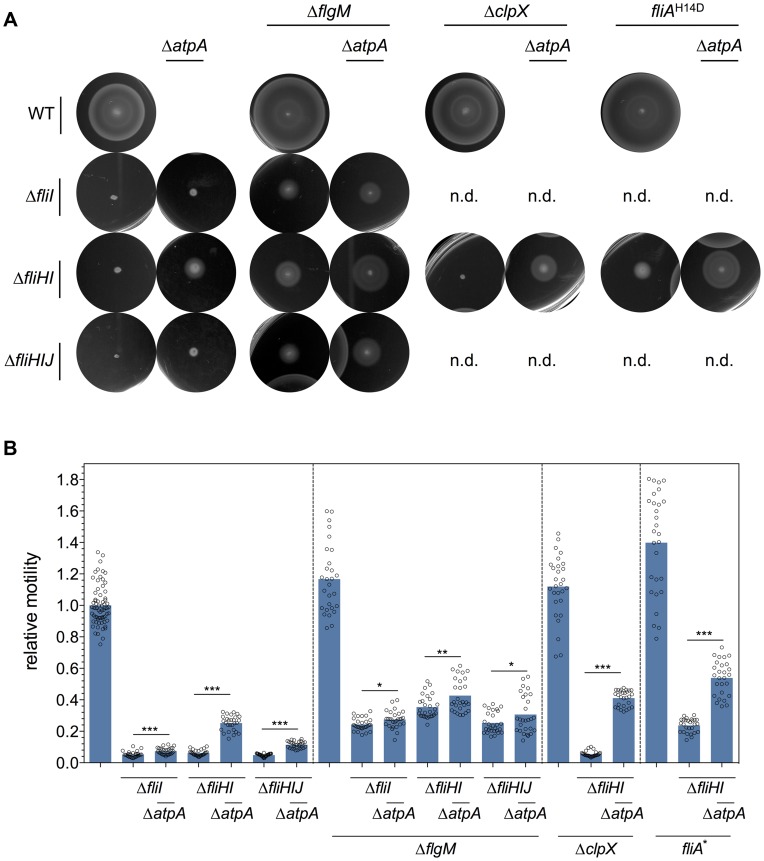
We constructed deletion mutants of clpX and atpA and tested the suppressor function in various fliHIJ deletion backgrounds by monitoring their swimming behavior in soft agar plates (Fig. 2A and Fig. S2). The absence of the FOF1 ATPase restored motility of a fliI deletion mutant to about 5% of the wildtype, of a fliHI deletion mutant to about 25% of the wildtype and of a fliHIJ deletion mutant to about 11% of the wildtype (Fig. 2B). In an otherwise wildtype background, deletion of atpA resulted in a pronounced growth defect (Fig. S3), however did not affect the free-swimming velocity (Fig. S4).
Figure 2. Motility of fliHIJ mutants is increased by mutations in ΔatpA, ΔflgM, ΔclpX and fliA H14D.
Null mutations in atpA, flgM, clpX, and the more stable FliAH14D variant increased motility of fliHIJ mutant strains in a swimming motility assay using 0.3% soft agar plates. (A) Representative soft agar motility plates after 4.5 hours incubation at 37°C of the FliC-phase locked wildtype and fliHIJ mutant strains. n.d., not determined. (B) Quantified relative motility of fliHIJ mutant strains. The diameter of the motility swarm relative to the wildtype was measured after 4.5 hours incubation. Biological replicates are shown as individual data points. Data were analyzed by the Student's t test. Stars indicate significantly different motility (*, P<0.05; **, P<0.01; ***, P<0.001).
As noted above, the absence of ClpXP protease components affects flagellar class 2 and class 3 gene expression by preventing proteolytic degradation of the flagellar master regulatory protein complex FlhDC. Similarly, a deletion of the FlgM anti-σ28 factor, or a FlgM-resistant σ28 mutant (FliAH14D) results in an increase of flagellar class 3 gene expression. We thus analyzed the motility of clean deletion mutants of clpX and flgM, and the fliA H15D allele. When combined with deletions of the fliHIJ ATPase complex components, the flgM deletion and the FlgM-resistant σ28 mutant raised motility levels up to 24–35% of the wildtype. The clpX deletion, however, had no apparent effect on motility when deleted alone. Next, the flgM, clpX, and fliA H15D mutations were combined with a null mutation of the FOF1 ATPase as described above. Interestingly, the atpA mutation had only a slight effect on the motility of fliI, fliHI and fliHIJ mutants in the flgM null background, which could be explained by impaired coordination of flagellar Class 3 gene expression with the assembly state of the flagellum. However, the apparent PMF increase of the atpA mutation did substantially increase motility in the clpX null and fliA H15D backgrounds to approximately 41–54% of the wildtype (Fig. 2B and Fig. S2). We thus concluded that a 20% increase in PMF by inactivation of the ATP synthase [29], [30] substantially enhances fT3SS export capability and thereby bypasses the FliHIJ ATPase requirement for flagellar function. We next reasoned that an artificial induction of an additional PMF-draining system would antagonize the suppressor function of the atpA null strain in the FliHIJ ATPase mutant backgrounds, which we tested by inducing the PMF-dependent inner membrane tetracycline/proton antiporter (TetA). Induction of the TetA efflux pump counteracted the suppressor phenotype of the atpA deletion of various fliHIJ deletion mutants in a motility assay (Fig. S5). We conclude that high levels of TetA tetracycline/proton antiporter activity drained the PMF and thereby negatively affected protein export via the fT3SS.
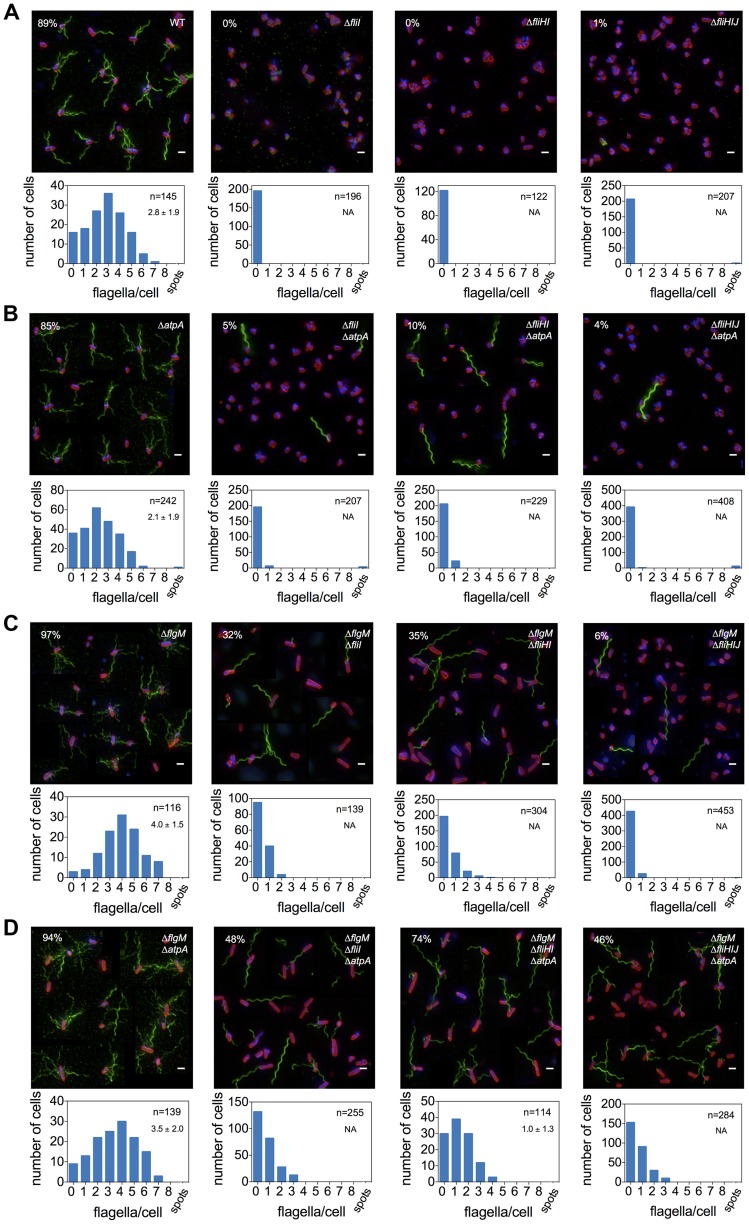
The type-III secretion apparatus is a PMF-powered protein exporter and we reasoned that increased PMF due to the atpA deletion provided additional energy to overcome the requirement of the flagellar ATPase complex for flagellar assembly. Flagellar filament assembly will only occur in the presence of a functional and efficiently working fT3SS. Deletion mutants of fliI, fliHI and fliHIJ were non-flagellated (Fig. 3A). The atpA deletion resulted in a small increase in the frequency of bacteria that produced functional flagella in a fliI and fliHIJ deletion strain up to approximately 4% of the population. However, the absence of atpA in a fliHI deletion mutant increased the frequency of flagella formation to about 10% of the bacteria (Fig. 3B). There appears to exist an important bottleneck in flagellar assembly that was overcome by the increase in PMF in the atpA deletion mutants. We observed formation of only one, singular filament in cases where flagellar assembly could proceed beyond completion of a hook-basal body complex. It has been shown previously that under wildtype conditions about 90% of detectable HBBs had a filament attached. Thus, virtually every secretion competent HBB had switched to late substrate secretion mode [32]. Since we observed only single flagellar filaments of wildtype lengths in our fliHI mutants (Fig. 3B and Fig. S6), it is a possibility that a localization mechanism exists that preferentially targets late flagellar substrates to a secretion system that is in late-type secretion mode.
Figure 3. Frequency of flagellar filament formation of fliHIJ mutants is increased in atpA and flgM null backgrounds.
The absence of the flagellar ATPase subunits FliH, FliI and FliJ results in a non-flagellated phenotype (A). Additional deletions in atpA (B) and flgM (C) substantially increase the frequency of flagellar filament formation of fliI, fliHI and fliHIJ mutant strains. Flagellar formation in the flgM null background is further enhanced by combination with the atpA mutation (D). Top: A montage of representative fluorescent microscopy images is shown. Flagellar filaments were stained using anti-FliC immunostaining and detected by FITC-coupled secondary antibodies (green), DNA was stained using Hoechst (blue) and cell membranes using FM-64 (red). Scale bar 2 µm. The percentage of cells with at least one filament is presented in the upper left corner. Bottom: Histogram of counted flagellar filaments per cell body. Number of counted cells and average number of filaments per cell +/− standard deviation based on Gaussian non-linear regression analysis is given in the upper right hand corner.
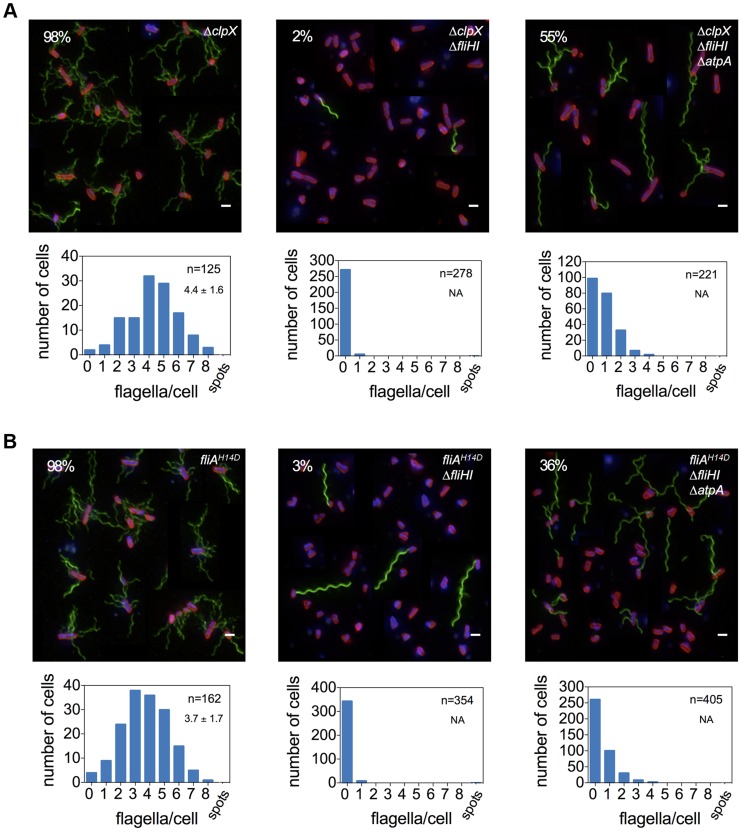
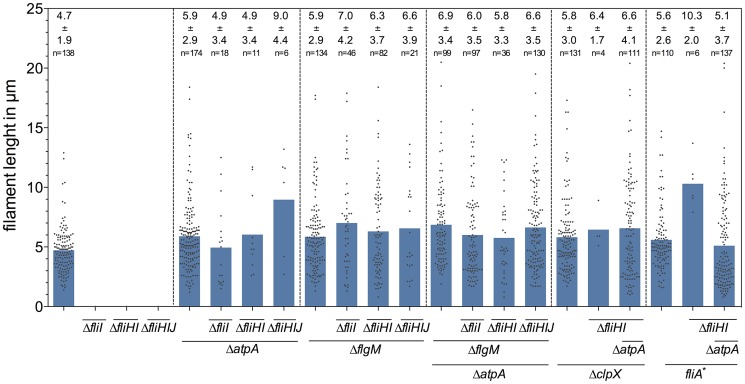
The cytoplasmic C-ring of the flagellum (composed of FliG, FliM, FliN) is essential for efficient localization of flagellar secretion substrates to the vicinity of the export gate by functioning as an affinity site for the localization of substrate/ATPase complexes. We have previously shown that a defect in the C-ring structure can be bypassed by providing an excess of secretion substrate [28]. Mutations in flgM, clpX and the fliA H14D (FlgM-bypass) allele result in increased secretion substrate levels. We tested the effects of increased substrate levels on secretion via the flagellar type-III secretion apparatus in the absence of the FliHIJ ATPase complex. In various fliHIJ mutant backgrounds null mutations of flgM, clpX or fliA H14D increased the frequency of bacteria able to produce at least one flagellum up to 35% of the population (Fig. 3C, Fig. 4). The deletion of flgM and atpA enhanced the frequency of flagellar assembly to a maximum of 74% of the population in the fliHI mutant background (Figs. 3C and 3D). The absence of the ATP synthase increased flagellar filament formation in the clpX or fliA H14D backgrounds to 55% and 36%, respectively (Figs. 4A and 4B). A significant increase in secreted flagellin was also observed in the flgM, clpX or fliA H14D mutant backgrounds under conditions when the PMF was apparently elevated by the atpA deletion (Fig. 5). We additionally measured the lengths of flagellar filaments visualized by flagellin immunostaining (Figs. 3 and 4) and observed that the average lengths of flagellar filaments of the various fliHIJ mutants is as long or longer than the wildtype filament lengths. The average lengths of filaments were also increased in an otherwise wildtype background if an excess of secretion substrates (e.g. by deleting the negative regulator flgM) was provided or the PMF was increased by deletion of the atpA subunit (Fig. 6).
Figure 4. Frequency of flagellar filament formation of a fliHI mutant strain is increased in clpX null and fliAH14D backgrounds.
A deletion in clpX (A) and the more stable fliAH14D variant (B) increase the frequency of flagellar filament formation in a fliHI mutant strain. Flagellar formation is further enhanced by combination with an atpA mutation. Top: A montage of representative fluorescent microscopy images is shown. Flagellar filaments were stained using anti-FliC immunostaining and detected by FITC-coupled secondary antibodies (green), DNA was stained using Hoechst (blue) and cell membranes using FM-64 (red). Scale bar 2 µm. The percentage of cells with at least one filament is presented in the upper left corner. Bottom: Histogram of counted flagellar filaments per cell body. Number of counted cells and average number of filaments per cell +/− standard deviation based on Gaussian non-linear regression analysis is given in the upper right hand corner.
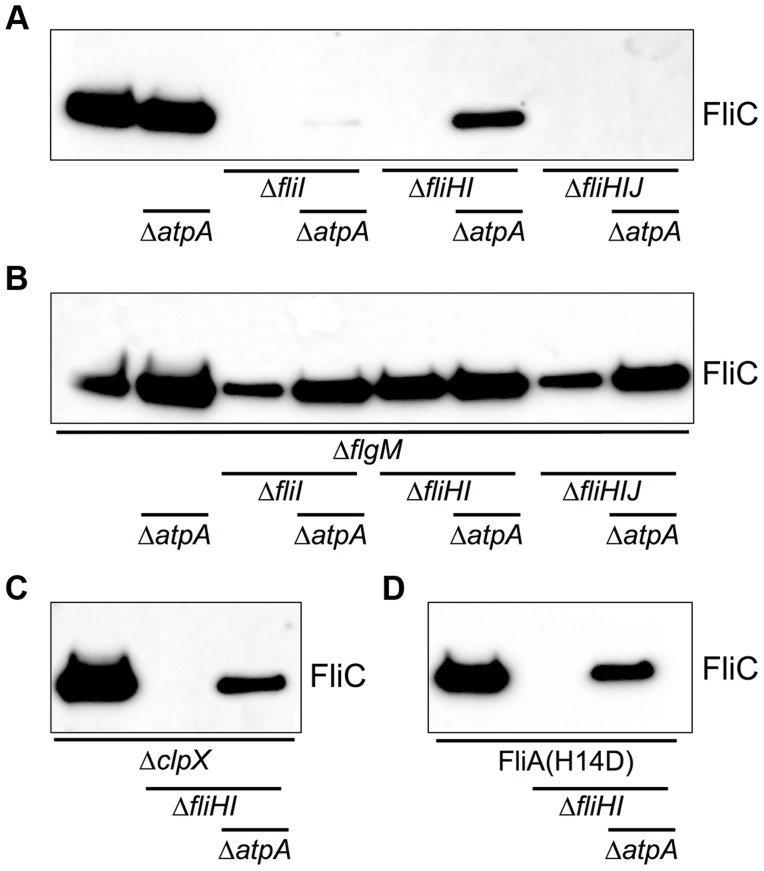
Figure 5. Flagellin protein secretion is restored in the absence of FliHIJ ATPase components by ΔatpA, ΔflgM, ΔclpX, and fliAH14D mutations.
Secreted FliC flagellin protein was analyzed by anti-FliC immunostaining in the FliC-phase locked wildtype and fliHIJ mutant strains. (A) Wildtype and fliHIJ mutants in combination with ΔatpA. (B) Wildtype and fliHIJ mutants in combination with ΔflgM and ΔatpA. (C) Wildtype and fliHI deletion mutant in combination with ΔclpX and ΔatpA. (D) Wildtype and fliHI deletion mutant in combination with fliAH14D and ΔatpA.
Figure 6. Lengths of flagellar filaments in fliHIJ mutants.
Plot showing the lengths of individual flagellar filaments of the fliHIJ mutants visualized by anti-FliC immunostaining in Figure 3 and Figure 4. The average lengths of flagellar filaments +/− standard deviation and the number of measured filaments are presented in the upper part of the graph.
In a complementary experiment, we tested the possibility that an increase in PMF (by deletion of atpA) or an excess of secretion substrates (by deletion of flgM) would allow for flagellar protein export in FlhA mutants where proton-flow through the export apparatus is blocked [33]. As shown in Fig. S6, no secretion of flagellin was detected under either condition, demonstrating that the proton flux through FlhA is essential for flagellar type-III secretion in contrast to ATP hydrolysis by the FliHIJ complex.
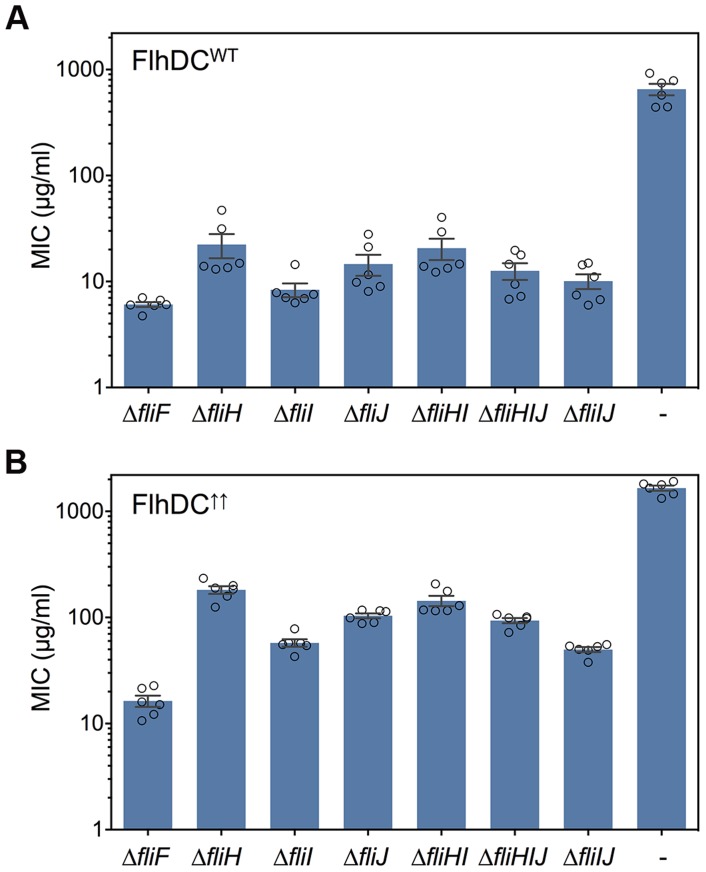
It has been suggested that the regulatory subunit of the ATPase complex, FliH, blocks the entrance gate of the secretion channel in the absence of its cognate FliI ATPase subunit [15], [16], [34]. In order to dissect the contribution of individual components of the FliHIJ complex to the type-III export process across the cytoplasmic membrane, we utilized a hook protein - β-lactamase based reporter system (FlgE-Bla). In the absence of the proximal rod subunits FlgB and FlgC, the FlgE-Bla fusion protein is secreted into the periplasm [28], [35], conferring quantifiable resistance to Ampicillin. We analyzed the export capability of various single and combination mutants of fliH, fliI and fliJ by determining the minimal inhibitory concentration (MIC) against Ampicillin. A deletion of the inner membrane scaffold protein, FliF, was used as a negative control and the MIC of the fliF mutant represents the basal level of Ampicillin resistance in this assay. Single deletion mutants of fliH and fliJ substantially increased the MIC about 3.5-fold compared to the fliF control strain (Fig. 7A). This is consistent with the previously reported leaky-motility phenotype of fliH and fliJ mutants [22], [26]. A deletion mutant of fliI also displayed increased MIC values, albeit significantly less than either fliH or fliJ null strains. Interestingly, if the fliI mutation was combined with either the fliH or fliHJ, the MIC values reached the level of the single fliH or fliJ null strains. Importantly, if only fliH was retained and both fliI and fliJ were deleted, the export capability was reduced to the level of the single fliI deletion. Under excess FlgE-Bla reporter construct conditions, secretion was also restored in the fliI single deletion mutant as MIC levels were 3.5-fold higher compared to the fliF control (Fig. 7B). The MIC values of the fliH and fliJ single mutants were 11 and 6.5-fold increased, respectively. Similar as under physiological FlgE-Bla substrate conditions, the MIC levels were increased 9 and 6-fold in fliHI or fliHIJ combinatory mutations to the levels of the single fliH and fliJ mutants. If only FliH was retained (ΔfliIJ), the MIC values were decreased to the level of the single fliI mutant. These results demonstrate that FliH functions as an inhibitor of the type-III export process. However, significant export can occur in the absence of both the negative regulator FliH and the ATPase FliI if excess substrate is provided, further adding evidence to our results that fT3SS can efficiently occur without ATP hydrolysis by FliI.
Figure 7. FliH functions as a negative regulator of type-III protein translocation.
Export of FlgE-Bla fusion protein into the periplasm was analyzed in various fliHIJ deletion strains. All strains additionally harbored a deletion of the proximal rod genes (ΔflgBC6557) and the FlgE-Bla fusion protein under its native promoter (flgE6569::bla). (A) Minimal inhibitory concentration (MIC) values with flagellar genes expressed at normal levels. (B) Summary of MIC values with flagellar genes expressed at elevated levels due to a PflhD promoter-up mutation (PflhD* = (P1+P4 -10 TATAAT)). The error bars represent the standard error of the mean (SEM) and biological replicates are shown as individual data points.
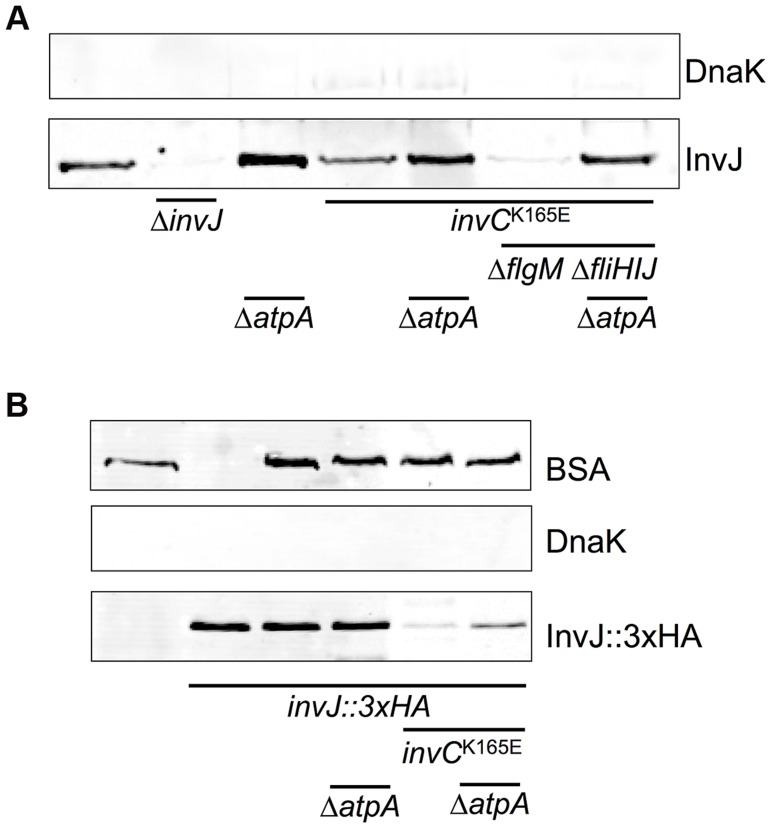
The ATPase InvC is associated with the vT3SS encoded by Spi1 and essential for Spi1 vT3SS function and virulence [17], [36]. Previously, loss-of-function mutations in InvC have been isolated that specifically target membrane association, oligomerization and catalytic activity of the ATPase [37]. Here, we employed the catalytically-inactive InvCK165E mutant to elucidate the role of ATPase activity in protein secretion via the Spi1 vT3SS. The InvCK165E mutant strain secreted significantly less InvJ substrate protein, however the defect in secretion was bypassed by a null mutation in the atpA subunit of ATP synthase (Fig. 8A). Importantly, the fT3SS-associated ATPase FliI did not complement InvC function, as a fliHIJ deletion in the InvCK165E ΔatpA mutant strain did not abolish InvJ secretion. Similar results were obtained using a 3×HA-tagged InvJ substrate protein (Fig. 8B).
Figure 8. Protein secretion via the vT3SS in a catalytically-inactive ATPase mutant strain is rescued by deletion of atpA.
Secretion of the Spi1 vT3SS substrate InvJ or a 3×HA tagged InvJ variant. (A) Secreted InvJ protein in the wildtype, ΔinvJ, ΔatpA, the catalytically-inactive Spi1 ATPase invCK165E, invCK165E ΔatpA, invCK165E ΔflgM, and invCK165E ΔflgM ΔfliHIJ ΔssaN ΔatpA mutant strains. Detection of DnaK protein was included as a cell lysis control. (B) Levels of secreted InvJ::3×HA protein in the wildtype, ΔinvJ::3×HA, ΔinvJ::3×HA ΔatpA, ΔinvJ::3×HA invCK165E, and ΔinvJ::3×HA invCK165E ΔatpA mutant strains. 288 ng BSA was added to each supernatant fractions (except for lane 2) and served as a precipitation control. DnaK protein served as a cell lysis control.
Discussion
The results demonstrate that ATPase activity is universally dispensable for protein export via bacterial type-III secretion systems. We propose that the function of the T3SS-associated ATPase complex is to deliver secretion substrates to the export apparatus and ensure efficient substrate unfolding, which was first reported by Akeda and Galán [17]. Either increased substrate production or increased PMF overcomes the requirement for the ATPase complex (Fig. S7). However, increased PMF or substrates do not bypass the requirement for proton flux through FlhA in contrast to the dispensability of ATP hydrolysis by the FliHIJ complex (Fig. S6).
In summary, our data demonstrate that efficient type-III protein export occurs in the absence of the ATP hydrolyzing components; therefore it is reasonable to assume that the export gate itself has a primary function in substrate unfolding and translocation. We speculate that the function of the ATPase complex in substrate unfolding and chaperone release is secondary to assure a highly efficient export process. Our data would suggest that chaperone release and unfolding occur inefficiently without ATPase activity, but is catalyzed by FliHIJ. Increasing PMF thus shifts the equilibrium of the spontaneous process.
The observation that export of a FlgE-Bla reporter construct was similarly increased in both the fliHI and fliJ mutant strains indicated that the FliHI and FliJ components of the ATPase complex acted in different pathways of the type-III export process. A possible scenario would be that FliJ – which interacts with FlhA of the export gate – activates the efficient Δψ-driven type-III protein export as suggested by Minamino et al. [21] (see also Fig. S7). FliH binds to both the ATPase FliI and the cytoplasmic C-ring, thereby localizing secretion substrates to the export gate for efficient unfolding by FliI prior to a Δψ or ΔpH-driven translocation process. In the absence of FliJ, the inner membrane export apparatus can still function as a less-efficient ΔpH-driven protein-proton antiporter and the secretion process is facilitated by substrate unfolding via the FliHI complex. The non-essential substrate unfolding is not occurring in a fliHI mutant strain, however the export gate operates in the highly-efficient Δψ export mode, which might be able to energize substrate unfolding as well.
The switch from early to late (flagellin-like) substrate secretion is a bottleneck in the process of flagellar bioassembly. The importance of this step in flagellar bioassembly was evidenced by the fact that we observed assembly of single flagellar filaments of wildtype lengths and longer in our fliHIJ deletion mutants (Fig. 6). It has been demonstrated before that the inner membrane ring complex, the C-ring and integral membrane components of the fT3SS assemble normally in the absence of the fliHIJ complex [38], [39]. Thus, the fliHIJ mutants analyzed here presumably assemble a physiological number of secretion system complexes. Only upon completion of the hook-basal-body structure, the secretion apparatus undergoes its secretion substrate specificity switch. In the absence of the FliHIJ complex, flagellar protein export is strongly impaired and thus HBB completion and the switch to filament-type secretion is a rare event. However, there appears to exist a mechanism for localization of the available late flagellar substrates to the single hook-basal-body that is in a late-export-competent state since we observed filament lengths that were as long or longer than the wildtype in our fliHIJ mutants (Fig. 6). An increase in PMF then likely assists in efficient export of late secretion substrates and thereby overcomes the requirement of the FliHIJ ATPase complex for late substrate secretion – similar to the related scenario where excess substrates bypass the requirement of the FliHIJ ATPase complex.
Our findings also have important implications for the evolution of the bacterial flagellum and type-III secretion systems. As mentioned above, the cytoplasmic components of the type-III secretion system share strong homology to components of the FOF1 ATP synthase and it has been proposed that the flagellum was derived from a proto FOF1 ATP synthase where ATP hydrolysis might have energized the export process. The present results would indicate that a proto F1-ATPase was added to a primordial type-III export system (which might have been proton-powered) with the evolutionary benefit of facilitating the export process. This made the bacterial type-III secretion apparatus to the highly efficient protein-export system of the contemporary flagellum and injectisome.
In summary, type-III protein export is a process of great complexity and involves a substantial number of checkpoints to ensure the correct order of export. A reasonable, general proposal for the export mechanism of bacterial type-III secretion systems is that the respective ATPase complexes bind to substrate proteins, shuttle the substrates to the base of the growing structure, energize chaperone release and substrate unfolding in an ATP-dependent manner and present the substrate to the membrane-embedded export apparatus components for efficient PMF-dependent secretion.
Materials and Methods
Bacterial strains, plasmids and media
All bacterial strains used in this study are listed in Table 1. Cells were grown in either lysogeny broth (LB) [40] or TB broth (1% Tryptone and 0.5% NaCl). Motility agar was prepared as described before [28]. The generalized transducing phage of Salmonella enterica servovar Typhimurium P22 HT105/1 int-201 was used in all transductional crosses [41].
Table 1. List of Salmonella enterica servovar Typhimurium LT2 strains used in this study.
| Strain number | Relevant genotype | Reference |
| TH437 | wildtype | J. Roth |
| TH6232 | Δhin-5717::FRT | [47] |
| TH8208 | Δhin-5717::FRT fliA*5225(H14D) | Lab collection |
| TH8419 | ΔinvJ::FKF | Lab collection |
| TH9949 | flgE6569::bla ΔflgBC655 | [35] |
| TH11801 | ΔatpA::tetRA | [15] |
| TH12465 | flgE6569::bla ΔflgBC655 ΔfliF7387 | This study |
| TH12472 | flgE6569::bla ΔflgBC655 ΔfliH7394 | This study |
| TH12473 | flgE6569::bla ΔflgBC655 ΔfliI7395 | This study |
| TH12474 | flgE6569::bla ΔflgBC655 ΔfliJ7396 | This study |
| TH12475 | flgE6569::bla ΔflgBC655 ΔfliHI7397 | This study |
| TH12476 | flgE6569::bla ΔflgBC655 ΔfliHIJ7398 | This study |
| TH12477 | flgE6569::bla ΔflgBC655 ΔfliIJ7399 | This study |
| TH13867 | Δhin-5717::FRT ΔfliI7364 | This study |
| TH13868 | Δhin-5717::FRT ΔfliHI7366 | This study |
| TH13869 | Δhin-5717::FRT ΔfliHIJ7367 | This study |
| TH14002 | Δhin-5717::FRT clpX1::Tn10dCm ΔfliHI7366 | This study |
| TH14009 | Δhin-5717::FRT ΔflgM5628::FRT ΔfliI7364 | This study |
| TH14010 | Δhin-5717::FRT ΔflgM5628::FRT ΔfliHI7366 | This study |
| TH14011 | Δhin-5717::FRT ΔflgM5628::FRT ΔfliHIJ7367 | This study |
| TH14017 | Δhin-5717::FRT fliA*5225 (H14D) | This study |
| TH14018 | Δhin-5717::FRT fliA*5225 (H14D) ΔfliHI7366 | This study |
| TH14019 | Δhin-5717::FRT fliA*5225 (H14D) ΔfliHIJ7367 | This study |
| TH14129 | Δhin-5717::FRT ΔfliHI7366 ΔclpX | This study |
| TH14130 | Δhin-5717::FRT clpX1::Tn10dCm ΔfliHI7366 atp (Mot+) | This study |
| TH14184 | ΔfliHI7366 Δhin-5717::FRT ΔclpX ΔatpA::tetRA | This study |
| TH14260 | ΔflgM5628::FRT ΔfliHI7366 Δhin-5717::FRT ΔatpA::tetRA | This study |
| TH14261 | fliA*5225 (H14D) ΔfliHI7366 Δhin-5717::FRT ΔatpA::tetRA | This study |
| TH14292 | ΔfliHI7366 Δhin-5717::FRT ΔatpA::tetRA | This study |
| TH14826 | Δhin-5717::FCF ΔflgM5628::FRT | This study |
| TH15426 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) | This study |
| TH15427 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) ΔfliF7387 | This study |
| TH15430 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) ΔfliI7395 | This study |
| TH15431 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) ΔfliHIJ7398 | This study |
| TH15925 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) ΔfliJ7396 | This study |
| TH16031 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) ΔfliH7394 | This study |
| TH16032 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) ΔfliHI7397 | This study |
| TH20154 | invC314 (K165E) | This study |
| TH20637 | invC314 ΔatpA::tetRA | This study |
| TH20708 | invC314 ΔflgM5628::FRT ΔfliHIJ7367 ΔssaN110 | This study |
| TH20709 | invC314 ΔflgM5628::FRT ΔssaN110 ΔfliHIJ7367 ΔatpA::tetRA | This study |
| TH20714 | invJ320::3×HA (SAGASA linker on both sides of 3×HA; between aa140 and 141) | This study |
| TH20750 | invJ320::3×HA ΔatpA::tetRA | This study |
| TH20830 | invC314 invJ321::3×HA (SAGASA linker on both sides of 3×HA; between aa140 and 141) | This study |
| TH20831 | invC314 invJ322::3×HA (SAGASA linker on both sides of 3×HA; between aa140 and 141) ΔflgM5628::FRT ΔfliHIJ7367 ΔssaN110 | This study |
| TH20832 | invC314 invJ321::3×HA ΔatpA::tetRA | This study |
| TH20833 | invC314 invJ322::3×HA ΔflgM5628::FRT ΔfliHIJ7367 ΔssaN110 ΔatpA::tetRA | This study |
| EM404 | Δhin-5717::FRT ΔflgM5628::FRT | This study |
| EM405 | Δhin-5717::FRT ΔatpA::tetRA | This study |
| EM406 | Δhin-5717::FRT ΔfliI7364 ΔatpA::tetRA | This study |
| EM407 | Δhin-5717::FRT ΔfliHIJ7367 ΔatpA::tetRA | This study |
| EM408 | Δhin-5717::FRT ΔflgM5628::FRT ΔfliI7364 ΔatpA::tetRA | This study |
| EM409 | Δhin-5717::FRT ΔflgM5628::FRT ΔfliHIJ7367 ΔatpA::tetRA | This study |
| EM415 | Δhin-5717::FRT ΔflgM5628::FRT ΔatpA::tetRA | This study |
| EM417 | Δhin-5717::FRT ΔclpX | This study |
| EM1236 | flgE6569::bla ΔflgBC655 PflhD7793 (P1+P4 -10 TATAAT) ΔfliIJ7399 | This study |
| EM1959 | Δhin-5717::FRT flhA22345 (D208A) ΔatpA::tetRA | This study |
| EM1960 | Δhin-5717::FRT flhA22346 (D208E) ΔatpA::tetRA | This study |
| EM1961 | Δhin-5717::FRT flhA22347 (D208K) ΔatpA::tetRA | This study |
| EM2037 | Δhin-5717::FRT flhA22345 (D208A) ΔflgM5628::FKF | This study |
| EM2038 | Δhin-5717::FRT flhA22346 (D208E) ΔflgM5628::FKF | This study |
| EM2039 | Δhin-5717::FRT flhA22347 (D208K) ΔflgM5628::FKF | This study |
Protein secretion assay
Secretion of FliC and InvJ into the culture supernatant was analyzed as described previously [32], [42]. For analysis of InvJ secretion, cultures were grown under Spi1-inducing conditions (high-osmolarity (0.3M NaCl, final) and low-oxygen (without agitation)) [43]. Secreted proteins were separated from the cellular fraction and precipitated using a final concentration of 10% Trichloroacetic acid (TCA). Protein levels of cellular and secreted fractions were analyzed by SDS polyacrylamide gel electrophoresis and immunoblotting. Antibodies against FliC (rabbit), InvJ (rabbit, kind gift of Sam Miller, University of Washington), hemaglutinin (mouse, Pierce), bovine serum albumin (mouse, Pierce) and DnaK (mouse, Abcam) were used for detection.
Minimal inhibitory concentration assay (FlgE-Bla secretion)
The minimal inhibitory concentration against ampicillin resulting from the export of hook-β-lactamase (FlgE-Bla) fusion proteins was analyzed as described by Lee et al. [35] with minor modifications. Briefly, cultures were grown at 37°C in a 96-well plates in LB media to mid-log phase. An aliquot was transferred in LB media supplemented with increasing concentrations of fresh ampicillin. Cells were grown for 4.5 hours at 37°C in a shaking incubator before absorbance was measured at OD600. The minimal inhibitory concentration values were determined as follows using R for Mac OS X and a modified version of package ‘stinepack’. Relative growth values were calculated by normalization of the measured OD600 values of any ampicillin (Amp) concentration to the zero Amp control. Next, a Stineman interpolation was used for a smooth curve fit of the relative growth values and root finding was performed to determine the MIC by calculating the interception with a threshold set at 5% of residual growth of the zero Amp control.
Fluorescent microscopy
Fluorescent microscopy analysis was performed as described before [32]. Briefly, logarithmically growing bacteria were applied to a poly-L-lysine treated coverslip well. Bacterial cells were fixed by addition of final 2% formaldehyde and 0.2% glutaraldehyde. Flagella were stained using polyclonal anti-FliC antibodies (rabbit) and anti-rabbit conjugated Alexa Fluor 488 secondary antibodies (Invitrogen). DNA staining was performed using Hoechst (Invitrogen) and cell membranes were stained using FM-64 (0.5 mg ml−1, Invitrogen). Images were collected with optical Z sections every 100 nm using an Applied Precision optical sectioning microscope and deconvolved using softWoRx v.3.4.2 (Applied Precision). The pixel data of individual Z sections of the deconvolved images were projected on a single plane using the Z-project tool (settings: maximal intensity) of ImageJ v.1.6.0 [44].
Free swimming assay
Bacterial cultures were grown in TB media at 37°C and aliquots were mixed with TB media containing 10% (final) Ficoll 400 (Sigma) and applied to a microscopic chamber consisting of a microscope slide and coverslip spaced by two strips of double sided adhesive tape for free swimming speed analysis. Free-swimming bacterial cells were observed close to the coverslip surface and at least two 30-second movies from different field of views were recorded. Movies were imported into ImageJ v.1.6.0 and the trajectories of several hundred cells were determined by the “2D ParticleTracker” ImageJ plug-in (http://www.mosaic.ethz.ch/Downloads/ParticleTracker, [45]). The median velocities of at least 100 cells were calculated using a custom-made [R] script as described by [46].
Supporting Information
Mutations in atp locus restore motility of a fliHI mutant strain. (A) Representative soft agar motility plate of motility suppressors of the non-motile fliHI mutant. Exemplary motility swarms of the wildtype TH6232 (WT), the parental fliHI strains TH13868 (ΔfliHI) and TH14002 (ΔfliHI clpX), and the originally isolated suppressor mutant in the atp locus TH14130 (ΔfliHI clpX atp) are shown. The mutation in the atp locus restored motility of the fliHI deletion strain. Motility plates were incubated at 37°C for 4.5 hours before imaging. The clpX mutation alone does not increase motility after 4.5 hours incubation but note that the original suppressor mutants in clpXP were isolated after overnight incubation. (B) Quantified relative motility of the fliHIJ suppressor strains. The diameter of the motility swarm relative to the wildtype was measured after 4.5 hours incubation. Biological replicates are shown as individual data points.
(TIFF)
Quantification of motility of fliHIJ mutant strains. Representative images used for quantification of the swimming motility assay shown in Figure 2B. Swimming motility plates containing 0.3% agar were incubated for 4.5 hours at 37°C before imaging.
(TIFF)
Growth curves of the wildtype and ΔatpA mutant. Growth of the wildtype strain TH437 (WT) and ΔatpA strain TH11801 (ΔatpA) in lysogeny broth (A) and tryptone broth (B). An overnight culture was diluted 1∶100 in LB or TB medium in a 96-well plate and incubated at 37°C. Growth was monitored by measuring the OD600 every 15 minutes.
(TIFF)
Swimming speed of the ΔatpA mutant strain. Velocities of individual cells of the wildtype (TH6232) and the ΔatpA mutant strain (EM405) were determined in 30 min intervals throughout the growth curve starting at an optical density of approximately 0.4 (time = 0). Velocities of at least 100 individual cells are depicted as box diagrams with whiskers according to the Tukey method. The increase of the optical density (OD600) of both wildtype and ΔatpA mutant is shown in the inlet.
(TIFF)
Induction of PMF-draining TetA tetracycline/proton antiporter suppresses the restored motility of fliHIJ ΔatpA mutant strains. (A) Quantified relative motility of the wildtype (TH6232) and various fliHIJ atpA mutant strains ΔfliI (TH13867), ΔfliI ΔatpA (EM406), ΔfliHI (TH13868), ΔfliHI ΔatpA (TH14292), ΔfliHIJ (TH13869) and ΔfliHIJ ΔatpA (EM407) in the presence of the tetA inducer anhydrotetracycline. The atpA gene in strains EM406, TH14292 and EM407 was deleted using a tetRA resistance cassette. Expression of tetA was induced by addition of 1 µg/ml anhydrotetracycline (AnTc). Biological replicates are shown as individual data points. Data were analyzed by the Student's t test. Stars indicate significantly different motility (ns, non significant). (B) Representative soft agar motility plates after 4.5 hours incubation at 37°C in the presence of anhydrotetracycline.
(TIFF)
Mutations in ΔatpA or ΔflgM do not rescue flagellar protein export of FlhA Asp-208 mutants. Secreted FliC flagellin protein was analyzed by anti-FliC immunostaining in FliC-phase locked strains. The charged residue Asp-208 of FlhA has been previously implicated in proton flow through the export apparatus [33]. Increased levels of flagellar substrates and PMF were provided by a deletion of atpA or flgM, respectively. (A) Secretion of FliC under increased PMF conditions. Strains harbored a ΔatpA deletion and the wildtype flhA allele (EM405) or the flhA point mutations D208A (EM1959), D208E (EM1960) and D208K (EM1961). (B) Secretion of FliC under elevated substrate conditions. Strains harbored a ΔflgM deletion and the wildtype flhA allele (TH14826) or the flhA point mutations D208A (EM2037), D208E (EM2038) and D208K (EM2039).
(TIFF)
Model for the flagellar type-III protein export process. A schematic model of the flagellar type-III protein secretion process is presented as described in the text. The FliJ component of the ATPase complex interacts with FlhA of the membrane-embedded export apparatus and activates the efficient Δψ-driven type-III protein export [21]. Upper part: Under wildtype conditions, the FliHIJ ATPase complex binds to substrate proteins, shuttles the substrates to the base of the export apparatus, energizes chaperone release and substrate unfolding in an ATP-dependent manner and presents the substrate to the membrane-embedded export apparatus components for efficient proton motive force (PMF)-dependent secretion. The Δψ component of the PMF is utilized for the export process. Middle part of the figure: in a ΔfliH mutant, localization of secretion substrates to the export gate by binding of FliH to the C-ring is prevented. In a fliHI mutant strain, the non-essential substrate unfolding is not occurring. However, the membrane-embedded export apparatus operates in the highly-efficient Δψ export mode due to the presence of FliJ. Lower part of the figure: In the absence of FliJ (ΔfliIJ or ΔfliJ), the membrane components of the export apparatus can still function as a less-efficient ΔpH-driven protein-proton antiporter and the secretion process is facilitated by substrate unfolding via the FliHI complex in the ΔfliJ mutant strain.
(TIFF)
Acknowledgments
We are grateful to Nadine Körner (Helmholtz Centre for Infection Research, Braunschweig) for expert technical assistance, Takanori Hirano (University of Florida, Gainesville) for help in strain constructions, Alexander Böhm (LOEWE Center for Synthetic Microbiology, Marburg) for providing the [R] script to determine free swimming velocities, Kota Miura (European Molecular Biology Laboratory, Heidelberg) for the help with the ImageJ ParticleTracker 2D plugin and Tomas Johannesson (Icelandic Meteorological Office, Reykjavik, Iceland) and Dorothea Lindtke (University of Fribourg, Switzerland) for help with implementation of the [R] stinepack software for MIC determination. We thank members of the Hughes and Erhardt labs for useful discussions of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Swiss National Science Foundation grant FN-7626 subside n° 31003A_132947/1 and National Institute of Health grant GM056141 (to KTH) and Helmholtz Association Young Investigator grant n° VH-NG-932 and the People Programme (Marie Curie Actions) of the European Unions's Seventh Framework Programme grant n° 334030 (to ME). FDF acknowledges support by the President's Initiative and Networking Funds of the Helmholtz Association of German Research Centers (HGF) under contract number VH-GS-202. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Berg HC, Anderson RA (1973) Bacteria swim by rotating their flagellar filaments. Nature 245: 380–382. [DOI] [PubMed] [Google Scholar]
- 2. Chevance FFV, Hughes KT (2008) Coordinating assembly of a bacterial macromolecular machine. Nature Reviews Microbiology 6: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kearns DB (2010) A field guide to bacterial swarming motility. Nature Reviews Microbiology 8: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duan Q, Zhou M, Zhu L, Zhu G (2013) Flagella and bacterial pathogenicity. Journal of Basic Microbiology 53: 1–8.. [DOI] [PubMed] [Google Scholar]
- 5. Cornelis GR (2006) The type III secretion injectisome. Nature Reviews Microbiology 4: 811–825. [DOI] [PubMed] [Google Scholar]
- 6. Macnab RM (2004) Type III flagellar protein export and flagellar assembly. Biochimica et Biophysica Acta 1694: 207–217. [DOI] [PubMed] [Google Scholar]
- 7. Thomas DR, Francis NR, Xu C, DeRosier DJ (2006) The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. Journal of Bacteriology 188: 7039–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan F, Ohnishi K, Francis NR, Macnab RM (1997) The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Molecular Microbiology 26: 1035–1046. [DOI] [PubMed] [Google Scholar]
- 9. Claret L, Calder SR, Higgins M, Hughes C (2003) Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Molecular Microbiology 48: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Minamino T, Macnab RM (2000) FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Molecular Microbiology 37: 1494–1503. [DOI] [PubMed] [Google Scholar]
- 11. Gonzalez-Pedrajo B, Fraser GM, Minamino T, Macnab RM (2002) Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Molecular Microbiology 45: 967–982. [DOI] [PubMed] [Google Scholar]
- 12. Ibuki T, Imada K, Minamino T, Kato T, Miyata T, et al. (2011) Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nature Structural & Molecular Biology 18: 277–282. [DOI] [PubMed] [Google Scholar]
- 13. Imada K, Minamino T, Tahara A, Namba K (2007) Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proceedings of the National Academy of Sciences of the United States of America 104: 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pallen MJ, Bailey CM, Beatson SA (2006) Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases. Protein Sci 15: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT (2008) Energy source of flagellar type III secretion. Nature 451: 489–492. [DOI] [PubMed] [Google Scholar]
- 16. Minamino T, Namba K (2008) Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451: 485. [DOI] [PubMed] [Google Scholar]
- 17. Akeda Y, Galan JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911–915. [DOI] [PubMed] [Google Scholar]
- 18. González-Pedrajo B, Minamino T, Kihara M, Namba K (2006) Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Molecular Microbiology 60: 984–998. [DOI] [PubMed] [Google Scholar]
- 19. McMurry JL, Murphy JW, González-Pedrajo B (2006) The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45: 11790–11798. [DOI] [PubMed] [Google Scholar]
- 20. Bange G, Kümmerer N, Engel C, Bozkurt G, Wild K, et al. (2010) FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proceedings of the National Academy of Sciences of the United States of America 107: 11295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minamino T, Morimoto YV, Hara N, Namba K (2011) An energy transduction mechanism used in bacterial flagellar type III protein export. Nature communications 2: 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Minamino T, Chu R, Yamaguchi S, Macnab RM (2000) Role of FliJ in flagellar protein export in Salmonella . Journal of Bacteriology 182: 4207–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evans LDB, Stafford GP, Ahmed S, Fraser GM, Hughes C (2006) An escort mechanism for cycling of export chaperones during flagellum assembly. Proceedings of the National Academy of Sciences of the United States of America 103: 17474–17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, et al. (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science (New York, NY) 280: 602–605. [DOI] [PubMed] [Google Scholar]
- 25. Eichelberg K, Ginocchio CC, Galan JE (1994) Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. Journal of Bacteriology 176: 4501–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minamino T, Gonzalez-Pedrajo B, Kihara M, Namba K, Macnab RM (2003) The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. Journal of Bacteriology 185: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomoyasu T, Takaya A, Isogai E, Yamamoto T (2003) Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Molecular Microbiology 48: 443–452. [DOI] [PubMed] [Google Scholar]
- 28. Erhardt M, Hughes KT (2010) C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Molecular Microbiology 75: 376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jensen PR, Michelsen O (1992) Carbon and energy metabolism of atp mutants of Escherichia coli . Journal of Bacteriology 174: 7635–7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koch-Koerfges A, Kabus A, Ochrombel I, Marin K, Bott M (2012) Physiology and global gene expression of a Corynebacterium glutamicum ΔF(1)F(O)-ATP synthase mutant devoid of oxidative phosphorylation. Biochimica et Biophysica Acta 1817: 370–380. [DOI] [PubMed] [Google Scholar]
- 31. Martinez-Argudo I, Veenendaal AKJ, Liu X, Roehrich AD, Ronessen MC, et al. (2013) Isolation of Salmonella mutants resistant to the inhibitory effect of Salicylidene acylhydrazides on flagella-mediated motility. PLoS ONE 8: e52179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT (2011) An infrequent molecular ruler controls flagellar hook length in Salmonella enterica . The EMBO Journal 30: 2948–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hara N, Namba K, Minamino T (2011) Genetic Characterization of Conserved Charged Residues in the Bacterial Flagellar Type III Export Protein FlhA. PLoS ONE 6: e22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galán JE (2008) Energizing type III secretion machines: what is the fuel? Nature Structural & Molecular Biology. pp. 127–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee HJ, Hughes KT (2006) Posttranscriptional control of the Salmonella enterica flagellar hook protein FlgE. Journal of Bacteriology 188: 3308–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galan JE (2001) Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol 17: 53–86. [DOI] [PubMed] [Google Scholar]
- 37. Akeda Y, Galan JE (2004) Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. Journal of Bacteriology 186: 2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S (1992) Morphological pathway of flagellar assembly in Salmonella typhimurium . Journal of Molecular Biology 226: 433–446. [DOI] [PubMed] [Google Scholar]
- 39. Morimoto YV, Ito M, Hiraoka KD, Che YS, Bai F, et al. (2014) Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol 91: 1214–1226. [DOI] [PubMed] [Google Scholar]
- 40. Bertani G (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. Journal of Bacteriology 186: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanderson KE, Roth JR (1983) Linkage map of Salmonella typhimurium, Edition VI. Microbiol Rev 47: 410–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singer HM, Erhardt M, Steiner AM, Zhang M-M, Yoshikami D, et al. (2012) Selective purification of recombinant neuroactive peptides using the flagellar type III secretion system. mBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bajaj V, Lucas RL, Hwang C, Lee CA (1996) Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Molecular Microbiology 22: 703–714. [DOI] [PubMed] [Google Scholar]
- 44. Abramoff M, Magelhaes PJ, Ram SJ (2004) Image Processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- 45. Sbalzarini IF, Koumoutsakos P (2005) Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol 151: 182–195. [DOI] [PubMed] [Google Scholar]
- 46. Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, et al. (2010) Second Messenger-Mediated Adjustment of Bacterial Swimming Velocity. Cell 141: 107–116. [DOI] [PubMed] [Google Scholar]
- 47. Bonifield HR, Hughes KT (2003) Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. Journal of Bacteriology 185: 3567–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutations in atp locus restore motility of a fliHI mutant strain. (A) Representative soft agar motility plate of motility suppressors of the non-motile fliHI mutant. Exemplary motility swarms of the wildtype TH6232 (WT), the parental fliHI strains TH13868 (ΔfliHI) and TH14002 (ΔfliHI clpX), and the originally isolated suppressor mutant in the atp locus TH14130 (ΔfliHI clpX atp) are shown. The mutation in the atp locus restored motility of the fliHI deletion strain. Motility plates were incubated at 37°C for 4.5 hours before imaging. The clpX mutation alone does not increase motility after 4.5 hours incubation but note that the original suppressor mutants in clpXP were isolated after overnight incubation. (B) Quantified relative motility of the fliHIJ suppressor strains. The diameter of the motility swarm relative to the wildtype was measured after 4.5 hours incubation. Biological replicates are shown as individual data points.
(TIFF)
Quantification of motility of fliHIJ mutant strains. Representative images used for quantification of the swimming motility assay shown in Figure 2B. Swimming motility plates containing 0.3% agar were incubated for 4.5 hours at 37°C before imaging.
(TIFF)
Growth curves of the wildtype and ΔatpA mutant. Growth of the wildtype strain TH437 (WT) and ΔatpA strain TH11801 (ΔatpA) in lysogeny broth (A) and tryptone broth (B). An overnight culture was diluted 1∶100 in LB or TB medium in a 96-well plate and incubated at 37°C. Growth was monitored by measuring the OD600 every 15 minutes.
(TIFF)
Swimming speed of the ΔatpA mutant strain. Velocities of individual cells of the wildtype (TH6232) and the ΔatpA mutant strain (EM405) were determined in 30 min intervals throughout the growth curve starting at an optical density of approximately 0.4 (time = 0). Velocities of at least 100 individual cells are depicted as box diagrams with whiskers according to the Tukey method. The increase of the optical density (OD600) of both wildtype and ΔatpA mutant is shown in the inlet.
(TIFF)
Induction of PMF-draining TetA tetracycline/proton antiporter suppresses the restored motility of fliHIJ ΔatpA mutant strains. (A) Quantified relative motility of the wildtype (TH6232) and various fliHIJ atpA mutant strains ΔfliI (TH13867), ΔfliI ΔatpA (EM406), ΔfliHI (TH13868), ΔfliHI ΔatpA (TH14292), ΔfliHIJ (TH13869) and ΔfliHIJ ΔatpA (EM407) in the presence of the tetA inducer anhydrotetracycline. The atpA gene in strains EM406, TH14292 and EM407 was deleted using a tetRA resistance cassette. Expression of tetA was induced by addition of 1 µg/ml anhydrotetracycline (AnTc). Biological replicates are shown as individual data points. Data were analyzed by the Student's t test. Stars indicate significantly different motility (ns, non significant). (B) Representative soft agar motility plates after 4.5 hours incubation at 37°C in the presence of anhydrotetracycline.
(TIFF)
Mutations in ΔatpA or ΔflgM do not rescue flagellar protein export of FlhA Asp-208 mutants. Secreted FliC flagellin protein was analyzed by anti-FliC immunostaining in FliC-phase locked strains. The charged residue Asp-208 of FlhA has been previously implicated in proton flow through the export apparatus [33]. Increased levels of flagellar substrates and PMF were provided by a deletion of atpA or flgM, respectively. (A) Secretion of FliC under increased PMF conditions. Strains harbored a ΔatpA deletion and the wildtype flhA allele (EM405) or the flhA point mutations D208A (EM1959), D208E (EM1960) and D208K (EM1961). (B) Secretion of FliC under elevated substrate conditions. Strains harbored a ΔflgM deletion and the wildtype flhA allele (TH14826) or the flhA point mutations D208A (EM2037), D208E (EM2038) and D208K (EM2039).
(TIFF)
Model for the flagellar type-III protein export process. A schematic model of the flagellar type-III protein secretion process is presented as described in the text. The FliJ component of the ATPase complex interacts with FlhA of the membrane-embedded export apparatus and activates the efficient Δψ-driven type-III protein export [21]. Upper part: Under wildtype conditions, the FliHIJ ATPase complex binds to substrate proteins, shuttles the substrates to the base of the export apparatus, energizes chaperone release and substrate unfolding in an ATP-dependent manner and presents the substrate to the membrane-embedded export apparatus components for efficient proton motive force (PMF)-dependent secretion. The Δψ component of the PMF is utilized for the export process. Middle part of the figure: in a ΔfliH mutant, localization of secretion substrates to the export gate by binding of FliH to the C-ring is prevented. In a fliHI mutant strain, the non-essential substrate unfolding is not occurring. However, the membrane-embedded export apparatus operates in the highly-efficient Δψ export mode due to the presence of FliJ. Lower part of the figure: In the absence of FliJ (ΔfliIJ or ΔfliJ), the membrane components of the export apparatus can still function as a less-efficient ΔpH-driven protein-proton antiporter and the secretion process is facilitated by substrate unfolding via the FliHI complex in the ΔfliJ mutant strain.
(TIFF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.