Abstract
Genetic studies have suggested a functional link between cholesterol/sphingolipid metabolism and endocytic membrane traffic. Here we show that perturbing the cholesterol/sphingomyelin balance in the plasma membrane results in the massive formation of clusters of narrow endocytic tubular invaginations positive for N-BAR proteins. These tubules are intensely positive for sphingosine kinase 1 (SPHK1). SPHK1 is also targeted to physiologically occurring early endocytic intermediates, and is highly enriched in nerve terminals, cellular compartments specialized for exo-endocytosis. Membrane recruitment of SPHK1 involves a direct, curvature-sensitive interaction with the lipid bilayer mediated by a hydrophobic patch on the enzyme’s surface. The knockdown of SPHKs results in endocytic recycling defects, and a mutation that disrupts the hydrophobic patch of C. elegans SPHK fails to rescue the neurotransmission defects in loss-of-function mutants of this enzyme. Our studies support a role of sphingosine phosphorylation in endocytic membrane trafficking beyond the established function of sphingosine-1-phosphate in intercellular signaling.
Introduction
Endocytosis is a highly regulated membrane remodeling process that critically depends on the coordination of endocytic factors, their interaction with membrane lipids and metabolic changes in the bilayer. In particular, a proper balance between sterols and sphingolipids within the plasma membrane is required to maintain plasma membrane homeostasis and to support endocytosis. Sterols, which are intercalated among the acyl chains of phospholipids, help modulate membrane rigidity. Sphingolipids, a class of lipids that use sphingoid base as the backbone and that interact with sterols, also serve as structural components of membranes and help modulate their physical properties1, 2.
The involvement of sterols and sphingolipids in normal endocytic membrane trafficking is supported by many lines of evidence. Pharmacological perturbations of cholesterol by sequestration with filipin, or extraction with methyl-β–cyclodextrin (MβCD), impair several endocytic pathways3–5. In yeast, genetic perturbations of the metabolism of ergosterol, the yeast sterol, impair endocytosis6. Cholesterol has also been implicated in synaptic vesicle recycling7, 8, a process that critically relies on clathrin-mediated endocytosis.
Similarly, genetic studies revealed that sphingoid bases are required for endocytosis in yeast6, and mutations in enzymes of sphingolipid metabolism produce endocytic membrane traffic defects in metazoa. For instance, mutations in neutral ceramidase and in sphingosine kinase are responsible for defects in synaptic vesicle recycling at the neuromuscular junction in D. melanogaster9 and C. elegans10, 11, respectively. In addition, enzymes that regulate ceramide and sphingoid base homeostasis in flies are critical for the endocytic trafficking of rhodopsin and their absence results in the degeneration of photoreceptor cells12, 13.
Beyond a generic impact of perturbations of cholesterol and sphingolipid metabolism on endocytosis, a more specific genetic connection emerged in yeast between these lipids and the endocytic N-BAR domain-containing proteins, RVS161 and RVS167. N-BAR domains are membrane-binding modules that can generate and sense bilayer curvature14–16. RVS161 and RVS167, and their metazoan homologues amphiphysin and endophilin, coordinate constriction of endocytic necks with other events required for the endocytic reaction17–21. Forward genetic screens for suppressors of rvs161 or rvs167 mutants identified mutations in enzymes of sphingolipid metabolism22, 23. Subsequently, large-scale epistatic miniarray profile (E-MAP) studies extended these results by identifying additional enzymes of both sphingolipid and ergosterol metabolism that genetically interact with N-BAR proteins24.
Goal of this study was to shed lights on the mechanisms underlying the genetic link between sterol and sphingolipid metabolism and endocytic membrane traffic. We started by exploring the impact on N-BAR proteins of perturbing the cholesterol/sphingomyelin balance in the plasma membrane. These manipulations resulted in a robust redistribution of N-BAR proteins that correlates with the formation of massive narrow tubular plasma membrane invaginations. Strikingly, an enzyme of sphingolipid metabolism, sphingosine kinase 1 (SPHK1), was enriched on these membranes via a direct, curvature-sensitive interaction. We further observed that SPHK1 is enriched on physiologically occurring early endocytic intermediates and that defective SPHK function impairs endocytic recycling, pointing to a role of sphingosine phosphorylation in endocytosis.
Results
Acute perturbation of cholesterol or sphingomyelin induces massive N-BAR protein-positive endocytic tubular invaginations
Acute cholesterol extraction from cells with methyl-β-cyclodextrin (MβCD) results in the perturbation of clathrin-mediated endocytosis accompanied by formation of shallow clathrin-coated pits3, 4. To monitor the dynamics of this effect, we examined live cells expressing GFP-clathrin light chain (CLC) and endophilin 2-Ruby by TIRF microscopy (Fig. 1a–c). Endophilin is an endocytic adaptor recruited at the necks of late stage endocytic clathrin-coated pits, where it coordinates acquisition of bilayer curvature (via its BAR domain) with the recruitment of dynamin and synaptojanin (via its SH3 domain)18–20, two factors required for fission and uncoating respectively.
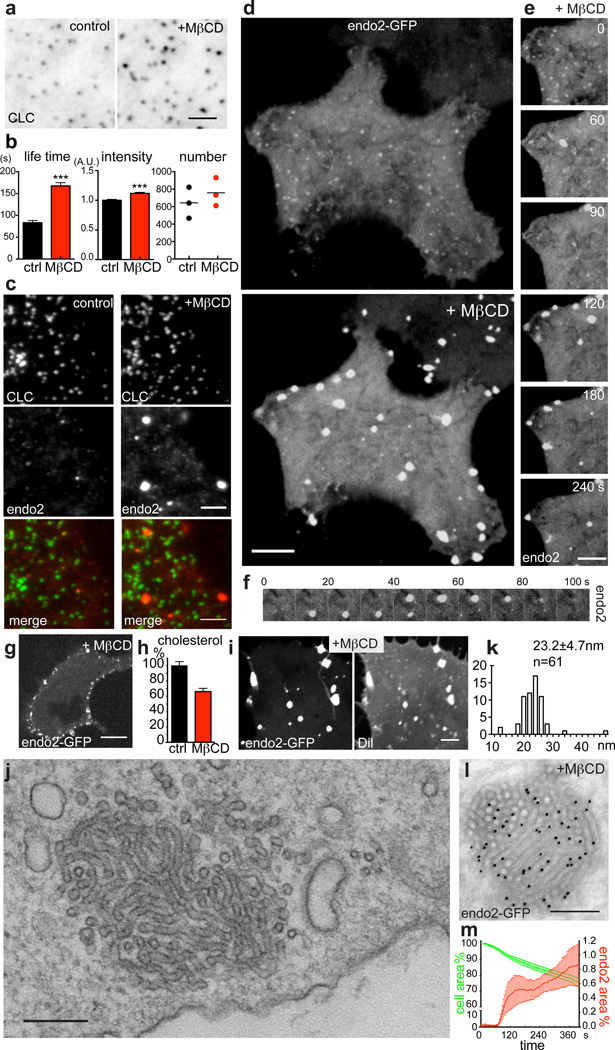
Figure 1. Acute perturbation of plasma membrane cholesterol induces massive endocytic tubular invaginations positive for N-BAR proteins.
a. TIRF images of a COS-7 cell expressing GFP-clathrin light chain (CLC) before (“control”, left) and after (“+MβCD”, right) 10 mM MβCD treatment at 37 °C. b. Statistical analysis of tracked clathrin-coated pits before (ctrl) and after (+MβCD) MβCD treatment (pooled results from three independent experiments). Mean lifetime: n=158 (ctrl) and 211 (+MβCD) pits. Mean fluorescence intensity: n=330 (ctrl) and 270 (+MβCD) pits. Error bars: standard error of the mean. [***] P < 0.0001, Student's t-test. Mean clathrin-coated pits number: n=3 (ctrl) and 3 (+MβCD) cells. c. TIRF images of a COS-7 cell expressing GFP-clathrin light chain and endophilin-2 Ruby before (“control”, left) and after (“+MβCD”, right) 10 mM MβCD treatment at 37 °C. d. Confocal images of HeLa cells expressing endophilin 2-GFP before (top) and after (bottom) MβCD treatment (bottom). e and f. Selected frames from a time series of the endophilin 2-GFP fluorescence from the cell shown in d. g. Confocal image of a middle section of a HeLa cell expressing endophilin 2-GFP after MβCD treatment. h. Measurement of cellular free cholesterol before and after MβCD treatment. n = 6 dishes Data are pooled from two independent experiments. Error bars: standard error of the mean. i. DiI staining of a cell expressing endophilin 2-GFP and treated with MβCD. j. Transmission electron microscopy image of a tubular membrane cluster formed during MβCD treatment. k. Histogram of tubule diameter. n=61 tubules. l. Anti-GFP immunogold labeling of a cell expressing endophilin 2-GFP and treated with MβCD. m. Average change of the footprint of cells (green) and of the percent areas (relative to the original footprints) occupied by endophilin 2 foci (red) during MβCD treatment. n = 7 cells. Data are pooled from 7 independent experiments. Error bars: standard error of the mean. Scale bar: 3 µm in a, c, and e; 5 µm in i; 10 µm in d and g; 200 nm in j and l.
Within minutes of MβCD treatment, a slower dynamics of the GFP-CLC spots (coated pits) was observed, consistent with a delay in the maturation of the pits (Fig. 1b). A corresponding modest increase in pit number was also detected (Fig. 1b). Surprisingly, a dramatic change in the localization and dynamics of endophilin 2-Ruby was also noted. In control cells, endophilin 2-Ruby accumulated transiently at hot spots (Fig. 1c, left), many of which represented late stage clathrin-coated pits. Within minutes from MβCD addition, endophilin 2-Ruby relocalized and clustered at multiple large foci that were up to one µm in size and did not coincide with clathrin-spots (Fig. 1c, right). Similar results were observed with endophilin 2-GFP (Fig. 1d and Supplementary movie S1). These foci appeared and disappeared in a dynamic fashion within minutes (Fig. 1e and f), but the rate of their formation progressively decayed until no new ones formed. The N-BAR domain of endophilin 2 fused to GFP (endo2ΔSH3-GFP) had a similar response to MβCD (Supplementary Fig. 1a), indicating that the SH3 domain is dispensable for this localization. Confocal images of middle sections of MβCD-treated cells revealed that large endophilin foci occurred exclusively near the cortical region (Fig. 1g). Analysis of cellular free cholesterol at 5 min after MβCD addition, i.e., when the majority of the endophilin foci had formed, showed an approximately 30% reduction (Fig. 1h), in agreement with previous reports25.
These effects of MβCD were robustly observed in multiple cell lines (Supplementary Fig. 1b and c) and were not an artifact due to endophilin overexpression, because endogenous endophilin 2 immunoreactivity underwent a similar redistribution (Supplementary Fig. 1d). An N-BAR domain-containing protein similar to endophilin, amphiphysin 2-GFP, co-clustered with endophilin 2-Ruby (Supplementary Fig. 1e) and accumulated into similar foci when expressed in endophilin null cells [mouse embryonic fibroblasts (MEFs) from endophilin triple KO mice20] (Supplementary Fig. 1f). Thus, endophilin marks the formation of some structures induced by cholesterol depletion, but is not needed for their formation.
MβCD extracts fatty acids and amphipathic molecules from the cells in addition to cholesterol26, and thus could cause effects independent of cholesterol. This possibility was excluded, as cholesterol add-back manipulations using cholesterol-MβCD complexes rescued the phenotype (Supplementary Fig. 1g). Moreover, a milder cholesterol extraction reagent, hydroxypropyl-β-cyclodextrin (HPβCD), resulted in a similar redistribution of endophilin (Supplementary Fig. 1h).
When the plasma membrane was prelabeled with the DiI lipid dye before MβCD treatment, the large endophilin foci were strongly positive for this dye (Fig. 1i), suggesting that they are sites of plasma membrane invaginations. Endophilin foci were also positive for chemical (TopFluor-PS)27 and genetically encoded (C2 domain of lactadherin–GFP)28 PtdSer probes (Supplementary Fig. 2a and b), consistent with the abundance of PtdSer in the plasma membrane. However, they were negative for makers of PI(4,5)P2 (the genetically encoded probe PHPLCδ and an anti-PI(4,5)P2 antibody) (Supplementary Fig. 2c and d), a phosphoinositide typically rapidly removed from membranes upon endocytosis, and for a marker of PI3P (the genetically encoded probe FYVEHrs) (Supplementary Fig. 2e), a signature phospholipid of early endosomes. Thus, they have the properties of very early endocytic intermediates.
The structure of the endophilin foci was investigated by electron microscopy (EM). This analysis revealed in MβCD-treated cells clusters of densely packed intertwined narrow membrane tubules (Fig. 1j), which in EM tomograms appeared to originate from the plasma membrane (Supplementary Movie S2). Their diameter was homogeneous (23.2 nm ± 4.7 nm) (Fig. 1k) and in the range of typical endophilin and amphiphysin coated tubules29, 30. Importantly, anti-GFP immuno-gold EM labeling of endophilin 2-GFP expressing cells revealed intense immunoreactivity on the tubular clusters, confirming their correspondence to the endophilin foci detected by immunofluorescence (Fig. 1l).
Formation of massive tubular plasma membrane invaginations should result in a decrease of the outer cell surface. Accordingly, scanning EM of mouse embryonic fibroblasts (MEFs), revealed a reduction of plasma membrane ruffles and filopodia upon MβCD treatment (Supplementary Fig. 2f), i.e., as expected from a reduction of the surface-to-volume ratio. The footprint of the cells was also reduced (Fig. 1m and Supplementary Fig. 2g) and the time course of this reduction correlated with the increase of the area occupied by endophilin foci in fluorescence images (Fig. 1m).
As formation of the clusters of membrane tubules was not dependent on endophilin (see above), endophilin may be recruited to the tubules via its bilayer curvature sensing properties. Alternatively, endophilin may participate in their formation, but such role may be overlapping, and thus not essential, with the role of other N-BAR proteins, such as amphiphysin (see above). Clathrin and dynamin were also dispensable for tubule formation, as endophilin foci occurred in cells where levels of clathrin heavy chain had been drastically reduced by RNAi and in cells lacking dynamin [dynamin triple KO MEFs31] (Supplementary Fig. 2h). In fact, endophilin foci formation in response to MβCD treatment was enhanced in cells lacking dynamin relative to control cells (Supplementary Fig. 2i), indicating that MβCD does not act by inhibiting dynamin-dependent membrane fission. Dynamin was concentrated at endophilin-GFP-positive foci via an SH3-dependent interaction (Supplementary Fig. 2j) (such accumulation did not occur with the overexpression of endo2ΔSH3-GFP [Supplementary Fig. 2j]), yet did not prevent tubule formation.
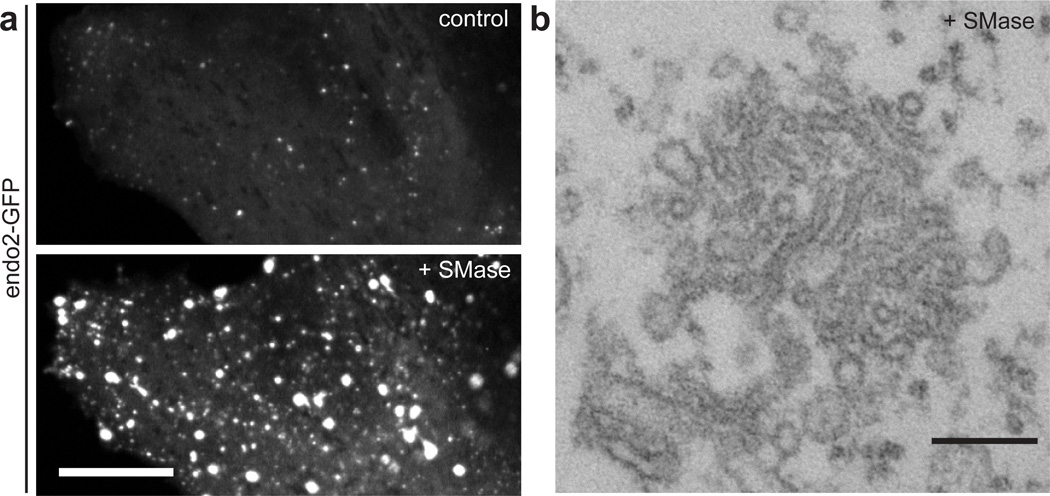
Cholesterol and sphingomyelin interact within the plasma membrane, and help define its physical properties32. We explored whether perturbing this partnership by hydrolyzing sphingomyelin, rather than by depleting cholesterol, also result in the tubular invaginations. Addition of purified bacterial sphingomyelinase (SMase) to endophilin2-GFP expressing cells produced a redistribution of GFP fluorescence similar to that produced by MβCD treatment and with a similar time-course (Fig. 2a). Likewise, EM revealed also in these cells the presence of clusters of small tubules (Fig. 2b).
Figure 2. Acute hydrolysis of plasma membrane sphingomyelin also induces massive endocytic tubular invaginations positive for N-BAR proteins.
a. Cell expressing endophilin 2-GFP before (“control”, top) and after (“+SMase”, bottom) sphingomyelinase (SMase) treatment (525 mU/ml) at 37 °C. b. Transmission electron microscopy image of a tubular membrane cluster induced by SMase treatment. Scale bar: 5 µm in a and 200 nm in b.
The sphingoid base modifying enzyme, sphingosine kinase 1 (SPHK1), is recruited to the tubular endocytic invaginations
In view of the strong genetic evidence linking sphingolipid metabolism to endocytic events, and in particular to the action of endocytic N-BAR proteins24 (Supplementary Fig. 3a), we explored the potential presence of sphingolipid metabolizing enzymes on the tubular endocytic intermediates generated by the above mentioned treatments that alter the cholesterol/sphingolipid balance.
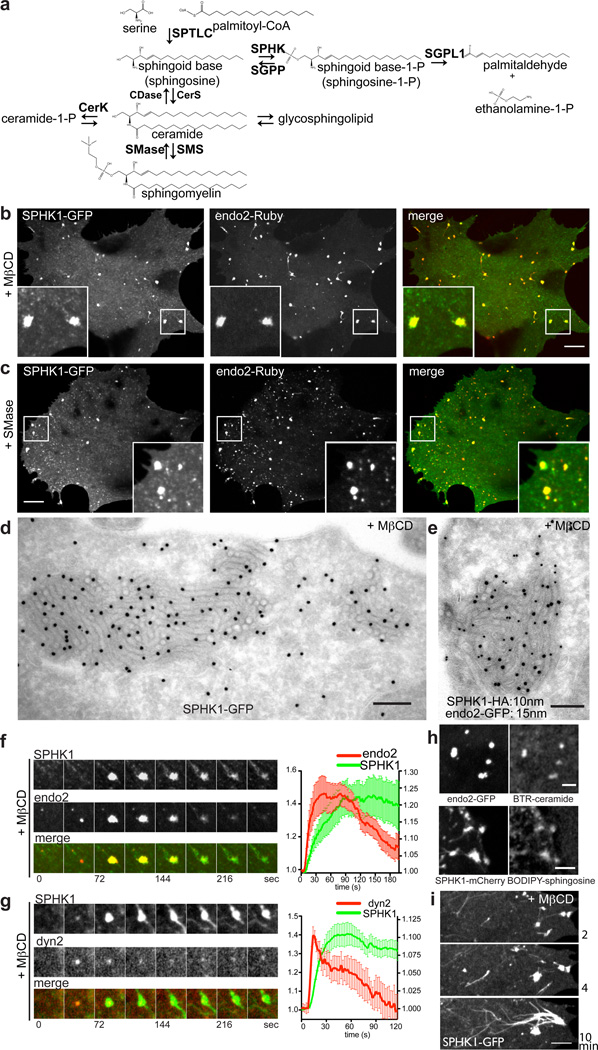
We examined the localization of GFP-fusions of several enzymes known to regulate sphingoid base levels (Fig. 3a and Supplementary Fig. 3b and c): neutral sphingomyelinase (SMPD3), neutral ceramidase (ASAH2), ceramide kinase (CerK), sphingosine kinase 1 (SPHK1), sphingosine-1-phosphate phosphatase 1 (SGPP1), sphingosine-1-phosphate lyase (SGPL1) and ceramide synthase 1 (CerS1). Strikingly, SPHK1, but not the other enzymes, was strongly enriched at the tubules induced by MβCD (Fig. 3b) or SMase treatment (Fig. 3c), where it colocalized with endophilin 2-Ruby. SPHK1 is a cytosolic kinase that specifically phosphorylates sphingoid bases to sphingoid base-1-phosphate. Interestingly, yeast sphingosine kinase (LCB4) is one of the enzymes that genetically interact with the RVS genes (LCB4 mutations suppress rvs161 and rvs167 mutations)24 (Supplementary Fig. 3a). Mammalian genomes encode two SPHKs, SPHK1 and SPHK2. FLAG-tagged SPHK2 was also present at endophilin foci (Supplementary Fig. 3d), consistent with a redundant function of the two genes in mice33. The enrichment of SPHK1-GFP at the tubular invaginations induced by MβCD was confirmed by EM with anti-GFP immunogold labeling (Fig. 3d). Co-immunogold labeling of cells co-expressing SPHK1-HA and endophilin 2-GFP further confirmed these results (Fig. 3e).
Figure 3. The sphingoid base modifying enzyme, sphingosine kinase 1 (SPHK1) is recruited to the tubular endocytic invaginations.
a. Sphingolipid metabolic pathway. SPTLC = serine palmitoyltransferase; SPHK = sphingosine kinase; SGPP = sphingosine-1-phosphate phosphatase; SGPL1 = sphingosine-1-phosphate lyase 1; CerS = ceramide synthase; CDase = ceramidase; SMS = sphingomyelin synthase; SMase = sphingomyelinase; CerK = ceramide kinase. b and c. GFP-tagged SPHK1 cosegregates into the clusters positive for Ruby-tagged endophilin 2 in cells treated with MβCD in b, or treated with SMase in c. d and e. Concentration of both SPHK1 and endophilin 2 on the endocytic tubular intermediates induced by MβCD treatment in a cell transfected with SPHK1-GFP alone in d or both SPHK1-HA and endophilin 2-GFP in e. Single or dual immunogold labeling with 15 nm gold particles for anti-GFP in c and e and 10 nm gold particles for anti-HA in e. f. Selected frames from a time series of a cluster colabeled for SPHK1-GFP and endophilin 2-Ruby (left). Fluorescence intensity of endophilin 2 (red) and SPHK1 (green) positive tubule clusters (n = 15 foci from 6 different cells) showing that recruitment of endophilin precedes SPHK1 (right). Pooled data from 6 independent experiments. Error bars: standard errors of the mean. g. Selected frames from a time series of a tubule cluster colabeled by SPHK1-GFP and dynamin 2-RFP (left). Fluorescence intensity of dynamin 2 (red) and SPHK1 (green) positive tubule clusters (n = 16 foci from 6 different cells) showing that recruitment of dynamin precedes SPHK1 (right). Pooled data from 6 independent experiments. Error bars: standard error of the mean. h. Tubule clusters formed after MβCD treatment are positive for fluorescent ceramide (BODIPY-TR-ceramide, top) and fluorescent sphingosine (BODIPY-sphingosine, bottom). i. Confocal images of a cell expressing SPHK1-GFP 2 min (top), 4 min (middle) and 10 min (bottom) after MβCD treatment. SPHK1 strongly labels filament-like structures originated from the clusters. Scale bar: 10 µm in b and c; 200 nm in d and e; 3 µm in h; and 5 µm in i.
Live-cell imaging revealed that recruitment of SPHK1 lagged slightly behind formation of endophilin-positive foci (Fig. 3f and g). However, SPHK1 accumulated at tubule clusters independent of endophilin, as SPHK1 persisted at these foci after the loss of endophilin (Fig. 3f) and SPHK1 foci still formed in endophilin triple KO cells (Supplementary Fig. 3e). Accordingly, no physical interaction between the two proteins was observed. Fluorescent sphingomyelin metabolites, BODIPY-TR-ceramide and BODIPY-sphingosine, accumulated at tubule clusters (Fig. 3h), raising the possibility that the corresponding endogenous metabolites may facilitate SPHK1 recruitment. SPHK1-positive foci eventually disassembled via the emergence from them of SPHK1 filament-like structures (most likely bundles of tubules) (Fig. 3i).
Recruitment of SPHK1 to physiologically occurring endocytic intermediates
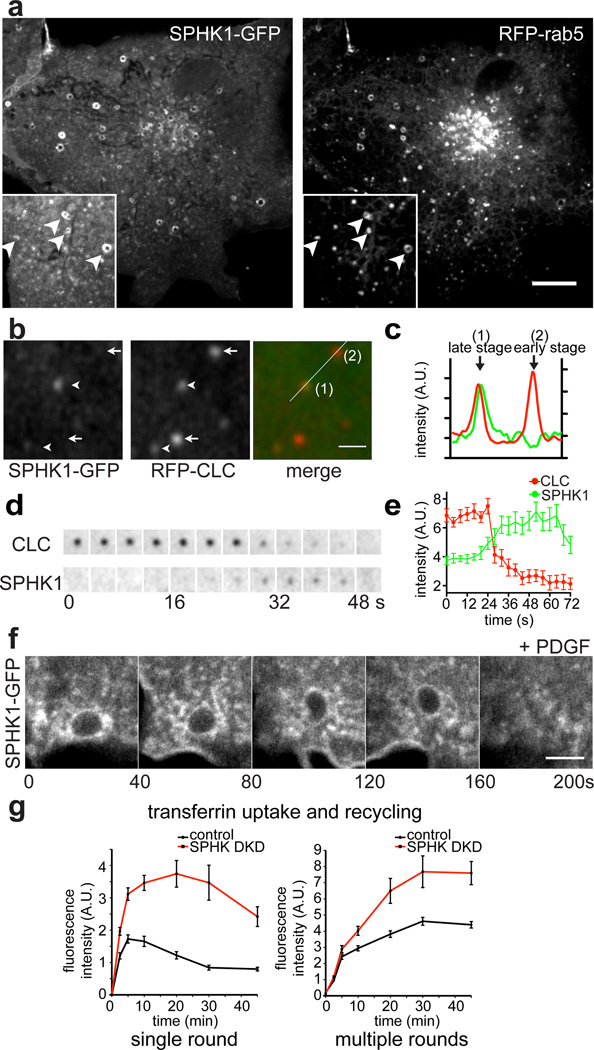
SPHK1 was shown to have a primarily cytosolic localization and to be recruited to the plasma membrane in response to pharmacological stimuli34, 35. SPHK1 was also reported to be localized on endosomes36 and phagosomes37, 38. Our results shown above, i.e., the recruitment of SPHK1 to “exaggerated” early endocytic intermediates induced by experimental perturbations, support the possibility that physiologically occurring early endocytic structures represent one of its sites of action. We revisited the subcellular targeting of SPHK1 under control conditions using SPHK1-GFP and observed an enrichment of this protein both on Rab5-positive early endosomes (Fig. 4a) and on even earlier endocytic intermediates. For example, SPHK1 was present at a subset (62%) of endocytic clathrin-coated pits (Fig. 4b and c), and at such pits the SPHK1-GFP signal peaked at late stages, when the clathrin signal had already started to decay (Fig. 4d and e). As the clathrin signal dims in parallel with the endophilin signal19, 39, even at these physiologically occurring sites SPHK1 recruitment lagged behind the endophilin recruitment, as observed at MβCD-induced endophilin foci. In addition, SPHK1 was also recruited to macropinosomes induced by PDGF stimulation (Fig. 4f) and neighboring membranes.
Figure 4. Recruitment of SPHK1 to physiologically occurring endocytic intermediates.
a. SPHK1-GFP is localized on Rab5 (RFP-Rab5) positive endosomal structures. b – e. SPHK1 is recruited to very late stage clathrin-coated endocytic pits. b. Confocal image of a COS-7 cell expressing SPHK1-GFP and RFP-clathrin light chain (CLC) showing that SPHK1 is present at a subset of endocytic clathrin-coated pits (arrowheads), but not others (arrows). c. Intensity profile of SPHK1 (green) and clathrin (red) fluorescence along the line drawn in b. d. Selected frames from a time series of a clathrin-coated pit showing that SPHK1 is recruited to the very late stage during clathrin-mediated endocytosis. e. Mean fluorescence intensity measurement of SPHK1-GFP and RFP-CLC. Pooled data from five independent experiments. n = 15 clathrin-coated pits from 5 different cells. Error bars: standard error of the mean. f. SPHK1-GFP is recruited to macropinosomes induced by PDGF stimulation. Neighboring endocytic membranes are also positive for the kinase. g. Double knockdown (DKD) of SPHK1 and SPHK2 in HeLa cells causes defects in transferrin recycling. Fluorescence intensity of Alexa Fluor 594-transferrin in control and DKD cells were calculated from projected Z-stack images. Numbers of cells counted were more than 30 for each time point. The exact n values are included in the Methods. Data are from a single experiment and representative of three different experiments Error bars: standard error of the mean. Scale bar: 10 µm in a, 3 µm in b, and 5 µm in f.
A function of SPHK1 in the early endocytic system was supported by loss-of-function experiments in HeLa cells. As shown in Figure 4g, RNAi-induced knockdown of SPHK1 and SPHK2 (Supplementary Figure 4) resulted in an accumulation of internalized transferrin relative to controls, both in a single round assay (uptake and release of prebound transferrin) and in a multiple rounds assay (uptake and release in the continued presence of transferrin), indicating a defect in recycling. An accumulation of internalized transferrin upon SPHK1 KD was also previously observed in a genome-wide RNAi screen in HeLa cells40.
These observations suggest that the accumulation of SPHK1 on the tubular structures produced by depletion of cholesterol or sphingomyelin reflects a coupling of SPHK1 function to early endocytic events that occurs under physiological conditions.
SPHK1 directly binds to negatively charged membranes with a preference for high positive curvature
Previous studies suggested that both protein-protein41, 42 and protein-lipid interactions43, 44 contribute to the membrane recruitment of SPHK1, a cytosolic enzyme. Given the strong enrichment of SPHK1 on narrow tubular membranes, we tested the possibility that such recruitment may be mediated by direct bilayer binding and positive curvature sensing properties.
C-terminally tagged human SPHK1 (either SPHK1-FLAG or SPHK1-GFP-FLAG) was purified to near homogeneity from extracts of transfected Expi293T cells by anti-FLAG affinity chromatography followed by size-exclusion chromatography (Fig. 5f and Supplementary Fig. 5a). The purified protein was enzymatically active as shown by an in vitro kinase assay involving sphingosine and 32P-ATP as substrates, which revealed an activity of approximately 106 pmol/mg/min (Supplementary Fig. 5b), consistent with previous reports45. SPHK1-FLAG bound to liposomes containing negatively charged lipids (PS or an acidic lipids mixture), but not to liposomes containing neutral lipids only (Fig. 5a). Moreover, when SPHK1-GFP-FLAG was tested for binding to giant unilamellar vesicles (GUVs labeled by a lipid dye) from which narrow tubules had been pulled, it became selectively enriched relative to lipids on the narrow tubules (Fig. 5b), although it did not change the membrane curvature (Supplementary Fig. 5c), suggesting that SPHK1 senses, but does not generate high bilayer curvature (Fig. 5c) under the experimental conditions tested.
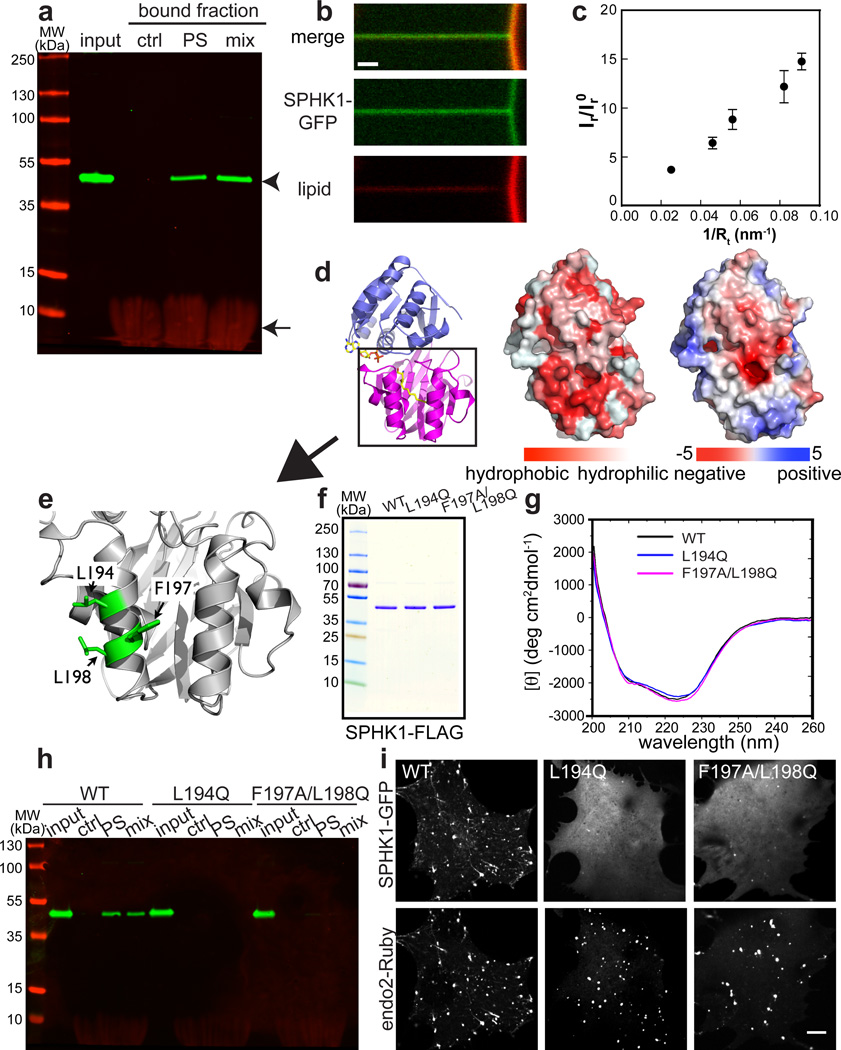
Figure 5. SPHK1 directly binds to negatively charged membranes with a preference for high positive curvature.
a. Liposome flotation assay demonstrating that purified SPHK1 does not bind neutral liposomes (control, ctrl)[PE:PC:chol = 45:50:5], but does bind negatively charged liposomes that contain PS (PS) [PE:PC:PS:chol = 45:10:45:5] or a mixture of acidic phospholipids (mix)[PE:PC:PS:PA:PI(4)P:PI(4,5)P2:chol = 25:10:30:10:10:10:5]. Arrowhead indicates SPHK1-FLAG and arrow indicates fluorescent lipids. b. Curvature-dependent sorting of SPHK1-GFP on an artificial lipid bilayer. SPHK1-GFP is enriched relative to a membrane lipid (red, Texas red-DHPE) on a membrane tether pulled from a low curvature giant unilamellar vesicle (GUV). c. Mean ratio between green (SPHK1-GFP) and red (Texas red-DHPE) fluorescence intensities from the cross-sectional area of the tether, normalized to the two fluorescence intensities on the GUVs, for varying membrane tension (and thus membrane curvature). n = 6 pulled tubules. Each tubule represents one independent experiment, and data from 6 measurements were pooled. Error bars: standard error of the mean. d. Crystal structure of human SPHK1 (PDB ID 3VZB). Ribbon representation (left), surface hydrophobicity (middle) and electrostatic surface (right) are shown. In the ribbon representation, the N-terminal lobe is shown in blue and the C-terminal lobe in magenta. The bound substrate and ADP (superposed from PDB ID 3VZD) are also shown. e. Zoomed-in view of the framed region in d, highlighting the three mutated, surface-exposed and hydrophobic residues, L194, F197 and L198 (green), which are key components of the hydrophobic patch shown in the middle panel of d. f. Coomassie blue-stained SDS-PAGE showing purified SPHK1-FLAG (WT), SPHK1L194Q-FLAG (L194Q) and SPHK1F197A/L198Q-FLAG (F197A/L198Q). g. CD spectrum of purified SPHK1-FLAG (WT), SPHK1L194Q-FLAG (L194Q) and SPHK1F197A/L198Q-FLAG (F197A/L198Q) showing correct folding of the mutant fusion proteins. h. SPHK1L194Q-FLAG (L194Q) and SPHK1F197A/L198Q-FLAG (F197A/L198Q) do not bind either neutral liposomes (ctrl) or negatively charged liposomes (PS and mix). i. Confocal images of cells co-expressing endophilin 2-Ruby (bottom) and either SPHK1WT-GFP, SPHK1L194Q-GFP or SPHK1F197A/L198Q –GFP as indicated, after MβCD treatment. Scale bar: 1 µm in b and 10µm in h.
The recently published crystal structure of human SPHK146 provided an opportunity to predict potential binding interfaces. SPHK1 is represented nearly exclusively by the kinase module, which is characterized by an N-terminal ATP binding lobe (blue in Fig. 5d, left) and a C-terminal substrate recognition lobe (magenta in Fig. 5d, left). Inspection of surface hydrophobicity demonstrates an evolutionarily conserved hydrophobic patch in the substrate recognition lobe (Fig. 5d, middle). The patch corresponds to one of the two helices that were proposed to function as a gate to control the flux of the substrate sphingosine into the interior of the protein (Fig. 5e). This may explain why the L194Q mutation (Fig. 5e) in this patch was previously shown to impair catalytic function41, although leucine 194 is not part of the catalytic site. Such result had previously been attributed to misfolding of the mutant protein41, but we did not find evidence for this possibility based on CD spectroscopy of purified SPHK1L194Q (Fig. 5f and g). A plausible explanation is that the L194Q mutation may impair access to the substrate. As this hydrophobic patch is surrounded by some positively charged residues (Fig. 5d, right), it may represent a lipid bilayer-binding interface with hydrophobic residues partially penetrating the membrane. This interpretation was supported by the diffuse cytosolic distribution of SPHK1L194Q and of a SPHK1 mutant that harbors two other mutations in the same hydrophobic patch, SPHK1F197A, L198Q (Fig. 5i). Both mutants folded correctly (Fig. 5g), but failed to bind to acidic liposomes in vitro (Fig. 5h) and were not recruited to the tubular membrane invaginations induced by cholesterol extraction in living cells (Fig. 5i).
A SPHK1 mutant that does not localize to tubular invaginations is non-functional in vivo
To determine whether the properties that mediate the recruitment of SPHK1 to the tubular invaginations have a physiological significance, we next investigated whether perturbation of the hydrophobic patch rescues the absence of SPHK1 in a functional assay. In neurons, SPHK1 is preferentially targeted to nerve terminals, a compartment highly specialized for the endocytic recycling of synaptic vesicles, as shown by immunocytochemistry47 and by the accumulation of SPHK1-GFP in presynaptic terminals of cortical neurons in vitro47 (and Fig. 6a), or of retinal photoreceptors in vivo (Fig. 6b). The single SPHK1 in C. elegans, cSPHK-1, is also concentrated at synaptic sites along axons, and its absence results in lower sensitivity to aldicarb, indicating a defect in neurotransmitter secretion10, 48. Interestingly, cSPHK-1 is primarily localized at periactive zones of synapses, which are sites of high endocytic activity10. In cSPHK-1, leucine 194 of the hydrophobic patch is replaced by another hydrophobic amino acid, valine 268. Supporting a critical importance of the patch, cSPHK-1V268Q-GFP did not exhibit the same highly punctate localization as cSPHKWT-GFP in axons (Fig. 6c and d). Importantly, while cSPHK-1WT-GFP (Fig. 6e) and even human SPHK1 (Supplementary Fig. 6) rescued neurotransmission in cSPHK-1 mutant worms, cSPHK-1V268Q-GFP did not (Fig. 6e).
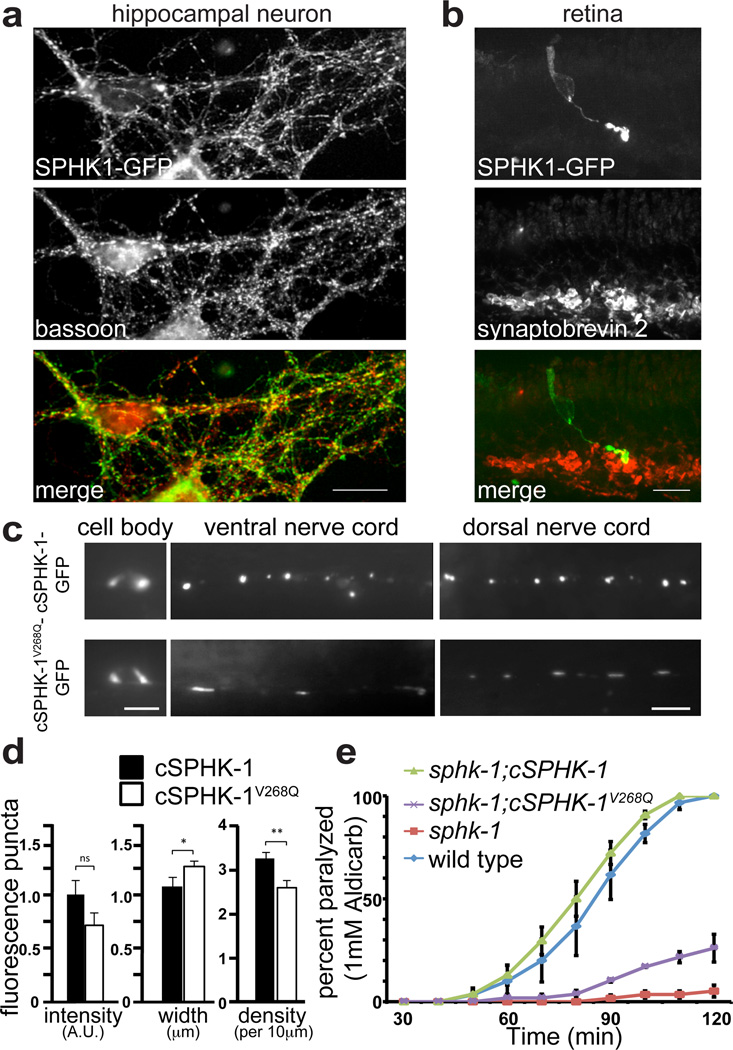
Figure 6. SPHK1 is concentrated at synapses and mutations that abolish membrane binding do not rescue synaptic transmission defects of loss-of-function mutants.
a. SPHK1-GFP is targeted to presynaptic terminals in cultured hippocampal neurons as shown by colocalization with immunoreactivity for Bassoon, a marker of pre-synaptic nerve terminals. b. SPHK1 is selectively enriched at the presynaptic terminal of photoreceptor cells in mouse retina. Z-projection confocal image of a mouse retina slice containing a SPHK1-GFP expressing rod cell (as a result of electroporation). The retina was fixed and immunostained for the synaptic vesicle marker synaptobrevin 2 to reveal synapses of the outer plexiform layer where the axon ending of the rod cell terminates. c – e. Analysis of worm sphingosine kinase (cSPHK-1). c. The localization of cSPHK-1WT-GFP or of the predicted membrane binding mutant cSPHK-1V268Q-GFP in the cell body (left), ventral (middle) and dorsal nerve cord (right) of motor neurons of C. elegans. d. Quantification of parameters of the fluorescence spots in the axon terminals (intensity, width and density) in wild-type animals expressing WT and mutant cSPHK-1. For cSPHK-1-GFP, n= 321 synapses from 17 worms, and for cSPHK-1V268Q-GFP, n=387 synapses from 23 worms. Each animal is an independent experiment, and data from independent experiments were pooled. Error bars: standard error of the mean. [ns] not significant; [*] P < 0.05; [**] P < 0.005, Student's t-test. e. Time-course of the onset of paralysis of the indicated worm strains upon exposure to the acetylcholine esterase inhibitor aldicarb (1 mM). sphk-1; cSPHK-1 and sphk-1;cSPHK-1V268Q stand for sphk-1 null mutants expressing full length worm SPHK-1-GFP or SPHK-1V268Q-GFP cDNA in neurons, respectively. A total of 60 worms were counted from each plate. Percentages were averaged at each time point per genotype and plotted graphically. n = 3 plates. The analysis was repeated two times, with two different transgenic lines. Data shown are from a single experiment. Error bars: standard error of the mean. Scale bar: 10 µm in a, 20 µm in b, and 10 µm in c.
Discussion
Our study shows that lowering cholesterol or sphingomyelin levels in the plasma membrane results in its dramatic remodeling, with the formation of massive tubular endocytic intermediates. These findings emphasize the critical importance of an appropriate cholesterol/sphingolipid balance in the control of plasma membrane integrity and cell surface area. N-BAR proteins and SPHK1 are highly enriched on the tubular endocytic structures formed under these conditions. Together with evidence for the presence of SPHK1 on physiologically occurring endocytic membranes, and for a defect of endocytic recycling in cells lacking SPHKs, these findings point to a link between sphingosine metabolism and early stages of endocytosis.
The reciprocal interactions of cholesterol and sphingomyelin help determine bilayer rigidity and generate microdomains that control protein localization and function1, 2. We speculate that perturbing the balance of these two lipids affects the membrane either directly by changing its physical properties49–51, or indirectly by modifying protein-lipid interactions. These changes, in turn, increase the propensity of the membrane to be deformed by the numerous proteins with curvature generating properties that are present in the cytosol15, 16, 52. These include BAR domain-containing endocytic proteins, as demonstrated by the accumulation of amphiphysin and endophilin, which are normally selectively concentrated at the short necks of endocytic pits17–19, 21, 39, on the tubules. Under physiological conditions equilibrium between these factors likely plays an important role in regulating cell surface dynamics. Sequestration of N-BAR proteins on the tubules may contribute to the reported accumulation of shallow clathrin-coated endocytic pits in cholesterol-extracted cells3, 4.
The massive recruitment of SPHK1 at the tubule clusters revealed a link between these “exaggerated” endocytic intermediates and sphingolipid metabolism and helped us discover a physiologically occurring relation between endocytosis and SPHK1 recruitment. Membrane curvature sensing53 of SPHK1 may play a role in its recruitment, as we have found that SPHK1 binds negatively charged lipid bilayers with curvature sensing properties. However, the delay of its recruitment relative to endophilin suggests the importance of additional factors. One factor could be the accumulation of its substrate, sphingosine, in the membrane. The propensity of SPHK1 to bind the bilayer may facilitate its access to the substrate and allow it to remain membrane bound through multiple catalytic cycles.
Exogenous sphingomyelinase, which generates ceramide, likely has the dual effect of altering the physicochemical state of the plasma membrane and of producing an accumulation of sphingosine, a catabolic product of ceramide. Depletion of cholesterol may achieve the same effect indirectly. By destabilizing cholesterol-sphingomyelin microdomains, MβCD may trigger internalization of excess sphingomyelin, thus activating the cascade that results in upregulated levels of sphingosine.
The metabolism of sphingomyelin requires a cooperation of enzymes located on intracellular membranes and at the cell surface54. There is evidence, however, for a selective concentration of sphingomyelin at the plasma membrane55, thus suggesting mechanisms to enrich it at this membrane and to remove it and its metabolites from internalized membranes. While the building blocks of sphingomyelin are synthesized in the ER and at the Golgi complex54, at least a pool of sphingomyelin is generated directly at the plasma membrane by a plasma membrane-localized sphingomyelin synthase (SMS2)56. Likewise, a neutral sphingomyelinase, which hydrolyzes sphingomyelin to ceramide57, 58 and a neutral ceramidase, which hydrolyzes ceramide to sphingosine59, 60 are also present at the cell surface. We speculate that endocytosis may correlate with the conversion of at least a pool of sphingomyelin to sphingosine, which can then flip across the membrane and be converted to sphingosine-1-phosphate by SPHKs. As endocytic recycling is affected in the absence of SPHKs, this metabolic cascade may not be simply a byproduct of membrane traffic but also impact endocytic trafficking.
Sphingosine-1-phosphate can then translocate to other membranes, such as the ER, through the cytosol as it is moderately aqueous soluble61. In the ER it can be dephosphorylated by sphingosine-1-phosphate phosphatase (SGPP1)62 and recycled for sphingomyelin resynthesis, or irreversibly degraded by sphingosine 1-phosphate lyase (SGPL1)63 to provide precursors for phospholipid synthesis64. A pool of sphingosine-1-phosphate may also translocate across membranes and function as an extracellular signaling molecule via the binding to a group of G-protein coupled receptors (GPCRs)65. The signaling action of sphingosine-1-phosphate via GPCRs is currently the most established function of this metabolite in mammals. However, as these GPCRs are not expressed in lower organisms, where SPHK and sphingosine-1-phosphate are present, it is possible that the most fundamental role for sphingosine-1-phosphate may relate to membrane dynamics.
METHODS
Cell culture
HeLa cells, COS-7 cells, Ptk cells and HEK293T cells were purchased from ATCC (Manassas, VA). Primary fibroblasts were isolated from 16–18 days embryos by the 3T3 method66. Endophilin 1, 2 and 3 triple KO mouse fibroblasts20 and dynamin 1, 2 and 3 triple conditional KO mouse embryonic fibroblasts18, 31 were generated according to previous protocols from our laboratory. Cells were grown in DMEM + 10% fetal bovine serum with penicillin/streptomycin supplement. Cell culture reagents were from Life Technologies (Carlsbad, CA).
For MβCD treatment, MβCD stock solution (5 × concentration) was prepared immediately before the experiments and used at 37 °C (10 mM final concentration) in imaging buffer (10 mM HEPES pH7.4, 120 mM NaCl, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2). For rescue experiments, a solution of cholesterol–methyl-β-cyclodextrin was prepared immediately before the experiments and added to cells at 800 µg/mL final concentration. Bacterial sphingomyelinase was added to cells at a final concentration of 500 mU/mL at 37 °C. PDGF was added to serum-starved (overnight) cells at the final concentration of 20 ng/mL in imaging buffer. Fluorescent lipid dyes were added for 1 min at 37 °C to cells plated on 35 mm glass-bottom dishes (MatTek) at the concentration recommended by the manufacturers. Cells were then briefly rinsed with imaging buffer before live-cell imaging. All cell-based experiments have been repeated at least three times.
RNAi
siRNA-dependent knockdown of clathrin heavy chain in mouse fibroblasts was performed as described67. For SPHK1 and SPHK2 knockdown, HeLa cells were transfected with siRNAs using Lipofectamine RNAi MAX (Life Technologies, Carlsbad, CA) and cultured for 72 h prior to analysis. siRNAs (HSC.RNAI.N182965.12.1 and HSC.RNAI.N182965.12.2 for human SPHK1; HSC.RNAI.N020126.12.2 and HSC.RNAI.N020126.12.3 for human SPHK2; NC1 negative control duplex as a control) were from Integrated DNA Technologies (IDT, Coralville, IA). Knockdown efficiency was evaluated by qPCR (Supplementary Fig. 4). For this purpose, total RNA was extracted using the RNeasy MiniKit (QIAGEN) and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). The resulting cDNA was subjected to real-time qPCR with Fast SYBR Green Master Mix (Life Technologies). Measurements were normalized to an internal control (ribosomal protein S26 mRNA), and results were presented as relative mRNA levels.
Plasmids
The following plasmids were kind gifts: GFP-Clathrin light chain (GFP-CLC) (James Keen, Thomas Jefferson University, Philadelphia, PA), PM-GFP (Tobias Meyer, Stanford University, Stanford, CA), GFP-PHPLCδ (Antonella De Matteis, Telethon Institute of Genetics and Medicine, Naples, Italy) and GFP-2xFYVEHrs (Harald Stenmark, Oslo University, Oslo, Norway). mRFP-clathrin light chain (RFP-CLC), endophilin 2-GFP, endophilin 2-Ruby, endophilin 2ΔSH3-GFP, GFP-amphiphysin II, dynamin 2-RFP and RFP-Rab5 were previously generated in our laboratory. C2lact-GFP was from addgene (#22852). GFP-SMPD3 (human) plasmid was from GeneCopoeia (cat No. EX-U1166-M29). Protein-coding cDNA clones corresponding to mouse ASAH2 missing the N-terminal 19 aa (clone ID 40018929), mouse SGPL1 (clone ID 4212293), human CerS1 (clone ID 6192817), mouse CerK (clone ID 6400872), mouse SPHK2 (clone ID 4987298) and human SPHK1 (clone ID 5213270) were from Thermo Fisher Scientific (Waltham, MA). The full length ASAH2 sequence was amplified by PCR using primers including the sequence encoding the N-terminal 19 aa (which were missing from the commercial plasmid) and subcloned into the pEGFP-N1 vector to obtain the ASAH2-GFP plasmid. SGPL1 and CerK were subcloned into the pEGFP-N1 vector to generate the SGPL1-GFP and the CerK-GFP plasmids, respectively. CerS1 was subcloned to generate the CerS1-oxGFP plasmid, where oxGFP tag is a version of GFP optimized for expression in the ER lumen (kind gift from Erik Snapp, Albert Einstein College of Medicine). Human SGPP1 plasmid was a kind gift from Lina Obeid (Stony Brook School of Medicine) and the plasmid was subcloned into the pEGFP-C1 vector to generate the GFP-SGPP1 plasmid. SPHK2 was subcloned into the pEGFP-N1 vector to generate SPHK2-GFP. As GFP tagged SPHK2 were prone to degradation in cells, a smaller tag, FLAG-tagged SPHK2 (SPHK2-FLAG), was generated from SPHK2-GFP by swapping the sequence of GFP with the nucleotide sequence encoding the FLAG tag. SPHK1 was subcloned into the pEGFP-N1 and pmCherry-N1 vectors, respectively, to generate C-terminal GFP or mCherry fusions. SPHK1-HA, SPHK1-FLAG, and SPHK1-GFP-FLAG constructs were generated from SPHK1-GFP by either swapping the sequence of GFP with the nucleotide sequence encoding the HA or FLAG tags, or inserting the nucleotide sequence encoding the FLAG tag at the C-terminal side of GFP (before the stop codon) using the in-Fusion HD cloning Kit (Clontech). GFP-tagged or FLAG-tagged mutant constructs of SPHK1, SPHK1L194Q and SPHK1F197A/F198Q were generated from SPHK1-GFP or SPHK1-FLAG plasmid using the QuikChange II XL Mutagenesis Kit (Agilent Technologies). The pCAG-SPHK1-GFP plasmid was generated by subcloning SPHK1-GFP into the pCAG-EGFP-N1 vector containing the chicken β-actin promoter.
Antibodies
Antibodies were obtained from the following sources: anti-Bassoon antibody (clone SAP7F407, Cat. No. ADI-VAM-PS003, Enzo Life Sciences, Farmingdale, NY, dilution 1:200 for immunofluorescence), anti-synaptobrevin 2 antibody (clone 69.1, Cat. No. 104 211, Synaptic Systems, Göttingen, Germany, dilution 1:400 for immunofluorescence), anti-HA antibody (clone 3F10, Cat. No. 11867423001, Roche, dilution 1:500 for immuno-gold labeling), anti-GFP antibody (Cat. No. A-11122, Life Technologies, Carlsbad, CA, dilution 1:100 for immuno-gold labeling), anti-FLAG antibody (clone M2, Cat. No. F3165, Sigma-Aldrich, St. Louis, MO, dilution 1:1000 for western blot), and anti-PI(4,5)P2 antibody (Cat. No. Z-A045, mouse IgM in ascites clone 2C11, Echelon Biosciences, Salt Lake City, UT, dilution 1:100 for immunofluorescence). Rabbit anti-endophilin 2 antibodies (dilution 1:50 for immunofluorescence) were generated in our laboratory20.
Other reagents
BODIPY-sphingosine was a gift from Robert Bittman (Queens College of CUNY Flushing, NY). Reagents purchased from commercial sources: methyl-β-cyclodextrin, cholesterol-methyl-β-cyclodextrin (cholesterol-water soluble), 2-hydroxypropyl-β-cyclodextrin and sphingomyelinase from Bacillus cereus (Sigma-Aldrich, St. Louis, MO); Dil and BODIPY-TR-ceramide, 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine triethylammonium salt (TR-DHPE)(Life Technologies, Carlsbad, CA); TopFluor PS, brain sphingosine, egg PC, liver PE, cholesterol from sheep wool, brain PS, egg PA, brain PI(4)P, brain PI(4,5)P2, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)-2000] (Avanti Polar Lipids, Alabaster, AL); and ATP, [γ-32P] (PerkinElmer, Waltham, MA). Electron microscopy reagents were purchased from Electron Microscopy Sciences.
Immunofluorescence
Immunofluorescence was performed according to standard procedures67. Samples were imaged with a Zeiss Axioplan 2 microscope using a Plan-Apochromatic 40× objective and a Hamamatsu ORCA II digital camera (Bridgewater, NJ) under the control of MetaMorph software (Molecular Devices, Sunnyvale, CA) or by spinning disk confocal microscopy. Immunofluorescence staining using the anti-PI(4,5)P2 antibody was performed according to Hammond et al.68.
Transferrin internalization
Control and SPHKs double KD cells were plated on 12 mm glass coverslips at a density of 50,000 cells/coverslip one day before assay. Cells were depleted of unlabeled transferrin by incubating them with serum-free DMEM for 1 h at 37 °C. For the “single round” uptake assay, cells were chilled with ice-cold DMEM for 10 min, labeled with 10µg/mL Alexa Fluor 594-transferrin (Life Technologies, Carlsbad, CA) in DMEM for 1 h, and briefly rinsed with DMEM always on ice. Coverslips were then transferred to pre-warmed (37 °C) DMEM and incubated at 37 °C for different times (2.5, 5, 10, 20, 30, and 45 min) to allow uptake and release of transferrin. For the “multiple rounds” uptake assay, cells were transferred (without pre-incubation on ice) to pre-warmed (37 °C) DMEM containing 10µg/mL Alexa Fluor 594-transferrin and incubated at 37 °C for different times. In the case of both assays, at the end of each incubation cells were immediately transferred to pre-chilled DMEM and then further processed on ice as follows: rinses in DMEM; stripping of surface-bound transferrin with three 2 min incubations with cold acid buffer (0.2M Acetic Acid pH 2.5, 0.2 M NaCl); washes in PBS and fixation with cold 4% formaldehyde in 0.1 M sodium phosphate, pH 7.2 for 3 min on ice and further fixation at room temperature for 15 min. Cells were washed with PBS and mounted for microscopic analysis. Fluorescence images were acquired by spinning disk confocal microscopy�at 500 nm intervals in the z-axis (5�µm total depth). The resulting z-stack of images was collapsed to a single image by the average intensity projection method in ImageJ software (version 1.43u, NIH). After background correction, whole cell fluorescence signals was quantified and analyzed. For single round assay, n = 32, 34, 33, 36, 32, and 31 (control); n=38, 35, 34, 33, 31, and 34 (DKD) were quantified for time point at 2.5, 5, 10, 20, 30, and 45 min, respectively. For multiple rounds assay, n = 36, 30, 35, 31, 32 and 37 (control); n = 33, 33, 30, 33, 33, and 31 (DKD) were quantified for time point at 2.5, 5, 10, 20, 30, and 45 min, respectively.
Cell transfection
Fibroblasts and hippocampal neuronal cultures were electroporated with Amaxa Nucleofector using solution R/program A-24 and mouse neuron solution/program O-03 (Lonza, Basel, Switzerland). HeLa and COS-7 cells were transfected with Lipofectamine 2000 (Life Technologies, Carlsbad, CA).
Live cell imaging
Spinning disc confocal microscopy and total internal reflection fluorescence microscopy were performed as described previously67.
Clathrin-coated pit tracking and analysis
TIRF images of COS-7 cells expressing GFP-CLC before and after MβCD treatment were analyzed using an enhanced version of the tracking method implemented in MATLAB69. Endocytic pits with lifetime of at least 12 s were selected. Only clathrin-coated pits that appeared and disappeared either before or after the addition of MβCD containing solutions (i.e. with the entire life-time in the control or in the MβCD condition) were used for quantification and analyzed separately. Statistical significance was calculated by two-tailed Student’s t-test. For the analysis of the presence of SPHK1 at endocytic clathrin coated pits, 53 clathrin-coated pits from 5 different live cells were counted.
Cholesterol measurement
Cells were plated in 6-well plates in sextuplicate the day before the assay. MβCD stock solution was added to the cells and incubated for 5 min at 37 °C in imaging buffer. After one rinse in PBS, cells were directly lysed in lysis buffer [150 mM NaCl, 20 mM Tris, 1% Triton X-100, and 1 mM EDTA, pH 7.4, supplemented with an EDTA-free protease inhibitor cocktail tablet (cOmplete)]. Free cholesterol in the lysate was measured using an enzyme-coupled assay kit (Amplex Red Cholesterol Assay Kit, Life Technologies, Carlsbad, CA). Fluorescence was detected using a 96-well black clear-bottom plate using a Tecan Infinite M1000 microplate reader. The relative cholesterol concentration for each sample was normalized to protein concentration (BCA Protein Assay Reagent) (Thermo Fisher Scientific) (n = 6 for control and MβCD-treated cells).
Electron microscopy
Cells expressing endophilin 2-GFP were plated on 35 mm gridded, glass-bottom MatTek dishes at 30% confluency one day before the assay. MβCD-treated (5 min) or sphingomyelinase-treated (10 min) cells and the corresponding coordinates on the grid were imaged using spinning disc confocal microscopy while fixing cells with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer. Cells were then post-fixed in 1% OsO4, 1.5% K4Fe(CN)6, 0.1 M sodium cacodylate buffer, en bloc stained with 0.5% uranyl magnesium acetate, dehydrated, and embedded in Embed 812. Ultrathin sections were observed with a Philips CM10 microscope at 80 kV and images were taken with a Morada 1000×1000 CCD camera (Olympus).
Immunogold labeling for HA or GFP tags
Cells were fixed with a mixture of 2% formaldehyde and 0.125% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, and processed for ultracryomicrotomy as described previously70. Ultrathin sections were immuno-labeled with primary antibodies followed by protein A coupled to 10 or 15 nm gold particles.
Electron tomography
The plastic sections (200–250 nm) were placed in a Fischione 2020 single-tilt holder (PA, USA), and examined in a Tecnai TF20 transmission electron microscope (FEI, The Netherlands) operated at 200 kV (Yale Center for Cellular and Molecular Imaging, Yale University). The single-tilt series were collected at every 1° over a ± 60° range using a FEI Eagle 4K × 4K CCD camera. Images were aligned using gold particles, the final tomograms were reconstructed using R-weighted algorithm of the IMOD software71.
Scanning electron microscopy
Control and MβCD-treated (5 min) cells were fixed with 4% formaldehyde, 2.5% glutaraldehyde. Samples were post fixed in 2% osmium tetroxide in sodium cacodylate buffer and dehydrated through an ethanol series to 100%. Samples were dried using a Polaron critical point dryer using liquid carbon dioxide as transitional fluid. Coverslips were glued to aluminum stubs, and sputter coated with 20nm gold using an EMS (Electron Microscopy Science) sputter coater. The samples were viewed and digital images acquired in an FEI ESEM between 5 –15 kV at a working distance of 12mm.
Protein expression and purification
SPHK1-FLAG, SPHK1L194Q-FLAG, and SPHK1F197A/L198Q-FLAG, and SPHK1-GFP-FLAG were expressed in suspension cultures of Expi293F cells (Life Technologies, Carlsbad, CA) for three to five days. Cells were lysed by freeze-thawing in buffer containing 20 mM Tris, pH 7.4, 250 mM NaCl, 10% glycerol, and cOmplete EDTA-free protease inhibitor cocktail (Roche). FLAG tagged fusion proteins were purified on anti-FLAG M2 affinity gel (Sigma-Aldrich, St. Louis, MO) and eluted with 3 × FLAG peptides (Sigma-Aldrich, St. Louis, MO) before further purification using size-exclusion chromatography.
Sphingosine kinase activity assay
Kinase activity of purified SPHK1 was assayed as described previously45. Purified SPHK1 (2 µL) was incubated with 180 µL assay buffer (25 mM Tris-HCl, pH 7.4, 10% glycerol, 250 mM NaCl, 1 mM DTT), 10 µL 1 mM sphingosine from a 1 mM stock, and the solution was vortexed before adding 10 µL [γ-32P]-ATP containing ATP mixture. The reaction mixture was incubated for 30 min at 37 °C. The 1 mM sphingosine stock was prepared by adding methanol-dissolved brain sphingosine (Avanti Polar Lipids, Alabaster, AL) into a 4 mg/mL fatty acid-free BSA solution in PBS. The solution was vortexed and sonicated to generate a fine suspension (i.e., cloudy but with no obvious particles). The sphingosine solution was found to be stable for 1 month when stored at –20 °C. The radioactive ATP mixture was prepared immediately before experiments by adding 20 µL 32P-ATP to 180 µL cold ATP solution (20 mM ATP in 0.2 M MgCl2). The kinase reaction was terminated by adding 20 µL of 1 M HCl and the lipid product was extracted and separated by TLC analysis in running buffer (8:2:1:2, v/v, mixture of 1-butanol:methanol:acetic acid:H2O). The plate was exposed in phosphoimager screens for 18–20 h before scanning.
Liposome flotation assay
Natural lipids (Avanti Polar Lipids, Alabaster, AL) and trace amounts of Texas Red-DHPE (Life Technologies, Carlsbad, CA) were mixed into borosilicate glass tubes at the indicated molar percent and dried under N2 gas, and further dried in the vacuum overnight. The next day, lipids were hydrated in assay buffer (20 mM Tris pH7.4, 150 mM NaCl, 2mM DTT), and liposomes were generated by freeze-thawing. The liposome suspension (50 µL of 2 mM) and 5 µg purified proteins were mixed with assay buffer to obtain a 100 µL mixture in a 5 × 41 mm ultracentrifuge tube (ultra-clear, No. 344090, Beckman Coulter) and incubated at room temperature for 10 min. The mixture was further mixed with 50 µL assay buffer and 150 µL of 80% w/v Nycodenz solution in assay buffer (final Nycodenz solution = 40%), overlaid with 250 µL 30%w/v Nycodenz and another 80 µL assay buffer. The gradients were centrifuged in a swing bucket SW55-Ti rotor at 48,000 rpm for 3 hours at 4 °C to allow liposomes and the bound proteins to float to the top layer. Lipid fractions were collected from the top and solubilized in SDS-PAGE loading sample buffer. 1/10 of the input and 1/3 of the collected top fractions were loaded on the gels, which were processed by western blotting with fluorescent antibodies. Antibody (green) and lipid (red) fluorescence was visualized by an Odyssey infrared imaging system. Liposome flotation experiments were repeated more than three times.
Curvature sorting of SPHK1 on tubular membranes
Curvature-sorting experiments were carried out as previously described72. Briefly, giant unilamellar vesicles (GUVs) [65% DOPC, 34% DOPS, 0.3% Texas Red-DHPE and 0.7% DSPE-PEG(2000)-biotin] were made by electros welling73. GUV dispersions were then diluted and incubated with 500 nM SPHK1-GFP in assay buffer (2 mM Tris, pH 7.4, 30 mM NaCl) containing polystyrene beads at room temperature for approximately 15 min. Subsequently, the sample was added into a measurement chamber mounted onto a confocal laser scanning microscope system. Two micropipettes were inserted into the chamber to aspirate a bead and a GUV, respectively, and a nanometer radius tether was pulled between the bead and the GUV. The quantification of SPHK1-GFP sorting was achieved by recording the confocal xz scans of the cross-section areas of the tethers for varying membrane tension (and thus curvature). Streptavidin-conjugated polystyrene beads with diameter of 6 µm were purchased from Polysciences (Warrington, PA). Micropipettes (World Precision Instruments, Sarasota, FL) were manufactured as described72.
Primary sequence analysis of sphingosine kinase and crystal structure visualization
Primary sequences corresponding to sphingosine kinases from different species were obtained from UniProt database, and the sequences were aligned using ClustalW server. The crystal structure of hSPHK1 (PDB ID 3VZB) was rendered in Pymol74 with ADP superposed from 3VZD (PDB ID) to show the active site. Hydrophobic surfaces are colored based on a normalized hydrophobicity scale using Color_h script75, and the electrostatic surface potential was calculated using APBS software76.
Circular dichroism (CD) spectrometer experiment
A Chirascan Circular Dichroism spectrometer (Applied Photophysics, Surrey, UK) was used to examine the conformation of purified SPHK1-FLAG, SPHK1L194Q-FLAG and SPHK1F197A/L198Q-FLAG at a concentration of 0.2 mg/mL. Measurements were carried out in 20 mM Tris pH 7.4, 150 mM NaCl, 0.5 mM TCEP, 10% glycerol at 20 °C.
In vivo experiment in mouse retina
In vivo electroporation to transfect mouse retinal progenitors at P0 with plasmid DNA (pCAG-SPHK1-GFP) was performed as previously described77 in three mice. Retinae were harvested one to two months after electroporation and dissected under a fluorescent microscope (Leica, MZFL III) to select GFP-positive area. No statistical method was used to predetermine sample size. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment. Animal care and use was carried out in accordance with our institutional guidelines.
Experiments in C. elegans
All behavioral and microscopy experiments were performed on young adult hermaphrodites. sphk-1(ok1097) was provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The wild type reference strain was N2 Bristol. sphk-1(ok1097) was outcrossed 10 times before use in experiments. The following transgenic strains were also used (strain no. [extrachromosomal array; plasmid]): OJ889 [vjEx338; pJC35],JPC01 [jcaEx01; pJC152] and OJ2111 [vjEx804; pDS449].
The SPHK-1V268Q-GFP variant was generated by Quikchange PCR (Stratagene) from sphk-1WT cDNA. Human and C. elegans sphingosine kinase cDNAs were subcloned and expressed under the pan neuronal promoter, psnb-1, to generate the following expression constructs: pJC152 [psnb-1::sphk-1V268Q-gfp], pJC35 [psnb-1::sphk-1WT-gfp], and pDS449 [psnb-1::humanSPHK1]. Microscopy and behavioral analysis were performed as described previously10, 11. For analysis of neurotransmission, animals were tested for their sensitivity to aldicarb (Bayer CropScience, Ventura CA), an inhibitor of acetylcholinesterase. The percentages were averaged at each time point per genotype and plotted graphically. For each experiment, the genotype was blind to the scorer.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shawn Ferguson, Adam Frost and Tobias Walther for discussion and advice, Jeremy Baskin for thorough reading of the manuscript, Frank Wilson, Louise Lucast, Lijuan Liu and Heather Czapla for outstanding technical support, Morven Graham, Xinran Liu and Stacy Wilson for help with microscopy experiments, members of T. Walther, T. Melia, and C. Burd laboratories (Yale University) for help with lipid experiments. We also acknowledge the help of the Yale Center for Cellular and Molecular Imaging and Yale Center for Genomics and Proteomics. This work was supported in part by grants from the NIH (NS36251, DK45735 and DA018343 to P.D.C., GM097552 to T.B., and NS071085 to D.S.) and from the Ellison Medical Foundation to P.D.C.
Footnotes
AUTHOR CONTRIBUTIONS
H.S. and P.D.C. designed the experiments and wrote the manuscript; H.S. performed the experiments. Experimental work was also contributed by F.G. (electron microscopy), Y.W. (electron microscopy), J.C. and D.S. (C. elegans experiments), C.Z. (curvature sorting), I.M. (neuronal experiment), Y.K. (retina experiment), and X.W. (CD spectroscopy).
COMPETING INTERESTS
The authors declare no competing financial interests.
Reference
- 1.Breslow DK, Weissman JS. Membranes in balance: mechanisms of sphingolipid homeostasis. Molecular cell. 2010;40:267–279. doi: 10.1016/j.molcel.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodal SK, et al. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subtil A, et al. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci U S A. 1999;96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigismund S, et al. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Hannich JT, Umebayashi K, Riezman H. Distribution and functions of sterols and sphingolipids. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiele C, Hannah MJ, Fahrenholz F, Huttner WB. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol. 2000;2:42–49. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- 8.Dason JS, Smith AJ, Marin L, Charlton MP. Vesicular sterols are essential for synaptic vesicle cycling. J Neurosci. 2010;30:15856–15865. doi: 10.1523/JNEUROSCI.4132-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohrbough J, et al. Ceramidase regulates synaptic vesicle exocytosis and trafficking. J Neurosci. 2004;24:7789–7803. doi: 10.1523/JNEUROSCI.1146-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan JP, Hu Z, Sieburth D. Recruitment of sphingosine kinase to presynaptic terminals by a conserved muscarinic signaling pathway promotes neurotransmitter release. Genes Dev. 2012;26:1070–1085. doi: 10.1101/gad.188003.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JP, Sieburth D. Localized sphingolipid signaling at presynaptic terminals is regulated by calcium influx and promotes recruitment of priming factors. J Neurosci. 2012;32:17909–17920. doi: 10.1523/JNEUROSCI.2808-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acharya U, Acharya JK. Enzymes of sphingolipid metabolism in Drosophila melanogaster. Cell Mol Life Sci. 2005;62:128–142. doi: 10.1007/s00018-004-4254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonamine I, et al. Sphingosine kinases and their metabolites modulate endolysosomal trafficking in photoreceptors. J Cell Biol. 2011;192:557–567. doi: 10.1083/jcb.201004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallop JL, McMahon HT. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem Soc Symp. 2005:223–231. doi: 10.1042/bss0720223. [DOI] [PubMed] [Google Scholar]
- 16.Shen H, Pirruccello M, De Camilli P. SnapShot: membrane curvature sensors and generators. Cell. 2012;150:1300, 1300 e1301–1300 e1302. doi: 10.1016/j.cell.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson SM, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera RM, Zoncu R, Lucast L, De Camilli P, Toomre D. Two synaptojanin 1 isoforms are recruited to clathrin-coated pits at different stages. Proc Natl Acad Sci U S A. 2006;103:19332–19337. doi: 10.1073/pnas.0609795104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milosevic I, et al. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 2011;72:587–601. doi: 10.1016/j.neuron.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin-and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 22.Desfarges L, et al. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast. 1993;9:267–277. doi: 10.1002/yea.320090306. [DOI] [PubMed] [Google Scholar]
- 23.Morgan J, et al. Altering sphingolipid metabolism in Saccharomyces cerevisiae cells lacking the amphiphysin ortholog Rvs161 reinitiates sugar transporter endocytosis. Eukaryot Cell. 2009;8:779–789. doi: 10.1128/EC.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguilar PS, et al. A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nat Struct Mol Biol. 2010;17:901–908. doi: 10.1038/nsmb.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zidovetzki R, Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim Biophys Acta. 2007;1768:1311–1324. doi: 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunaldi K, Huang N, Hamilton JA. Fatty acids are rapidly delivered to and extracted from membranes by methyl-beta-cyclodextrin. J Lipid Res. 2010;51:120–131. doi: 10.1194/jlr.M900200-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kay JG, Koivusalo M, Ma X, Wohland T, Grinstein S. Phosphatidylserine dynamics in cellular membranes. Mol Biol Cell. 2012;23:2198–2212. doi: 10.1091/mbc.E11-11-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 29.Farsad K, et al. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mim C, et al. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park RJ, et al. Dynamin triple knockout cells reveal off target effects of commonly used dynamin inhibitors. J Cell Sci. 2013;126:5305–5312. doi: 10.1242/jcs.138578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramstedt B, Slotte JP. Sphingolipids and the formation of sterol-enriched ordered membrane domains. Biochim Biophys Acta. 2006;1758:1945–1956. doi: 10.1016/j.bbamem.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 33.Mizugishi K, et al. Essential role for sphingosine kinases in neural and vascular development. Mol Cell Biol. 2005;25:11113–11121. doi: 10.1128/MCB.25.24.11113-11121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson KR, Becker KP, Facchinetti MM, Hannun YA, Obeid LM. PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. Extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA) J Biol Chem. 2002;277:35257–35262. doi: 10.1074/jbc.M203033200. [DOI] [PubMed] [Google Scholar]
- 35.ter Braak M, et al. Galpha(q)-mediated plasma membrane translocation of sphingosine kinase-1 and cross-activation of S1P receptors. Biochim Biophys Acta. 2009;1791:357–370. doi: 10.1016/j.bbalip.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi S, et al. Identification and characterization of RPK118, a novel sphingosine kinase-1-binding protein. J Biol Chem. 2002;277:33319–33324. doi: 10.1074/jbc.M201442200. [DOI] [PubMed] [Google Scholar]
- 37.Kusner DJ, et al. The localization and activity of sphingosine kinase 1 are coordinately regulated with actin cytoskeletal dynamics in macrophages. J Biol Chem. 2007;282:23147–23162. doi: 10.1074/jbc.M700193200. [DOI] [PubMed] [Google Scholar]
- 38.Thompson CR, et al. Sphingosine kinase 1 (SK1) is recruited to nascent phagosomes in human macrophages: inhibition of SK1 translocation by Mycobacterium tuberculosis. Journal of immunology. 2005;174:3551–3561. doi: 10.4049/jimmunol.174.6.3551. [DOI] [PubMed] [Google Scholar]
- 39.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collinet C, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 41.Sutherland CM, et al. The calmodulin-binding site of sphingosine kinase and its role in agonist-dependent translocation of sphingosine kinase 1 to the plasma membrane. J Biol Chem. 2006;281:11693–11701. doi: 10.1074/jbc.M601042200. [DOI] [PubMed] [Google Scholar]
- 42.Jarman KE, Moretti PA, Zebol JR, Pitson SM. Translocation of sphingosine kinase 1 to the plasma membrane is mediated by calcium- and integrin-binding protein 1. J Biol Chem. 2010;285:483–492. doi: 10.1074/jbc.M109.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahelin RV, et al. The mechanism of membrane targeting of human sphingosine kinase 1. J Biol Chem. 2005;280:43030–43038. doi: 10.1074/jbc.M507574200. [DOI] [PubMed] [Google Scholar]
- 44.Delon C, et al. Sphingosine kinase 1 is an intracellular effector of phosphatidic acid. J Biol Chem. 2004;279:44763–44774. doi: 10.1074/jbc.M405771200. [DOI] [PubMed] [Google Scholar]
- 45.Olivera A, Spiegel S. Sphingosine kinase. Assay and product analysis. Methods Mol Biol. 1998;105:233–242. doi: 10.1385/0-89603-491-7:233. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, et al. Molecular basis of sphingosine kinase 1 substrate recognition and catalysis. Structure. 2013;21:798–809. doi: 10.1016/j.str.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 47.Kajimoto T, et al. Involvement of sphingosine-1-phosphate in glutamate secretion in hippocampal neurons. Mol Cell Biol. 2007;27:3429–3440. doi: 10.1128/MCB.01465-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 49.Hao M, Mukherjee S, Maxfield FR. Cholesterol depletion induces large scale domain segregation in living cell membranes. Proc Natl Acad Sci U S A. 2001;98:13072–13077. doi: 10.1073/pnas.231377398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishimura SY, Vrljic M, Klein LO, McConnell HM, Moerner WE. Cholesterol depletion induces solid-like regions in the plasma membrane. Biophysical journal. 2006;90:927–938. doi: 10.1529/biophysj.105.070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vrljic M, Nishimura SY, Moerner WE, McConnell HM. Cholesterol depletion suppresses the translational diffusion of class II major histocompatibility complex proteins in the plasma membrane. Biophysical journal. 2005;88:334–347. doi: 10.1529/biophysj.104.045989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirchhausen T. Bending membranes. Nat Cell Biol. 2012;14:906–908. doi: 10.1038/ncb2570. [DOI] [PubMed] [Google Scholar]
- 53.Antonny B. Mechanisms of membrane curvature sensing. Annual review of biochemistry. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 54.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 55.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu B, Hassler DF, Smith GK, Weaver K, Hannun YA. Purification and characterization of a membrane bound neutral pH optimum magnesium-dependent and phosphatidylserine-stimulated sphingomyelinase from rat brain. J Biol Chem. 1998;273:34472–34479. doi: 10.1074/jbc.273.51.34472. [DOI] [PubMed] [Google Scholar]
- 58.Tani M, Hannun YA. Neutral sphingomyelinase 2 is palmitoylated on multiple cysteine residues. Role of palmitoylation in subcellular localization. J Biol Chem. 2007;282:10047–10056. doi: 10.1074/jbc.M611249200. [DOI] [PubMed] [Google Scholar]
- 59.El Bawab S, Bielawska A, Hannun YA. Purification and characterization of a membrane-bound nonlysosomal ceramidase from rat brain. J Biol Chem. 1999;274:27948–27955. doi: 10.1074/jbc.274.39.27948. [DOI] [PubMed] [Google Scholar]
- 60.Tani M, Iida H, Ito M. O-glycosylation of mucin-like domain retains the neutral ceramidase on the plasma membranes as a type II integral membrane protein. J Biol Chem. 2003;278:10523–10530. doi: 10.1074/jbc.M207932200. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Pacios M, et al. Sphingosine-1-phosphate as an amphipathic metabolite: its properties in aqueous and membrane environments. Biophysical journal. 2009;97:1398–1407. doi: 10.1016/j.bpj.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mao C, Wadleigh M, Jenkins GM, Hannun YA, Obeid LM. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J Biol Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 63.Bandhuvula P, Saba JD. Sphingosine-1-phosphate lyase in immunity and cancer: silencing the siren. Trends Mol Med. 2007;13:210–217. doi: 10.1016/j.molmed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 64.Nakahara K, et al. The Sjogren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Molecular cell. 2012;46:461–471. doi: 10.1016/j.molcel.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 65.Lee MJ, et al. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279:1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
Reference
- 66.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen H, et al. Constitutive activated Cdc42-associated kinase (Ack) phosphorylation at arrested endocytic clathrin-coated pits of cells that lack dynamin. Mol Biol Cell. 2011;22:493–502. doi: 10.1091/mbc.E10-07-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) The Biochemical journal. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang L, Shen H, De Camilli P, Duncan JS. Tracking clathrin coated pits with a multiple hypothesis based method. Med Image Comput Comput Assist Interv. 2010;13:315–322. doi: 10.1007/978-3-642-15745-5_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- 71.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of structural biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 72.Zhu C, Das SL, Baumgart T. Nonlinear sorting, curvature generation, and crowding of endophilin N-BAR on tubular membranes. Biophysical journal. 2012;102:1837–1845. doi: 10.1016/j.bpj.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mathivet L, Cribier S, Devaux PF. Shape change and physical properties of giant phospholipid vesicles prepared in the presence of an AC electric field. Biophysical journal. 1996;70:1112–1121. doi: 10.1016/S0006-3495(96)79693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schrodinger, LLC The PyMOL Molecular Graphics System. 2010 [Google Scholar]
- 75.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. Journal of molecular biology. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 76.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.