Abstract
Purpose
To compare the precision of measuring the pennation angle and fiber length in the Vastus Lateralis (VL) using two distinctly different diffusion tensor imaging sequences.
Materials and Methods
We imaged the thigh of ten normal subjects on a 3T MR imager with twice refocused spin echo (TRSE) and stimulated echo (STEAM) DTI-MRI techniques. Both techniques took the same total acquisition time, employed the same diffusion weighting and gradient directions. Using the diffusion tensor images produced by each sequence muscle fiber bundles were tracked from the aponeurosis by following the first eigenvector of the diffusion tensor. From these tracks we calculated the pennation angle and fiber length.
Results
The STEAM acquisition resulted in significantly higher SNR, lower ADC, higher FA values and longer fibers than the TRSE. Although no difference in the pennation angle between the two acquisitions was found, the TRSE sequence had a significantly greater within subject dispersion in the pennation angle of tracked fibers which may indicate a reduction in the coherence of fiber bundles.
Conclusion
Diffusion tensor imaging of muscle using a STEAM acquisition resulted in significant improvements in the SNR and FA, resulting in tracking a larger number of muscle fiber bundles over longer distances and with less within subject dispersion.
Keywords: Quadriceps muscle, Stimulated echo, Muscle Fibers, Fractional Anisotropy, Pennation angle
Diffusion tensor imaging (DTI) has been used extensively both clinically [1] and for research [2] to map white matter fiber tracts in the central nervous system (CNS). In this application DTI takes advantage of the anisotropic structure of axons, which permits faster diffusion of water molecules parallel to and slower diffusion perpendicular to the axon. DTI has also been applied, although to a much lesser extent, for defining the micro structure of other tissues which also have an inherent structural anisotropy [3]. For example, skeletal muscles consist of bundles of individual muscle fibers, which are in turn comprised of multiple parallel protein bands called myofibrils. This protein configuration and the overall cellular geometry give muscle fibers a highly anisotropic structure. Consequently, DTI can be used to reveal the orientation of muscle fibers [4, 5].
The value of these analyses is derived partly from the observation that the spatial arrangement of muscle fibers with respect to the muscle's line of action is directly related to the muscle's ability to produce force [6, 7]. In particular, two structural phenomena, the pennation angle and muscle fiber length, are integral for assessing the ability of a muscle to generate force. Estimating muscle fiber structure through DTI in fact may be easier than deriving aspects of axonal structure because the muscle fiber diameters, which are approximately 10-90 μm, match the diffusion length within the typical diffusion time of between 10- 50 ms [8, 9]. This could be particularly helpful when assessing changes in muscle fibers as a result of injury or training, where measurements cannot currently be readily made non-invasively over the whole muscle. While there is growing interest in applying these techniques to muscle, data are still lacking regarding the effect of different DTI pulse sequences on the derived muscle fiber length and pennation angle.
The application of DTI to muscle presents a number of unique challenges that require adaptation of the twice refocused spin echo (TRSE) MR pulse sequences commonly used in the brain. One factor is the smaller T2 to T1 ratio in muscle versus the white matter in the CNS [10]. One consequence of this is that the signal-to-noise ratio (SNR) in diffusion weighted images is poorer in muscle compared to the brain [11]. An additional challenge, more important to imaging muscle than the CNS, is minimizing the contribution to the image of adipose tissue that surrounds and infiltrates the muscle. Because diffusion weighted images are typically acquired with echo planar imaging (EPI) techniques, the off-resonance protons in fat lead to displacement of the fat in the image so that it overlaps the muscle of interest [5]. The result will be a bias in the estimates of the diffusion coefficients and added variability to the estimates of the eigenvector directions [12, 13]. Because of the much smaller amount of fat in the scalp, this problem is not nearly as significant for imaging the brain. Lastly, localized eddy current-induced fields which result from the prolonged high gradient pulses to encode diffusion can further degrade the geometric fidelity and general image quality [11, 14]. These well recognized challenges have encouraged the exploration of alternative diffusion encoding strategies from the more commonly used TRSE, echo planar acquisitions typically used with DTI.
To deal with these challenges some investigators have explored using a single-echo spin echo acquisition similar to the original Stejskal-Tanner technique but with a modified gradient design to reduce eddy current generation [14-16]. Within a TRSE technique alternative bipolar gradient encoding schemes have been investigated to minimize the compromise in image quality and SNR [17]. Bipolar gradient schemes have reduced geometric distortion but at the expense of slightly longer TE than monopolar encoding schemes [18] [14]. A few groups have used a Stimulated Echo Acquisition Mode (STEAM) sequence, wherein the diffusion encoding occurs during the mixing time (TM) [8, 19]. As only T1 recovery occurs during TM, it is possible to increase TM to achieve an adequate b value while simultaneously reducing TE. The lengthened diffusion encoding time also reduces the need for high amplitude gradients, thus reducing the induced eddy currents that spatially distort the images. While in limited studies both techniques have been shown to work in muscle, a comparison between the two pulse sequences, particularly for quantitative fiber tracking applications, is lacking in the literature. Such a comparison will be of value to clinicians and researchers alike to help them optimize or choose the sequence that would work best for their particular application.
Previous studies that have evaluated either a STEAM or a TRSE sequence for imaging muscle have used different field strengths (1.5 T and 3.0T), have compared the sequences in-vivo in human muscle, run simulations, or used excised brain tissue [5, 11, 14]. Thus, the purpose of this study was to compare a TRSE DTI sequence to a STEAM DTI sequence on the Vastus Lateralis (VL) of healthy human participants.
MATERIALS AND METHODS
The comparison of TRSE and STEAM techniques in muscle imaging will likely be strongly affected by the SNR they generate. For the STEAM sequence the SNR varies as
| (1) |
The signal from the TRSE acquisition varies as
| (2) |
The sequence parameters TE and TR will typically be different for the two techniques and will depend upon the diffusion weighting chosen. The mixing time TM is, of course, unique to the STEAM sequence. The factor of ½ in Eq. (1) is a consequence of the formation of a stimulated echo. Steidle et al. and Karampinos et al. have demonstrated that beyond some intermediate diffusion weighting, the reduction in TE in STEAM benefits the SNR so significantly that it more than compensates for the factor of two reduction in SNR [11, 20]. One offsetting factor in the SNR for STEAM is that the inclusion of the mixing time increases the minimum TR and so reduces the available time for multiple excitations to contribute to an average. This leads to a reduction in SNR which, must be weighed against any increase in SNR arising from reduced TE. This factor is included in the √N terms in both Eqs. (1) and (2). Our own simulations of the SNR behavior of the two sequences based on Eqs. (1) and (2) and including the effects of averaging multiple acquisitions within a constrained total acquisition time recapitulate the basic results predicted by Steidle et al. and Karampinos et al. That is, STEAM is predicted to have a larger SNR beyond a b-weighting of approximately 200 s/mm2 [11, 20]. The exact value for b above which STEAM has superior SNR depends somewhat on the assumed relaxation times for muscle, the specifics of the pulse sequences used and the number of slices acquired. Nevertheless, the simulations point the way to achieving the best performance of the two sequences (a STEAM acquisition with a longer TR and fewer excitations than the TRSE acquisition which should have a shorter TR and more excitations). With these optimal acquisition parameters, the simulation predicts an approximately 33% increased SNR from STEAM than from a TRSE acquisition.
Lastly, for STEAM there will be a reduction in the amplitude of the signal from fat because of the exp(-TM/T1) decay term in Eq. (1). At 3.0 T, fat has a T1= 382 ms while for muscle T1= 1377 ms [21]. Because of the much shorter T1 for fat we can anticipate a greater reduction in its signal compared to muscle which will in turn reduce the contamination of the muscle signal from any superimposed fat. This benefit will not accrue to the TRSE images.
MRI
Prior to starting the study all subjects reviewed and signed an informed consent document approved by the University Institutional Review Board. The participants in the study were 25±7.3 years old and included 7 females and 3 males. Following the informed consent, subjects were imaged on a 3T MAGNETOM Trio, a Tim System (Siemens AG, Healthcare Sector, Erlangen, Germany) MR imager. They lay supine, feet-first, with a small bolster placed under their knees. The bolster helped to reduce lower back pain that would have resulted from keeping their legs fully extended. Flexing the subject's knees also slightly elongated and straightened the muscle fibers in the VL. The MR signal was received from a multi-element, phased array flexible body coil wrapped around the subject's upper thigh and centered over the VL. Once in the magnet, subjects were asked to shift their body slightly so the leg being imaged was centered in the magnet, thereby improving the B0 field homogeneity over the region of interest. Imaging began with a series of localizers to determine the position and orientation of the subject's leg and the superior and inferior extent of the VL in order to place the imaging sections uniformly along the muscle. Images were acquired perpendicular to the thigh at an axial-oblique orientation of approximately 10° matching the slight flexion of the knee in the magnet bore. For the TRSE vs. STEAM comparison, we acquired a single packet of eleven slices using each sequence; each slice was 6 mm thick with no gap between slices. Thus a packet spanned a distance of 66 mm. Before the acquisition of each packet we acquired one gradient echo image set in the same location as the diffusion weighted images. We used these images to place the volume of interest (VOI) to perform a careful shimming using a combination of automated and manual shim adjustments. The shim box was sized and oriented to include primarily muscle and exclude subcutaneous fat. Over the VOI the full width at half maximum peak height (FWHM) value after shimming typically was on the order of 30 Hz. In the same position as the diffusion images we acquired a field map using a double-echo, gradient echo technique. These images were used in the subsequent analysis to correct for spatial distortions arising from magnetic field inhomogeneities.
Both DTI acquisitions used Siemens prototype implementations. This was particularly necessary in the case of the STEAM DTI acquisition but also in the case of the TRSE acquisition, which used an alternative gradient encoding scheme to minimize the TE. The single-shot EPI acquisition parameters were TR/TE=3000/64.8 ms with number of signal excitations (Nex) of 4. This choice of parameters reflects a compromise between eddy current suppression and a desire to keep TE short. The STEAM acquisition had TR/TE=4000/36.4 ms with Nex=3. Also for the STEAM sequence, the following parameters were set: mixing time TM=173.0 ms, gradient separation Δ=185.8 ms and a gradient duration of δ=5.4ms. Both diffusion sequences used the same gradient direction table with 27 gradient directions at b= 500 s/mm2 and 4 repetitions at b=0 [22]. Both sequences used a GRAPPA factor of 2, had the same FOV = 192 mm and acquisition matrix of 96 × 96 producing image voxels with dimensions of 2 × 2 × 6 mm3 . The EPI phase encoding direction was posterior-to-anterior so as to shift subcutaneous fat away from the VL muscle in the images. Both sequences had the same total acquisition time of 6:00 min. For the STEAM technique the effect of mixing time on the diffusion tensor was assessed by varying TM from 42.5 to 173 ms for a single acquisition of a single slice in a subset of six subjects. In addition to the diffusion tensor images, high-resolution proton-density and T2-weighted fast spin echo images (1 × 1 mm2 in-plane) were acquired in order to assist with visualizing the boundaries of the muscle during data analysis. These images were acquired at the same slice positions and field of view as the two DTI data sets.
Fat suppression was achieved through a combination of techniques. First the acquisition was preceded with an adiabatic SPAIR suppression pulse. Second, the sequences alternated the polarity of the slice select gradients so that fat excited in the first pulse was not fully refocused in subsequent pulse. The image intensity of fat and muscle was measured for both techniques in the b=0 images on six slices on two of the volunteers.
Diffusion tensor calculation and fiber tracking
Eddy-current induced distortions in the diffusion weighted images were corrected by registering these images to the b=0 images as an integral part of image reconstruction rather than during data postprocessing. The saved EPI images were corrected for geometric distortion based on the gradient echo field maps and using FUGUE from the FSL software package (FMRIB, Oxford, UK). Following image registration and distortion correction, all subsequent steps were performed using custom routines in Matlab (The MathWorks Inc., Natick, MA) software. The diffusion tensor was computed from the full b-matrix information for each image voxel above a predefined SNR threshold, and values for the fractional anisotropy (FA), apparent diffusion coefficient (ADC), axial and radial diffusivities were extracted along with the direction of the principal eigenvector.
The fiber tracking was performed following previously established techniques [23-25]. First the border of the VL and its deep aponeurosis were traced separately from the b=0 image of each individual slice by an investigator (BN) who has had over 2 years of experience tracing images. The b=0 images from the STEAM acquisition were used in seeding the fiber origins for both methods since they had the best signal-to-noise properties. Also, using a common aponeurosis seeding mesh allows for a more meaningful and direct comparison of fibers tracked from the two DTI data acquisition methods. The high-resolution proton density images provided visual guidance but were not used directly for manual tracing of muscle and aponeurosis, thus circumventing the potential issue of misregistration with the DTI data. The digitized aponeurosis positions were smoothed and interpolated within and between slices to form a three-dimensional mesh consisting of 66 rows × 90 columns. The mesh points of intersection were then used as seed points to track muscle fibers. To ensure that fiber tracking would begin in the muscle itself rather than in the low-SNR region of the aponeurosis, the mesh was shifted by one pixel width. Fiber tracking was then performed by following the direction of greatest diffusion. The length of each step was chosen at 2 mm, equal to the in-plane pixel dimensions, and at each point along the track the direction vector of the next step was estimated by nearest-neighbor interpolation from known voxel values. Fibers were terminated if they left the side of the muscle, or had an FA of less than 0.10 or greater than 0.75, or a curvature greater than 45°. A low-FA threshold value of 0.10 was chosen to ensure that a sufficient number of fibers would be tracked for the lower-SNR TRSE technique to allow a comparison.
The pennation angle and fiber length were then calculated from the tracked fibers as described previously [26]; [25]. Pennation angle was calculated separately for each fiber tract as the average angle between the normal vector to the plane of the aponeurosis at the point of fiber tract insertion and the position vectors of the first five points along the length of the fiber tract. Smoothing of the plane of the aponeurosis was used to obtain robust estimates of the direction of the normal vector at each mesh point. The length of each tract was determined by summing the distance between each successive pair of points along the fiber tract, starting at the aponeurosis of fiber insertion and ending at the muscle border.
Statistical Analyses
Summary measures of SNR characteristics of the underlying data and of the DTI metrics for each of the two techniques were obtained for each study participant by averaging across voxels within the manually traced VL muscle masks. Voxels of low signal within a band along the aponeurosis in each image slice were excluded. Signal and noise amplitudes were measured in the b=0 images. The noise standard deviation was estimated within the muscle ROI itself from paired differences among the replicate b=0 images [27] . Because of the four (4) repetitions at b=0 for each acquisition, the use of paired image differences cancels out inherent fluctuations in image intensity across the muscle thus allowing for a reliable estimate of the noise level. The mean pennation angle and fiber length values were determined by Jacobian-weighted summation across individual fibers rather than as the algebraic mean. The Jacobian reflects the non-uniformity of the seeding mesh along the aponeurosis surface and allows for a weighting of the metrics by the actual aponeurosis area represented by the individual fiber tracts. (It should be noted that the number of tracks used for calculation of the mean fiber length is much smaller than the total number of fibers tracked, since only a fraction of the fibers reach the muscle border within the 66 mm length of the packet of image slices.) Within-subject fiber coherence was assessed by measuring the variability in pennation angle among individual fibers after the fitting (and removal) of gross trends in overall fiber direction. (For this assessment of variability, fiber orientation was measured in the laboratory reference system rather than with respect to the plane of the aponeurosis.) Summary statistics (means, standard deviations) were calculated for each condition, paired t-tests were used to compare the two sequences and correlations between SNR and the other outputs were made using Pearson product moment correlation coefficients. In order to assess the effect of SNR on muscle fiber tracking within each technique separately, the number of signal excitations (Nex) used for calculation of the diffusion tensor was varied from 1 to 4 for TRSE and from 1 to 3 for STEAM.
RESULTS
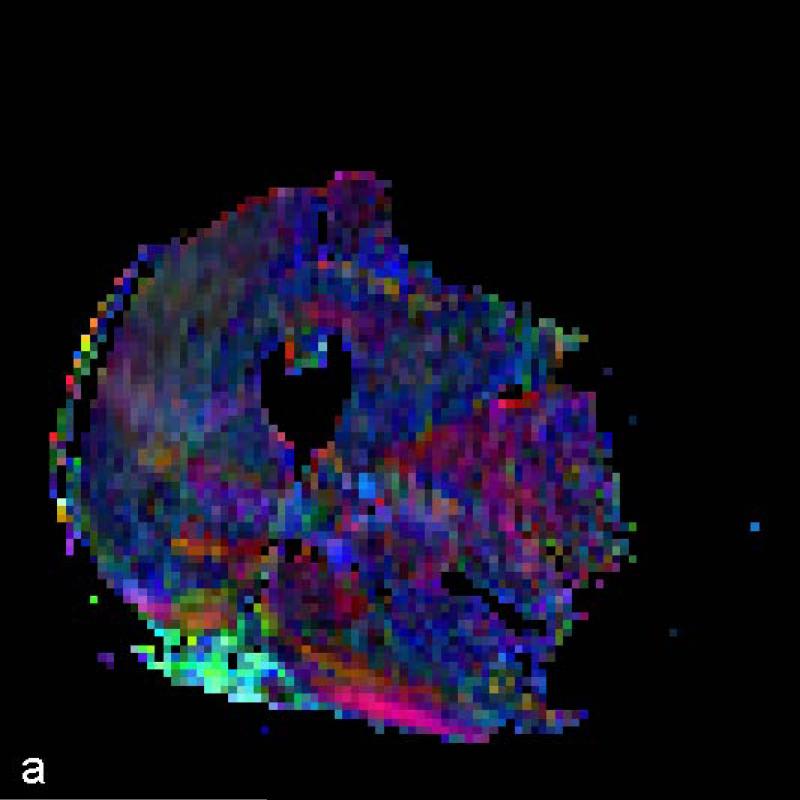
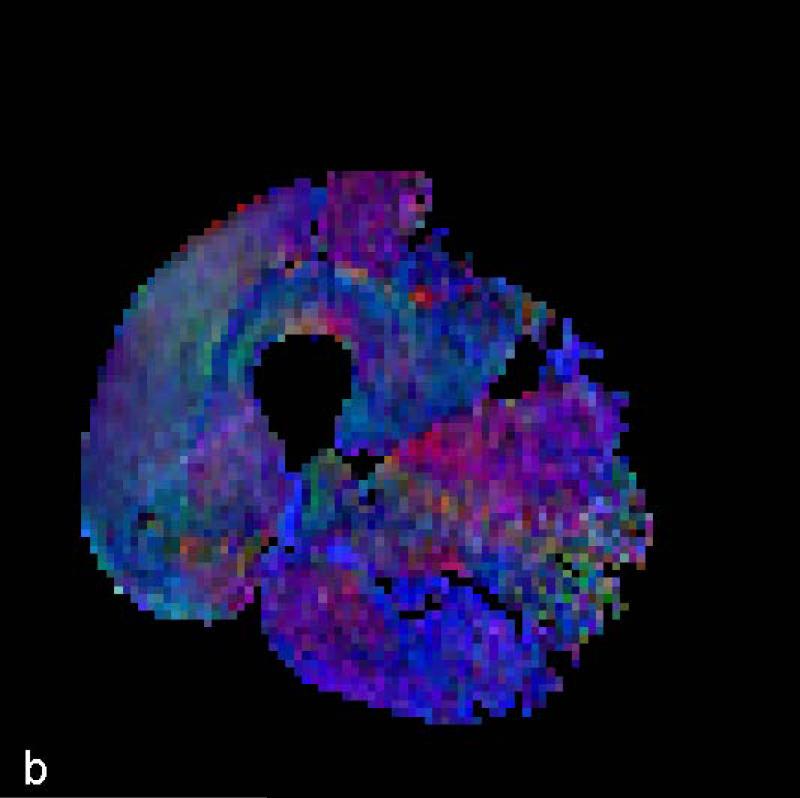
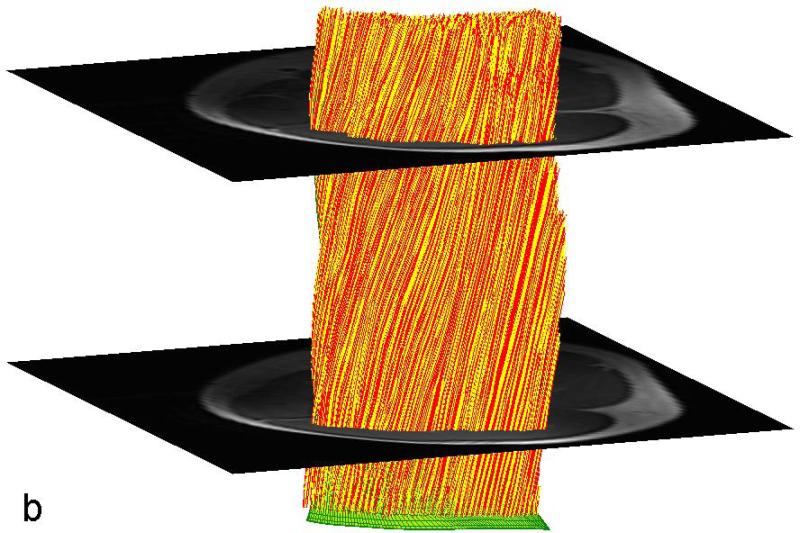
The data for each subject are presented for the STEAM sequence in Table 1 and for the TRSE sequence in Table 2. Typical color-coded fractional anisotropy (FA) images along with the b=0 image for one of the eleven slices acquired using both techniques in one volunteer are shown in Fig. 1. When comparing the two sequences we found that the STEAM acquisition resulted in a significantly greater SNR, lower ADC and higher FA values than the TRSE sequence (Table 3). The difference in SNR between the two techniques is evident in the noisier FA maps derived from the TRSE sequence (see Fig. 1). There was also a significant difference in the mean estimated fiber length, but not the mean pennation angle, between the two sequences (Table 3). While there was a significant difference in radial diffusivity between TRSE and STEAM, the measure was not correlated among subjects, indicative of poor reliability of this parameter estimate for the TRSE technique. Likewise, the subjects’ SNR values for the two techniques were uncorrelated. In comparison, the FA and axial diffusivity values as well as fiber length for the two techniques were highly correlated among subjects. Lastly, the pennation angle associated with the TRSE sequence had a significantly greater within subject standard deviation (3.09±0.58 vs. 2.10±0.64, p<0.001) than the STEAM sequence, indicative of a reduction in coherence of tracked muscle fibers. Figure 2 illustrates the fiber tracking results for the two techniques in the same volunteer. The more random and less coherent fiber orientations are evident for the TRSE acquisition.
Table 1.
Individual estimates of SNR, FA, ADC, Axial and Radial diffusivity, total number of number of fiber tracts propagated, average pennation angle and average length of tracked fiber bundles per subject derived from the STEAM acquisition.
| Subject | SNR | FA | ADC | Axial diffusivity | Radial diffusivity | Fiber Number | Pennation Angle | Fiber length |
|---|---|---|---|---|---|---|---|---|
| 1 | 38.1 | 0.33 | 1.21 | 1.67 | 0.98 | 5932 | 11.2 | 24.5 |
| 2 | 38.0 | 0.29 | 1.22 | 1.64 | 1.01 | 5940 | 16.5 | 41.1 |
| 3 | 41.2 | 0.29 | 1.32 | 1.76 | 1.10 | 5911 | 14.3 | 39.2 |
| 4 | 43.9 | 0.25 | 1.24 | 1.60 | 1.06 | 5856 | 16.4 | 32.7 |
| 5 | 36.8 | 0.28 | 1.23 | 1.62 | 1.03 | 5885 | 20.2 | 33.9 |
| 6 | 29.9 | 0.23 | 1.23 | 1.54 | 1.07 | 5699 | 21.5 | 27.0 |
| 7 | 36.8 | 0.31 | 1.21 | 1.65 | 0.99 | 5925 | 15.1 | 26.8 |
| 8 | 34.8 | 0.34 | 1.25 | 1.75 | 1.01 | 5940 | 17.4 | 39.0 |
| 9 | 46.5 | 0.31 | 1.26 | 1.72 | 1.02 | 5914 | 17.3 | 26.0 |
| 10 | 45.4 | 0.37 | 1.21 | 1.74 | 0.94 | 5849 | 14.6 | 18.4 |
Table 2.
Individual estimates of SNR, FA, ADC, Axial and Radial diffusivity, total number of number of fiber bundles tracked, average pennation angle and average length of tracked fiber bundles per subject derived from the TRSE acquisition.
| Subject | SNR | FA | ADC | Axial diffusivity | Radial diffusivity | Fiber Number | Pennation Angle | Fiber length |
|---|---|---|---|---|---|---|---|---|
| 1 | 31.3 | 0.25 | 1.52 | 1.93 | 1.31 | 5829 | 9.5 | 18.4 |
| 2 | 39.6 | 0.20 | 1.53 | 1.87 | 1.36 | 5639 | 19.5 | 40.5 |
| 3 | 34.7 | 0.23 | 1.62 | 2.02 | 1.41 | 5755 | 13.8 | 30.8 |
| 4 | 27.7 | 0.18 | 1.57 | 1.88 | 1.42 | 5551 | 15.1 | 30.3 |
| 5 | 25.4 | 0.21 | 1.51 | 1.87 | 1.33 | 5673 | 13.9 | 32.6 |
| 6 | 29.5 | 0.22 | 1.40 | 1.72 | 1.24 | 5087 | 17.5 | 16.8 |
| 7 | 26.5 | 0.26 | 1.46 | 1.88 | 1.26 | 5759 | 16.8 | 29.1 |
| 8 | 30.2 | 0.26 | 1.44 | 1.85 | 1.24 | 5671 | 13.5 | 33.1 |
| 9 | 35.1 | 0.21 | 1.54 | 1.90 | 1.36 | 5818 | 15.1 | 21.0 |
| 10 | 38.0 | 0.27 | 1.54 | 1.98 | 1.32 | 5810 | 17.9 | 18.9 |
Figure 1.
Color FA maps and b=0 image at the mid-thigh level on one volunteer derived from (a) TRSE, (b) STEAM and (c) STEAM (b=0).
Table 3.
Average values for SNR, FA, ADC, Axial and Radial diffusivity, the number of fiber tracks and average pennation angle and fiber track length comparison between STEAM and TRSE. Additionally the results of the correlation between the two sequences and the p value (last column) of the Student's t-test comparison between the two techniques for each derived quantity are given.
| STEAM | TRSE | P value of Correlation | t value | P value between sequences | |
|---|---|---|---|---|---|
| SNR | 39.1 ±5.20 | 31.8±4.80 | 0.214 | 4.35 | 0.002 |
| FA | 0.30 ±0.04 | 0.23±0.03 | 0.012 | 7.82 | 0.000 |
| ADC | 1.24 ±0.03 | 1.51±0.06 | 0.149 | 16.0 | 0.000 |
| Axial diffusivity | 1.67±0.07 | 1.89±0.08 | 0.010 | 13.1 | 0.000 |
| Radial diffusivity | 1.02±0.04 | 1.33±0.07 | 0.234 | 15.2 | 0.000 |
| Fiber Number | 5885±73 | 5659±220 | 0.002 | 4.39 | 0.002 |
| Pennation Angle | 16.4±2.94 | 15.3±2.84 | 0.238 | 1.20 | 0.262 |
| Fiber length | 30.8±7.49 | 27.2±7.88 | 0.001 | 2.90 | 0.018 |
Figure 2.
Fiber tracking results on the same volunteer derived from the primary diffusion eigen-vector. The tracks in (a) were derived from the TRSE acquisition while those in (b) were derived from the STEAM acquisition. Shown in green is the seeding mesh of the aponeurosis.
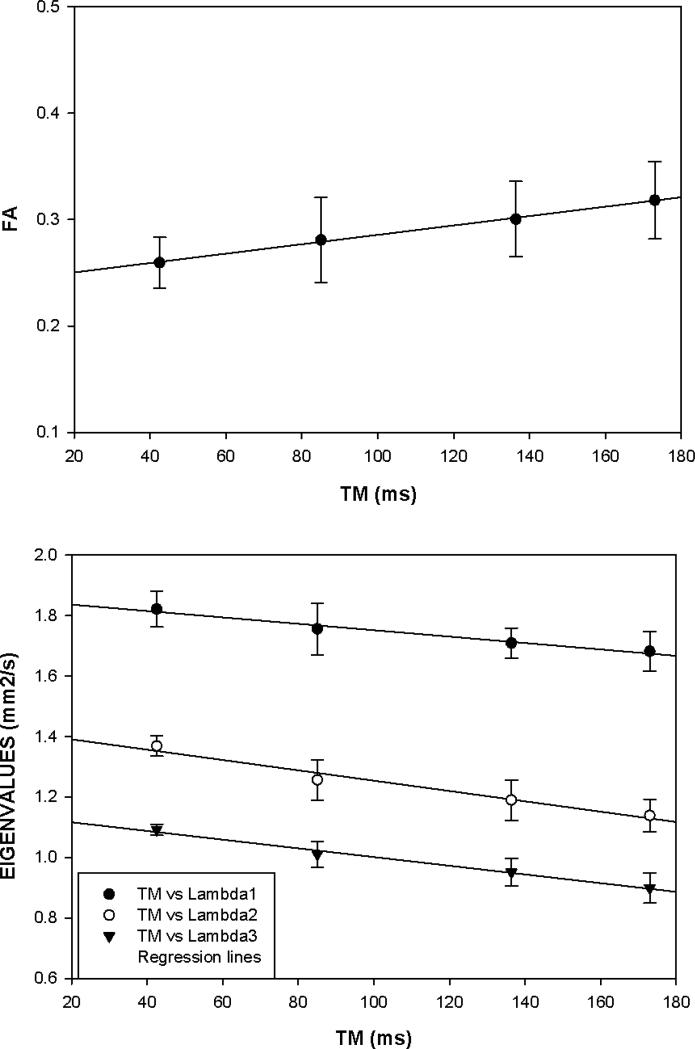
The effects of varying TM from 42.5 to 173 ms can be seen in Figure 3 which illustrates the dependence of the diffusion tensor metrics for the VL muscle ROI mask within a single slice. The eigenvalues decrease with increasing TM, with the transverse components λ2 and λ3 decreasing at a faster rate than the longitudinal λ1 leading to an overall increase in FA. The significantly shorter diffusion time for the TRSE (Δ=24.8 ms, i.e. the time duration between the start of diffusion encoding and the start of diffusion decoding) compared with that for STEAM (Δ=185.8 ms) is responsible for a considerable amount of the difference in the measured FA between the two sequences. Using the data from Figure 3 we can predict an FA reduction of 0.072 from the difference in diffusion times between the two acquisition techniques. This value is identical to the difference between the average FA (FASTEAM = 0.30, FATRSE = 0.23) derived from the ten volunteers. Similarly, the difference in diffusion times would explain a difference in the axial diffusivity of -0.16 mm2/s and in the radial diffusivity of -0.23 mm2/s for TRSE relative to STEAM. With this correction the diffusivities derived from the TRSE images would equal 1.73 mm2/s for the axial and 1.09 mm2/s for the radial diffusivity, values which are within 3.8% and 6.9% respectively of the axial and radial diffusivities of 1.67 mm2 and 1.02 mm2/s derived from the STEAM acquisition.
Figure 3.
a) Diffusion eigenvalues, λ1, λ2 and λ3 and B) average fractional anisotropy, FA as a function of the mixing time TM in the STEAM acquisition. Values plotted are averaged over the VL in one slice in six subjects. The error bars represent ± 1 standard deviation from the six subjects.
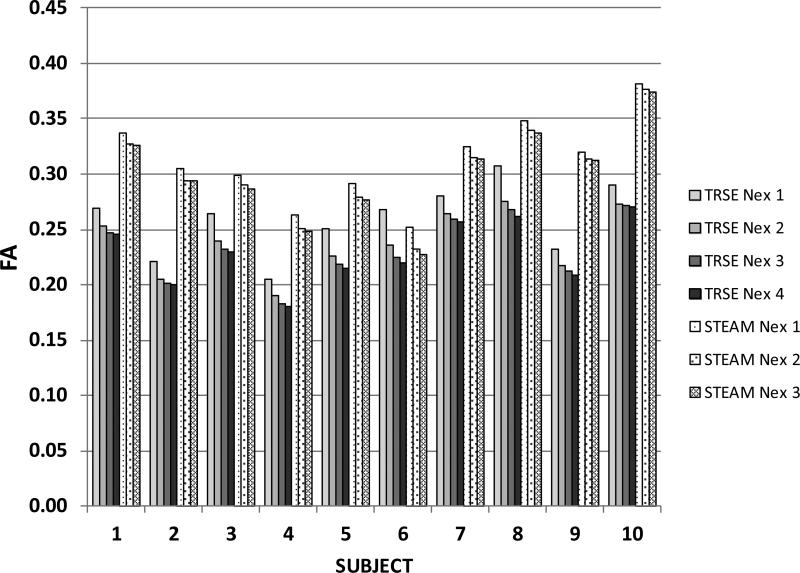
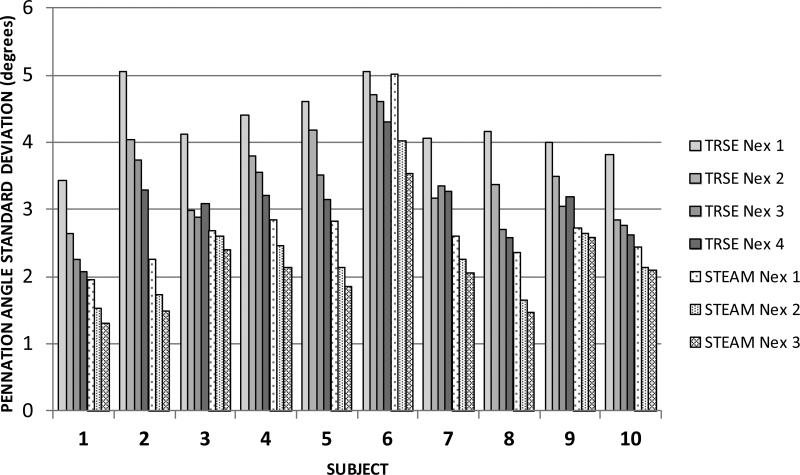
Figure 4 shows the effect of number of signal excitations on the FA. The SNR in the b=0 images followed the expected dependence (not shown). Figure 5 shows the effect of Nex on the within-subject variability in the fiber pennation angle. The variability is consistently less for STEAM than for the TRSE technique, even though the SNR may be lower for TRSE in some subjects and for some combinations of number of excitations. Lastly, the average difference in contrast between muscle and fat relative to the signal intensity for muscle, i.e. (Sm-Sf)/Sm, was 40% larger for the STEAM acquisition compared to the TRSE acquisition which demonstrates the improvement in contrast between muscle and fat for the STEAM acquisition. This improved fat suppression in STEAM arises from the exp(-TM/T1) term in Eq. (1) and because T1 for fat is much lower than for muscle.
Figure 4.
Illustrating the dependence of FA on the number of DTI signal averages for the TRSE and STEAM techniques . The values in Table 1 represent the plotted bars for Nex=4 for the TRSE and Nex=3 for the STEAM.
Figure 5.
Illustrating the dependence of variability in the measurement of fiber angle on the number of DTI signal excitations for TRSE and STEAM techniques. The values in Table 1 record the values plotted for Nex=4 for the TRSE technique and Nex=3 for the STEAM technique.
DISCUSSION
There is growing interest in applying DTI-MRI to study the micro-structure of tissues such as muscle. However, using DTI-MRI in tissues other than the brain has a number of challenges that need to be overcome before routine application of the technique can begin. The purpose of this study was to compare two common approaches to study muscle structure using DTI-MRI. We found that the STEAM acquisition sequence had significantly higher SNR, FA, and lower ADC values than the TRSE acquisition.
These differences, particularly the larger SNR, translated into significantly more reconstructed fiber tracks and improved coherence in the tracks derived from the STEAM acquisitions. A range of values for SNR ratio of the b=0 images of 25-60 has been reported as the minimum needed to reconstruct muscle fibers [13, 27]. The average SNR values for both the STEAM (39.1) and TRSE sequence (31.8) were above this lower threshold. To a great extent the difference in SNR is expected based on our predictions for the two techniques. Equations (1) and (2) predict a ratio of the SNR at b=500 s/mm2 of 1.39 while the measured ratio was 1.23. The difference could arise from differences between the actual and the assumed relaxation times of muscle. Comparing the two techniques, the largest source of the higher SNR in the STEAM sequence is the reduced TE afforded by the sequence structure. The anticipated improvement in the contrast between fat and muscle as a consequence of the decay of the fat signal during the mixing time was observed in the images produced by the STEAM acquisition. The reduction in the signal from fat presents an additional benefit to STEAM as it reduces the bias to the signal in muscle from superimposed signal from fat and so improves the fidelity of the muscle diffusion weighted measurements.
To study the potential effect of SNR on FA for each of the techniques separately, we varied the number of excitations used for calculation of the diffusion tensor. The observed but perhaps counterintuitive slight increase in FA value that accompanies a reduction of SNR is consistent with results from prior simulation studies [13, 27]. Not captured by the mean within the muscle ROI mask, however, is the voxel-to-voxel variability in FA value; this noisiness is evident in the spatial FA maps and leads to greater variability in fiber orientation (Fig. 2). The FA values in the current study were significantly higher and ADC values lower in the STEAM sequence as compared to the TRSE sequence, although the higher FA in the STEAM sequence were not directly associated with the improved SNR. In addition, even though the SNR from the STEAM sequence in the current study was lower than a previous report on DTI in the thigh, the FA values between our study and the previous study were similar [24]. Also, these results are consistent with another study which reported adjusting the diffusion time may be an important factor to consider in DTI acquisitions of muscle [11]. More recently, Sigmund et al. have argued that longer diffusion times are necessary when imaging muscle versus the brain to account for the longer fiber length of muscle [28]. An alternative view was offered by Kim et al., who showed that that very long diffusion times will reduce SNR; they advocated a trade-off between diffusion time and SNR to achieve optimal results [8].
The differences between sequences propagated to fiber length measurements but not the pennation angles. The values for the pennation angle of the Vastus Lateralis were similar to another paper using a single echo SE sequence on the Vastus Lateralis of the thigh [24]. Because the FA values were lower in the TRSE sequence we had to lower the minimum FA value down to 0.1 to track a sufficient number of fibers until they exited the muscle. The lower FA coupled with a lower SNR resulted in the fibers being more variably oriented. This was evidenced by a greater within-subject standard deviation of the fiber orientation for the TRSE sequence than the STEAM sequence. The detrimental effect of low SNR on fiber coherence as reflected by greater within-subject variation in the fiber angle estimate was evident for each DTI method separately as well. A similar effect of increased uncertainty in the fiber orientation with decreasing SNR has been demonstrated in a Monte Carlo simulation study [13]. This likely carried over to the determination of fiber length as well, resulting in greater premature early termination of fibers in the TRSE vs. STEAM sequence. Interestingly, the values achieved for fiber length though are lower than what has been previously reported from cadaveric dissection [29]. There were several potential reasons for this difference. As we were primarily interested in comparing the two sequences, only one 66 mm packet was taken at mid-thigh. The termination of the fibers was determined as they exited the side of the ROI, thus we likely did not have a sufficiently long VOI to track the physiological length of each fiber. Other potential differences between previous cadaveric dissections and our measurements could be attributed to partial volume effects and having to shift 1 pixel or 2 mm off the aponeurosis to start tracking. However, the results allowed us to compare the fiber tracking between two commonly used diffusion tensor acquisitions.
This study is not without limitations. First we were unable to exactly replicate TRSE sequences used in previous studies because of differences in the hardware of the imager platforms such as RF receiver coils, gradient strength and slew rates and also differences in software such as differences in pulse sequences available from different vendors. Thus the echo time in the TRSE sequence used was slightly longer than that used in other DTI studies on muscle. While this will affect the SNR directly, reducing TE will also result in a lower FA because of the shorter diffusion time. The higher SNR achieved by STEAM coupled with a higher FA makes tracking muscle fiber bundles easier.
In conclusion, within the same total imaging time, the STEAM sequence produced significantly higher SNR, FA and lower ADC values. Using the diffusion tensor images derived from the STEAM acquisition, we could track significantly more muscle fiber bundles with less within subject dispersion in estimating the pennation angle. While both sequences were able to successfully reconstruct fibers, the STEAM sequence offers greater sensitivity to detect subtle differences in FA and fiber orientation. This improved sensitivity might be valuable when using DTI-MRI to detect muscle injury or the effects of exercise for training or rehabilitation.
Acknowledgement
Grant support: NIH/NIAMS R01 AR050101, UL1TR000117, K23AR062069
REFERENCES
- 1.Le Bihan D, Mangin JF, Poupon C, et al. Diffusion tensor imaging: concepts and applications. J Magn Reson Imaging. 2001;13:534–46. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- 2.Xue R, van Zijl PC, Crain BJ, Solaiyappan M, Mori S. In vivo three-dimensional reconstruction of rat brain axonal projections by diffusion tensor imaging. Magn Reson Med. 1999;42:1123–7. doi: 10.1002/(sici)1522-2594(199912)42:6<1123::aid-mrm17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 3.Le Bihan D, Mangin JF, Poupon C, et al. Diffusion tensor imaging: concepts and applications. J Magn Reson Imaging. 2001;13:534–46. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- 4.Damon BM, Ding Z, Anderson AW, Freyer AS, Gore JC. Validation of diffusion tensor MRI-based muscle fiber tracking. Magn Reson Med. 2002;48:97–104. doi: 10.1002/mrm.10198. [DOI] [PubMed] [Google Scholar]
- 5.Sinha S, Sinha U, Edgerton VR. In vivo diffusion tensor imaging of the human calf muscle. J Magn Reson Imaging. 2006;24:182–90. doi: 10.1002/jmri.20593. [DOI] [PubMed] [Google Scholar]
- 6.Lieber RL, Fridén J. Functional and clinical significance of skeletal muscle architecture. Muscle & Nerve. 2000;23:1647–1666. doi: 10.1002/1097-4598(200011)23:11<1647::aid-mus1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 7.Lieber RL, Ward SR. Skeletal muscle design to meet functional demands. Phil. Trans. R. Soc. B. 2011;366:1466–1476. doi: 10.1098/rstb.2010.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Chi-Fishman G, Barnett AS, Pierpaoli C. Dependence on diffusion time of apparent diffusion tensor of ex vivo calf tongue and heart. Magnetic Reson Med. 2005;54:1387–96. doi: 10.1002/mrm.20676. [DOI] [PubMed] [Google Scholar]
- 9.Polgar J, Johnson MA, Weightman D, Appleton D. Data on fibre size in thirty-six human muscles. An autopsy study. J Neurol Sci. 1973;19:307–18. doi: 10.1016/0022-510x(73)90094-4. [DOI] [PubMed] [Google Scholar]
- 10.Schick F, Eismann B, Jung WI, Bongers H, Bunse M, Lutz O. Comparison of localized proton NMR signals of skeletal muscle and fat tissue in vivo: two lipid compartments in muscle tissue. Magn Reson Med. 1993;29:158–67. doi: 10.1002/mrm.1910290203. [DOI] [PubMed] [Google Scholar]
- 11.Steidle G, Schick F. Echoplanar diffusion tensor imaging of the lower leg musculature using eddy current nulled stimulated echo preparation. Magn Reson Med. 2006;55:541–8. doi: 10.1002/mrm.20780. [DOI] [PubMed] [Google Scholar]
- 12.Williams SE, Heemskerk AM, Welch EB, Li K, Damon BM, Park JH. Quantitative effects of inclusion of fat on muscle diffusion tensor MRI measurements. J Magn Reson Imaging. 2013:1292–1297. doi: 10.1002/jmri.24045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damon BM. Effects of image noise in muscle diffusion tensor (DT)-MRI assessed using numerical simulations. Magn Reson Med. 2008;60:934–44. doi: 10.1002/mrm.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finsterbusch J. Double-spin-echo diffusion weighting with a modified eddy current adjustment. Magn Reson Imaging. 2010;28:434–40. doi: 10.1016/j.mri.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Finsterbusch J. Eddy-current compensated diffusion weighting with a single refocusing RF pulse. Magn Reson Med. 2009;61:748–54. doi: 10.1002/mrm.21899. [DOI] [PubMed] [Google Scholar]
- 16.Stejskal E, Tanner J. Spin diffusion measurements: Spin echoes in the presence of a time-dependent field gradient. J. Chem Phys. 1965;42:288–292. [Google Scholar]
- 17.Feiweier T. International Society of Magnetic Resonance Imaging. Montreal, Canada: 2011. Bipolar Diffusion Encoding with Implicit Spoiling of Undesired Coherence Pathways. [Google Scholar]
- 18.Reese NB, Bandy WD. Use of an inclinometer to measure flexibility of the iliotibial band using the Ober test and the modified Ober test: differences in magnitude and reliability of measurements. J Orthop Sports Phys Ther. 2003;33:326–30. doi: 10.2519/jospt.2003.33.6.326. [DOI] [PubMed] [Google Scholar]
- 19.Tanner J. Use of the stimulated echo in NMR diffusion studies. J. Chem Phys. 1970:52. [Google Scholar]
- 20.Karampinos DC, Banerjee S, King KF, Link TM, Majumdar S. Considerations in high-resolution skeletal muscle diffusion tensor imaging using single-shot echo planar imaging with stimulated-echo preparation and sensitivity encoding. NMR in Biomed. 2012;25:766–778. doi: 10.1002/nbm.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bazelaire CM, Duhamel GD, Rofsky NM, Alsop DC. MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology. 2004;230:652–9. doi: 10.1148/radiol.2303021331. [DOI] [PubMed] [Google Scholar]
- 22.Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magnetic Reson Med. 1999;42:515–25. [PubMed] [Google Scholar]
- 23.Heemskerk AM, Sinha TK, Wilson KJ, Ding Z, Damon BM. Quantitative assessment of DTI-based muscle fiber tracking and optimal tracking parameters. Magn Reson Med. 2009;61:467–72. doi: 10.1002/mrm.21819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kan JH, Heemskerk AM, Ding Z, et al. DTI-based muscle fiber tracking of the quadriceps mechanism in lateral patellar dislocation. J Magn Reson Imaging. 2009;29:663–70. doi: 10.1002/jmri.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lansdown DA, Ding Z, Wadington M, Hornberger JL, Damon BM. Quantitative diffusion tensor MRI-based fiber tracking of human skeletal muscle. J Appl Physiol. 2007;103:673–81. doi: 10.1152/japplphysiol.00290.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heemskerk AM, Sinha TK, Wilson KJ, Ding Z, Damon BM. Quantitative assessment of DTI-based muscle fiber tracking and optimal tracking parameters. Magnetic Resonance Medicine. 2009;61:467–72. doi: 10.1002/mrm.21819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froeling M, Nederveen AJ, Nicolay K, Strijkers GJ. DTI of human skeletal muscle: the effects of diffusion encoding parameters, signal-to-noise ratio and T2 on tensor indices and fiber tracts. NMR in Biomed. 2013;26:1339–1352. doi: 10.1002/nbm.2959. [DOI] [PubMed] [Google Scholar]
- 28.Sigmund EE, Sui D, Ukpebor O, et al. Stimulated echo diffusion tensor imaging and SPAIR T(2) -weighted imaging in chronic exertional compartment syndrome of the lower leg muscles. J Magn Reson Imaging. 2013 doi: 10.1002/jmri.24060. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker I, Baxter GD, Woodley SJ. The vastus lateralis muscle: An anatomical investigation. Clin Anat. 2010;23:575–585. doi: 10.1002/ca.20974. [DOI] [PubMed] [Google Scholar]