Abstract
We examined the genotype-phenotype interactions of Cyp51+/− mice carrying one functional allele of lanosterol 14α-demethylase from cholesterol biosynthesis. No distinct developmental or morphological abnormalities were observed by routine visual inspection of Cyp51+/− and Cyp51+/+ mice and fertility was similar. We further collected a large data-set from female and male Cyp51+/− mice and controls fed for 16 weeks with three diets and applied linear regression modeling. We used 3 predictor variables (genotype, sex, diet), and 39 response variables corresponding to the organ characteristics (7), plasma parameters (7), and hepatic gene expression (25). We observed significant differences between Cyp51+/− and wild-type mice in organ characteristics and blood lipid profile. Hepatomegaly was observed in Cyp51+/− males, together with elevated total and low-density lipoprotein cholesterol. Cyp51+/− females fed high-fat, high-cholesterol diet were leaner and had elevated plasma corticosterone compared to controls. We observed elevated hepatocyte apoptosis, mitosis and lipid infiltration in heterozygous knockouts of both sexes. The Cyp51+/− females had a modified lipid storage homeostasis protecting them from weight-gain when fed high-fat high-cholesterol diet. Malfunction of one Cyp51 allele therefore initiates disease pathways towards cholesterol-linked liver pathologies and sex-dependent response to dietary challenge.
Introduction
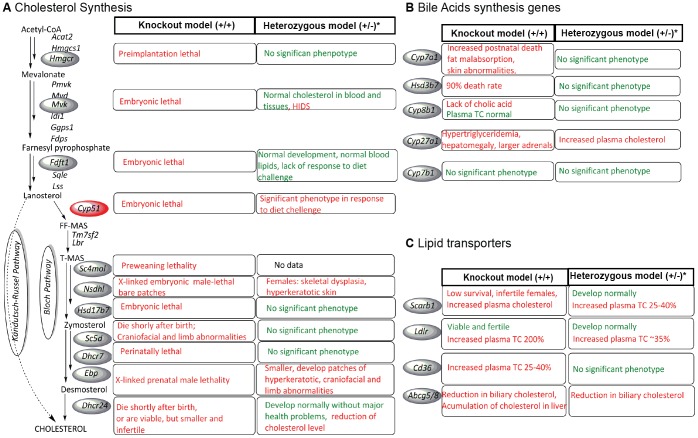
Cholesterol, an essential compound of cell membranes, regulates permeability, fluidity, and membrane signaling capacity [1], is a precursor of steroid hormones and bile acids, and plays an important role in cell proliferation [2], [3]. Cholesterol originates from two sources – the dietary intake (30–50%) and de novo synthesis (50–70% in men) [4]. Its abnormal blood concentration leads to the increased risk of heart diseases and brain strokes. Thus, regulation on the cellular level and on the level of the entire organism is essential [5]. The lipid homeostasis is performed mainly by the liver, the major organ of lipid clearance [6] and synthesis. Almost 40% of the cholesterol is synthesized in the murine liver [7], and the pathway is well conserved in mammals. The loss of function of genes from cholesterol synthesis, metabolism or transport results in lethality or other serious conditions, where the severity of the phenotype depends on the position of gene in the pathway [8], [9], [10], [11]. Most murine studies focus on the complete knockout models that are unlikely to be found in humans, due to the lethal developmental phenotype, while mice heterozygous for the cholesterol-linked genes seldom present a distinct phenotype (Figure 1). However, the cholesterol homeostasis in humans exhibits examples where abnormalities manifest with the heterozygous variants, such as in the genes of cholesterol synthesis (HMGCR, DHCR7, DHCR24 and CYP51A1), where polymorphisms associate with preterm delivery or low birth weight [12], [13], [14].
Figure 1. Characteristics of knock-out and heterozygous knock-out mouse models.
Figure shows mouse characteristics in the cholesterol synthesis (A), bile acid synthesis pathways (B) and lipid transporters (C). In red are marked disease phenotypes, while in green are marked mild/no phenotype. *The heterozygous knock-out models often were not thoroughly investigated. Gene names and references to literature describing mouse models: Acat2 acetyl-coenzyme A acetyltransferase 2; Hmgcs1 3-hydroxy-3-methylglutaryl-coa synthase 1; Hmgcr 3-hydroxy-3-methylglutaryl-coa reductase [55]; Pmvk phosphomevalonate kinase; Mvd diphosphomevalonate decarboxylase; Mvk mevalonate kinase [56]; Idi1 isopentenyl-diphosphate delta isomerase 1; Ggps1 geranylgeranyl diphosphate synthase 1; Fdps farnesyl diphosphate dynthase; Fdft1 farnesyl-diphosphate farnesyltransferase 1 [57]; Sqle squalene epoxidase; Lss lanosterol synthase; Cyp51 lanosterol 14-alpha-demethylase [17], Tm7sf2 C-14 sterol reductase; Lbr lamin-B receptor; Sc4mol methylsterol monooxygenase 1*(WTSI); Nsdhl NAD(P) dependent steroid dehydrogenase-like [58], [59]; Hsd17b7 hydroxysteroid (17-beta) dehydrogenase 7 [60]; Sc5d lathosterol oxidase [61]; Dhcr7 7-dehydrocholesterol reductase [62]; Ebp emopamil binding protein (sterol isomerase) [63]; Dhcr24 24-dehydrocholesterol reductase [41], [64]; Cyp7a1 cholesterol 7-alpha-monooxygenase [65], [66]; Hsd3b7 3 beta-hydroxysteroid dehydrogenase type 7 [67]; Cyp8b1 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase [68]; Cyp27a1 Sterol 26-hydroxylase [69]; Cyp7b1 25-hydroxycholesterol 7-alpha-hydroxylase [70]; Scrab1 scavenger receptor class b, member 1 [71]; Ldlr low density lipoprotein receptor [72]; Cd36 fatty acid translocase [73], [74]; Abcg5 ATP-binding cassette sub-family g member 5; Abcg8 ATP-binding cassette sub-family g member 8 [75].
The focus of our study is lanosterol 14α-demethylase CYP51, a cytochrome P450 from the cholesterol biosynthesis pathway. In humans, the CYP51A1 shows low nucleotide variability compared to other genes of the pathway and other related cytochrome P450 genes [15]. The mouse Cyp51 gene is 89% identical to the human counterpart [16]. The complete knockout of Cyp51 is embryonically lethal in mice [17]. In humans homozygous CYP51A1 dysfunctions have not been detected so far, as they probably spontaneously abort in early development. Cyp51 is likely not essential for normal spermatogenesis [18] even if the products of lanosterol demethylation might serve as signaling sterols [19]. In humans, CYP51A1 was hemizygously deleted in a family with cerebral cavernous malformations [20], and the gene was proposed as a candidate for the cause of pediatric cataracts [21]. The CYP51A1 heterozygous common variant (rs6465348) associates with the decreased birth weight in preterm babies and with the changed lipid profile in pregnant women [12]. Due to the crucial role of cholesterol synthesis for organisms’ integrity, the reported associations of CYP51A1 polymorphisms with brain function or early development likely represent only a portion of potential malformations caused by dysfunction of CYP51.
To assess the global role of decreased expression of Cyp51 by 50%, we investigated a large colony of Cyp51 heterozygous knockout (Cyp51+/−) mice of both sexes challenged with three diets differing in the fat and/or cholesterol content. The statistical modeling revealed novel associations between the Cyp51 genotype, the nutrition and the lipid homeostasis. Those factors, together with histopathology pinpointed to the liver as the most prominent disease target organ.
Materials and Methods
Animals
Heterozygous males Cyp51+/− (B6.129SV-Cyp51<tm1.1Bfro>) were obtained from the Department of Animal Science, Biotechnical Faculty, University of Ljubljana and mated with C57BL/6JOlaHsd females (Harlan, Italy). The experiments were performed at the Medical Experimental Center, Medical Faculty, University of Ljubljana. All the animals of over 95% of C57BL/6J background were housed 3–7 per cage (825 cm2 floor space) on Lignocel ¾ bedding material (Germany) in a 22–23°C room, 55±10% humidity and a 12 h light/dark cycle (illumination between 07.00 am −07.00 pm) with free access to food and acidified tap water (pH = 3). The experiment was approved by the Ethical Committee for laboratory animals of the Republic of Slovenia and Veterinary Administration of the Republic of Slovenia granted permit numbers 34401-31/2011/4 and 34401-29/2011/3. The experiment was conducted in accordance with the Directive 2010/63/EU on the protection of animals used for scientific purposes, as well as in accordance with National Institutes of Health guidelines for work with laboratory animals.
Diet regime
Litters were weaned at age of 3 weeks, genotyped as described [17] and randomly distributed to one of the diets for period of 16 weeks (8–15 mice/group; each group represents mice of specific gender and genotype on one of the diets):
LFnC (Low Fat no Cholesterol diet) (Item no. 1324 LFnC GmbH & Co. KG, Germany), a control diet with no cholesterol, supplying 2844 kcal/kg where 65% of the calories derives from carbohydrates, 24% from proteins and 11% from fat.
HFnC (High Fat no Cholesterol diet) – High Fat Clinton-Cybulsky rodent diet without cholesterol (D12106C, Research Diets INC, USA) supplying 4056 kcal/kg where 40.0% of the calories derives from carbohydrates, 20.0% from proteins and 39.9% from fat.
HFC (High Fat diet with Cholesterol) - High Fat Clinton-Cybulsky rodent diet with 1.25% cholesterol (D12108C, Research Diets INC, USA) supplying 4056 kcal/kg where 40.0% of the calories derives from carbohydrates, 20.0% from proteins and 39.9% from fat.
Organ and blood collection
A total of 142 mice were sacrificed after a 5 h fast with cervical dislocation during the light period (11.30 am −2.30 pm) to limit diurnal variability. During the autopsy the mice were weighed; organs were removed, trimmed of adipose tissue, weighed and patho-morphologically examined. For the histology, left lateral lobe (lobus sinister lateralis) was fixed in 4% formalin and embedded in paraffin, sectioned (5 µm) and stained with hematoxylin and eosin. The rest of the liver was cut into small slices, snap frozen in liquid nitrogen and stored at −80°C for further analyses. Immediately after cervical dislocation, the blood was taken from the right ventricle, collected into lithium-heparin coated tubes, and centrifuged for 15 minutes at 4°C at 3000×g; plasma was collected, snap frozen in liquid nitrogen, and stored at −80°C.
Histology
Liver sections of each mouse were scored individually for different parameters such as size, shape and polymorphism of the hepatocytes, nuclei and nucleoli, granulation of the cytoplasm, presence of inflammation, cholestasis, steatosis, apoptosis, mitosis, and potential abnormalities of bile ducts. Histological scoring was performed on hematoxylin and eosin stained sections in a blinded fashion by a pathologist, with a scoring system for steatosis as follows: no steatosis was assigned when less than 5% of hepatocytes contained lipid droplets, mild steatosis when 5–25% of hepatocytes contained lipid droplets, moderate when 25–50% of hepatocytes contained lipid droplets, severe steatosis when lipid droplets were present in 50–75% of hepatocytes, and very severe when lipid droplets were present in more than 75% of hepatocytes. Apoptoses and mitoses were counted on the whole liver section. Liver samples were taken in the same way for every mouse (vertical cut in the middle of the right lobe) to exclude variability due to technical preparation of the sample. Apoptotic cells were recognized by characteristic morphological features, such as chromatin condensation, cell shrinkage, and nuclear fragmentation [22]. Mitotic figures were identified according to the criteria proposed by van Diest et al [23]. Only cells with clear morphological features of metaphase, anaphase, and telophase were counted, avoiding apoptotic and hyperchromatic nuclei.
Blood parameters
Plasma was analyzed by the Veterina Le Marechal d.o.o, Ljubljana by Architect Ci 8200 (Abbott Corporation, USA) using methods recommended by The International Federation of Clinical Chemistry and Laboratory Medicine (IFCC) and the Clinical and Laboratory Standards Institute (CLSI), with original or recommended reagents: Total Cholesterol (TC) with Architect/Aeroseth 7D62 (Abbott), High Density Lipoprotein Cholesterol (HDL) with Architect/Aeroseth 3K33-20 (Abbott), Triglycerides (TG) with Architect/Aeroseth 7D74 (Abbott) and Free Fatty Acids (FFA)/Non-Esterified Fatty Acids NEFA-HR (2) 436–91995 (Wako Chemicals GmbH). Non-HDL cholesterol (LDL) was calculated using the Friedewald equation [24]: LDL [mmol] = TC [mmol]-HDL [mmol]-(TG [mmol])/2.2).
Corticosterone was measured in plasma (5 animals/group, in duplicates) by a Corticosterone (Rat/Mouse) ELISA kit (EIA-5186, DRG Instruments GmbH) according to the manufacturer’s instructions. Testosterone measurements in the males’ plasma were measured with the Testosterone (Rat/Mouse) ELISA kit (EIA-5179, DRG Instruments GmbH) according to the manufacturer’s instructions. The average absorbance of samples was used for curve-fitting with Four Parameter Logistics using the ReaderFit software (https://www.readerfit.com/).
Gene expression analyses
The total RNA was extracted from 30 mg of liver using the QuickGene RNA tissue kit SII (FujiFilm). RNA concentration was measured with NanoDrop ND-1000 and the quality (RNA Integrity Number) checked on RNA chips (Agilent). The total RNA (3 µg) in water was treated with amplification grade DNase I (Roche) by standard protocol and used immediately for the cDNA synthesis with the SuperScript III cDNA Synthesis Kit (Invitrogen). Real-time qPCR was performed with the LightCycler 480 using SYBR Green I Master according to instructions (Roche Applied Science). Primers were designed using the Primer3 software in a way that they would span over the intron and validated with melting and standard curve analyses (Table S1). The reaction was performed in a 5 µl volume using 3.75 ng of cDNA, 1.15 µl of water, 2.5 µl of SYBR Green I Master 2×, 0.3 µl of each 2.5 mM primers with the thermocycling program: 95°C for 10 min, then 95°C for 10 s, 60°C for 30 s, and 72°C for 5 s for 45 cycles plus a dissociation step (60–95°C). The relative expression was calculated as previously described [25]. Rplp0, Utp6c and Eif2a were selected for normalization as previously described [26].
Western Blot Analysis
The proteins were isolated from liver that was homogenized in lysis buffer (30 mg of tissue in 1 mL: 20 mM TRIS pH 7.5, 150 mM NaCl, 1% NP-40, 10% glycerol, 5 mM EDTA, 1 mM PMSF and protease inhibitor cocktail Complete Roche) and incubated on an orbital shaker for 1 h at 4°C followed by 15 min centrifugation at 4°C and 12000 rpm. The protein concentration was measured with Pierce BCA Protein Assay kit (Thermo Scientific). The samples were separated by 12% SDS-PAGE and electro-transferred onto a PVDF membrane (Millipore, Eschborn, Germany). The membranes were blocked with 5% (w/v) nonfat dry milk in PBST containing 0.05% or 0.1% (v/v) Tween 20 for 1 h at room temperature. The membranes were then incubated with the appropriate primary antibodies (self-made rabbit polyclonal anti-CYP51 antibodies against the peptide QRLKDSWAERLDFNPDRY, 1∶2000; anti-HMGCR, 1∶1000, ABS229, Millipore, Billerica, MA, USA; anti-NSDHL, 1∶250, 15111-1-AP, Proteintech Group, Inc., Chicago, IL, USA; anti-TM7SF2, 1∶250, 12033-1-AP, Proteintech Group, Inc., Chicago, IL, USA; anti-GAPDH antibodies, 1∶5000, G9545, Sigma Aldrich, St. Louis, MO, USA). Visualization was performed by goat anti-rabbit IgG peroxidase conjugate (1∶5000, A0545, Sigma Aldrich, St. Louis, MO, USA) with 5% nonfat dry milk in PBST and chemiluminescence recorded.
Statistical analysis
The data was processed and analyzed using different R [27] and Bioconductor [28] packages. Linear regression modeling was employed using three predictor variables corresponding to genotype (Cyp51+/− and Cyp51+/+), sex (Female and Male) and diet (LFnC, HFnC and HFC) and 39 response variables corresponding to organ characteristics (7 variables), plasma parameters (7 variables), and hepatic gene expression (25 variables).
The package car [29] was used to estimate univariate normalizing transformation of the response variables by Box-Cox power transformation family on the predictors. The Box-Cox transformation of data was necessary, as gene expression results rarely present normal distribution and they were transformed with parameter λ close to 0, which is similar to logarithmic transformation. The transformed response variables were modeled using a linear modeling approach and empirical Bayes smoothing to the standard errors by the limma package [30]. A linear model was fitted using three predictor variables and their pairwise interactions.
Benjamini-Hochberg's method for controlling the false discovery rate (FDR) [31] was used to determine statistically significant response variables at significance level of α = 0.1. The significance level of α = 0.1 was chosen, due to small number of animals analyzed in each group. We wished to assess diet/gender/genotype effects on multiple and heterogeneous levels (i.e. globally). As this is (to our best knowledge) the first study of such kind, we have chosen a rather relaxed level of statistical significance (α = 0.1) in order not to miss more subtle, but important effects. The parameters that reached statistical difference close to α = 0.1 will be discussed as the trend and tendencies, but the main focus of the manuscript were differences significant at the level of α = 0.05. Differences between genotypes were assessed across all the mice as well as separately for each diet, sex and their combinations. HFnC and HFC diets were compared to the LFnC diet across all the mice as well as separately for each sex and genotype. Interaction between the diets and genotypes were estimated across all the mice, as well as separately for each sex. Figure S1 presents the hierarchical tree for assessing the Cyp51 genotype effect. Particular contrasts within the tree allow also studies of the diet – genotype interactions. The numbers of animals analyzed within each contrast (organs characteristics, plasma parameters, liver histology and hepatic gene expression) are shown in Table S2.
Results
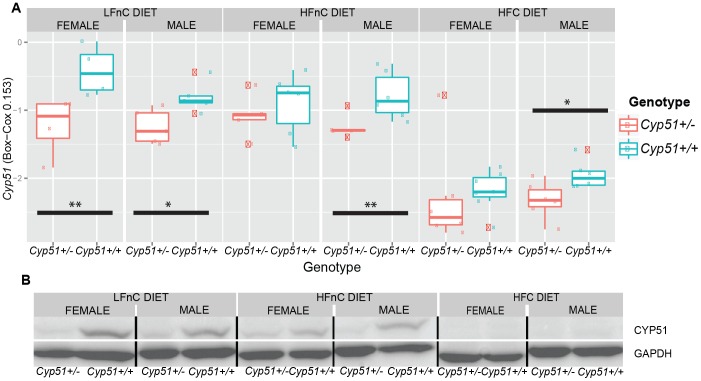
The Cyp51+/− animals on the standard laboratory diet (LFnC) develop normally based on standard visual inspection and as expected they reproduce normally [18]. The Cyp51+/− heterozygotes show a decreased expression of Cyp51 mRNA and protein compared to the Cyp51+/+ controls on each diet (Figure 2), indicating that Cyp51 is transcribed from both alleles with no compensatory mechanisms in Cyp51+/− mice. In wild types, the expression is higher on the two diets with no cholesterol (LFnC and HFnC compared to HFC), irrespective of the presence of excess dietary fats (HFnC) and is expectedly repressed on the high-fat, high-cholesterol (HFC) diet due to the cholesterol feedback regulation. A thorough investigation of the blood lipid panel, liver histology, and gene expression by linear regression modeling revealed other significant differences between Cyp51+/− mice and wild types.
Figure 2. The expression of Cyp51 (A) on mRNA and (B) protein levels (western blot analysis).
The global effect of the Cyp51+/− genotype in males and in females
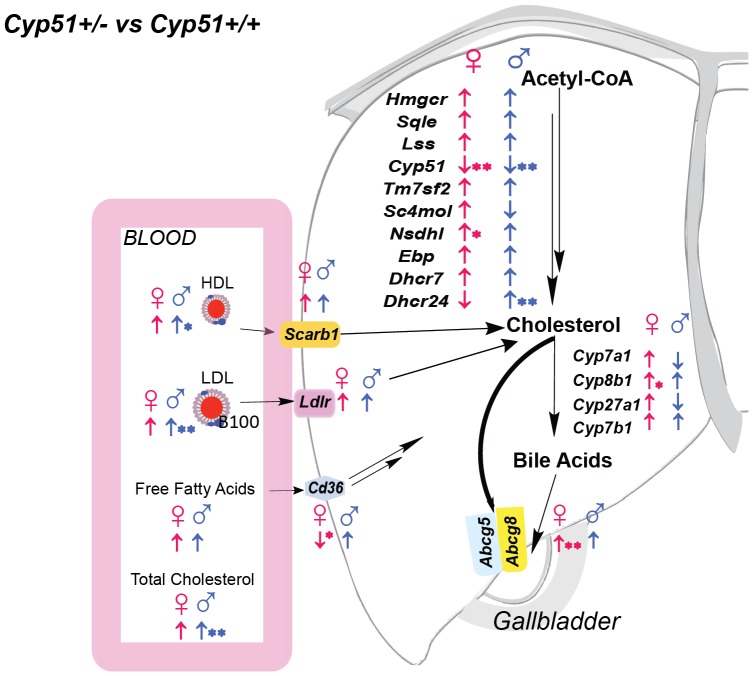
Initially we evaluated the effect of Cyp51 genotype in sex-matched animals regardless of the feeding regime. This can simulate a random population where feeding is not controlled. The measured values of the body and organ weights, plasma lipids, steroid hormones and relative gene expression are shown in Table S3 with statistically significant features summarized in Table 1 and Figure 3. Cyp51+/− males had significantly increased plasma total cholesterol, HDL and LDL cholesterol (20–35% higher than controls) and present hepatomegaly (Table 1). On the other hand, Cyp51+/− females remained leaner than wild-type controls, with the plasma lipid profile similar to that of controls. The hepatic gene expression is summarized in Figure 3. As expected, the Cyp51+/− mice exhibited a decreased expression of Cyp51 and a trend of increased expression of majority of other cholesterogenic genes. The observed sexual differences (Nsdhl increased in females, Dhcr24 increased in males) require further investigation. On top of that, Cyp51+/− females had significantly increased expression of Abcg8 transporter, responsible for the cholesterol clearance to the bile, as well as the tendency of the increased expression of Cyp8b1 from alternative bile acid synthesis, and slightly decreased expression of fatty acid translocase Cd36. These changes in gene expression could aid in maintaining the healthier blood lipid profile of Cyp51+/− females compared to the males. The results from pooled population where mice ate either of the diets suggest that Cyp51 haploinsufficiency is more harmful for males than females.
Table 1. Significant differences in gross phenotype between Cyp51+/− and Cyp51+/+ mice.
| Females Cyp51+/− | Females Cyp51+/+ | Males Cyp51+/− | Males Cyp51+/+ | |||||||
| Mean | SEM | Mean | SEM | P value | Mean | SEM | Mean | SEM | P value | |
| FAT TISSUE/BWa , b | 0.048 | 0.002 | 0.059 | 0.004 | **0.033 | 0.065 | 0.004 | 0.065 | 0.005 | 0.984 |
| LIVER/BWa , b | 4.25 | 0.058 | 4.38 | 0.075 | 0.326 | 4.51 | 0.124 | 4.13 | 0.091 | **0.029 |
| HDL-CHOLESTEROL [mM] | 1.43 | 0.115 | 1.28 | 0.076 | 0.369 | 1.98 | 0.103 | 1.66 | 0.091 | * 0.069 |
| LDL-CHOLESTEROL [mM] | 1.09 | 0.117 | 0.91 | 0.102 | 0.127 | 1.52 | 0.168 | 1.13 | 0.135 | **0.029 |
| TOTAL CHOLESTEROL [mM] | 2.92 | 0.234 | 2.58 | 0.172 | 0.194 | 3.92 | 0.255 | 3.18 | 0.212 | **0.029 |
BW – body weight;
organ weights/body weight ratios are given in organ weight[g]*100/body weigh [g].
*p<0.1, **p<0.05.
Figure 3. The global effect of Cyp51 haploinsufficiency.
The genotype effect shown on blood lipid profile (n = 8–15) and hepatic gene expression analyses in Cyp51+/− mice compared to Cyp51+/+ matched with sex (n = 4–7) and blood lipid profile differences. The direction of arrows indicate (↑) up-regulation or (↓) down-regulation of expression in heterozygous females (♀) and males (♂). The direction of arrows shows also a trend of change in genes that did not reach statistical significance. *(p<0.1); **(p<0.05). Results are joined for all diets.
Liver histology
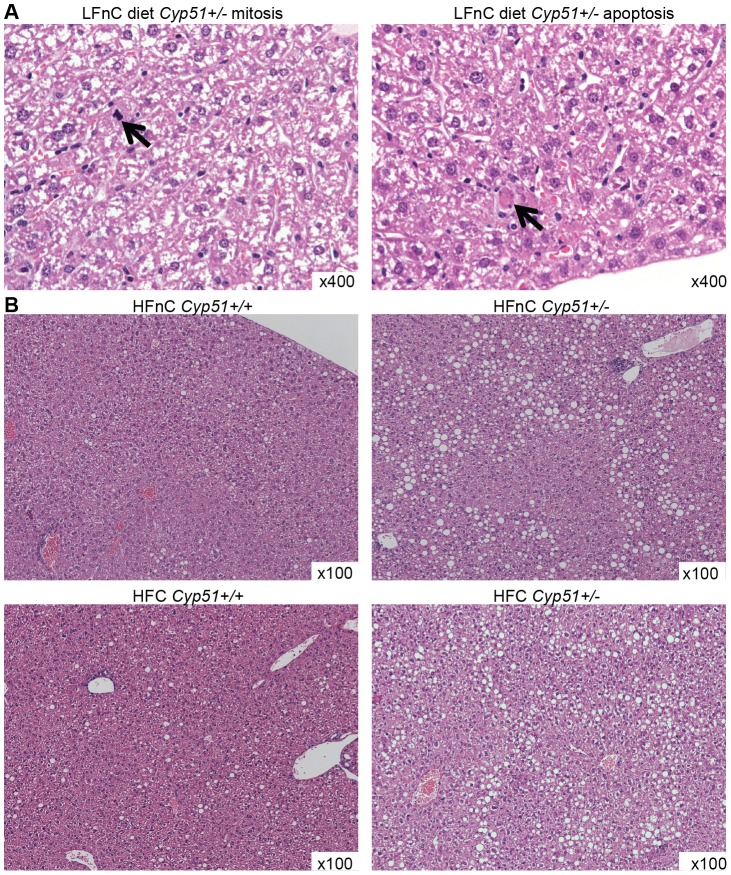
While the global model suggested hepatomegaly only in Cyp51+/− males, the histopathology shows similar features in livers of both sexes, thus results are not separated according to the sex (Table 2 and Figure 4). On the standard laboratory diet (LFnC), livers of the Cyp51+/− mice show frequent apoptoses and mitoses compared to the wild-type mice. The apoptoses and mitoses were still present in Cyp51+/− mice on the high fat diets with or without cholesterol, yet less frequent than before, suggesting that dietary lipids partially rescued this phenotype. However, both high-fat diets in Cyp51+/− mice resulted in a moderate steatosis compared to mild steatosis of wild-types, indicating that whole body CYP51 and/or normal cholesterol synthesis may play a role in the hepatic accumulation of the fats.
Table 2. Summary of the liver histology.
| Histopathology | LFnCa | HFnCb | HFCc | |||
| Cyp51+/− | Cyp51+/+ | Cyp51+/− | Cyp51+/+ | Cyp51+/− | Cyp51+/+ | |
| Apoptosis, mitosis | Frequent(A = 25; M = 8)d | No(A = 4; M = 0)d | Rare(A = 7; M = 2)d | No(A = 1; M = 0)d | Rare(A = 4; M = 1)d | No(A = 4; M = 0)d , e |
| Steatosis | No | No | Yes moderate | Yes mild | Yes moderate | Yes mild |
LFnC – low fat without cholesterol diet;
HFnC – high fat without cholesterol diet;
HFC – high fat diet with cholesterol;
A – apoptosis, M – mitosis (number of observed events);
5 instead of 6 animals were analyzed.
Figure 4. Liver histology, hematoxylin and eosin stain.
A Mitotic (left) and apoptotic cells with hyper-eosinophilic cytoplasm containing fragments of dense basophilic nuclear material (right) in Cyp51+/− mice on LFnC diet; original magnification x400. B Mild focal steatosis in Cyp51+/+ mice and moderate steatosis in Cyp51+/− mice on HFnC and HFC diets; original magnification x100.
Are Cyp51+/− males at a higher disease risk on high-fat high-cholesterol diet?
In this part, we investigated how mice with one missing Cyp51 allele react to either a low-fat or high-fat diet, with or without cholesterol. On LFnC standard laboratory diet (low-fat, cholesterol-free), both Cyp51+/− and wild-types matched by sex had similar body and organ weights, as well as the plasma lipid levels (Table 3). In addition to the significantly reduced Cyp51 in heterozygotes of both sexes, Abcg5 transporter, which plays a role in the clearance of cholesterol to the bile, was increased in both sexes and Dhcr24 only in males (Figure S2 A).
Table 3. Significant differences between Cyp51+/− and Cyp51+/+ mice on each diet separately.
| LFnC Females | LFnC Males | HFnC Females | HFnC Males | HFC Females | HFC Males | |||||||
| parameter | FC | P.value | FC | P.value | FC | P.value | FC | P.value | FC | P.value | FC | P.value |
| Body Weight | 0.97 | 0.954 | 1.04 | 0.808 | 1.00 | 0.993 | 1.05 | 0.883 | 0.87 | **0.039 | 1.03 | 0.712 |
| Fat Tissue/BWa , b | 1.00 | 0.954 | 1.00 | 0.911 | 0.84 | 0.466 | 0.95 | 0.893 | 0.63 | **0.006 | 1.09 | 0.712 |
| Heart/BWa , b | 0.95 | 0.954 | 1.04 | 0.828 | 1.07 | 0.589 | 0.94 | 0.835 | 1.11 | *0.085 | 0.95 | 0.617 |
| Spleen/BWa , b | 1.03 | 0.954 | 1.17 | 0.808 | 1.09 | 0.993 | 0.94 | 0.966 | 1.00 | 0.578 | 0.79 | *0.080 |
| Liver/BWa , b | 0.95 | 0.954 | 1.04 | 0.808 | 1.02 | 0.993 | 1.01 | 0.982 | 0.95 | 0.351 | 1.23 | **0.001 |
| Corticosterone | 0.86 | 0.954 | 0.77 | 0.808 | 0.87 | 0.993 | 0.86 | 0.997 | 2.22 | *0.085 | 0.60 | 0.617 |
| Sqle | 0.66 | 0.134 | 1.19 | 0.702 | 1.38 | 0.263 | 1.48 | 0.132 | 2.50 | **0.014 | 1.00 | 0.822 |
| Cyp51 | 0.41 | **0.003 | 0.60 | *0.070 | 0.78 | 0.435 | 0.57 | **0.048 | 1.57 | 0.644 | 0.60 | *0.090 |
| Tm7sf2 | 0.80 | 0.151 | 1.10 | 0.506 | 1.29 | 0.108 | 1.08 | 0.662 | 1.47 | *0.060 | 1.13 | 0.633 |
| Nsdhl | 0.79 | 0.288 | 1.16 | 0.677 | 1.81 | **0.006 | 1.18 | 0.410 | 1.83 | **0.035 | 1.11 | 0.788 |
| Dhcr7 | 0.89 | 0.507 | 0.75 | 0.496 | 1.71 | *0.080 | 1.19 | 0.566 | 0.78 | 0.732 | 1.11 | 0.768 |
| Dhcr24 | 0.90 | 0.573 | 1.50 | **0.028 | 0.95 | 0.662 | 1.29 | *0.098 | 1.03 | 0.949 | 0.93 | 0.843 |
| Cyp8b1 | 1.00 | 0.860 | 1.03 | 0.908 | 1.29 | 0.362 | 1.06 | 0.651 | 2.00 | **0.011 | 1.04 | 0.663 |
| Cyp27a1 | 1.02 | 0.709 | 0.85 | 0.317 | 1.16 | 0.381 | 1.07 | 0.798 | 1.59 | **0.009 | 1.09 | 0.767 |
| Cyp7b1 | 1.00 | 0.566 | 1.46 | 0.251 | 1.67 | 0.181 | 1.09 | 0.935 | 3.00 | **0.018 | 0.97 | 0.787 |
| Cd36 | 1.32 | 0.420 | 1.00 | 0.894 | 0.92 | 0.667 | 1.00 | 0.632 | 0.68 | *0.063 | 1.39 | 0.194 |
| Abcg5 | 2.25 | *0.073 | 2.00 | *0.069 | 1.67 | 0.272 | 0.60 | 0.159 | 0.57 | 0.235 | 0.74 | 0.338 |
FC – fold change calculated as a ratio between average values for Cyp51+/− mice and average values for Cyp51+/+ mice.
BW – body weight;
organ weights/body weight ratios are given in organ weight[g]*100/body weigh [g].
On the diet with no cholesterol but with high fats (HFnC) there was similarly no changes in the body, organ weights and plasma lipids between Cyp51+/− and wild-type mice of both sexes. Sexual dimorphism was observed at the mRNA level of cholesterol synthesis genes. The majority of measured genes from this pathway were up-regulated in Cyp51+/− mice even if different genes show statistical significance in males and in females – the overexpression of Nsdhl and Dhcr7 in females and again Dhcr24 in males (Figure S2 B). A tendency of high NSDHL protein level was observed in both sexes. (Figure S3). From these data, we can deduce that the Cyp51+/− genotype is not immediately harmful for animals if at the high fat, cholesterol-free diet.
A dramatic influence on Cyp51+/− mice was observed on the high fat diet with 1.25% of cholesterol (HFC). The Cyp51+/− females remain significantly leaner compared to wild-types, with lower fat tissue/body weight ratio, and lower body weight, with increased plasma corticosterone (Table 3). The Cyp51+/− males have significantly enlarged livers compared to the wild-types. The expression of several genes from the cholesterol and bile acid synthesis is significantly increased in Cyp51+/− females compared to their controls. In females, the mRNA levels of Tms7Sf2 and Nsdhl were significantly higher in Cyp51+/− mice compared to controls (Figure S2 C), and TMS7SF2 also on the protein level, while the NSDHL protein dropped below the detection threshold (Figure S3 and Figure S4). This can be explained by the overall repression of cholesterol synthesis on the high-fat diet with cholesterol compared to other diets (Table S3). Interestingly, it appears that Cyp51+/− females repress cholesterol synthesis less effectively than males. From other pathways we noted a decreased expression of fatty acid translocase Cd36 in Cyp51+/− females, but not in males (Figure S2 C).
To further investigate the sexual dimorphism, we analyzed the Cyp51 genotype-sex interactions (Figure S5), which confirmed the significant differences in organ characteristics and in the expression of Cd36. The significant differences were found also in the expression of Cyp27a1 and Cyp7b1 from the alternative bile acid synthesis pathway, where Cyp51+/− females exhibited increased expression. Cyp27a1 is involved in bile acid synthesis pathway, but can also metabolize sterol intermediates from the cholesterol synthesis, including lanosterol [32]. The Cyp7b1 may also be involved in the development of atherosclerosis [33], oxysterol metabolism, and sex hormone synthesis. The elevated expression of this gene can play a role in the protective effect on cholesterol levels in Cyp51+/− females. On the other hand, the increase of Lpl expression was determined in the Cyp51+/− males, which should not be the case in the livers of adult animals [34] and can be indicative of an early phases of metabolic syndrome.
The analysis of diet-genotype interactions confirmed observed sexual dimorphisms, resulting in different patterns of gene expression in males and females. The cholesterogenic genes Nsdhl, Sqle, Cyp51 and Sc4mol increase in Cyp51+/− females when fed with HFnC diet compared to LFnC diet and decrease with cholesterol supplementation (HFC), whereas they are decreased in wild-type females when fed with either of the high-fat diets. On the protein level, we confirmed the decrease in NSDHL expression in Cyp51+/− females on HFC diet, and the decrease in its expression in wild-type females on both high-fat diets. The expression of Nsdhl, Sqle, and Tm7sf2 was still increased in lean Cyp51+/− females fed HFC compared to their wild-type counterparts, suggesting that the feedback regulation of their expression might be less efficient. The high-fat feeding triggered differential expression of Abcg5 transporter in Cyp51+/− males compared to the wild types, whereas in females more diet-genotype interactions were observed. The analysis of interactions did not discover differences in expression of nuclear receptors, leaving the mechanism of the observed phenotype unclear.
The Cyp51 genotype interactions with the diet: cholesterol or other dietary fats?
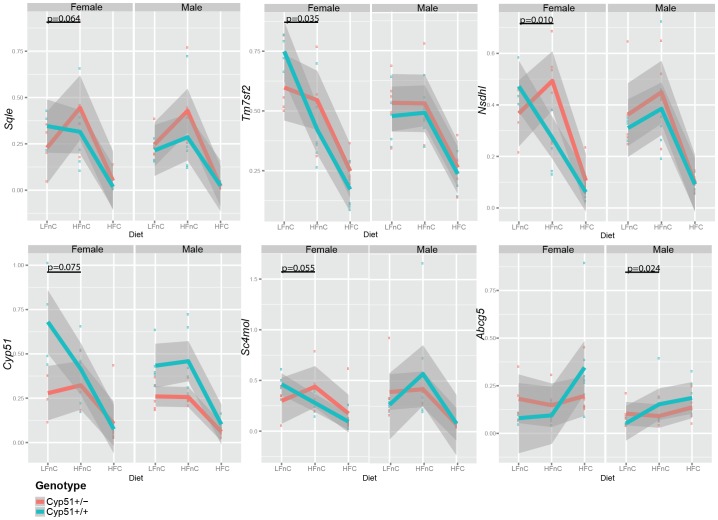
We questioned how the dietary fats affect the Cyp51+/− genotype is affected, in the absence of cholesterol (HFnC vs LFnC) or in the presence of cholesterol (HFC vs HFnC), which is the usual case in high-fat nutrition. The major impact was observed for the cholesterol and lipid homeostasis genes, which is shown in the expression plots (Figure 5).
Figure 5. The diet-genotype interactions (effect of high fat feeding).
Plots showing the change in expression levels in males and females, both Cyp51+/− (red) and Cyp51+/+ (blue). Grey color indicates a linear smooth of 0.95 confidence intervals. The significant interactions between factors (Cyp51 genotype and diet) that can be understood as the differences between slopes of lines are indicated by their p values.
The LFnC and HFnC diets were without significant amounts of cholesterol (<0.01%), so there is a need for the increased cholesterol synthesis. However, females and males adapt to this condition differently. Only females with the Cyp51+/−genotype increase the expression of genes from cholesterol synthesis on HFnC diet, while the adaptation is already present in wild-type males (Figure 5, Table S4). The difference is observed also at the level of Abcg5 (from direct clearance of cholesterol), where only the wild-type males, but not females increase the expression on high fat load (comparing LFnC and HFnC) (Figure 5, Table S4).
The cholesterol feedback regulation does not differ majorly between the Cyp51+/− and wild-type mice in the presence of dietary cholesterol, which is shown in an interaction study between Cyp51+/− vs Cyp51+/+ genotypes and high-fat diets with and without cholesterol (HFnC vs HFC). Results indicate that irrespective of the Cyp51 genotype, mice reacted similarly to cholesterol supplementation in the high-fat milieu (Table S4). Differences in the expression of Ldlr in females and Lpl in males were significant, but remained in the same direction in both genotypes (Figure S6, Table S4).
Discussion
The genotype-phenotype correlation is a complex feature, where genes interacting with the environment can lead to less predictable phenotypes even in single-gene knockout models. To assess the global role of the decreased expression of Cyp51 from cholesterol synthesis, we investigated mice with one functional Cyp51 allele since the inactivation of both alleles led to the embryonically lethal phenotype [17]. We applied a controlled environment which mimics the human eating habits - mice of both sexes were on diets for 16 weeks and not only 1 to 8 weeks as used in many rodent studies [35], [36], including these from the Mouse Phenome Database (http://phenome.jax.org). A large data-set from the Cyp51+/− mice and corresponding controls was collected and analyzed by linear regression modeling taking into account the sexual dimorphism, which is characteristic for cholesterol homeostasis in rodents and humans [36], [37], [38].
It appears on first glance, the Cyp51 heterozygous knockout has a negligible effect, because mice seem to develop normally. A deeper analysis revealed significant changes in the phenotype on the organ and metabolic level. Overall, the Cyp51+/− males seem to be more affected than Cyp51+/− females, presenting hepatomegaly and an increased plasma lipid profile. The liver histology showed apoptotic and mitotic events in the Cyp51+/− mice of both sexes and on all diets (less severe with the high-fat), which was not observed in the wild-type mice and should not be found in livers of healthy animals. The hepatic apoptoses and mitoses are indication of pathological events in cells (such as toxicity, increased oxidative stress, increased accumulation of intermediates, inflammation, etc.), and they are biomarkers for severity of nonalcoholic fatty liver disease (NAFLD) [39]. The Cyp51+/− females were leaner than wild-type littermates and did not show significant changes in the blood lipid profile compared to wild-types. They also did not show hepatomegaly, but displayed hepatic apoptoses, mitoses and increased lipid infiltration on high-fat diets, which may relate to a group of difficult to diagnose cases of “lean NAFLD” [40]. The extrapolation of metabolic data obtained from murine models directly to humans is not always straightforward. Additionally, the mRNA levels do not necessarily reflect the protein levels and enzymes activity. In our study we found a good correlation between mRNA and protein levels for CYP51 and NSDHL (and partially also for TM7SF2). This is true also for HMGCR where the expression did not differ significantly between the genotypes at the mRNA or protein levels (Figure S7). The liver is a major organ of cholesterol synthesis in mice, but in primates the synthesis mainly occurs in extra hepatic tissues. However, murine models proved to be a great tool investigating inborn errors of cholesterol and bile acid synthesis and phenotypes relate well to human individuals [9], [10]. It is thus plausible to speculate that human individuals of both sexes with functional polymorphisms in the CYP51A1 are at increased risk to develop liver pathologies.
Our results indicate that the diminished expression of Cyp51 from cholesterol synthesis was more harmful for males than for females. The male predominance was observed also for Dhcr24 as another cholesterol synthesis gene, where the viable Dhcr24−/− knockout males showed a more severe phenotype than females [41]. However, in our case, the female Cyp51+/− phenotype resembles the ‘lean NAFLD’ with increased corticosterone on the high-fat diet, which might represent another risk factor for rapid development of fatty liver disease. Additionally, the expression of genes from cholesterol synthesis in Cyp51+/− females appears to be more sensitive to the high-fat diet challenge. It appears that females (even Cyp51 wild-types) are sensing lack of cholesterol better than males (especially on the high fat diet) increasing the expression of cholesterol synthesis genes more efficiently than males, which improves the flux through the cholesterol synthesis pathway towards cholesterol [36].
The expression of cholesterogenic genes was shown to be sex-biased; however, data reported in literature are inconsistent [36], [42], [43]. We identified higher expression of several genes in females including Srebp2, Hmgcr, Lss and Ebp from cholesterol synthesis (Table S5), which is in line with Gatti et al [43] and relates well with Lorbek et al [36]. However, in our study we investigated sexual dimorphism in context of Cyp51 genotype and we uncovered novel differences at the level of expression of cholesterogenic genes. Knockout of one Cyp51 allele led to a reduced expression of the CYP51 protein in both sexes. The Cyp51 haploinsufficiency triggered the increased expression of Dhcr24 in males and Nsdhl in females. DHCR24 metabolizes different precursor sterols, including lanosterol which is the major substrate of CYP51 [44]. NSDHL accumulates on the lipid droplets where it is involved in storage of neutral lipids, such as triacylglycerol and cholesterol esters [45] and possibly contributes to the cholesterol clearance in Cyp51+/− females. The Nsdhl gene is located on the X chromosome and possibly due to a presence of a single copy cannot be overexpressed in males, but it seems to play an important role in the cholesterol homeostasis of Cyp51+/− females. The leaner Cyp51+/− females showed the increased expression of Cyp8b1 from alternative pathway of bile acid synthesis and Abcg8 from transport of cholesterol to bile suggesting increased cholesterol clearance. The decreased adipose tissue correlates with a healthier blood lipid profile in Cyp51+/− females and decreased expression of Cd36 accounting for the protective effect from cardiovascular diseases [46], [47] in Cyp51+/females, but not in males. The mechanism of observed sex differences cannot be explained by our current experimental design, but one possible explanation for sex differences could be in the interaction of lowered Cyp51 expression with female sex hormones [48], [49]. The sex hormone hypothesis is supported by the study of Romero-Aleshire et al [50] demonstrating that in the mouse menopause model, even without a high-fat diet treatment, progression into menopause caused a significant increase in the cholesterol levels indicating that estrogens might indeed play a role.
The global model focusing on differences observed only due to Cyp51 genotype show that Cyp51+/− males had significantly increased total cholesterol and LDL cholesterol compared to Cyp51+/+ mice, while Cyp51+/− females managed to maintain blood lipids on similar level as wild-types, possibly due to the increased clearance via Abcg5/8 transporters or modified lipids storage. Apparently, the diminished expression of Cyp51 was still sufficient for cholesterol synthesis, even in the absence of cholesterol in the diet, but resulted in increased mRNA of other cholesterogenic genes and that further led to dysregulation of lipid homeostasis. Heterozygous mice showed increased plasma cholesterol concentration, increased hepatic lipid storage and signs of metabolic stress (indicated by hepatic apoptotic and mitotic events in both sexes and increased corticosterone in females). The mRNA levels of nuclear receptors could not explain this phenotype and the mechanism of dysregulation of lipid homeostasis is yet to be discovered. The decrease in Cd36 and increase in gene expression from the bile acid synthetic pathway on HFC diet likely contributed to better management of the healthy blood lipid profile in Cyp51+/− females, but significantly increased corticosterone might represent an independent fatty liver risk factor [51]. Corticosterone is a stress hormone that, when elevated, can lead to changes in metabolism of carbohydrates, proteins, and lipid homeostasis, resulting also in the leaner phenotype in mice fed with high-fat diet [52]. The increased corticosterone does not always affect cholesterol levels in rodents [53], which was also the case in Cyp51+/− females. Because Cyp51+/− females were housed in the same manner as wild-types at all diet regimes, the increased corticosterone in Cyp51+/− HFC group suggests metabolic stress that could be a consequence of the loss of one Cyp51 allele.
Conclusions
The heterozygous mutations leading to the decreased protein levels or enzyme activity often result in mild phenotypes that can be easily overlooked. The cumulative effects of single mutations, which may cause very mild phenotypes independently, together with the environmental factors, carry important information about causes of complex diseases. It seems that much of the genetic control of common diseases is due to rare variants with a strong impact on disease risk in individual patients [54]. Our work shows that even heterozygous loss of function of one Cyp51 allele from the cholesterol synthesis leads to significant phenotype differences in male and female mice, where Cyp51+/− males are more affected by harmful cholesterol lipid profile and hepatomegaly, but also Cyp51+/− females reveal risk factors of the fatty liver disease. It is thus plausible to assume that the etiology of liver diseases differs between males and females, where cholesterol synthesis represents a crucial point of sexual dimorphism.
Supporting Information
Outline of the statistical analysis.
(TIF)
The genotype effect on hepatic gene expression and blood lipids for (A) low fat, cholesterol-free diet; (B) high fat, cholesterol-free diet; and (C) High fat with 1.25% of cholesterol added diet. The direction of arrows indicate (↑) up-regulation or (↓) down-regulation of expression in heterozygous females (♀) and males (♂). * indicates p<0.1 and **p<0.05.
(TIF)
The expression of Nsdhl (A) on mRNA and (B) protein levels (western blot analysis) **p<0.05.
(TIF)
The expression of Tm7sf2 (A) on mRNA and (B) DHCR14 protein levels (western blot analysis) *p<0.1.
(TIF)
The significant interactions between sex and Cyp51 genotype in mice fed with the high-fat diet with 1.25% of cholesterol - the organ characteristics and the hepatic gene expression. The interactions in organ characteristics/blood profile were not observed on cholesterol-free diets. The Dhcr24 and Abcg8 were significantly different (p = 0.06 and 0.07 respectively) on the LFnC diet and Abcg5 reached significance (p = 0.08) on the HFnC diet (data not shown).
(TIF)
The significant interactions between the Cyp51 genotype and two high-fat diets with and without cholesterol.
(TIF)
The expression of Hmgcr (A) on mRNA and (B) protein levels (western blot analysis) **p<0.05.
(TIF)
The list of primers used for RT-PCR analysis.
(DOCX)
Number of animals analyzed within each contrast; A – body characteristics B – plasma lipids profile C – hepatic gene expression, D – liver histology.
(DOCX)
The average values (Mean) for weights, lipid profile and gene expression and standard error means (SEM) for global model (regardless diets) and for each diet separately.
(DOCX)
The interactions between diets and genotypes. Left panel shows the significant interactions between two cholesterol-free diets and Cyp51 genotype (indicated p values). The directions of arrows indicate the directions of changes from LFnC diet to HFnC diet, stars indicate statistically significant changes (** p<0.05 and * p<0.1). The right panel shows the significant interactions (p values indicated) between two high-fat diets and Cyp51 genotype. The directions of arrows indicate directions of changes for HFnC to HFC diet and stars indicate statistically significant changes.
(DOCX)
The sexual dimorphism – differences between females and males expressed as FC (females mean/males mean). *p<0.1; **p<0.05.
(DOCX)
Acknowledgments
We would like to thank Ksenija Kodra for her work with the animal husbandry, Marjan Kuznik, Helena Klavzar, Rok Kosir and Ursula Prosenc Zmrzljak for technical assistance at euthanasia.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by FP7 FightingDrugFailure ITN Marie Curie grant #238132 and Slovenian Research Agency program grant P1-0104, grant P1-0010 and project J7-4053. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Goluszko P, Nowicki B (2005) Membrane cholesterol: a crucial molecule affecting interactions of microbial pathogens with mammalian cells. Infection and immunity 73: 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernandez C, Martin M, Gomez-Coronado D, Lasuncion MA (2005) Effects of distal cholesterol biosynthesis inhibitors on cell proliferation and cell cycle progression. Journal of lipid research 46: 920–929. [DOI] [PubMed] [Google Scholar]
- 3. Singh P, Saxena R, Srinivas G, Pande G, Chattopadhyay A (2013) Cholesterol biosynthesis and homeostasis in regulation of the cell cycle. PloS one 8: e58833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dietschy JM (1984) Regulation of cholesterol metabolism in man and in other species. Klinische Wochenschrift 62: 338–345. [DOI] [PubMed] [Google Scholar]
- 5. Goedeke L, Fernandez-Hernando C (2012) Regulation of cholesterol homeostasis. Cellular and Molecular Life Sciences 69: 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turley SD, Spady DK, Dietschy JM (1995) Role of liver in the synthesis of cholesterol and the clearance of low density lipoproteins in the cynomolgus monkey. Journal of lipid research 36: 67–79. [PubMed] [Google Scholar]
- 7. Dietschy JM, Turley SD (2002) Control of cholesterol turnover in the mouse. The Journal of biological chemistry 277: 3801–3804. [DOI] [PubMed] [Google Scholar]
- 8. Porter FD, Herman GE (2011) Malformation syndromes caused by disorders of cholesterol synthesis. Journal of lipid research 52: 6–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lorbek G, Lewinska M, Rozman D (2012) Cytochrome P450s in the synthesis of cholesterol and bile acids–from mouse models to human diseases. The FEBS journal 279: 1516–1533. [DOI] [PubMed] [Google Scholar]
- 10. Horvat S, McWhir J, Rozman D (2011) Defects in cholesterol synthesis genes in mouse and in humans: lessons for drug development and safer treatments. Drug metabolism reviews 43: 69–90. [DOI] [PubMed] [Google Scholar]
- 11. Ikonen E (2006) Mechanisms for cellular cholesterol transport: defects and human disease. Physiological reviews 86: 1237–1261. [DOI] [PubMed] [Google Scholar]
- 12. Lewinska M, Zelenko U, Merzel F, Golic Grdadolnik S, Murray JC, et al. (2013) Polymorphisms of CYP51A1 from cholesterol synthesis: associations with birth weight and maternal lipid levels and impact on CYP51 protein structure. PloS one 8: e82554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steffen KM, Cooper ME, Shi M, Caprau D, Simhan HN, et al. (2007) Maternal and fetal variation in genes of cholesterol metabolism is associated with preterm delivery. Journal of perinatology: official journal of the California Perinatal Association 27: 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bream EN, Leppellere CR, Cooper ME, Dagle JM, Merrill DC, et al. (2013) Candidate gene linkage approach to identify DNA variants that predispose to preterm birth. Pediatric research 73: 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lewinska M, Zmrzljak UP, Rozman D (2013) Low nucleotide variability of CYP51A1 in humans: meta-analysis of cholesterol and bile acid synthesis and xenobiotic metabolism pathways. Acta chimica Slovenica 60: 875–883. [PubMed] [Google Scholar]
- 16. Debeljak N, Horvat S, Vouk K, Lee M, Rozman D (2000) Characterization of the mouse lanosterol 14alpha-demethylase (CYP51), a new member of the evolutionarily most conserved cytochrome P450 family. Archives of biochemistry and biophysics 379: 37–45. [DOI] [PubMed] [Google Scholar]
- 17. Keber R, Motaln H, Wagner KD, Debeljak N, Rassoulzadegan M, et al. (2011) Mouse knockout of the cholesterogenic cytochrome P450 lanosterol 14alpha-demethylase (Cyp51) resembles Antley-Bixler syndrome. The Journal of biological chemistry 286: 29086–29097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keber R, Acimovic J, Majdic G, Motaln H, Rozman D, et al. (2013) Male germ cell-specific knockout of cholesterogenic cytochrome P450 lanosterol 14alpha-demethylase (Cyp51). Journal of lipid research 54: 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keber R, Rozman D, Horvat S (2013) Sterols in spermatogenesis and sperm maturation. Journal of lipid research 54: 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muscarella LA, Guarnieri V, Coco M, Belli S, Parrella P, et al. (2010) Small deletion at the 7q21.2 locus in a CCM family detected by real-time quantitative PCR. Journal of biomedicine & biotechnology 2010: 854737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aldahmesh MA, Khan AO, Mohamed JY, Hijazi H, Al-Owain M, et al. (2012) Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes. Genetics in medicine: official journal of the American College of Medical Genetics 14: 955–962. [DOI] [PubMed] [Google Scholar]
- 22. Geske FJ, Gerschenson LE (2001) The biology of apoptosis. Human pathology 32: 1029–1038. [DOI] [PubMed] [Google Scholar]
- 23. van Diest PJ, Baak JP, Matze-Cok P, Wisse-Brekelmans EC, van Galen CM, et al. (1992) Reproducibility of mitosis counting in 2,469 breast cancer specimens: results from the Multicenter Morphometric Mammary Carcinoma Project. Human pathology 23: 603–607. [DOI] [PubMed] [Google Scholar]
- 24. Friedewald WT, Levy RI, Fredrickson DS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clinical chemistry 18: 499–502. [PubMed] [Google Scholar]
- 25. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kosir R, Acimovic J, Golicnik M, Perse M, Majdic G, et al. (2010) Determination of reference genes for circadian studies in different tissues and mouse strains. BMC molecular biology 11: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Team RDC (2013) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 28. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome biology 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox J, Weisberg S (2011) An R companion to applied regression. Thousand Oaks, Calif.: SAGE Publications. xxii, 449 p. p. [Google Scholar]
- 30.Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman VC, S Dudoit, R Irizarry, W Huber, editor. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer. 397–420. [Google Scholar]
- 31. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 57: 289–300. [Google Scholar]
- 32. Pikuleva I, Javitt NB (2003) Novel sterols synthesized via the CYP27A1 metabolic pathway. Archives of biochemistry and biophysics 420: 35–39. [DOI] [PubMed] [Google Scholar]
- 33. Bjorkhem I, Andersson O, Diczfalusy U, Sevastik B, Xiu RJ, et al. (1994) Atherosclerosis and sterol 27-hydroxylase: evidence for a role of this enzyme in elimination of cholesterol from human macrophages. Proceedings of the National Academy of Sciences of the United States of America 91: 8592–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang H, Eckel RH (2009) Lipoprotein lipase: from gene to obesity. American journal of physiology Endocrinology and metabolism 297: E271–288. [DOI] [PubMed] [Google Scholar]
- 35. Henkel AS, Anderson KA, Dewey AM, Kavesh MH, Green RM (2011) A chronic high-cholesterol diet paradoxically suppresses hepatic CYP7A1 expression in FVB/NJ mice. Journal of lipid research 52: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lorbek G, Perse M, Horvat S, Bjorkhem I, Rozman D (2013) Sex differences in the hepatic cholesterol sensing mechanisms in mice. Molecules 18: 11067–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Marinis E, Martini C, Trentalance A, Pallottini V (2008) Sex differences in hepatic regulation of cholesterol homeostasis. The Journal of endocrinology 198: 635–643. [DOI] [PubMed] [Google Scholar]
- 38. Wang X, Magkos F, Mittendorfer B (2011) Sex differences in lipid and lipoprotein metabolism: it's not just about sex hormones. The Journal of clinical endocrinology and metabolism 96: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wieckowska A, Zein NN, Yerian LM, Lopez AR, McCullough AJ, et al. (2006) In vivo assessment of liver cell apoptosis as a novel biomarker of disease severity in nonalcoholic fatty liver disease. Hepatology 44: 27–33. [DOI] [PubMed] [Google Scholar]
- 40. Younossi ZM, Stepanova M, Negro F, Hallaji S, Younossi Y, et al. (2012) Nonalcoholic fatty liver disease in lean individuals in the United States. Medicine 91: 319–327. [DOI] [PubMed] [Google Scholar]
- 41. Wechsler A, Brafman A, Shafir M, Heverin M, Gottlieb H, et al. (2003) Generation of viable cholesterol-free mice. Science 302: 2087. [DOI] [PubMed] [Google Scholar]
- 42. Yang X, Schadt EE, Wang S, Wang H, Arnold AP, et al. (2006) Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome research 16: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gatti DM, Zhao N, Chesler EJ, Bradford BU, Shabalin AA, et al. (2010) Sex-specific gene expression in the BXD mouse liver. Physiological genomics 42: 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Acimovic J, Rozman D (2013) Steroidal triterpenes of cholesterol synthesis. Molecules 18: 4002–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caldas H, Herman GE (2003) NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Human molecular genetics 12: 2981–2991. [DOI] [PubMed] [Google Scholar]
- 46. Kavanagh IC, Symes CE, Renaudin P, Nova E, Mesa MD, et al. (2003) Degree of oxidation of low density lipoprotein affects expression of CD36 and PPARgamma, but not cytokine production, by human monocyte-macrophages. Atherosclerosis 168: 271–282. [DOI] [PubMed] [Google Scholar]
- 47. Itabe H, Ueda M (2007) Measurement of plasma oxidized low-density lipoprotein and its clinical implications. Journal of atherosclerosis and thrombosis 14: 1–11. [DOI] [PubMed] [Google Scholar]
- 48. Pellegrini M, Pallottini V, Marin R, Marino M (2014) Role of sex hormone estrogen in prevention of lipid disorder. Current medicinal chemistry. [DOI] [PubMed] [Google Scholar]
- 49. Trapani L, Violo F, Pallottini V (2010) Hypercholesterolemia and 3-hydroxy-3-methylglutaryl coenzyme A reductase regulation in aged female rats. Experimental gerontology 45: 119–128. [DOI] [PubMed] [Google Scholar]
- 50. Romero-Aleshire MJ, Diamond-Stanic MK, Hasty AH, Hoyer PB, Brooks HL (2009) Loss of ovarian function in the VCD mouse-model of menopause leads to insulin resistance and a rapid progression into the metabolic syndrome. American journal of physiology Regulatory, integrative and comparative physiology 297: R587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. D'Souza A M, Beaudry JL, Szigiato AA, Trumble SJ, Snook LA, et al. (2012) Consumption of a high-fat diet rapidly exacerbates the development of fatty liver disease that occurs with chronically elevated glucocorticoids. American journal of physiology Gastrointestinal and liver physiology 302: G850–863. [DOI] [PubMed] [Google Scholar]
- 52. Chuang JC, Cui H, Mason BL, Mahgoub M, Bookout AL, et al. (2010) Chronic social defeat stress disrupts regulation of lipid synthesis. Journal of lipid research 51: 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ricart-Jane D, Rodriguez-Sureda V, Benavides A, Peinado-Onsurbe J, Lopez-Tejero MD, et al. (2002) Immobilization stress alters intermediate metabolism and circulating lipoproteins in the rat. Metabolism: clinical and experimental 51: 925–931. [DOI] [PubMed] [Google Scholar]
- 54. Cirulli ET, Goldstein DB (2010) Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nature Reviews Genetics 11: 415–425. [DOI] [PubMed] [Google Scholar]
- 55. Ohashi K, Osuga J, Tozawa R, Kitamine T, Yagyu H, et al. (2003) Early embryonic lethality caused by targeted disruption of the 3-hydroxy-3-methylglutaryl-CoA reductase gene. The Journal of biological chemistry 278: 42936–42941. [DOI] [PubMed] [Google Scholar]
- 56. Hager EJ, Tse HM, Piganelli JD, Gupta M, Baetscher M, et al. (2007) Deletion of a single mevalonate kinase (Mvk) allele yields a murine model of hyper-IgD syndrome. Journal of inherited metabolic disease 30: 888–895. [DOI] [PubMed] [Google Scholar]
- 57. Tozawa R, Ishibashi S, Osuga J, Yagyu H, Oka T, et al. (1999) Embryonic lethality and defective neural tube closure in mice lacking squalene synthase. The Journal of biological chemistry 274: 30843–30848. [DOI] [PubMed] [Google Scholar]
- 58. Cunningham D, Spychala K, McLarren KW, Garza LA, Boerkoel CF, et al. (2009) Developmental expression pattern of the cholesterogenic enzyme NSDHL and negative selection of NSDHL-deficient cells in the heterozygous Bpa(1H)/+ mouse. Molecular genetics and metabolism 98: 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Caldas H, Cunningham D, Wang X, Jiang F, Humphries L, et al. (2005) Placental defects are associated with male lethality in bare patches and striated embryos deficient in the NAD(P)H Steroid Dehydrogenase-like (NSDHL) Enzyme. Molecular genetics and metabolism 84: 48–60. [DOI] [PubMed] [Google Scholar]
- 60. Jokela H, Rantakari P, Lamminen T, Strauss L, Ola R, et al. (2010) Hydroxysteroid (17beta) dehydrogenase 7 activity is essential for fetal de novo cholesterol synthesis and for neuroectodermal survival and cardiovascular differentiation in early mouse embryos. Endocrinology 151: 1884–1892. [DOI] [PubMed] [Google Scholar]
- 61. Krakowiak PA, Wassif CA, Kratz L, Cozma D, Kovarova M, et al. (2003) Lathosterolosis: an inborn error of human and murine cholesterol synthesis due to lathosterol 5-desaturase deficiency. Human molecular genetics 12: 1631–1641. [DOI] [PubMed] [Google Scholar]
- 62. Fitzky BU, Moebius FF, Asaoka H, Waage-Baudet H, Xu L, et al. (2001) 7-Dehydrocholesterol-dependent proteolysis of HMG-CoA reductase suppresses sterol biosynthesis in a mouse model of Smith-Lemli-Opitz/RSH syndrome. The Journal of clinical investigation 108: 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Derry JM, Gormally E, Means GD, Zhao W, Meindl A, et al. (1999) Mutations in a delta 8-delta 7 sterol isomerase in the tattered mouse and X-linked dominant chondrodysplasia punctata. jderry@immunex.com. Nature genetics 22: 286–290. [DOI] [PubMed] [Google Scholar]
- 64. Mirza R, Hayasaka S, Takagishi Y, Kambe F, Ohmori S, et al. (2006) DHCR24 gene knockout mice demonstrate lethal dermopathy with differentiation and maturation defects in the epidermis. The Journal of investigative dermatology 126: 638–647. [DOI] [PubMed] [Google Scholar]
- 65. Erickson SK, Lear SR, Deane S, Dubrac S, Huling SL, et al. (2003) Hypercholesterolemia and changes in lipid and bile acid metabolism in male and female cyp7A1-deficient mice. Journal of lipid research 44: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 66. Ishibashi S, Schwarz M, Frykman PK, Herz J, Russell DW (1996) Disruption of cholesterol 7alpha-hydroxylase gene in mice. I. Postnatal lethality reversed by bile acid and vitamin supplementation. The Journal of biological chemistry 271: 18017–18023. [DOI] [PubMed] [Google Scholar]
- 67. Shea HC, Head DD, Setchell KD, Russell DW (2007) Analysis of HSD3B7 knockout mice reveals that a 3alpha-hydroxyl stereochemistry is required for bile acid function. Proceedings of the National Academy of Sciences of the United States of America 104: 11526–11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li-Hawkins J, Gafvels M, Olin M, Lund EG, Andersson U, et al. (2002) Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. The Journal of clinical investigation 110: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Repa JJ, Lund EG, Horton JD, Leitersdorf E, Russell DW, et al. (2000) Disruption of the sterol 27-hydroxylase gene in mice results in hepatomegaly and hypertriglyceridemia. Reversal by cholic acid feeding. The Journal of biological chemistry 275: 39685–39692. [DOI] [PubMed] [Google Scholar]
- 70. Li-Hawkins J, Lund EG, Turley SD, Russell DW (2000) Disruption of the oxysterol 7alpha-hydroxylase gene in mice. The Journal of biological chemistry 275: 16536–16542. [DOI] [PubMed] [Google Scholar]
- 71. Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, et al. (1997) A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proceedings of the National Academy of Sciences of the United States of America 94: 12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE, et al. (1993) Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. The Journal of clinical investigation 92: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, et al. (1999) A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. The Journal of biological chemistry 274: 19055–19062. [DOI] [PubMed] [Google Scholar]
- 74. Febbraio M, Guy E, Coburn C, Knapp FF Jr, Beets AL, et al. (2002) The impact of overexpression and deficiency of fatty acid translocase (FAT)/CD36. Molecular and cellular biochemistry 239: 193–197. [PubMed] [Google Scholar]
- 75. Yu L, Hammer RE, Li-Hawkins J, Von Bergmann K, Lutjohann D, et al. (2002) Disruption of Abcg5 and Abcg8 in mice reveals their crucial role in biliary cholesterol secretion. Proceedings of the National Academy of Sciences of the United States of America 99: 16237–16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Outline of the statistical analysis.
(TIF)
The genotype effect on hepatic gene expression and blood lipids for (A) low fat, cholesterol-free diet; (B) high fat, cholesterol-free diet; and (C) High fat with 1.25% of cholesterol added diet. The direction of arrows indicate (↑) up-regulation or (↓) down-regulation of expression in heterozygous females (♀) and males (♂). * indicates p<0.1 and **p<0.05.
(TIF)
The expression of Nsdhl (A) on mRNA and (B) protein levels (western blot analysis) **p<0.05.
(TIF)
The expression of Tm7sf2 (A) on mRNA and (B) DHCR14 protein levels (western blot analysis) *p<0.1.
(TIF)
The significant interactions between sex and Cyp51 genotype in mice fed with the high-fat diet with 1.25% of cholesterol - the organ characteristics and the hepatic gene expression. The interactions in organ characteristics/blood profile were not observed on cholesterol-free diets. The Dhcr24 and Abcg8 were significantly different (p = 0.06 and 0.07 respectively) on the LFnC diet and Abcg5 reached significance (p = 0.08) on the HFnC diet (data not shown).
(TIF)
The significant interactions between the Cyp51 genotype and two high-fat diets with and without cholesterol.
(TIF)
The expression of Hmgcr (A) on mRNA and (B) protein levels (western blot analysis) **p<0.05.
(TIF)
The list of primers used for RT-PCR analysis.
(DOCX)
Number of animals analyzed within each contrast; A – body characteristics B – plasma lipids profile C – hepatic gene expression, D – liver histology.
(DOCX)
The average values (Mean) for weights, lipid profile and gene expression and standard error means (SEM) for global model (regardless diets) and for each diet separately.
(DOCX)
The interactions between diets and genotypes. Left panel shows the significant interactions between two cholesterol-free diets and Cyp51 genotype (indicated p values). The directions of arrows indicate the directions of changes from LFnC diet to HFnC diet, stars indicate statistically significant changes (** p<0.05 and * p<0.1). The right panel shows the significant interactions (p values indicated) between two high-fat diets and Cyp51 genotype. The directions of arrows indicate directions of changes for HFnC to HFC diet and stars indicate statistically significant changes.
(DOCX)
The sexual dimorphism – differences between females and males expressed as FC (females mean/males mean). *p<0.1; **p<0.05.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.