Abstract
BACKGROUND
Fluorescence in situ hybridization (FISH) to identify specific DNA target sequences in the nuclei of nondividing cells of numerous solid neoplasms has contributed to the introduction of molecular cytogenetics as a useful adjunct to cytology, leading recently to the “marriage” of the 2 disciplines. Numerous cancer molecular markers can now be investigated using different technical approaches, at both the gene and expression levels, in biopsies of various suspected cancers, including differentiated thyroid carcinoma. The limited amount of bioptic material is often insufficient to carry out multiple tests, and optimizing handling of the biopsy is desirable. METHODS We have developed a home-brew tetracolor break-apart probe able to simultaneously identify the 2 most common genetic alterations in differentiated thyroid carcinoma: RET/PTC variants in papillary thyroid carcinoma and PAX8/PPARg fusion and variants in follicular thyroid carcinoma. RESULTS The probe had 100% specificity, 99.5% sensitivity, and ≥3% cutoff. The probe was tested on RET/PTC and PAX8/PPARg RT-PCR positive controls, and feasibility was assessed in 368 thyroid nodule fine-needle aspirations (FNA). In the latter analysis, 24 FNAs had split RET signal, and 9 had split PPARg signal. FISH analysis of available surgically removed nodules confirmed the sensitivity of FISH in detecting abnormal clones and oligoclones. CONCLUSIONS The home-brew tetracolor probe showed high feasibility, optimizing the use of the biological material in relation to the available molecular tests and maximizing the FISH experimental and slide-scoring times. This probe may be considered an alternative to RT-PCR when recovery and quality of RNA amplification from FNA are insufficient. Cancer (Cancer Cytopathol) 2014;122:377–385. © 2014 The Authors. Cancer Cytopathology published by Wiley Periodicals, Inc. on behalf of American Cancer Society. This is an open access article under the terms of the Creative Commons Attribution-NonCommercial-NoDerivs License, which permits use and distribution in any medium, provided the original work is properly cited, the use is non- commercial and no modifications or adaptations are made.
Keywords: thyroid cancer, FISH, RET/PTC, PAX8/PPARg, FNA
INTRODUCTION
The “marriage of cytology and cytogenetics” has recently been celebrated, as reported by Dal Cin and coworkers: “Dearly Beloved, we are gathered here to join in happy matrimony…Cytology and Cytogenetics?&”1 To ensure a long-lasting marriage, it is of importance to continuously improve and innovate both the cytological side, especially through the immunohistochemical approach,2,3 and the cytogenetic side. In this view, refining the fluorescence in situ hybridization (FISH) probe panel suitable for targeting specific DNA regions involved in changes (such as gene loss, amplification, gene fusion) is crucial both because of the implications for cancer research and for the efficient use of FISH as an adjunct to cytology. Many commercially available probes are today routinely used in cytology and histology, mainly as ancillary tools in the diagnosis of soft-tissue tumors.4 This approach is also gaining favor for analysis of tumors of epithelial origin5; we recently discussed the pros and cons of FISH in papillary thyroid nodules.6 Papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC), which account for more than 80% of all thyroid cancers, represent 2 histotypes showing definite and distinct morphological patterns. However, in a number of cases the morphological figure may be ambiguous and diagnosis not straightforward, especially in cytological preparations in which the tissue architecture of the gland is lost. In these cases, molecular-based methodologies may be a valuable adjunct. Progress in our understanding of the molecular pathways implicated in the pathobiology of PTC and FTC variants has revealed that several genetic changes play a material role in the thyroid carcinogenic process. Mutations of BRAFV600E, rearrangements of the tyrosine kinase receptor genes RET or TRK, or mutations in RAS have been identified in approximately 70% of PTCs. Mutations of RAS or PAX8/PPARg rearrangements have been identified in up to 75% of FTCs. The utility of molecular screening for these markers in routine fine-needle aspiration (FNA) has been asserted.7 It was demonstrated that more than 8% of nodules with indeterminate cytological diagnosis bear genetic alterations, including RET and PPARg rearrangements.7 Moreover, indeterminate cytology lacking the most frequent genetic alterations are less likely to be malignant (6% to 28% risk),8 indicating a benefit of the combination of cytological evaluation and molecular analysis in the management of patients with thyroid nodules. Until recently, the genetic alterations within and among PTC and FTC were largely considered to be mutually exclusive.9 However, Guerra and coworkers10 recently reported concomitant BRAFV600E mutation and RET rearrangement in approximately 19% of the nodules studied. A rare case with a RET/PTC rearrangement and simultaneous PAX8/PPARg rearrangement was also reported.11 The use of highly sensitive methods (such as Southern blot on reverse transcriptase-polymerase chain reaction [RT-PCR]) has been crucial in discovering the occurrence of sporadic oligoclones with RET rearrangements in histologically benign lesions, whereas, the occurrence of consistent clones remains exclusive to PTC.12 On the other hand, the meaning of PPARg rearrangement in benign nodules is still unclear, although studies in transgenic models suggest that lesions carrying this anomaly may be precursors of PAX8/PPARg-positive FTC.13 These observations have both pathobiological and clinicopathological significance and call for establishing precise diagnostic cutoff values. The simultaneous investigation by FISH of the integrity of RET and PPARg genes (indicative of RET/PTC and PAX8/PPARg or variant fusions, respectively) would improve thyroid FNA characterization by maximizing the use of available biological material and would reduce the time required for performing FISH and scoring slides. This optimization is desirable, in part, because of the emergence of numerous promising molecular tests that are suited to FNA.14 Moreover, the ability to analyze concurrently the integrity of these 2 genes would help to identify cases with a dual mutation and would facilitate determining their clonal relationship.
To this end, we have developed a tetracolor break-apart cocktail probe in which 5′ and 3′ flanking sequences of the RET and PPARg genes are labeled with 4 different fluorophores. To assess the feasibility of tetracolor FISH, the probe was tested on 368 FNA and 13 corresponding surgically removed nodules.
MATERIALS AND METHODS
Tetracolor Break-Apart Probe Preparation
A total of 10 bacterial artificial chromosome (BAC) clones were selected from the University of California Santa Cruz Human Genome Browser (http://genome.cse.ucsc.edu) and were obtained from the BACPAC Resources Center, Children’s Hospital Oakland Research Institute (Oakland, Calif.; http://www. chori.org/). Selected BAC clones flanking the 5′ and 3′ of RET (located at 10q11.2)6 and PPARg (located at 3p25)15 are reported in Figure 1. The 2 proximal and 2 distal RET BAC clones span 272.9 and 351.5 kb, respectively; the 3 proximal and 3 distal PPARg BAC clones span 609.7 and 528.4 kb, respectively (Fig. 1).
Figure 1.
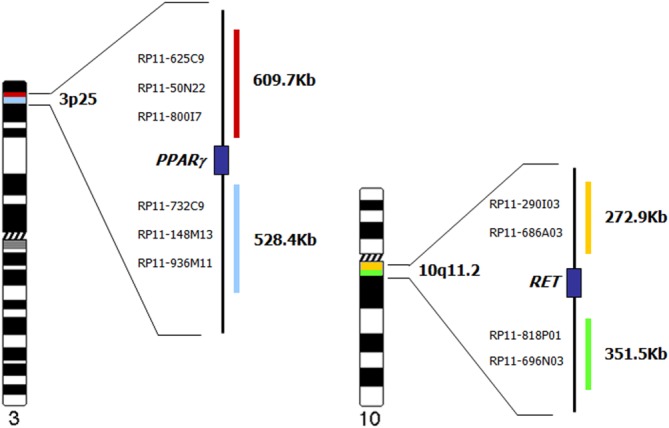
Schematic representation of the proximal and distal positions of the BAC clones used in the preparation of the gold/green RET and red/aqua PPARg premix for the tetracolor probe setup.
BACs were expanded, and DNA was isolated by a HiPure Plasmid Midiprep Kit (Invitrogen, Carlsbad, Calif.) and labeled by nick translation according to the manufacturer’s instructions. Four different fluorophores (Abbott Molecular/Vysis, Downers Grove, Ill) were used to prepare the tetracolor break-apart mix: proximal and distal RET probes were labeled with Spectrum Gold dUTP and Spectrum Green dUTP, respectively; proximal and distal PPARg probes were labeled with Spectrum Aqua dUTP and Spectrum Red dUTP, respectively, according to the manufacturer’s instructions. The labeled DNA from all 10 BAC clones was pooled to yield a probe with strong FISH signals.
Fluorescence In Situ Hybridization
The tetracolor probe was hybridized on cytogenetic and cytological preparations using 10 μL of probe per slide (final probe concentration 40 ng/μL). Hybridization was performed on a HYBrite unit (Abbott Molecular/Vysis). Denaturation was performed for 5 minutes at 75°C, followed by overnight hybridization at 37°C. Posthybridization washes were performed at 60°C in 0.1× SSC 3 times for 5 minutes. Slides were counterstained with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride). Preparations were examined on an Olympus BX41 epifluorescence microscope equipped with a charge-coupled device camera (Cohu, San Diego, Calif.) interfaced with the CytoVision system (software version 3.9; Applied Imaging, Pittsburgh, PA). Cytogenetic preparations (metaphases and nuclei) from phytohemagglutinin-stimulated lymphocyte cultures (in DMEM/F12 medium with additives; Gibco/Life Technologies, Milan, Italy) derived from apparently healthy male volunteers were used to establish the analytical specificity (percentage of FISH signal located at the correct chromosomal position, RET at 10q11.2 and PPARg at 3p25) and sensitivity (percentage of metaphases or nuclei with correct FISH signal pattern) of the tetracolor probe (Fig. 2a).
Figure 2.
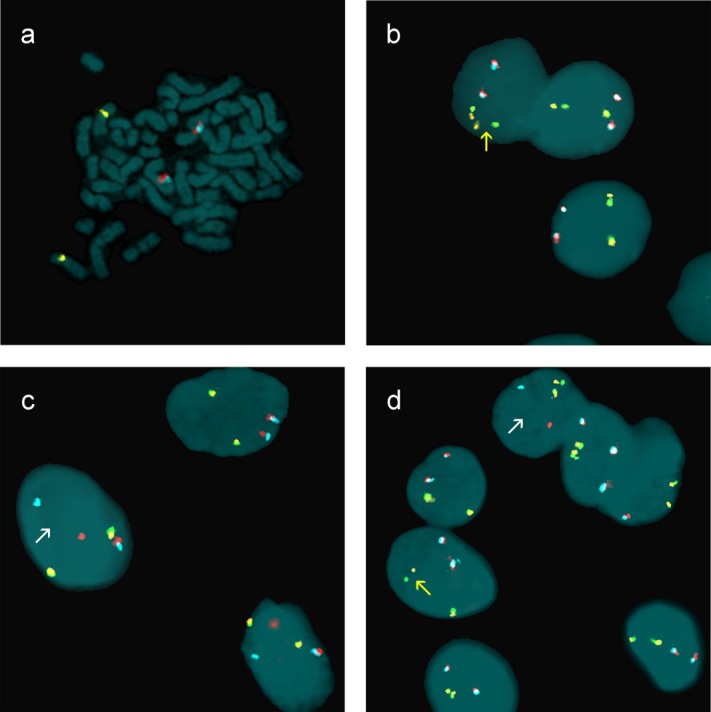
FISH with a home-brew tetracolor probe for identification of RET and PPARg breaks. (a) Specificity of the tetracolor mix: gold/green RET gene, located at 10q11.2, and red/aqua PPARg gene, located at 3p25. (b) Nuclei of FNA (nodule 375) showing broken RET: break-apart of 1 FISH gold/green signal (arrow). (c) Nuclei of FNA (nodule 265) showing broken PPARg: break-apart of 1 FISH red/aqua signal (arrow). (d) Nuclei of a control case with both broken RET and PPARg: break-apart of 1 FISH gold/green (yellow arrow) signal and 1 FISH red/aqua (white arrow). Nuclei and chromosomes are counterstained with DAPI.
Nuclei from formalin-fixed, paraffin embedded (FFPE) tissue samples, obtained as described6 from the apparently tumor-free contralateral lobe of 3 PTCs and 3 FTCs negative for RET/PTC and PAX8/PPARg by conventional RT-PCR, were used for establishing the probe cutoff value. This source was chosen to ensure that no tumor tissue was present in the sample: a 30-μm section, consecutive to the 4-μm section used for the histological investigation, was used for nuclei isolation.
Nuclei from PTCs and FTCs positive for RET/PTC and PAX8/PPARg alterations by conventional RT-PCR were used to confirm the probe efficiency.
For specificity analysis, metaphases and nonoverlapping nuclei with 4 visible color signals were scored. Each fluorescence signal was singularly captured, a pseudocolor was assigned to each, and the acquired images were superimposed. Merged images were edited with the Applied CytoVision FISH program. Fifty metaphases were scored to assess probe specificity. To establish probe sensitivity, the metaphase findings were integrated with a scoring of 600 interphase nuclei. Signal pattern interpretation was as follows: 2 unsplit gold/green and 2 unsplit red/aqua FISH signals were interpreted as unbroken RET and PPARg genes; 1 split gold/green FISH signal plus 1 unsplit gold/green signal and 2 unsplit red/aqua FISH signals were interpreted as 1 broken RET; and 1 split red/aqua FISH signal plus 1 unsplit red/aqua signal and 2 unsplit gold/green FISH signals were interpreted as 1 broken PPARg.
To ensure the feasibility of using a home-brew tetracolor FISH probe in a clinical setting, the probe was tested on 368 cytologically classified FNA samples retrieved from our file of thyroid smears maintained at −30°C. In detail, during ultrasound-guided FNA a minimum of 5 direct smears were prepared. All smears except 1 (or more if available) were fixed in alcohol and used for cytological analysis. The remaining smears were air-dried, treated with a hypotonic solution (KCl 0.075M) for 5 minutes at 37°C, and fixed with methanol acetic (3:1) solution for 10 minutes at room temperature. Slides were air-dried and checked under contrast-phase microscopy at low magnification (20×). The presence of approximately 150 nuclei, including both grouped (at least 5 consistent groups) and spread, was the cutoff for eligibility. Small nuclei, associated with lymphocytic infiltration, were not considered. This procedure usually ensured a recovery of a minimum of 75 nuclei with FISH signal. Thawed slides were aged 24 hours at room temperature and, prior to being used for FISH, were pretreated at room temperature in acetic acid 70% (v/v) for 15 minutes, in phosphate-buffered saline (PBS) balanced solution for 5 minutes, and in a graded ethanol series (70%–90%–95%–100%) for 5 minutes each. Preparations were digested with pepsin solution (10 mg in 100 mL of 0.01 N HCl; Sigma Aldrich, Milan, Italy) for 3 minutes at 37°C, rinsed in PBS, and dehydrated in the graded ethanol series. Of the 368 cytologically classified FNA samples, 281 have been previously reported, and previous data were available from an analysis of RET integrity obtained with a dual-color FISH probe.6 According to the British Thyroid Association, the Italian Society of Anatomic Pathology, and diagnostic cytology consensus, cytological diagnosis assigned these FNA specimens to the following cytological classes: Thy 1 (20 nodules), Thy 2 (236 nodules), Thy 3 (75 nodules), Thy 4 (17 nodules), and Thy 5 (20 nodules); see Table1.
Table 1.
Tetracolor FISH in FNA samples: RET and PPARg rearrangements
| FNA | Cytology | FISH Broken RET | FISH Broken PPARg |
|---|---|---|---|
| 20 | Thy 1 | 0 | 1 |
| 236 | Thy 2 | 9 | 6 |
| 75 | Thy 3 | 9 | 2 |
| 17 | Thy 4 | 1 | 0 |
| 20 | Thy 5 | 5 | 0 |
For each sample, a minimum of 75 and a maximum of 300 nonoverlapping nuclei were scored. The study was carried out in accordance with review board–approved protocols and in compliance with the Helsinki Declaration (http://www.wma.net/en/30publications/10policies/b3). Written informed consent was obtained from all patients before FNA. None of the patients of this pilot study was advised to undergo surgery on the basis of FISH results. Moreover, 13 samples were available for the FISH postsurgical control study on touch preparations or isolated nuclei from FFPE, obtained as described.6
RESULTS
Fluorescence In Situ Hybridization and Probe Validation
The FISH analysis on lymphocytes established that the probe exhibited 100% specificity for both RET and PPARg and 99.5% sensitivity for RET and 99.8% for PPARg. Taking into account the lower value, we consider the tetracolor probe specificity to be 100% and sensitivity to be 99.5%.
To establish the cutoff value, the probe was then tested on 1500 nuclei isolated from FFPE samples of apparently normal thyroid tissue. The cutoff value for correctly identifying normal thyroid samples, calculated as the beta inverse function,16 was split FISH signals in 2.4% of cells for RET and 0% for PPARg. On this basis, a sample was considered negative for broken RET when a split FISH signal was observed in <3% of nuclei. A sample was also considered negative for broken PPARg when a split FISH signal was observed in <3% of nuclei (arbitrarily set because no broken PPARg were observed in controls). Conversely, a sample was considered positive if a broken signal for RET or PPARg was observed in ≥3% of nuclei. Moreover, as already noted,6 the RET break-apart FISH signal was observed in ≥3% and <6.8% of nuclei only in single isolated scattered cells, whereas the RET break-apart FISH signal was observed in ≥6.8% of nuclei among grouped nuclei.
Probe efficiency was confirmed on previously published RT-PCR positive controls.11,17 In 1 control case of PTC that was positive for RET/PTC1 and negative for PAX8/PPARg, we found by FISH that 19% of nuclei had broken RET and intact PPARg. In 1 control case of FTC that was negative for RET/PTC and positive for PAX8/PPARg, we found that 21% of nuclei had broken PPARg and intact RET. In 1 control case of PTC follicular variant positive for both RET/PTC3 and PAX8/PPARg, we confirmed the rearrangements of RET (in 30% of nuclei) and PPARg (in 14% of nuclei).
Detection of RET and PPARg Rearrangement in Cytological and Histological Samples
We found a total of 33 FNAs (9%) with broken genes, 24 with broken RET (Fig. 2b), and 9 with broken PPARg (Fig. 2c): broken RET was observed in 9 Thy 2 (3.8%), 9 Thy 3 (12%), 1 Thy 4 (5.9%), and 5 Thy 5 (25.0%) FNAs; broken PPARg was present in 1 Thy 1 (5%), 6 Thy 2 (2.5%), and 2 Thy 3 (2.6%) FNAs. No FNA exhibited a dual rearrangement (Table1) similar to the one found in the control case (Fig. 2d).
Of 18 surgically removed nodules, 13 were available for the FISH study. All these were RET-positive FNAs. The surgical samples positive for PPARg unfortunately were not available for FISH analysis. In a previous report,6 we set a 6.8% cutoff for RET clonal rearrangement, which was associated with malignancy. In general, this threshold was confirmed in the present series, that is, RET rearrangements were observed in the corresponding tumor cells (Table2). Moreover, these 13 samples were negative for PPARg rearrangement, confirming the results obtained in FNA. Of 28 surgically removed Thy 3 FISH-negative samples (2.6%) histology was available for 28 nodules: 21 benign and 7 malignant (2 PTC, 2 FTC, 3 PTC-follicular variant).
Table 2.
FISH Results on Cytological and Histological Preparations Using the Tetracolor Break-Apart Probe
| Nodule ID | FNA | Tissue | |||
|---|---|---|---|---|---|
| Cytology | FISH Broken RET | FISH Broken PPARg | Histology | FISH Broken RET | |
| 532 | Thy 1 | neg | pos | Adm goiter | NA |
| 1 | Thy 2 | posOC | neg | No surgery | |
| 16 | Thy 2 | posOC | neg | No surgery | |
| 20 | Thy 2 | posOC | neg | FTA | neg |
| 34 | Thy 2 | neg | pos | No surgery | |
| 40 | Thy 2 | posOC | neg | No surgery | |
| 80 | Thy 2 | neg | pos | No surgery | |
| 169 | Thy 2 | neg | pos | No surgery | |
| 334 | Thy 2 | posOC | neg | No surgery | |
| 294 | Thy 2 | posC | neg | No surgery | |
| 362 | Thy 2 | posOC | neg | No surgery | |
| 380 | Thy 2 | neg | pos | No surgery | |
| 411 | Thy 2 | neg | pos | No surgery | |
| 490 | Thy 2 | posOC | neg | No surgery | |
| 519 | Thy 2 | posOC | neg | No surgery | |
| 535 | Thy 2 | neg | pos | No surgery | |
| 9 | Thy 3 | posOC | neg | Benign | neg |
| 26 | Thy 3 | posOC | neg | PTC-FV | neg |
| 32 | Thy 3 | posOC | neg | No surgery | |
| 36 | Thy 3 | posOC | neg | Benign | neg |
| 82 | Thy 3 | posOC | neg | Benign | neg |
| 265 | Thy 3 | neg | pos | PTC-FV | NA |
| 323 | Thy 3 | posOC | neg | Benign | neg |
| 375 | Thy 3 | posOC | neg | microPTC | posC |
| 413 | Thy 3 | neg | pos | Adm goiter | NA |
| 488 | Thy 3 | posOC | neg | Mltn goiter | NA |
| 542 | Thy 3 | posOC | neg | PTC-FV | NA |
| 60 | Thy 4 | posC | neg | PTC | posC |
| 13 | Thy 5 | posOC | neg | PTC-WLV | neg |
| 228 | Thy 5 | posC | neg | PTC | posC |
| 260 | Thy 5 | posC | neg | PTC | posC |
| 284 | Thy 5 | posC | neg | PTC | posC |
| 295 | Thy 5 | posOC | neg | PTC | neg |
Adm goiter, adenomatous goiter; FTA, follicular thyroid adenoma; NA, not available; pos, positive, break ≥3%, no threshold set for PPARg clonal change (see discussion); posC, positive, clonal change (break ≥6,8%); posOC, positive, oligoclonal change (break ≥3% and <6,8%); PTC, papillary thyroid carcinoma; PTC-FV, PTC–follicular variant; Mltn goiter, multinodular goiter; PTC-WLV, PTC–Whartin-like variant.
Lesion with occult PTC; number of patients = 368.
DISCUSSION
Different kinds of genetic alterations, as well as epigenetic phenomena, have been identified that are now considered important information for classification, prognosis, and treatment of thyroid cancer. Among commonly observed changes are mutations, gene amplification, chromosome translocations leading to chimeric genes, chromosome gains, and promoter methylation.14 Consequently, considering all these features, the impact of gene alteration testing on the diagnosis and management of patients with thyroid cancer is increasing, particularly in view of more accurate cancer risk stratification of lesions with an indeterminate presurgical diagnosis.8 Currently, several molecular approaches are available to investigate thyroid tumor cell pathobiology at different levels (gene, chromosome, gene expression). These include CGH array,18 DNA sequencing,7 polymerase chain reaction (conventional, multiplex, quantitative), FISH, immunohistochemistry, proteomics,19 and the recently introduced mass spectrometry.20 The integration of results obtained by these technologies has been invaluable in clarifying genetic alterations associated with thyroid cancer and in interpreting the key roles of impaired signaling pathways. One of the main aspects highlighted by the molecular studies is the presence of intratumoral heterogeneity in the distribution of the genetic changes,21 for example, a mixture of cells with and without rearrangements. This finding may dispute the tumorigenic role attributed to the most common thyroid carcinoma–related mutations, confining them to a secondary role in tumor initiation but possibly maintaining a key role in the final diagnosis, with clinical and treatment implications.22 In fact, the biological and clinical meaning of finding or not finding multiple changes in the same or separate clones might be different, especially if such heterogeneity affects morphological aspects. As the presence of dual mutations in the same nodule is an emerging finding, understanding the reciprocal role of concurrent impaired genes that are involved in the same or different signaling pathways is challenging.10 Among the above-mentioned techniques, only FISH enables identifying gene fusion within a single nucleus and permits localizing chimeric genes within the same cell or in different cells. In this regard, the probe we have developed has an added value, allowing determination of the prevalence of simultaneous RET and PPARg rearrangements in differentiated thyroid cancer variants. In the present series, we found 33 cases with RET or PPARg rearrangement. PPARg rearrangement was found in Thy 1-Thy 3 classes but not in Thy 4 or Thy 5. Absence of PPARg rearrangement in Thy 5 cases is expected because PTCs are usually found in this class.
We recently reported the importance of FISH as an adjunct to cytology in thyroid nodule management, highlighting the challenge associated with interpretation of results and the need for precise laboratory FISH cutoffs.6 Interpretation of FISH results (far from being “counting dots”) can be rather complex, and the pathologist and the cytogeneticist should work in concert to reach a conclusive result. It is a fact that oligoclonal foci of molecularly abnormal epithelial cells without morphologic abnormalities can be detected throughout the thyroid epithelium, and the finding of isolated single sporadic FISH RET-positive cells in FNA might reflect this pattern. Accumulating knowledge on the prevalence of RET and PPARg rearrangements in thyroid nodules would lead to a better understanding not only of the follicular cell carcinogenetic process but also of the diagnostic and prognostic potential of these markers. The clinical implication of RET activation in sporadic cells is still debated, and it is not known if a phenomenon of field cancerization23 and a different capability of individual cells to respond to growth-regulating signals from neighboring cells are responsible for the expansion of a “cell patch” with RET rearrangement. Nevertheless, although considered fully benign, careful follow-up is indicated for FNA with benign classification (Thy 2) and RET rearrangement ascertained by Southern blot on RT-PCR because they grow rapidly.12 According to Marotta et al,12 FISH is, at present, the most appropriate method for detecting clonal changes with a view toward refining indeterminate cytology. In our view,6 FNA containing grouped cells with an identical abnormal FISH pattern is indicative of an appreciable clonal growth, whereas sporadic cells with the same abnormal pattern possibly indicate an oligoclonal process. Translating FISH analysis of thyroid FNA into the clinical setting will be a future step; at the moment, the International System for Human Cytogenetic Nomenclature24 does not contain definitions dedicated to FISH results for clones, oligoclones, clonal evolution, and unrelated clones. In the present series, excluding nodule 375, with a histological diagnosis of microcarcinoma and sporadic change (possibly from FNA bias), 71% of Thy 3 nodules with RET-positive scattered cells proved to be benign, confirming the appropriateness of a ≥6.8% cutoff for malignancy. Because of the unavailability of surgical material for FISH, any effort to correlate the presence of sporadic cells with PPARg rearrangement and the final diagnosis was impaired. Nevertheless, we would point out that a Thy 3 cytological sample with broken PPARg in 16% of nuclei had a diagnosis of follicular variant of PTC, a variant often exhibiting this anomaly. Further study will be needed to confirm the hypothesis that PPARg rearrangements may be precursors of PPARg-rearranged positive FTC, as suggested by analysis of a transgenic mouse model.13 Our findings indicate the feasibility of optimizing molecular cytogenetics in presurgical thyroid material, improving and supporting the use of FISH as an adjunct to cytology. Early cytogenetic studies have characterized different cytogenetic subclasses of differentiated thyroid tumors.25 FISH analysis revealed a higher rate of thyroid tumors with clonal cytogenetic deviations than conventional cytogenetics.26 Rapidly increasing the use of molecular approaches for analyzing FNA material is occurring,7,14 especially in the effort to better characterize nodules with indeterminate cytological diagnosis.8 Although the management of approximately 30% of thyroid cancers remains problematic because they do not show common mutations detectable by the currently available molecular tests,27 molecular tests may be informative in most nodules with indeterminate cytology. The use of ancillary techniques on direct-smear aspirates represents a significant evolution for cytopathologic techniques.28 Data correlating clonal/oligoclonal expansion of gene-rearranged cells with clinical outcomes in thyroid neoplasms need to be explored and improved because both RET/PTC and PAX8/PPARg protein products are attractive diagnostic targets.29,30 FNA provides a unique chance for presurgical diagnosis of a thyroid nodule. Assessing the presence of the corresponding chimeric genes in presurgical samples is becoming unavoidable. RT-PCR is a good routine methodology for this purpose; however, recovery and quality of RNA amplification from FNA may be a limiting factor. In this case, the tetracolor probe may overcome this limitation, although further studies are needed to ascertain whether and to what extent FISH technology may improve the diagnostic accuracy of indeterminate (Thy 3) cytology. As for other tumors1 such as renal tumors,31 the application of FISH to FNA has been reported to be significant for tumor subclassification. In this regard, collecting as much information as possible on the status of RET and PPARg in thyroid FNA and comparing the data with the morphological patterns and clinical outcomes would provide a deeper and more focused understanding of the diagnostic and prognostic value of these rearrangements in thyroid lesions.
FUNDING SOURCES
This work was partially supported by the Italian Ministry of University and Research (PRIN grant No 2007MBW5M7), and Fondazione Banco Sardegna (Grant No 1282/2010-1132).
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures.
REFERENCES
- Dal Cin P, Qian X, Cibas ES. The marriage of cytology and cytogenetics. Cancer Cytopathol. 2013;121:279–290. doi: 10.1002/cncy.21270. [DOI] [PubMed] [Google Scholar]
- Rossi ED, Martini M, Capodimonti S. Diagnostic and prognostic value of immunocytochmistry and BRAF mutation analysis on liquid-based biopsies of thyroid neoplasms suspicious for carcinoma. Eur J Endocrinol. 2013;168:853–859. doi: 10.1530/EJE-13-0023. [DOI] [PubMed] [Google Scholar]
- Zimmermann AK, Camenisch U, Rechsteiner MP, Bode-Lesniewska B, Rössle M. Value of immunohistochemistry in the detection of BRAFV600E mutations in fine-needle aspiration biopsies of papillary thyroid carcinoma. Cancer Cytopathol. 2014;122:48–58. doi: 10.1002/cncy.21352. [DOI] [PubMed] [Google Scholar]
- Lindeman NI, Dal Cin P, Cheng L, Zhang DY. Molecular Genetic Pathology. New York: Humana Press; 2008. Molecular testing for solid tumors; pp. 467–495. [Google Scholar]
- Cheng L, Eble JN. Molecular Surgical Pathology. New York: Springer; 2013. [Google Scholar]
- Caria P, Dettori T, Frau DV. Assessing RET/PTC in thyroid nodule fine-needle aspirates: the FISH point of view. Endocr Relat Cancer. 2013;20:527–536. doi: 10.1530/ERC-13-0157. [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Wald AI, Roy S, Durso MB, Nikiforov YE. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1852–E1860. doi: 10.1210/jc.2013-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov YE, Ohori NP, Hodak SP. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: a prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab. 2011;96:3390–3397. doi: 10.1210/jc.2011-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra A, Zeppa P, Bifulco M, Vitale M. Concomitant BRAFV600E mutation and RET/PTC rearrangement is a frequent occurrence in papillary thyroid carcinoma. Thyroid. 2013 doi: 10.1089/thy.2013.0235. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Caria P, Dettori T, Frau DV. Simultaneous occurrence of PAX8-PPARg and RET-PTC3 rearrangements in a follicular variant of papillary thyroid carcinoma. Am J Surg Pathol. 2012;36:1415–1420. doi: 10.1097/PAS.0b013e318264bdd6. [DOI] [PubMed] [Google Scholar]
- Marotta V, Guerra A, Sapio MR, Vitale M. RET/PTC rearrangement in benign and malignant thyroid diseases: a clinical standpoint. Eur J Endocrinol. 2011;165:499–507. doi: 10.1530/EJE-11-0499. [DOI] [PubMed] [Google Scholar]
- Kim CS, Zhu X. Lessons from mouse models of thyroid cancer. Thyroid. 2009;19:1317–1331. doi: 10.1089/thy.2009.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–1069. doi: 10.1016/S0140-6736(13)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA, Fletcher JA, Cibas ES, Caulfield C, Allard P, Kroll TG. Molecular detection of PPARγ rearrangements and thyroid carcinoma in preoperative fine-needle aspiration biopsies. Endocr Pathol. 2008;19:166–174. doi: 10.1007/s12022-008-9036-0. [DOI] [PubMed] [Google Scholar]
- Wiktor AE, Van Dyke DL, Stupca PJ. Preclinical validation of fluorescence in situ hybridization assays for clinical practice. Genet Med. 2006;8:16–23. doi: 10.1097/01.gim.0000195645.00446.61. [DOI] [PubMed] [Google Scholar]
- Frau DV, Lai ML, Caria P. Trisomy 17 as a marker for a subset of noninvasive thyroid nodules with focal features of papillary carcinoma: cytogenetic and molecular analysis of 62 cases and correlation with histological findings. J Clin Endocrinol Metab. 2008;93:177–181. doi: 10.1210/jc.2007-0970. [DOI] [PubMed] [Google Scholar]
- Unger K, Malisch E, Thomas G. Array CGH demonstrates characteristic aberration signatures in human papillary thyroid carcinomas governed by RET/PTC. Oncogene. 2008;27:4592–4602. doi: 10.1038/onc.2008.99. [DOI] [PubMed] [Google Scholar]
- Dom G, Tarabichi M, Unger K, Thomas G. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br J Cancer. 2012;107:994–1000. doi: 10.1038/bjc.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainini V, Pagni F, Garancini M. An alternative approach in endocrine pathology research: MALDI-IMS in Papillary Thyroid Carcinoma. Endocr Pathol. 2013;24:250–253. doi: 10.1007/s12022-013-9273-8. [DOI] [PubMed] [Google Scholar]
- Unger K, Zitzelsberger H, Salvatore G. Heterogeneity in the distribution of RET/PTC rearrangements within individual post-Chernobyl papillary thyroid carcinomas. J Clin Endocrinol Metab. 2004;89:4272–4279. doi: 10.1210/jc.2003-031870. [DOI] [PubMed] [Google Scholar]
- Vitale M. Intratumor BRAFV600E heterogeneity and kinase inhibitors in the treatment of thyroid cancer: a call for participation. Thyroid. 2013;23:517–519. doi: 10.1089/thy.2012.0614. [DOI] [PubMed] [Google Scholar]
- Braakhuis BJ, Tabor MP, Kummer JA. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- Shaffer LG, McGowan-Jordan J, Schmid M, editors. An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: S. Karger; 2013. [Google Scholar]
- Caria P, Vanni R. Cytogenetic and molecular events in adenoma and well-differentiated thyroid follicular-cell neoplasia. Cancer Genet Cytogenet. 2010;203:21–29. doi: 10.1016/j.cancergencyto.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Drieschner N, Rippe V, Laabs A. Interphase fluorescence in situ hybridization analysis detects a much higher rate of thyroid tumors with clonal cytogenetic deviations of the main cytogenetic subgroups than conventional cytogenetics. Cancer Genet. 2011;204:366–374. doi: 10.1016/j.cancergen.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Hassell LA, Gillies EM, Dunn ST. Cytologic and molecular diagnosis of thyroid cancers: is it time for routine reflex testing? Cancer Cytopathol. 2012;120:7–17. doi: 10.1002/cncy.20186. [DOI] [PubMed] [Google Scholar]
- Knoepp SM, Roh MH. Ancillary techniques on direct-smear aspirate slides: a significant evolution for cytopathology techniques. Cancer Cytopathol. 2013;121:120–128. doi: 10.1002/cncy.21214. [DOI] [PubMed] [Google Scholar]
- Ye L, Santarpia L, Gagel RF. The evolving field of tyrosine kinase inhibitors in the treatment of endocrine tumors. Endocr Rev. 2010;31:578–599. doi: 10.1210/er.2009-0031. [DOI] [PubMed] [Google Scholar]
- McIver B, Grebe SK, Eberhardt NL. The PAX8/PPAR gamma fusion oncogene as a potential therapeutic target in follicular thyroid carcinoma. Curr Drug Targets Immune Endocr Metabol Disord. 2004;4:221–234. doi: 10.2174/1568008043339802. [DOI] [PubMed] [Google Scholar]
- Roh MH, Dal Cin P, Silverman SG, Cibas ES. The application of cytogenetics and fluorescence in situ hybridization to fine-needle aspiration in the diagnosis and subclassification of renal neoplasms. Cancer Cytopathol. 2010;118:137–145. doi: 10.1002/cncy.20077. [DOI] [PubMed] [Google Scholar]


