Abstract
The DNA of E. coli contains 19,120 6-methyladenines and 12,045 5-methylcytosines in addition to the four regular bases and these are formed by the postreplicative action of three DNA methyltransferases. The majority of the methylated bases are formed by the Dam and Dcm methyltransferases encoded by the dam (DNA adenine methyltransferase) and dcm (DNA cytosine methyltransferase) genes. Although not essential, Dam methylation is important for strand discrimination during repair of replication errors, controlling the frequency of initiation of chromosome replication at oriC, and regulation of transcription initiation at promoters containing GATC sequences. In contrast, there is no known function for Dcm methylation although Dcm recognition sites constitute sequence motifs for Very Short Patch repair of T/G base mismatches. In certain bacteria (e.g., Vibrio cholerae, Caulobacter crescentus) adenine methylation is essential and in C. crescentus, it is important for temporal gene expression which, in turn, is required for coordinating chromosome initiation, replication and division. In practical terms, Dam and Dcm methylation can inhibit restriction enzyme cleavage; decrease transformation frequency in certain bacteria; decrease the stability of short direct repeats; are necessary for site-directed mutagenesis; and to probe eukaryotic structure and function.
INTRODUCTION
DNA methylation in bacteria is most often thought of in its role to protect DNA from restriction endonucleases. In addition to this role, however, studies in Escherichia coli, Salmonella enterica serovar Typhimurium (referred to as S. enterica hereafter) and Caulobacter crescentus have shown that methylated bases have other biological functions. In these cases, the methylated bases are not part of a restriction/modification system and the enzymes that produce them are often referred to as orphan or solitary DNA methyltransferases. The postreplicative DNA methylation produced by these enzymes superimposes on the primary DNA sequence secondary information that has significance for DNA transactions such as transcription, transposition, initiation of chromosome replication, mRNA utilization and prevention of mutations by DNA repair. These alterations are brought about in two ways, the first being simply a change in the steady-state level of the methyltransferase either up or down from normal. The second mechanism is through the configuration of the nucleotide sequence subject to methylation; it can exist as symmetrically methylated, unmethylated or two possible hemi-methylated arrangements. The details about the changes in DNA transactions through alteration of methyltransferase levels or state of methylation sequences form the bulk of this review.
This review is an updated version of that which appeared in the last print edition of Escherichia coli and Salmonella: Cellular and Molecular Biology [1]. Several reviews of DNA methylation have appeared recently that are considered complementary to this article [2–5].
METHYLATED BASES IN DNA
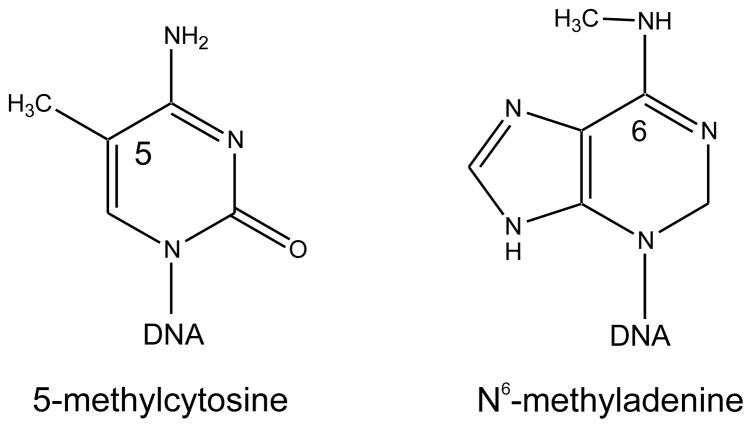
The DNA of E. coli K-12 contains two modified bases; 6-methyl-adenine (6-meAde, Fig. 1) and 5-methylcytosine (5-meCyt, Fig. 1). About 1.5% of all adenines (19,120 in GATC) and 0.75% of all cytosines (12,045 in CCWGG) in the chromosome are methylated, and the modifications occur in specific sequences resulting from the action of three DNA methyltransferases (Table 1). The EcoK adenine methyltransferase, which is encoded by the hsd (host specificity) genes, is part of the classical EcoK restriction/modification system described in detail in EcoSal chapter 4.4.6. The gene for a fourth methyltransferase, YhdJ, is not expressed under laboratory growth conditions [6].
FIGURE 1.
Structures of 5-methylcytosine and N6-methyladenine.
TABLE 1.
DNA methyltransferases in E. coli K-12
| Gene(s) | Modification methyltransferase | Recognition sequencea | Number in genomeb | Restriction endonucleasesc |
|---|---|---|---|---|
| hsdSM | M.EcoK | -AAC(N6)GTCG- | 595 | EcoKI |
| dam | Dam | -GATC- | 19,120 | DpnI, DpnII, Sau3A |
| dcm | Dcm | -CCWGG- | 12,045 | EcoRII, BstNI |
| yhdJ | YhdJ | -ATGCAT- | 839 | NsiI |
The base in boldface is modified.
All M.EcoK sites are methylated. For Dam and Dcm, most sites are methylated but this number also includes hemimethylated and unmethylated sites. YhdJ methylation is below the limit of detection at this sequence.
Restriction enzymes recognize the same sequence as the methyltransferases. EcoKI cuts only if the sequence is unmethylated; DpnI cleaves only if the sequence is methylated; Sau3A cuts regardless of the state of methylation. EcoRII cleaves only at unmethylated sites and BstNI cutting is not affected by methylation.
The Dam (DNA adenine methyltransferase) enzyme, which modifies GATC sequences, forms over 99% of the 6-meAde in E. coli DNA, since strains lacking this enzyme contain only the contribution expected from the EcoK enzyme [7, 8]. The Dcm (DNA cytosine methyltransferase) protein, methylating CC(A/T)GG sites, is responsible for all the 5-meCyt in DNA, since none of this modified base can be detected in cells deleted for the dcm gene (2). A dam dcm hsdS mutant contains no detectable modified bases in DNA, indicating that such bases are not essential for E. coli viability [8].
Although dam gene function is not essential for viability of E. coli in a wild-type background, it is required in recombination-deficient mutants such as recA, ruvABC, etc. [9]. The reason for this is explained below, in the dam-directed mismatch repair section. In contrast, in bacteria such as Vibrio cholerae, Dam methylation is an essential function although the reason for it has yet to be determined [10, 11].
Distribution of GATC Sequences in Chromosomal DNA
Analyses of E. coli DNA sequences [12–14] have indicated the following details about the GATC tetranucleotide. (i) It is represented, on average, once every 243 nucleotides, which is close to the 1/256 expected in a random base sequence. (ii) It is present at a higher than expected frequency in numerous chromosomal locations (e.g., dnaA, rpsP, metL, malP, rplS, xylB, gltX, and guaBA) in addition to oriC. The significance, if any, of this clustering is unknown for all these genes except dnaA and oriC (see Initiation of Chromosome Replication below). (iii) It is found more frequently in translated regions than in non-coding or non-translated regions, which is consistent with more frequent mismatch repair surveillance. In particular, rRNA- and tRNA-encoding genes exhibited the lowest GATC content of all genes examined. This deficiency may be correlated with unwanted palindromic secondary structures. (iv) Finally, the GATC tetranucleotide is never separated from another GATC sequence by more than 2 kb. This allows for dam-directed mismatch repair to occur over the whole genome, since it is less efficient at distances greater than 2 kb [15]. (See “DNA Mismatch Repair,” doi:10.1128/ecosalplus.7.2.5.)
The statistical data described above give the frequency of GATC sites in chromosomal DNA. These sites, however, can be present in unmethylated, hemimethylated, or fully methylated configurations. All GATC sites appear to be methylated in chromosomal and plasmid DNA isolated from E. coli by conventional standard methods and using restriction enzymes such as DpnI and DpnII to monitor methylation status. DpnI cleaves only at methylated sites, DpnII cleaves only at unmethylated sites, and Sau3AI (or BfuCI) will cut regardless of methylation status. Neither DpnI nor DpnII digests hemimethylated sequences [16, 17]. Techniques such as pulse-field gel electrophoresis of digested DNA and specific end-labeling procedures [18–21], however, indicate that the E. coli chromosome contains about 36 specific, unmethylated dam sites. The number and intensity of unmethylated sites in the chromosome vary depending on growth phase and growth rate, suggesting that the proteins which bind to them could be involved in gene expression or maintaining chromosome structure. The unmethylated dam sites appear to be mostly [18] or completely [22] modified in strains overproducing Dam, suggesting that the enzyme competes with other DNA binding proteins at these specific sites. Alternatively, at some GATC sites the increased Dam concentration may allow for modification of DNA structures (e.g., non-B-form DNA such as H-DNA [23]) relatively resistant to methylation at the normal cellular level of the enzyme. Palindromic structures containing GATCs are also relatively resistant to Dam methylation [24]. Alteration in helix stability by DNA methylation can be detected by abnormal migration of DNA fragments in denaturing gels [25]. Evidence for competition between Dam and other DNA binding proteins at several unmethylated sites has been obtained. These findings indicate that these sites are involved in regulation of gene expression, and they are discussed in more detail in that section below (Regulation of Gene Expression).
In addition to the unmethylated GATC sites discussed above, persistent hemimethylated sequences have been detected in the chromosome [26, 27]. These are distinct from the hemimethylated GATC sites which occur transiently immediately behind the replication fork due to the time lag in modifying new Dam methylation sites. The persistent hemimethylated sites are discussed in more detail below (Initiation of Chromosome Replication).
Single-molecule real-time deep DNA sequencing can identify modified bases and can, therefore, assign if a modified base in a particular motif is in a methylated, hemi-methylated or unmethylated state. In the pathogenic E. coli strain C227-11, 89.2%, 9.7% and 1.1% of GATC modifications were in methylated, hemi-methylated or unmethylated configurations [28]. Of 23 unmethylated sites tested, all were in non-coding regions and were near genes encoding the phosphotransferase system.
The results from E. coli strain C227-11, indicate that there is little “off-target” methylation by Dam at its normal cellular concentration. Single-molecule real-time deep DNA sequencing was also used to demonstrate that when Dam is overproduced from a multicopy plasmid, substantial methylation of A in GACC sequences occurs, as well as lower, but significant, modification in AATC, CATC and TATC sequences [29]. In vivo results derived from Dam overproduction should be treated with caution not only because of off-target GATC methylation, but also due to interference with SeqA (see “Initiation of chromosome replication”) and MutH action on hemi-methylated DNA (see “dam-directed mismatch repair”).
Dam methylation and DNA structure
Methylation of GATC sites can influence DNA structure because the methyl group protrudes into the major groove leading to duplex destabilization [30, 31]Alteration in helix stability by DNA methylation can be detected by abnormal migration of DNA fragments in denaturing gels (43). Dam methylation increases curvature of GATC-containing DNA as determined by electrophoretic mobility shift assays (52, 171). Non-B-form DNA such as H-DNA [165]) and palindromic structures containing GATCs are relatively resistant to methylation at the normal cellular level of Dam methyltransferase (3). Whether changes in curvature, especially in regions of high GATC content such as oriC, promote binding of proteins, such as SeqA, or play a role in origin function is not known. The destabilization of a methylated 13-bp duplex oligonucleotide is the basis for a fluorescence-based assay for Dam methyltransferase activity [32].
Distribution of CC(A/T)GG Sequences in Chromosomal DNA
Analyses of the Dcm recognition sequences, CCTGG and its complement CCAGG, indicated that these occur at a higher than expected frequency: every 385 bp instead of every 512 bp as predicted from random sequence [13]. As discussed below (VSP Repair), in stationary phase cells, the Dcm sequences are constantly subjected to cycles of deamination of 5-meCyt followed by repair of the resulting T-G mismatch and subsequent remethylation by Dcm. This cycling prevents the accumulation of C to T mutational changes in the chromosome although statistical data suggests such a drift has occurred albeit at a low frequency [33, 34].
The state of methylation at dcm sites can be monitored by digestion with EcoRlI, which cuts only if the sequence is unmethylated, and BstNI, which cleaves regardless of methylation status (Table 1). As is the case with dam sites, a small but undetermined number of unmethylated dcm sites have been detected in E. coli chromosomal DNA [18]. One assumes that there must also be hemimethylated dcm sites in chromosomal DNA, but their existence has not yet been demonstrated.
The investigation of unmethylated and hemimethylated dam and dcm sites in chromosomes has demonstrated the utility of this approach to identify regions of the chromosome with interesting biological features such as sequences that are bound by proteins which have regulatory functions. These kinds of studies should allow a functional dissection of the E. coli chromosome to be integrated with DNA sequence information.
Methylated dam and dcm Sequences in DNA of Other Bacteria and Phages
Methylated dam and dcm sites are found in most enterobacteria and the E. coli dam gene DNA hybridizes under stringent conditions to the DNA of other enterobacteria [35]. Methylated dam sites have also been detected in various gram-positive and gram-negative bacteria as well as in some archaebacteria [22]. Methylated dcm sites are found only in members of the Enterobacteriaceae (M. Lieb, personal communication). Strains with a dcm gene also contain the vsr gene (see VSP Repair).
There are many bacterial species (and all eukaryotes) which do not contain methylated dam and/or dcm sites and presumably have other unknown mechanisms to substitute for Dam and Dcm functions.
The GATC tetranucleotide in the genomes of several bacteriophages is present at lower than the expected frequency [14, 36]. The reason for this is not known although protection against host MutH endonuclease is often cited as a reason [37]. This seems very unlikely as phages such as fd, lambda and T7, which have undermethylated dam sites plaque on dam (mutH+) mutants with the same efficiency as wildtype [38]. Furthermore, the extent of methylation of the dam sequence in phages is also low. The probable reason for this is that methylation does not keep up with phage replication. For example, in phage lambda DNA, only about half the GATCs are methylated when propagated in wildtype bacteria but almost all are in wildtype bacteria over-expressing Dam [39].
Single-molecule real-time deep DNA sequencing has been used to analyze the “methylome” of Geobacter metallireducens, Chromohalobacter salexigens, Vibrio breoganii, Bacillus cereus, and two strains of Campylobacter jejuni [40]. The technique identified several known and new Type I, II and III restriction/modification motifs as well as methylated GATC sequences in C. salexigens and Vibrio breoganii, presumably produced by Dam homologs. This sequencing technology shows great promise in detecting modified bases in genomes although the detection of 5-methylcytosine was not possible for these studies, although it will be in the future (R. J. Roberts, New England Biolabs).
DNA METHYLATION GENES
The hsd genes which specify the EcoK methyltransferase map at 99 min on the genetic map and 4,615 kb on the physical map. The corresponding map locations for the dam gene are 76 min and 3,536 kb, those for the dcm gene are 44 min and 2,042 kb and the yhdJ gene at 73 min and 3,410 kb. The DNA methylation genes are thus unlinked to one another. All dam mutations, except one, are in a single complementation group and are recessive [41]. The exceptional mutation is very leaky, making it difficult to assign conclusively within the same complementation group. These genetic data suggest that no other functional dam methylation genes exist in E. coli. In the annotated E. coli genome, there is the damX gene which has no effect or relationship with dam methylation. This gene (b3388) was originally designated urf74.3 in the literature because its function was, and remains, unknown [42]. The origin or reason for the designation damX is unknown.
The dam Gene
The 834-bp dam gene is part of a transcriptional unit containing at least four genes [42, 43] and perhaps six or seven [44]. The locations of promoters and a transcriptional terminator which affect dam are shown in Fig. 2. Each promoter has been cloned individually, and the order of promoter strength is P2 > P1 > P3 > P4 > P5 [45]. Insertion of the cat (chloramphenicol acetyltransferase) coding sequence into the aroK gene reduces transcription across the dam gene by 70%, and a mini-Tn10 insertion in urf74.3 (damX; b3388) does so by 90% [43]. These data indicate that promoters P1 and P2 (situated about 3.5 kb upstream of dam) and P3 (located 2 kb upstream) are the most important for dam gene transcription. Promoters P1 through P4 all show the typical RNA polymerase sigma-70 recognition sequences.
FIGURE 2.
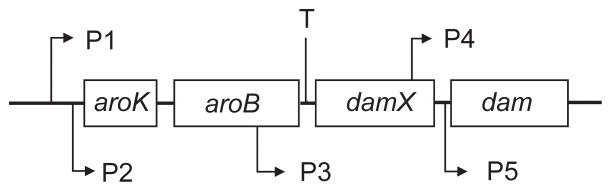
Organization of the dam transcriptional unit. The locations of promoters P1 through P5 are indicated, as is the transcription terminator (T) at the end of aroB. The major and growth-rate regulated promoter P2 is located 3.2 kb upstream of the dam gene.
Only promoter P2 has thus far been shown to be regulated [46]. This promoter is growth rate regulated by a mechanism distinct from that used for rRNA and tRNA gene promoters [45]. Transcription initiation from P2 is not affected by the stringent response, ribosomal feedback, or the level of Fis protein, all of which affect growth rate-dependent rRNA and tRNA promoters ([47]. Conversely, mutations in the cde (constitutive dam gene expression) gene, located at 15 min on the genetic map and 670 kb on the physical map, abolish growth rate regulation of the dam P2, but not rRNA growth rate-dependent, promoters [45]. The cde gene was subsequently shown to be identical to lipB [48], but the connection between lipoic acid biosynthesis and dam gene regulation remains unknown.
The rationale for growth rate regulation of the dam gene may be to correlate Dam levels with the amount of hemimethylated DNA close to the replication fork and at oriC. Cells growing with different doubling times synthesize DNA at different rates. To maintain the optimal level of hemimethylated DNA, the amount of Dam must be adjusted accordingly [45]. If dam gene expression were not regulated, too much or too little Dam would result in increased mutagenesis, asynchronous initiation of chromosomes, and alteration of the frequency of transposition.
Overproduction of Dam in E. coli alters the wildtype phenotype depending on the level of overproduction. At low level overproduction (about 10-fold), Dam out competes SeqA for GATC binding sites to produce a seqA phenocopy causing alterations in global gene transcription and chromosome initiation synchrony [49] (See Initiation of Chromosome Replication). Greater than 10-fold overproduction leads Dam to out compete the MutH protein for GATC sites as well resulting in a mutator phenotype due to inhibition of DNA mismatch repair [50, 51] (See dam-Directed Mismatch Repair). As described above, Dam overproduction leads to off-target methylation of GACC, AATC, CATC and TATC sequences [29].
Phage genomes often have an open reading frame encoding a putative Dam or Dam-like protein with signature motifs. For example, phage P1 and the T-even phages encode their own Dam methyltransferases which are expressed at some stage during their cell cycle. On the other hand, the lambda-like prophages in E. coli O157:H7 all have a putative dam gene which is not expressed under normal laboratory growth conditions. These are not pseudogenes because the E. coli O157:H7 VT2-Sa prophage derived dam homologue can be expressed to methylate GATC sequences [52]. When the host dam gene is eliminated from the chromosome of E. coli O157:H7, there is no detectable chromosomal GATC methylation indicating repression of the prophage dam genes [53]. The role of phage-encoded Dam in the life cycle remains obscure as is also the case for Vibrio phages [54].
The dcm Gene
The 1,419-bp dcm gene is overlapped at its 3′ end by the first six codons of the vsr gene, which is in a +1 register relative to dcm [55]. Such an overlap is uncommon in E. coli and in this case may serve to link the expression of these genes. Both genes appear to be transcribed into a single mRNA, and translation of vsr appears to be dependent upon translation of the upstream dcm coding sequence [55]. The mechanism by which this is achieved is not known. The location of the promoter(s) and its mode of regulation are also unknown. The possibility of growth rate regulation of dcm gene expression has not been tested. There is no obvious phenotype associated with the under- or overproduction of Dcm.
The yhdJ Gene
The yhdJ gene comprises 885 bp and is predicted to produce a protein of 294 amino acids with a predicted molecular weight of 33,397 Da. Annotation of the gene predicted that the protein contains conserved methyltransferase motifs for S-adenosyl-L-methionine (SAM) binding and catalysis. The order of these domains places YhdJ in the beta group of methyltransferases, along with CcrM and its homologs. E. coli YhdJ shares identity with the adenine methyltransferases M.AvaIII (55.8%) and CcrM from C. crescentus (34.3%). CcrM (cell cycle-regulated methyltransferase), is found in species of the alpha subdivision while Dam is present in the gamma subdivision of proteobacteria [2, 3]. Where it has been examined, CcrM is essential for viability, whereas Dam and its homologs are only essential in a few species [56]. In both E. coli and S. enterica, however, the yhdJ gene can be deleted without loss of viability and without any obvious phenotype [6]. In addition expression of the gene is below the level of detection in both E. coli and S. enterica under normal laboratory conditions of cultivation [6]. Nothing is known why this is so or how gene expression is regulated.
DNA METHYLTRANSFERASES
DNA methyltransferases transfer the methyl group from S-adenosyl-L-methionine (SAM) to specific residues in double-stranded DNA. Dam methyltransferase flips out the adenine residue from the DNA and modifies it [57, 58] and it is probable that the same basic mechanism is used for other E. coli methyltransferases. In E. coli, the substrate for Dam is GATC in hemimethylated DNA behind the replication fork. That is, the parental strand is methylated and methyl transfer occurs only onto the GATCs in the newly synthesized unmethylated strand.
Dam
The methylation of specific GATC sites in DNA of exponentially growing cells is rapid, occurring within the minimum time (about 1 min) allowed by the sensitivity of the method [26] for chromosomal DNA and 2–4 sec on plasmid DNA [59]. An already mentioned exception to this involves GATC sites in oriC and the dnaA promoter, which remain hemimethylated longer than other sites; this is due to the binding of SeqA (discussed in Initiation of Chromosome Replication).
Dam has been purified 3,000-fold and is a single polypeptide chain of 278 amino acids with an apparent molecular size of 32 kDa [60]. It has an Sw20, of 2.8S and a Stokes radius of 2.4 nm and exists in solution as a monomer. The enzyme has a turnover number of 19 methyl transfers per min (but see below) and an apparent Km of 3.6 nM for DNA. Double-stranded DNA is a better methyl acceptor than denatured DNA, and there is little difference in the rate of methylation between unmethylated and hemimethylated DNA. Dam transfers one methyl group per DNA binding event even when binding a fully unmethylated site (but see below).
Dam has been suggested to have two SAM-binding sites: a catalytic site, and one which increases specific binding to DNA perhaps as a result of an allosteric change in the protein [61]. DNA binding and/or methyl transfer is influenced by flanking sequence; the optimal sequence is 5′-GGGGATCAAG-3′ [62]. Dam is thought to bind the template and to slide processively along the DNA searching for substrate sequences [63, 64]. Further evidence for a sliding mechanism was the demonstration that in each binding event about 55 methylated GATC sites are formed before the enzyme dissociates from DNA [65](cf. previous paragraph and [60]). Processivity, however, is very dependent on sequence context [62]. An unmethylated GATC and its complement in duplex DNA are modified on both strands before the enzyme dissociates and this has been termed “intrasite processivity.” Intrasite methylation occurs only when GATCs are clustered and not separated by more than 400 bp [66].
In fast-growing bacteria there are 130 Dam molecules per cell in K-12 strains and 100 molecules per cell in B strains [67]. Each of these molecules would need to transfer 39 methyl groups per min to methylate all available GATC sites in a cell with a doubling time of 30 min. Dam is a substrate for the Lon protease [68] suggesting another possible regulatory mechanism in addition to growth rate-dependent transcriptional control [46].
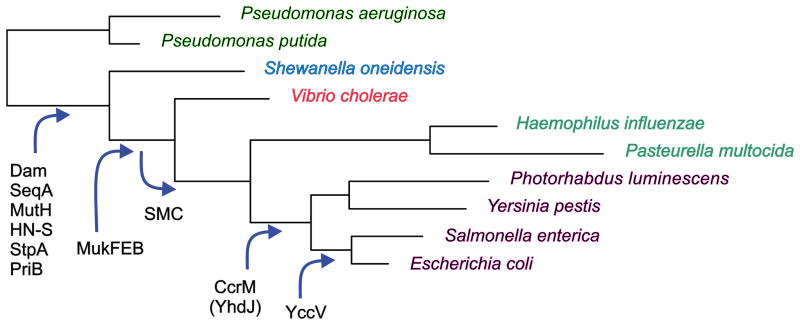
The Dam enzyme of E. coli is part of a family of methyltransferases that share nine amino acid sequence motifs [69] including the highly conserved -DPPY- that appears to be involved in SAM binding. Close relatives include the Dam proteins of phages P1, T1, T2 and T4, EcoRV, FokI, MboI, NlaIII and DpnII. The evolution of Dam appears to be a recent acquisition along with SeqA and MutH [2]. As shown in Fig. 3, Dam is present in one clade of bacteria which consists of the orders Enterobacteriales, Vibrionales, Aeromonadales, Pasteurellales, and Alteromonadales. Members of this clade share the following features. First, the dam gene is organized in an operon with aroK and aroB. Second, they have homologues to SeqA and MutH which have the same hemimethylated substrate as Dam. Third, they have separated the replication initiator gene (dnaA) from the origin of replication (oriC). Fourth, GATC sites are approximately 10-fold over-represented in their oriC and dnaA promoters. The phylogenetic distribution of Dam and its association with SeqA, MutH and other proteins has also been described using a different Web resource [70].
FIGURE 3.
Phylogeny of the Dam clade. TBLASTN searching was used with 4311 E. coli proteins against selected genomes with and without the dam gene. The genes encoding SeqA, MutH, HN-S, PriB and 75 other proteins were found in all selected genomes with the dam gene and in none of the genomes lacking it. Reproduced from [2] with permission from Elsevier Ltd.
The atomic structure of Dam complexed with DNA has been solved to 1.89 Å resolution in the presence of S-adenosyl-homocysteine [58]. The structure shows both non-specific backbone contacts and specific contacts with the GATC bases. Importantly, the aromatic ring of Y119 intercalates into the DNA between GA and TC thereby flipping the adenine into the enzyme’s active site. The unpaired T residue can adopt an intrahelical or extrahelical position. Four other important contacts are made: K9 to G, L122 and P134 to C and by R124 to T. These and flanking phosphate contacts by conserved residues (R95, N126, N132, and R137) position Dam on the DNA duplex.
An altered specificity Dam mutant has been obtained by extensive mutagenesis of the dam gene [71]. The variant methylates GATT, but not GATC sequences but its specific activity is only about 20% that of the parental enzyme. The alteration in specificity was facilitated by converting amino acids R124 and P134, which contact C and T respectively, to serine. Further studies along these lines may allow for conversion of GATC specificity to the off-target sequences, especially CACC.
Selective inhibitors of Dam have been sought for use as potential therapeutics because humans do not produce this enzyme. A high-throughput small molecule screen identified several lead compounds including those which appear to bind specifically to the allosteric sites of Dam [72]. Some compounds showed greater than 400-fold selectivity for Dam compared to murine DNA cytosine methyltransferase (Dnmt1). Dam activity can also be specifically inhibited by cyclic peptides (e.g., SGWYVRNM) at IC50s of 50–150 μM, concentrations which do not inhibit HhaI methyltransferase [73]. The mechanism of inhibition is not yet known. Several S-adenosyl-methionine analogues can inhibit Dam methyltransferase activity in the approximately 10 μM range [74].
Dcm
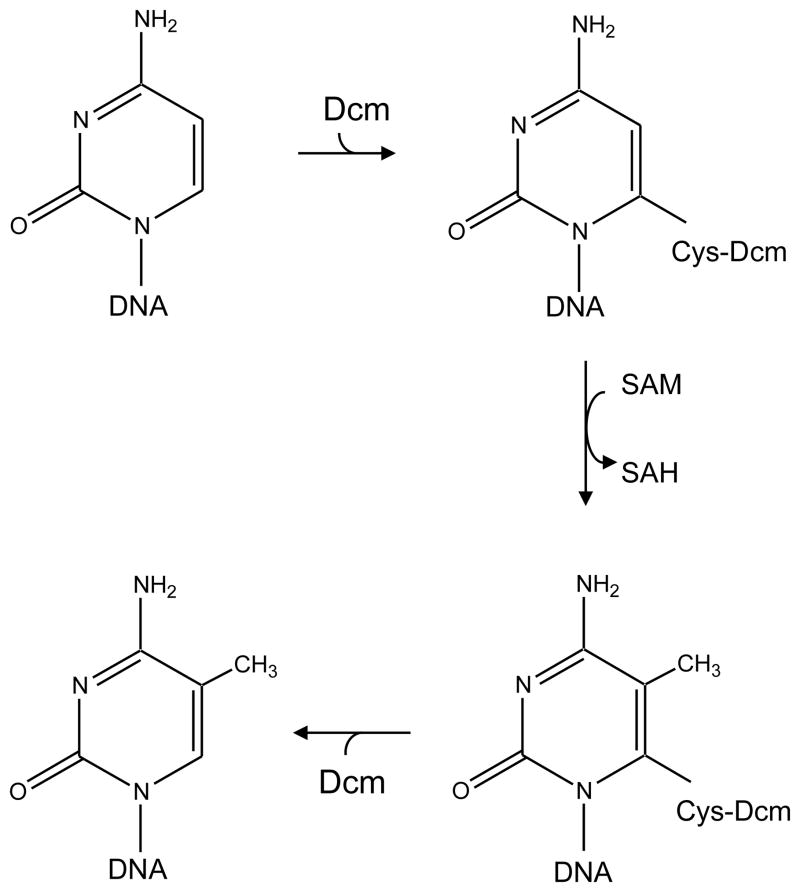
The purification and biochemical properties of the Dcm protein have not been reported, but from the DNA sequence, a 472-amino-acid protein of 53,465 kDa should be produced. Protein sequence comparisons indicate that, like other 5-meCyt methyltransferases, Dcm contains 10 conserved motifs including a Pro-Cys motif [75]. The cysteine residue is essential for catalysis but not for DNA binding, suggesting a mechanism of methyl transfer (Fig. 4) similar to that for thymidylate synthase [76], i.e., attack of the C-6 of cytosine by cysteine 177 of Dcm to activate the C-5 position for methylation [77, 78].
FIGURE 4.
Catalytic mechanism of methyl group transfer. Nucleophilic attack by cysteine-177 of Dcm at the C-6 position of cytosine leads to the formation of a covalent Dcm-DNA intermediate. This leads to activation of the C-5 position and transfer of the methyl group from SAM. The S-adenosyl-homocysteine (SAH) and the methylated cytosine are released from the covalent intermediate.
YhdJ
In E. coli over-expressing YhdJ, genomic DNA is protected from cleavage by NsiI while wild-type cellular DNA is not protected [6]. Similarly, partially purified YhdJ is able to protect DNA from NsiI cleavage in vitro. In vivo analysis of DNA isolated from the over-producing strain showed that the methylation occurred at the second or 3′ adenine in the NsiI recognition sequence, 5′-ATGCAT-3′. It is likely that this is, or at least contains, the YhdJ recognition sequence. CcrM methyltransferases play a key role in the initiation of chromosome replication but YhdJ over-production does not affect synchronization in dnaC2(Ts) bacteria [6]. This property together with its being a non-essential gene makes YhdJ distinct from other members of the CcrM family of methyltransferases.
BIOLOGICAL FUNCTION FOR 5-meCYT
To identify the biological role for cytosine methylation, mutant strains lacking this modified base in DNA were isolated [79, 80]. Unfortunately, no obvious phenotype has yet been found associated with the dcm mutations. In discussing possible functions for 5-meCyt, it is worth noting that in contrast to E. coli K- 12, E. coli B lacks the dcm gene.
dcm Mutations
The most widely used dcm allele, dcm-6 [80], is defective in both methylation and VSP repair (see below) and shows mutational changes in codons 26 and 45 compared to wild type [55]. The polar effect of the nonsense codon (TGA) at position 45 in dcm would most easily explain the effect on vsr. Mutations dcm-9 and dcm-10 are also Vsr− but dcm-1, dcm-4 and dcm-7 are Vsr+ (M. Lieb, personal communication).
In addition to dcm-6, the mec mutant allele of dcm [79] has been frequently employed, although the location of the mutation in the gene is not known. Two large deletions which remove dcm and additional genes have been shown to lack Dcm methylation [41, 81]. A site-directed deletion of the gene (JW1944) and its replacement with the kanamycin coding sequence is available from the Keio collection (http://ecoli.naist.jp).
VSP Repair
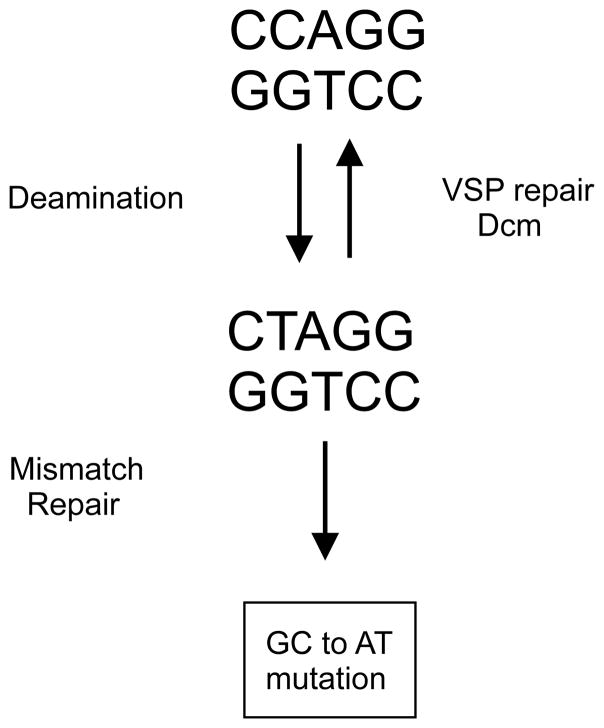
Spontaneous mutational hotspots for amber nonsense mutations occur in the lacI gene at the 5-meCyt residue in the Dcm recognition site CCAGG, altering it to CTAGG [82]. A similar result was obtained in the cI gene of phage lambda in growing bacteria [83] but the mutation frequencies at these hotspots were severely reduced in stationary phase bacteria [84]. When these amber mutations were used in genetic crosses, anomalous recombination frequencies were obtained [83] which led to the discovery of a very short patch (VSP) repair system correcting T-G mismatches in Dcm recognition sequences (Fig. 5) [85, 86].
FIGURE 5.
Deamination, repair and mutagenesis at a Dcm recognition site. Deamination of 5-meCyt in duplex DNA produces a T-G mismatch which is a substrate for Vsr endonuclease. After removal of the T residue, DNA polymerase I and DNA ligase reactions restore the original sequence which is re-methylated by Dcm. Failure to repair before DNA replication or if the MutHLS mismatch system acts on the mismatch before Vsr will produce a GC to AT mutation.
Such T-G mismatches can occur in non-replicating DNA by the deamination of 5-meCyt. This reaction is analogous to the deamination of cytosine to form a uracil-guanine mismatch, which is a substrate for uracil-N-glycosylase. In a similar manner, the T-G mismatch is a substrate for the strand- and sequence-specific Vsr endonuclease, followed by conventional DNA polymerase I-dependent excision repair [87] and finally by DNA ligase. VSP repair can thus be viewed as counteracting the potential mutagenic effects of 5-meCyt deamination (Fig. 5). As expected, in dcm mutants no mutational hotspots are detectable [82, 88].
The T-G mismatch resulting from cytosine deamination should also be recognized by the MutS protein of the dam-directed mismatch repair (MMR) system. This indeed is the case but surprisingly, it was found that inactivating MutS or its partner MutL reduced VSP repair by an order of magnitude [88]. This decrease may be related to the relative amounts of Vsr and the Mut proteins in logarithmic and stationary phase cells. In logarithmic phase cells, Vsr is not detectable while the Mut proteins are present at their normal concentration. In stationary phase cells, however, the endonuclease MutH protein (partners with MutS and MutL—see dam-Directed Mismatch Repair) concentration decreases three-fold [89] while the Vsr concentration is at its maximal [90]. In other words, during logarithmic growth, the Mut proteins are maximally efficient when replication errors are most likely while in stationary phase when DNA synthesis is minimal and 5-meCyt deaminations accumulate, VSP is most active.
One possible explanation for these results, is that MutS and MutL partner with MutH in growing cells but preferentially with Vsr in non-growing cells (both MutH and Vsr are endonucleases) [86]. Support for this idea is that Vsr interacts with MutL in a two-hybrid assay [91, 92]. One model for MutS translocation suggests that it forms loops [93] such that MutS and MutL are at the base of the loop and the mismatch is at the apex. In this case, the mismatch could be bound by another molecule of MutS in growing cells but in non-growing cells could be bound by Vsr [86]. An alternative model is that MutS and MutL enhance Vsr binding at the mismatch through an alteration of DNA secondary structure. Such a model was proposed based on the crystal structure of Vsr [94].
VSP repair is reduced in dam mutants and correlates with a reduction in the level of Vsr but not Dcm [95]. Since the dcm and vsr genes are co-transcribed, the regulation of vsr in is probably posttranslational. Discussion of this issue will be continued in the Post-Transcriptional Regulation section.
Protection from Restriction Endonucleases
Unmethylated dcm sites are substrates for the EcoRII restriction endonuclease [79], suggesting that one function may be protection of DNA from group N plasmids which produce these restriction endonucleases [13]. Upon transfer such plasmids into a naive cell, however, the cognate modification enzyme is produced before the restriction protein thereby affording protection even in the absence of Dcm. A role for Dcm in protection from restriction enzymes, therefore, seems problematic and in addition since some E. coli strains (e.g., strain B) do not possess the dcm gene it should have been eliminated from the biosphere by this model. It is of interest that the EcoRII methyltransferase shows about 70% amino acid sequence similarity with Dcm. Both enzymes methylate the same DNA sequence, and the function of M.EcoRII is known: it protects DNA from cleavage by EcoRII.
BIOLOGICAL FUNCTIONS FOR 6-meADE
Mutant strains lacking DNA adenine methylation were isolated in order to identify the role of this methylated base in cell metabolism. Unlike the dcm mutants, there are several phenotypic traits associated with dam mutants which have helped to define the multiple roles of 6-meAde in DNA metabolism.
dam Mutations
The most commonly used dam mutant alleles are dam-3, dam-4, dam-13::Tn9 (chloramphenicol resistance), and dam-16::KanR (Table 2). The mutational changes in dam-3 (Gly13Asp) and dam-4 (Gly12Glu) are surprising because these are not critical amino acids for DNA contacts in the co-crystal structure (see above). However, Gly12 makes a backbone phosphate contact between G and A in GATC and so the introduction of a negative charge might decrease enzyme binding. This explanation may also apply to Gly13.
TABLE 2.
E. coli K-12 dam alleles
| Allele | Type of mutation | Location (nt) | Reference |
|---|---|---|---|
| dam-3 | GC to AT transition, GGC (gly13) to GAC (asp) | 3,513,898 | [80] |
| dam-4 | GC to AT transition, GGG (gly12) to GAG (glu) | 3,513,901 | [259] |
| dam-13::Tn9 | Insertion of Tn9 | 3,513,725 – 3,513,726 | [260] |
| dam-16::Kan | Replacement of coding sequence | 3,513,240 – 3,513,773 | [261] |
| JW3350 (KanR) | Replacement of coding sequence | 3,513,102 – 3,513,933 | Keio collection [262] |
| dam-12::Mu | Insertion of MudII301(Ap, lac) | nd | [260] |
| dam-18::lacZ | lacZ fusion to dam at BamHI site at normal chromosomal location | 3,513,628 | [45] |
| dam-19::Kan | Insertion of EZ::TN<KAN-2> | nd | [263] |
Insertion and deletion alleles of dam have been isolated and characterized in Salmonella enterica. In general these have the same phenotypic properties as the E. coli mutants [96]. Other dam mutants were isolated on the basis of altered frameshift mutagenesis and the dam-1 allele confers properties similar to those of dam mutant alleles in E. coli [97].
The E. coli dam mutants exhibit a variety of phenotypic traits and other properties (Table 3). The bewildering array of differences compared to the wild type suggests that dam methylation and the level of Dam itself have multiple functions in the cell. These functions are correlated with three DNA transactions: DNA mismatch repair, regulation of gene expression and initiation of chromosome replication. For these transactions which are described in detail below, the amount of hemimethylated DNA trailing the replication fork is critical. Decreasing or increasing the level of hemimethylated DNA by using a Dam-overproducing plasmid or a dam mutant, respectively, profoundly alters the function involved.
TABLE 3.
Altered physiological properties of a dam mutant [1]
| Reduced Dam activity in vivo and in vitro, leading to a reduction of N-methyladenine in GATC sequences in DNA [80] |
| A mutator phenotype [7] |
| A hyper-recombination phenotype [259] |
| Alleviation of EcoK restriction [264] |
| Increased number of single-strand DNA strand breaks in dam lig (DNA ligase) cells [41] |
| Increased number of double-strand breaks in a dam recBC strain [107] |
| Increased sensitivity to UV light and certain chemicals [7, 109, 230, 255, 265–267] |
| Increased drug-induced mutagenesis [97] |
| Derepression of certain genes in the SOS regulon [110, 268] |
| Increased spontaneous induction of lysogenic phages [38, 241] |
| Inviability of dam mutant cells with mutations in recA, recB, recC, lexA, polA, priA or ruv [7, 107, 112, 268] |
| Increased precise excision and transposition of Tnl0 and other transposons [269] |
| Altered expression of certain chromosomal and non-chromosomal genes such as trpS, sulA, glnS, mom, dnaA, pap, traJ, finP, and tsp [4, 202, 205, 206] |
| Suppression of some dam phenotypes by second-site mutation in mutS, mutH, and mutL [99, 100] |
| Control of phage P1 DNA packaging into virions [219] |
| Asynchronous initiation of chromosome DNA replication [134] |
| Failure to support the growth of plasmids containing the E. coli origin (oriC) of chromosomal replication [270, 271] or the phage P1 ori [272] or those with the RepI replication protein [273] |
| Failure of Dam methylated plasmids to transform dam mutants at high efficiency [16] |
dam-Directed Mismatch Repair (see also module 7.2.5, “DNA Mismatch Repair”)
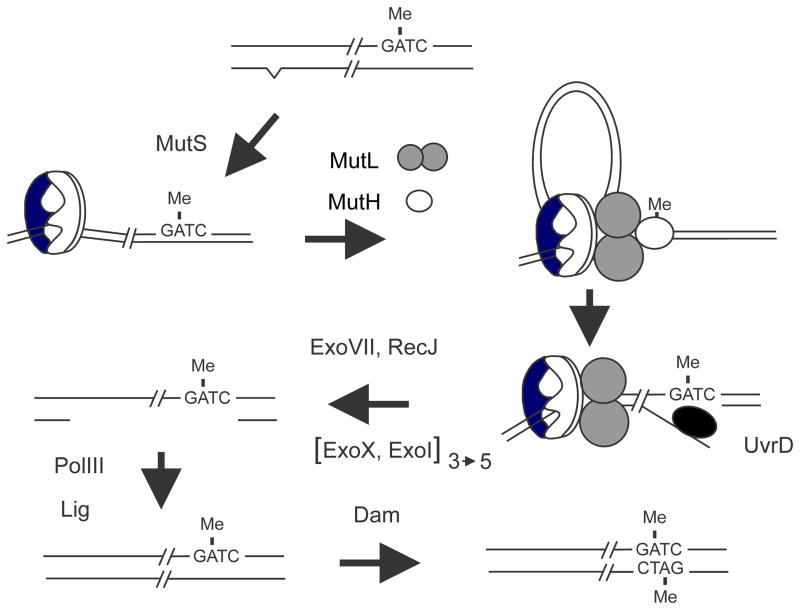
The most direct and convincing evidence for the involvement of dam methylation in mismatch repair comes from the use of in vitro-constructed heteroduplexes of phage lambda DNA [39]. Heteroduplexes containing a mismatched base pair were constructed with one strand methylated, both strands methylated, or neither strand methylated. The unmethylated strand was preferentially repaired in heteroduplexes containing one methylated and one unmethylated strand. If neither strand was methylated, repair occurred equally on both strands. No repair was observed when both strands were fully methylated [39]. These results indicate that the function of Dam methylation is to impart strand selectivity and that the role of the repair system in the wild type is to remove replication errors in the newly synthesized undermethylated DNA strand trailing the replication fork (Fig. 6). The errors are base mismatches or deletion/insertions of up to four nucleotides [98]. In dam mutants where strand discrimination is lost, mutations are introduced into the parental strand 50% of the time, thereby explaining the mutator phenotype. It has been repeatedly observed, however, that the mutation rate in dam bacteria is much less than 50% that of mutS, mutL, or mutH cells [99, 100]. This result has been interpreted to indicate that mismatch repair in dam cells may frequently lead to a lethal outcome [101]. As expected, the mutation spectrum of dam and mut strains is identical with AT to GC and GC to AT transitions and frameshift mutations predominating [95, 102–105].
FIGURE 6.
Dam-directed mismatch repair in E. coli. The top of the figure shows DNA immediately behind the replication fork in which the “old” top strand is methylated and the “new” strand is not and also contains a base mismatch (carat) created as a replication error. The mismatch is recognized and bound by MutS followed by recruitment of MutL and MutH to form a ternary complex. The formation of this complex is thought to involve DNA looping to bring the mismatch and a GATC sequence in close proximity but the details are unclear. In the ternary complex the latent nuclease activity of MutH is activated and it cleaves the new unmethylated strand 5′ to the GATC sequence. The nick created by MutH serves as an entry site for the UvrD helicase which unwinds the DNA exposing single-stranded DNA which is digested by one or more of the following exonucleases: RecJ, ExoVII, ExoX or ExoI. The exonuclease(s) used depends on the relative orientation of the mismatch to the GATC sequence; in the figure the direction of UvrD unwinding is 5′ to 3′ and so either ExoVII or RecJ or both are needed. If the mismatch was to the “right” of the GATC sequence, UvrD would unwind in the 3′ to 5′ direction and ExoX and/or ExoI would digest the single-stranded DNA. The gap created by nuclease digestion removes the mismatched base and is filled in by DNA polymerase III. The resulting nick is closed by DNA ligase and eventual Dam methylation precludes any further repair.
Further evidence for the role of Dam in strand discrimination is that dam mutants and wild-type cells overproducing Dam show a mutator phenotype [50, 51]. Unlike the wild type, where repair is confined to the newly replicated strand, dam mutants have lost strand discrimination and in addition to correcting mismatched bases in the new strand, mutations are introduced into the parental strand using the newly synthesized mutant strand as template. In Dam-overproducing cells, the high concentration of Dam greatly reduces the transient lifetime of hemimethylated GATCs in newly replicated DNA, thereby preventing mismatch repair and so the mutation rate increases.
Mismatch repair in Escherichia coli has two functions: correction of replication errors behind the replication fork and prevention of recombination between similar but not identical DNA sequences (“antirecombination”). To correct replication errors, the MutS protein binds to mismatches in DNA and recruits MutL and MutH resulting in activation of the latent endonuclease activity of MutH to produce a nick in the newly-synthesized unmethylated DNA strand 5′ to the G in a nearby GATC sequence. The UvrD helicase loads on the nicked DNA in a MutS- and MutL-dependent manner and begins to unwind single-strand DNA either in the 5′ to 3′ direction or the 3′ to 5′ direction depending on the orientation of the mismatch to the GATC sequence. The single-stranded DNA is digested either by ExoI, ExoVII or ExoX in the 3′ to 5′ direction or the RecJ or ExoVII in the 5′ to 3′ direction. The resultant gap is filled by the action of DNA polymerase III, and after the action of ligase the duplex DNA is methylated by Dam. Fully methylated DNA is not a substrate for mismatch correction.
The loss of directionality of mismatch repair in a dam mutant must be responsible for the formation of DNA single and double-strand breaks by mismatch repair [7, 106, 107]. The increase in the number of single-strand breaks in dam lig cells compared to dam bacteria indicates that most of the nicks created by MutH can be sealed by ligase [41].
The double-strand breaks created by mismatch repair are the basis for many of the phenotypes associated with dam listed in Table 3, such as inviability with inactive recombination genes, induction of prophages, induced SOS response, etc. The inviability of dam cells with recA, recBCD, ruvABC and priA indicates that double-stranded ends are formed and that the RecBCD pathway is required to repair mismatch repair-induced double-strand breaks [9]. There are two possible ways in which these breaks could be formed. First, a replication fork encountering a nick created by MutH would collapse according the model proposed by Kuzminov [108] and shown in Fig. 7A. This model requires that the mismatch repair system recognizes some kind of endogenous damage ahead of the replication fork. Wyrzykowski and Volkert [109] reported that base pairs containing oxidative lesions are subject to mismatch repair. Since these could occur ahead of an oncoming fork in dam cells, it might encounter the gap intermediate made during the mismatch repair reaction and lead to fork collapse. The RecBCD pathway would then restore the fork and the PriA pathway would reload the DnaB helicase followed by DNA polymerase III holoenzyme (“replication restart”).
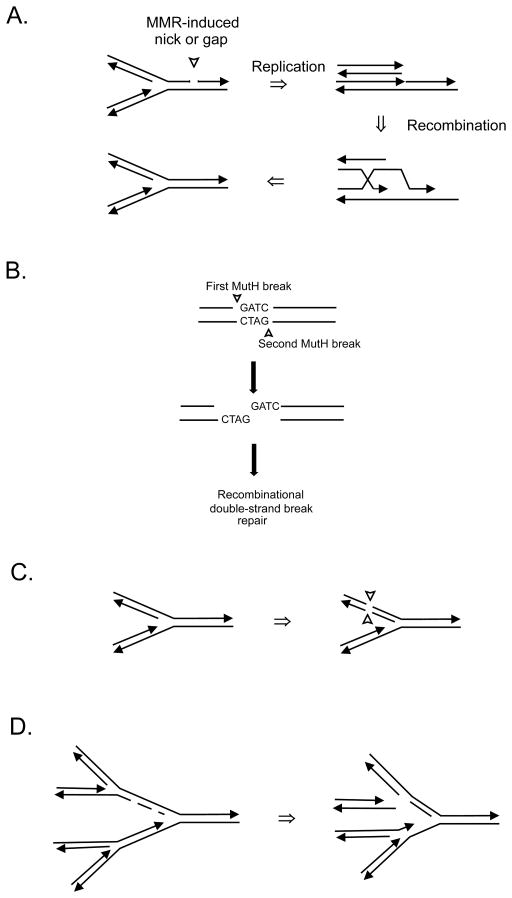
FIGURE 7.
Models illustrating double-strand break formation in a dam mutant. (A) A replication fork encounters a mismatch repair (MMR) intermediate of a nick or gap on one strand leading to replication fork collapse. The MMR intermediate could arise from the processing of endogenous DNA damage or from repair of a replication error from the previous replication. Recombination between daughter chromosomal arms can restore the fork which can then be loaded with the DnaB helicase and DNA polymerase III holoenzyme. (B) MutH nicking on opposite sides of the same GATC in non-replicating DNA produces a double-strand break which can be repaired using a sister chromosome. (C) MMR processing of a replication error either by action at the same GATC as in panel B or by overlapping excision tracts from GATCs on opposite strands producing a double-strand break that can be repaired using the daughter strands as template. (D) Mismatch repair-independent double-strand break formation. Asynchronous initiation of chromosome replication in a dam mutant could lead to two initiation events close together resulting in two closely spaced forks on each chromosomal arm. If the second fork catches up to the first, replication fork collapse occurs. The exposed double-stranded end becomes a substrate for RecBCD exonuclease which, when encountering a Chi site, loads RecA on single-stranded DNA thereby generating an SOS inducing signal.
In the second model, MutH nicks the GATC on the 5′ side of the G on both strands creating a double-strand break in non-replicating DNA (Fig. 7B). The same recombination proteins would be required to effect repair of the double-strand break with the exception of PriA as there should be no necessity for replication restart. However, if the double-strand break occurs immediately behind the fork (Fig. 7C) where mismatch repair normally acts, there might be fork disruption requiring replication restart. At present there is physical evidence for double-strand breaks in dam recBC cells but not in dam recBC mut bacteria [106, 107, 110]. The recBC mutation is necessary to stabilize double-stranded ends although neutral single-cell electrophoresis allowed visualization of double-strand breaks in a dam cell in the presence of RecBCD (Fig. 8) [110]. However, whether these breaks are dependent on replication (which would favor the first model) is not yet known.
FIGURE 8.
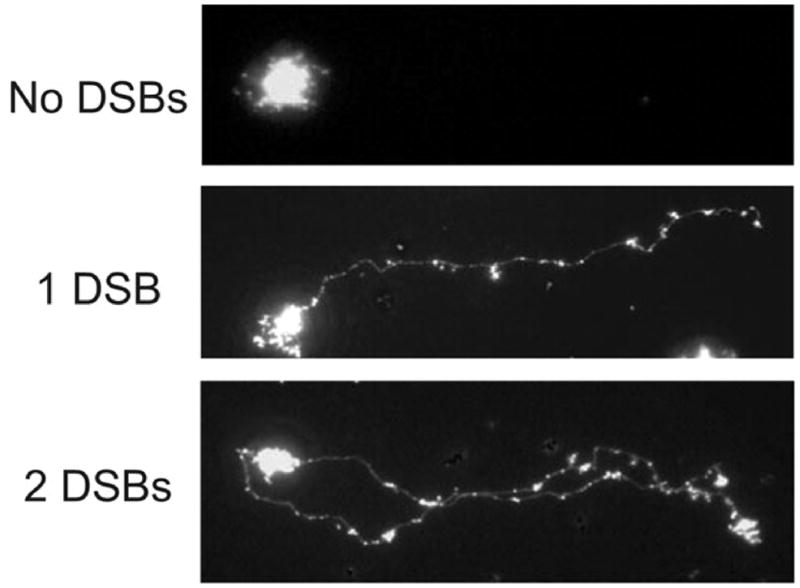
Double-strand breaks in an E. coli dam mutant detected by single-cell microgel electrophoresis showing disrupted cells with 0, 1 or 2 double-strand breaks. Reproduced from with permission from [110] copyright (2005) American Society for Microbiology.
It might be expected that the presence of double-strand breaks in dam cells is the likely explanation for the high basal level of the SOS response and induction of prophages. The SOS level in a population of dam cells is heterogeneous as measured by recA::gfp fusions [111] and this is probably a reflection of the stochastic nature of the SOS inducing signal. A dam lexA (Ind−) mutant is viable if RecA and RuvAB are supplied in trans from multicopy plasmids suggesting that these are the only necessary contribution from the SOS system [9]. Surprisingly, elimination of MutH, MutL, or MutS by mutation in dam cells does not reduce SOS regulon expression [112], indicating that an additional inducing signal must be generated although the nature of it is unknown but might be connected to asynchronous initiation of chromosome replication in dam bacteria (Fig. 7D) (see Regulation of Gene Expression).
Initiation of Chromosome Replication
Newly replicated DNA is hemimethylated and bound by SeqA
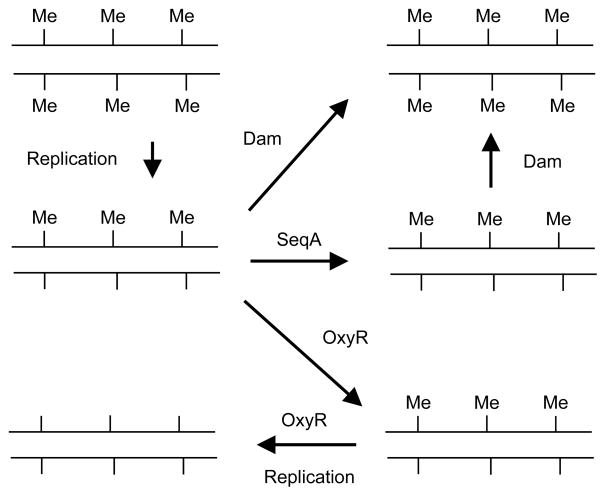
Replication of the fully methylated chromosome generates a transient wave of hemimethylated GATC sites (methylated on the parental strand, but not the daughter strand) behind the replication fork. Re-methylation of most chromosomal GATC sites takes place within 2–4 seconds after passage of the replication fork [59].
In the chromosomal replication origin, oriC, and the dnaA gene promoter, hemimethylated DNA persists for a large part of the cell cycle [26]. This is likely the result of the high density of GATC sites in these regions. There are 11 GATC sequences within the 245 bp minimal oriC, and eight GATC sequences within a 219 bp region covering dnaA promoters P1 and P2, which provides multiple binding sites for the SeqA protein [113]. SeqA consist of two functional domains. The N-terminus is responsible for oligomerization and the C-terminus for DNA binding [114, 115]. SeqA exists as a homodimer in solution [116, 117] and each dimer binds a pair of hemimethylated GATC sites separated by up to 31 bp [118, 119]. The SeqA protein also binds fully methylated DNA albeit with lower affinity [120]. It was proposed that dimers of SeqA are capable of oligomerizing into a left-handed helical structure (filament) with the hemimethylated DNA wrapped around it [116] thereby introducing negative supercoils [121]. The formation of a SeqA filament on hemimethylated oriC enables SeqA to bind hemimethylated GATC’sequences more tightly than the monomeric Dam enzyme and this may explain why oriC is sequestered for an extended period [116]. Support for this hypothesis comes from mutations that disrupt the SeqA dimer-dimer interface and hence prevents filament formation. Such mutations lead to a seqA mutant phenotype, despite the mutant protein retaining the ability to bind pairs of hemimethylated GATC’s as a dimer [116, 122]. Evidence also suggests that the DnaA protein assists SeqA in keeping oriC hemimethylated for a long time [123].
A number of chromosomal hemimethylated GATC sites in addition to oriC and the dnaA promoter are substrates for SeqA. Localization studies show that SeqA is localized in discrete foci dependent on ongoing replication [124, 125]. The foci most likely represent multimers of SeqA bound to hemimethylated DNA behind the replication fork [125, 126]. Cytological studies of SeqA foci further suggest that replication forks originating from the same origin are organized into replication factories [127].
Methylation and initiation synchrony
In fast growing cells the time required to replicate the bacterial chromosome exceeds the culture doubling time, and initiation of replication may take place one, two or even three generations prior to cell birth. Consequently, cells are born with replicating chromosomes and containing multiple origins of replication. Initiation of replication in such cells takes place only once at each origin within a short time interval of the cell cycle leading to synchronous initiation [128]. Cells therefore contain mainly 2n replication origins (where n = 0, 1, 2, 3, etc.).
Dam methylation, despite facilitating duplex opening in the oriC region [129], is not essential for initiation of replication. Rather, methylation of origin GATC sites are instrumental in maintaining initiation synchrony, because it allows for a discrimination between an old (uninitiated) origin and one that has recently been initiated.
Initiation at the first origin within a single cell is assumed to be set by binding of DnaA to one or more low-affinity sites within oriC (see below). The DnaA protein is furthermore assumed to be released from an origin upon initiation [130]. Following initiation, the two nascent and hemimethylated origins are rapidly bound by the SeqA protein, preferentially to two positions on either side of the IHF site [131] to prevent open complex formation [132, 133]. This sequestration renders the origin inaccessible to DnaA for about one third of a generation time and prevents immediate reinitiation [26, 113]. Consequently, the DnaA protein released from the first initiated origin in a cell will momentarily increase the DnaA/oriC ratio for the remaining fully methylated, i.e., “old” origins and their initiation will follow in a cascade-like manner (“the initiation cascade” [130]).
In dam mutant cells the situation is different. When replication has initiated at one of the cellular origins, the two newly synthesized origins are unmethylated, and will not be sequestered. They will therefore immediately re-bind the DnaA protein liberated at the initiation event, and bring the DnaA/origin ratio below the threshold level. This will prevent the occurrence of the initiation cascade. Because only a single origin is initiated at a particular time in the dam cells, the decrease in DnaA/origin ratio will not be as marked as in the wild-type cells, leading to a shorter time period for the build-up of initiation potential (accumulation of DnaA protein) for the next initiation. Consequently initiations in dam mutant cells are spread over the entire cell cycle, i.e. they are asynchronous [130].
In the absence of SeqA [113] or in the presence of high levels of Dam methyltransferase [134] origins are rapidly re-methylated, and re-initiations at the same origin occurs frequently, resulting in an asynchrony phenotype. This is also the case for origins that cannot be sequestered due to mutations in four or eight oriC GATC sites [135]. Density shift sedimentation analysis corroborates that the minimal inter-replication time, i.e., the time between two successive initiations from the same origin, is reduced only a few minutes in sequestration-deficient cells to about 0.6 generation relative to wild-type cells [136].
Cells where the initiation interval is extended, for example by oversupply of DnaA, also initiate asynchronously, and it was concluded that asynchronous initiation occurs when the initiation period is longer than origin sequestration [137, 138]. Asynchronous cells fail to discriminate between an old non-initiated origin from a recently initiated origin, and consequently origins compete for initiation factors and are randomly selected for initiation. This results in replication incompatibility, best demonstrated by the inability of these cells to stably maintain minichromosomes [138–140]. Amino acid starvation of E. coli cells results in induction of the “stringent response” mediated by guanosine tetra- and pentaphosphates. A consequence of this induction is the inhibition of chromosome replication intiation at oriC. This inhibition does not occur in seqA or dam mutant cells, suggesting that SeqA and Dam are required [141].
Methylation and once-per-cell cycle initiation of replication
In wild-type cells, initiations from the same oriC are separated by the culture doubling time on an average [136]. Sequestration of newly replicated hemimethylated origins is, despite immediate blocking of re-initiation, not sufficient to maintain once-per-cell cycle initiation at oriC. Sequestration lasts only about 1/3 of a generation time but provides a time interval during which other mechanisms operate to lower the intracellular concentration or the activity of the DnaA protein for initiation to occur one mass doubling later (for reviews see [142, 143]).
In E. coli DnaA is bound to the strong origin binding sites R1, R2 and R4 throughout the cell cycle [144]. Replication initiation commences with further binding of DnaA to the weaker recognition sites within the origin—that is, R2, R3, and R5, which are indifferent to the nucleotide-bound status of DnaA—and to I2, I3, tau1 and tau2 as well as 6-mer sites in the AT-rich region that are specific for DnaAATP [145–147]. The I1 box has been reported to bind DnaA associated with both ATP and ADP [145] or DnaAATP only[146].With the help of accessory proteins IHF, HU and DiaA [148, 149], a larger DnaA-DNA nucleoprotein complex is formed at oriC [150, 151] where both DnaAATP and DnaAADP participate [152–155]. Because many of the weaker DnaA binding sites are dispensable for proper timing and synchrony of initiation [156], it is thought that the DnaA filament is mainly anchored at high affinity binding sites such as R1 and R4. The DnaA-DNA complex promotes duplex opening in an adjacent AT-rich region. This open complex is stabilized by the binding of DnaAATP to specific 6 bp sequences in the single-stranded region [147]. The participation of DnaAATP in formation of the pre-RC complex and in stabilizing the open complex may explain why this configuration of the protein is limiting for initiation in vivo [157, 158]. Subsequently, the DnaA protein recruits the hexameric DnaB helicase associated with ATP-bound DnaC as a DnaB6DnaC6 complex to the single-stranded region of the open complex. DnaC loads the DnaB helicase on the open complex to promote further duplex opening to form the pre-RC stage. During this process, ATP is hydrolyzed and DnaC is released, and the transition to replication proceeds by the loading of two or three DNA polymerase III holoenzymes at the origin [159].
During the period where oriC is sequestered, the availability of active ATP-bound DnaA protein is lowered. Several mechanisms contribute to lowering both the amount and activity of DnaA available for initiation.
Because the dnaA gene is close to oriC, it is replicated soon after initiation. The dnaA gene promoter is, like oriC, rich in GATC sites and consequently sequestered and transcriptionally inactive for approximately the same fraction of the cell cycle as the origin [26]. The absence of de novo DnaA synthesis during sequestration where cell growth continues, contributes to lowering the amount of DnaA available for initiation when sequestration ends [160].
Another mechanism that contributes to lowering the amount of available DnaA while sequestration of the hemimethylated oriC persists is the generation of new DnaA binding sites outside the origin by replication. These serve to titrate DnaA protein away from oriC [161]. The most prominent of these DnaA binding sites, datA, is located about 470 kb away from oriC and replicated within the period of origin sequestration [162]. The datA region can bind several hundred molecules of DnaA [162]. Because datA contains five R-type DnaA boxes, it is believed to bind both DnaAATP and DnaAADP equally well although this has never been determined experimentally. Like sequestration of the dnaA promoter, titration of DnaA to non-oriC binding sites serves to lower the amount of the protein available for initiation.
Finally, the activity of DnaA is reduced by conversion of active DnaAATP to inactive DnaAADP by hydrolysis [163]. Two processes are described that contribute to this inactivation. One of these is a process called RIDA (Regulatory Inactivation of DnaA), which converts active DnaAATP to inactive DnaAADP by hydrolysis [163]. RIDA activity involves two proteins: The DnaA-related protein Hda [164] and the DNA-loaded beta clamp of the DNA polymerase III holoenzyme. In RIDA, the beta-subunit of Pol III forms a complex with the Hda and DnaA proteins, stimulating the ATPase activity of the latter to promote conversion of DnaAATP to the inactive DnaAADP [165]. RIDA is dependent on DNA-loaded beta-clamps, i.e., ongoing replication, and therefore the RIDA process is accelerated by an initiation event. However, because initiation of replication requires only a fraction of the DnaA protein to be ATP bound [152, 153] and because, newly synthesized DnaA protein is mainly ATP bound, an efficient turn-off of replication by RIDA is dependent on a period without de novo DnaA protein synthesis. This is provided by sequestration of the hemimethylated dnaA promoter.
A second mechanism for conversion of DnaAATP to DnaAADP involves the datA region in a process called DDAH (datA-dependent DnaA-ATP hydrolysis) [166]. DnaAATP binds to the DnaA-boxes in datA in an Integration Host Factor (IHF)-dependent manner, and the datA-DnaA-IHF complex promotes DnaAATP hydrolysis. This may seem contradictory as IHF is known to stimulate initiation of replication when it is bound to oriC prior to initiation [167, 168], but datA is bound after initiation has occurred. In this way the DDAH process assist RIDA in turning off unwanted initiations [166].
Origin sequestration-deficient cells are not able to maintain once-per-cell cycle initiation, despite being proficient in dnaA promoter sequestration, datA duplication and RIDA [113, 134, 136]. Therefore a period of time, where the origin cannot be initiated, is necessary to ensure a decrease in DnaAATP to a level that does not permit re-initiation in the same cell cycle. This time period is provided by the sequestration of hemimethylated origin. At the end of sequestration, when origins become available for re-initiation, the level of DnaAATP is below the threshold for initiation, and a period of growth is necessary to accumulate sufficient amounts of DnaAATP protein for the next round of initiation. This cell cycle-dependent variation in free DnaAATP within the cell represents the biological clock responsible for the constant inter-initiation period.
Methylation and nucleoid organization
The second function of SeqA in vivo is the organization of nascent nucleoids behind the replication fork. SeqA was found to bind numerous sites across the E. coli chromosome [169, 170] and fast growing E. coli cells contain multiple replication forks. Because SeqA binds new replication forks more readily than old ones [170], there is a preference for SeqA binding to origin proximal parts of the chromosome under these conditions. This may explain, or at least contribute to, the earlier observations that SeqA was found to bind the region surrounding the replication terminus, i.e., the ter macrodomain, poorly [169, 171, 172].
SeqA dimers bound to distant pairs of hemimethylated GATC sites can, concurrent with replication, organize nascent daughter chromosomes into nucleoid domains [116, 173] which may require also the ability of SeqA to generate positive supercoils [174]. Lack of SeqA leads to increased negative supercoiling [175]. SeqA is also involved in proper chromosome segregation as seqA mutants, SeqA overproducers and cells with non-sequesterable origins show gross defects in nucleoid positioning within the cell [135, 176, 177].
Two models have been proposed for segregation of newly replicated DNA as it leaves the replication factory. In the extrusion-capture model [178, 179], replication of DNA from a stationary factory pushes newly replicated DNA outwards. It has been proposed that SeqA could organize and channel hemimethylated DNA towards the positions of the new nucleoid [126, 179]). In the sister chromosome cohesion model, the newly replicated DNA is held together until late in the replication cycle. Upon release from cohesion, origins move rapidly from a central position towards the cell poles [180–182]. There is discrepancy in the literature regarding SeqA’s role in cohesion. In one study SeqA dimers were found not to bind separate DNA molecules [183] indicating that they act strictly in cis and therefore presumably are not involved in cohesion of sister chromosomes. Furthermore, cohesion between oriC sister copies was observed in dam mutant cells in vivo, and this argues against a role for Dam/SeqA in cohesion [184]. Sister chromosome cohesion, therefore, could result from catenation of daughter chromosomes behind the replication fork [185]. Interestingly, SeqA protein bound to hemimethylated DNA may play a role in decatenation as it interacts with, and stimulates, the decatenating activity of topoisomerase IV [122]. A recent report indicates SeqA dependent co-localization of newly replicated origins, especially during fast growth. In this study SeqA was capable of pairing newly replicated origins in vitro [186].
The role of DNA methylation for chromosome replication in other bacteria
The gamma-proteobacterium Vibrio cholerae is a close relative of E. coli. V. cholerae has its genome divided between two chromosomes. The origin of the larger chromosome I, oriCI, resembles E. coli oriC with respect to the positions of the DnaA binding sites, AT-rich region, GATC sites, etc., whereas oriCII of the smaller chromosome II, is different. oriCII carries a number of 11-bp or 12-bp iterons to which the chromosome II specific initiator protein, RctB, binds. Each of these iterons contains a GATC site [11]. Newly replicated oriCI and oriCII remain hemimethylated for an extended period when compared to other sites on the chromosome [187, 188]. Deletion of the seqA gene has opposite consequences for sequestration of oriCI and oriCII. SeqA deficiency results in a shortened sequestration period of oriCII, similar to what is observed for oriC in E. coli, but is unexpectedly long for oriCI [188]. SeqA deficiency resulted in overreplication of both chromosomes [188], whereas SeqA overproduction inhibits replication of both chromosomes [187].
Minichromosomes having either oriCI or oriCII replicate autonomously in wild-type E. coli cells but fail to transform dam mutants [11]. For oriCI, this indicates either an inability to replicate in the absence of Dam methylation or that incompatibility exists between the chromosomal oriC and the extrachromosomal oriCI, as previously described for E. coli minichromosomes in Dam-deficient cells [189]. In two independent studies, the oriC region of the E. coli chromosome was replaced with the V. cholerae oriCI region. In one study the dam gene was found to be dispensable for growth [188] whereas the other study found dam to be dispensable only if the overall level of negative supercoiling was increased [190]. This suggests that the genome organization around the origin may influence function and that care should be taken when analyzing origin function in a non-native context. However, in a V. cholerae strain carrying a fusion between chromosome I and II and with oriCI as sole replication origin, the dam gene could be deleted, which clearly shows that Dam methylation is not required for oriCI replication or any other processes essential for growth [191].
It is therefore conceivable that the inviability of V. cholerae dam mutants [10] results only from a failure to initiate replication at oriCII, which agrees well with the observation that oriCII minichromosomes transform and replicate as extrachromosomal elements in wild-type E. coli cells, but both fail to do so in dam mutant cells [11]. The initiation failure may result from difficulties in unwinding the unmethylated oriCII [25, 129].
The alpha-proteobacterium Caulobacter crescentus is dimorphic. Cell division results in a stalk cell and a swarmer cell. Initiation of replication takes place in the stalk cell immediately after cell division, whereas it is delayed in the swarmer cell until it differentiates into a stalk cell. The chromosomal replication origin covers about 1000 bp and contains five GANTC sequences; the substrate for the essential CcrM enzyme (Cell cycle-regulated DNA Methyltransferase) that methylates the A residue [192–194] (See “DNA Methylation in Caulobacter,” below). DnaA mediated replication initiation takes place on fully methylated DNA and overproduction of CcrM leads to additional initiations, suggesting that these are normally prevented from occurring on hemimethylated origins [193]. It is not clear why methylation of the origin is required for activity but this may result from altered origin structure or to facilitate origin opening or both [25, 129]. Newly replicated origins remain hemimethylated for most of the cell cycle because the CcrM enzyme is only present in late pre-divisional swarmer cells and after DNA replication is complete [193].
The dnaA gene of C. crescentus is also regulated by CcrM methylation. The dnaA gene is close to oriC and becomes hemimethylated at about the same time as the origin. When CcrM is present, the dnaA promoter becomes remethylated at about the same time as oriC, and because the fully methylated dnaA promoter is more active than its hemimethylated counterpart, the DnaA protein accumulates in swarmer cells to trigger a new round of initiation in the swarmer to stalk cell transition [195]. For reviews on C. cresentus chromosome replication, see [56, 196].
Regulation of Gene Expression
Since the state of GATC methylation (methylated, unmethylated, hemi-methylated) can affect specific binding of DpnI, DpnII, Sau3A (BfuCI), Dam, SeqA and MutH, then the presence of this tetranucleotide in promoter or regulatory sequences should also affect gene expression through inhibition or promotion of repressor or activator binding.
Regulation at methylated GATC sites
The P2 promoter region of the dnaA gene discussed above, for example, is fully active only in the completely methylated state, consistent with its biological role (25). Currently, this is the only example of this type of regulation.
Regulation at unmethylated GATC sites
In contrast to the lone dnaA example above, there is evidence that specific protein binding results in 36 unmethylated GATCs in the E. coli chromosome [19]. Nine of these are in the cyclic AMP binding protein (CAP)-binding sites preceding the mtlA, cdd, flhD, gcd, ycdZ, yffE, ppiA, proP, and srl (gut) operons [19], suggesting modulation of gene expression by Dam methylation through differential CAP binding. Other genes with GATCs that overlap with protein binding sites are: hrsA, kdgT (Fnr), pspA, yjdG (IHF), fep (Fur), carA (CarP, IHF), agn43 (flu) (OxyR), ppiA (Lrp, CAP), and yhiP (Lrp) [20, 21]. In only a few instances are there data to support specific binding of a regulatory protein either in vivo or in vitro. The level of methylation of the carbamoyl phosphate synthase (carA) gene promoter GATC was dependent upon cultural conditions; more methylation was detected when arginine and pyrimidines were present than in their absence, suggesting a possible regulatory effect. Protection of the yhiP GATC by Lrp was dependent on the presence of leucine [21].
Additional unmethylated GATC sites were found in the non-coding regions of rspA, ydjL, yahM, bhsA, yjdD, yhiP, yiaK, yidX, and yihU/V genes [20, 21] although their significance is not known. For all operons containing unmethylated GATCs it would be interesting to determine whether their level of expression changes in cells overproducing Dam. If so, it would suggest that methylation of the sites is important for regulating operon expression.
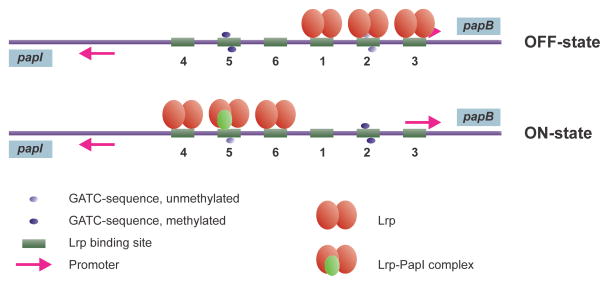
Studies on the pap operon have provided the most detailed evidence that the unmethylated GATCs are involved in controlling gene expression (reviewed in [197]). Pyelonephritis-associated pilus (Pap) expression is regulated by a phase variation mechanism in which individual cells either express pili (phase-on) or not (phase-off). When Pap pilus gene expression is in the phase-off state, GATC1028 is fully methylated and GATC1130 is unmethylated (Fig. 10). Conversely, in the phase-on state, the methylation state at these two sites is reversed. In a strain overproducing Dam, the transition of phase-off to phase-on is prevented, whereas in a dam mutant the opposite transition does not occur. The mechanism of phase variation is that Dam competes with the transcriptional activators Lrp and PapI such that Lrp is required for protection of GATC1130 and both Lrp and PapI are required for methylation protection of GATC1028 [197]. Other pilus systems also appear to be Dam controlled, although the evidence is not as complete as for pap [197].
FIGURE 10.
Switching at the pap promoter region. Lrp binds co-operatively to either Lrp binding sites 1–3 (in the OFF-state, non-piliated) or to the Lrp binding sites 4–6 (ON-state, piliated). Lrp binding site 3 overlaps the papB promoter and Lrp binding to site 3 inhibits transcription. Lrp binding sites 2 and 5 overlaps with GATC sites and Lrp binding to either site prevents methylation of that site by DamMT. Lrp binding to sites 1–3 mutually excludes binding to sites 4–6. When in OFF-state, each DNA replication produce one hemimethylated GATC site (in Lrp site 5) and one unmethylated GATC site (in Lrp site 2) and dissociate Lrp from its binding sites. The OFF-state is preserved by rebinding of Lrp to the same binding sites around the unmethylated Lrp binding site. A shift from phase OFF to ON may occur if PapI mediates Lrp binding to the hemimethylated Lrp site 5, followed by Lrp binding to sites 4 and 6. The shift is further stabilized by full methylation of site 2 by DamMT and conversion of the hemimethylated site 5 to unmethylated by subsequent DNA replications. This figure is adapted from [275] with permission from Cell Press.
Another example of proteins binding at specific GATCs leading to phase variation, is the agn43 (antigen 43 or flu) gene of E. coli which encodes an autotransporter protein that causes cells to aggregate and form precipitates in culture media [198]. The gene has three -GATC- sites in its promoter region and their methylation leads to full transcription initiation [199, 200]. During replication of the gene, the hemimethylated DNA can be bound by Dam or SeqA or OxyR (Fig. 11). Dam binding to the GATCs allows continued transcription and while SeqA binding is transient and favors its eventual replacement by Dam. OxyR binding, however, is tighter than SeqA and can exclude Dam so that after a second round of replication a fully unmethylated promoter region with bound OxyR is created and gene transcription cannot occur [201].
FIGURE 11.
Regulation of agn43 gene transcription in E. coli. The promoter region of the agn43 gene contains three GATC sequences which must be methylated (Me) for the gene to be expressed. Replication of the gene will cause transient hemimethylation allowing one of three proteins can bind this DNA. Dam action will methylated the GATCs on the new strand thereby preserving expression of the gene. SeqA can also bind but is easily displaced by Dam resulting in methylation and continued gene expression. OxyR binding prevents Dam action and after a second round of replication the GATCs are unmethylated and transcription is prevented. This modified figure is reproduced with permission from [276] copyright (2002) John Wiley and Sons Ltd.
Regulation at hemimethylated GATC sites
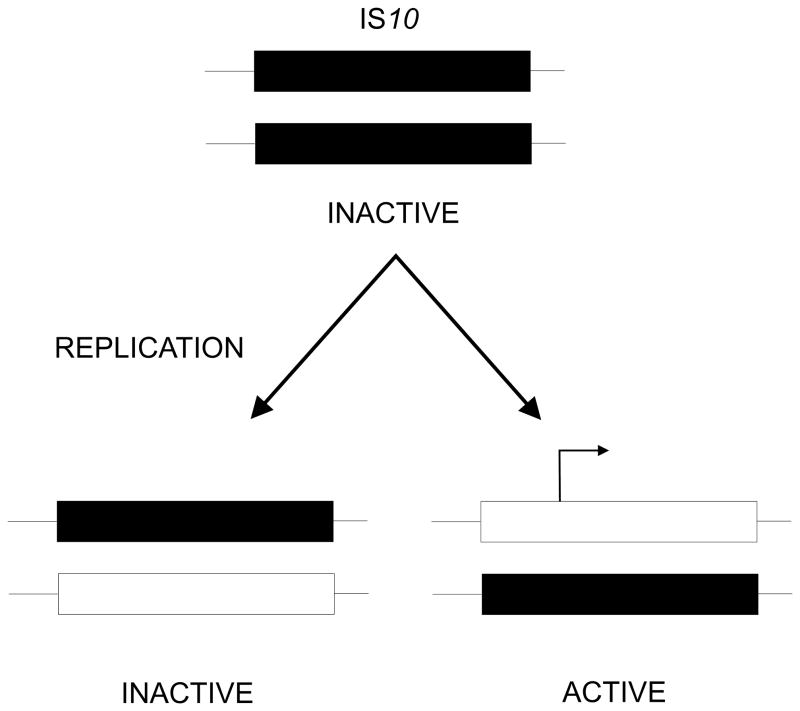
In addition to unmethylated sites, there is also evidence that hemimethylated GATC sites are important to control gene expression. The transposition frequency of Tn10 is directly related to the cellular concentration of Dam acting at two specific GATC sites in IS10 right [202]. Overproduction of Dam decreases transposition, whereas it is increased in a dam mutant. One of the GATC sites overlaps the -10 region of the transposase (tsp) promoter, while the other is near the inner end of IS10 in the target area for transposase action. In DNA that is not being replicated these sites are methylated and inert for transposition (Fig. 12). Upon replication, these sites become hemimethylated but only one of these is activated for transposition. The transposase promoter, in a wild-type strain, is active only in the configuration of methylated transposase coding strand and unmethylated non-coding strand.
FIGURE 12.
Activation of the tsp promoter in IS10. In wildtype bacteria both strands of IS10 are methylated (black rectangles). Upon replication, two hemimethylated forms are produced but only that with the methylated coding strand actively transcribes the tsp gene and moves to a new location while the inactive IS10 remains.
The coupling of transposase activation and action to hemimethylation means that transposition is repressed for most of the cell cycle but induced when the element is replicated. The asymmetry imposed at the replication fork means that only one of the two copies of the element can transpose. Hence one copy can remain in place while the other finds an alternative location. The coupling to replication helps to prevent the potentially deleterious effects of excessive transposition [202]. Other transposons such as Tn5 and Tn903 and the insertion element 1S3 also use Dam methylation to control transposition [203].
Several E. coli promoters have GATC sites in either the -10 or -35 regions. These include promoter regions for the sulA, trpS, trpR, tyrR, and glnS genes, and expression of these genes is increased in dam mutants compared to wild type (reviewed in references [204–206]). It is not known whether expression of these genes is increased in a hemimethylated configuration. However, even if it were the physiological role for a coupling of transcription of these genes to replication is not obvious. For trpR, one possibility is that since the trpR gene lies between oriC and the trp operon, a transient boost in trpR transcription might provide the increase in Trp repressor concentration needed when duplication of the operon occurs. However, subsequent experiments did not support this idea [207].
Microarray studies
Global gene expression comparing wildtype and dam mutants using microarrays has been measured in E. coli and S. enterica [49, 110, 208, 209]. The results are difficult to compare because different strain backgrounds, media, arrays and other experimental conditions were varied as well as the goals of the experiments. However, the up-regulation of SOS gene expression in the dam background was detected in each case and decreased motility in two studies. In addition, Oshima et al. [208] and Robbins-Manke et al. [110] confirmed the increase in transcription of several SOS genes by RT-PCR.
Oshima et al. [208] found that in addition to increased SOS regulon expression, genes involved in aerobic respiration, amino-acid and nucleotide metabolism were expressed at higher levels in the dam strain. Decreased signal was found for genes in anaerobic respiration, flagellar biosynthesis, chemotaxis and motility. Decreased motility of the dam mutant was demonstrated on motility plates. The increase or decrease in expression depended on the level of aeration. These results were interpreted in terms of altered binding of Fnr (fumarate nitrate reduction) and CRP (catabolite activator protein) in the promoter regions of affected genes. In addition to the array data, an increase in the steady-state level of a number of proteins was detected in dam cells by 2D-gel electrophoresis including YeaF (cell envelope), G3P1 (glyceraldehyde-3-phosphate dehydrogenase A) and GroEL (heat shock response) while a decreased level was noted for Pgk (phosphoglycerate kinase), SodA (superoxide dismutase) and G3P1 (under low aerobic conditions). Further filtering of these data has been reported [210].
The goal of the array experiments conducted by Løbner-Olesen et al. [49] was to compare transcription profiles in dam, seqA and Dam-overproducing wildtype cells. The striking result was that the profiles for the seqA mutant and the Dam-overproducing wildtype cells were very similar (Fig. 9). This result suggests that SeqA binds to the hemi-methylated region behind the fork but is prevented from doing so when Dam is present at a high concentration to reduce the amount of hemimethylated DNA. Since the absence of SeqA increases the superhelicity of chromosomal DNA [175], transcription can initiate at promoters not active at the physiological superhelical density of wildtype cells. There were few known genes in the dam strain, other than those belonging to the SOS regulon that showed significant differences in expression levels compared to wildtype. One of these was dnaA whose transcription level was moderately reduced confirming previous data obtained with lac fusions [211].
FIGURE 9.
Expression of individual genes as a function of position on the chromosome. All genes for which a significant signal was obtained were plotted relative to wild type as a function of position along the chromosome. The chromosome is linearized at a position directly opposite oriC. The replication origin has position 0 on the abscissa. (A) MG1655 seqA. (B) MG1655_pTP166 (Dam overproducer). (C) MG1655 dam-13::Tn9. Trendlines for the gene expression data are presented in A and B. All points above the trendline in panel A, i.e., genes that were derepressed in the seqA mutant are plotted as green dots, and all genes that were repressed in the seqA mutant as red dots. Expression data from individual genes in panels B and C have the same color assignment as in panel A (red and green dots). Reproduced from with permission from [49] and Copyright (2007) National Academy of Sciences, U.S.A.
Robbins-Manke et al. [110] investigated the transcription profiles of wildtype, dam, mutS and dam mutS cells as well as measuring the number of DNA double-strand breaks in each strain. As expected, there were many more double-strand breaks in the dam mutant compared to the wildtype, mutS and dam mutS bacteria. The array data indicated that the SOS regulon was expressed at a low level in wild-type cells but was at a higher level in the dam mutant. The dam mutS strain, however, still showed almost as much SOS regulon expression as the dam mutant and significantly more than the control mutS strain confirming a previous result showing elevated SOS expression in dam mut strains using recA::lac or sulA::lac fusions [112]. Expression of the SOS regulon genes, therefore, does not seem to correlate with mismatch repair-induced double-strand breaks in a dam background. Consequently, there must be a DNA substrate other than the processing of MutHLS-induced double-strand breaks that serves as an inducing signal (presumably RecA-covered single-stranded DNA) in dam cells although its nature remains unknown. One possibility, however, relates to the asynchronous initiation of replication in dam cells - if there are two initiation events close together perhaps the forks are so closely spaced that they run into each other producing replication fork collapse (Fig. 7D). The exposed double-stranded end becomes a substrate for RecBCD exonuclease which, when encountering a Chi site generates RecA-loaded single-stranded DNA thereby producing an SOS signal.
There were many genes showing increased or decreased expression in the dam background in the Robbins-Manke data set but a more restrictive filtering eliminated most of these except for the SOS genes and a few genes involved in translation [212].
Dam methylation affects virulence in S. enterica and some other bacteria (See Bacterial Virulence). A microarray analysis of wildtype and dam mutant transcripts in this strain identified increased SOS regulon expression in the dam strain just as in E. coli [209]. Invasion gene transcript levels in pathogenicity island SPI-1 were decreased in dam cells while those in the std fimbrial operon were increased. Altered expression patterns were found for certain flagellar (fliCD) and chemotaxis (cheR, STM3216) genes and for the lppB lipoprotein gene. The decreased expression of fli and che genes probably accounts for decreased motility on motility plates. The transfer operon (tra) genes of the conjugative virulence plasmid, pSLT, showed increased expression confirming previous lac fusion data (See Bacterial Virulence). All these changes in transcription probably help to explain the reduced motility, accumulation of Std protein in membranes and supernatants, envelope instability, and reduced virulence associated with the dam mutant. This study lays the foundation for further investigation into the complex regulation of the pathogenicity island and flagella genes.
Methylation-dependent gene expression and dam phenotypes
In Table 3, the properties of a dam mutant which can be explained by methylation-dependent gene expression include increased transposition by transposons and altered expression of chromosomal and plasmid genes. Since E. coli dam cells are viable, it follows that there are no essential genes whose expression are solely dependent on Dam methylation. Rather, Dam methylation can be viewed in the context of fine tuning basal levels of gene expression either by acting directly at GATCs in regulatory sequences or indirectly by affecting nucleoid structure or acting as a timing switch or in an epigenetic fashion.
Methylation-dependent gene expression in bacteriophage
Examples of gene expression modulated by Dam methylation have also been described in bacteriophage systems. The expression of the mom gene of bacteriophage Mu was found to be influenced by Dam methylation [213]. The mom gene, which is expressed late in the phage life cycle, encodes a modification enzyme which converts adenine to N6-carboxy-methyl adenine [214, 215]. The mom gene is non-essential but the mutant has a restricted host range suggesting that Mom’s action helps protect the phage DNA from host restriction systems [216]. Dam methylation prevents binding of the E. coli OxyR regulatory protein to a 43-bp region upstream of the phage Mu mom gene which contains three GATCs [213]. Although it was demonstrated that OxyR binding in vitro occurred on unmethylated (but not methylated) DNA substrates, it is probable that binding occurs at hemimethylated sites in vivo [217]. Once bound, the OxyR repressor prevents Dam from methylating the three critical GATC sites and prevents transcription initiation, perhaps by interfering with the action of the trans-activating C protein on RNA polymerase.
Cre is a bacteriophage P1 site-specific recombinase involved in the formation of covalently-closed circular DNA upon infection of the host. One of the cre promoters contains two GATCs in its -35 hexamer and its transcription is repressed by Dam [218]. The biological significance of this regulation is not known.
Control of Phage P1 DNA Packaging into Virions
Phage P1 encodes a dam gene which produces a Dam protein that is related to that of the E. coli host. P1 dam mutant phage growth is normal on a wild-type host but severely restricted on a dam mutant host [219]. The requirement for Dam by phage P1 is related to the mechanism of DNA encapsidation. Packaging begins at a fixed site (pac) on a concatemer and proceeds by a headful mechanism such that greater-than-genome-length units are packaged. This results in terminal redundancy (same DNA sequence at both ends), which allows the linear genome to circularize by recombination upon infection of the next host.
The pac cleavage site is flanked by seven GATCs in a region of 162 bp. In vitro and in vivo experiments indicate that cleavage of both strands occurs only on fully Dam methylated pac regions [219]. Since the phage requires concatemeric DNA for packaging and because the phage proteins necessary for pac cleavage are synthesized early in the life cycle, there must be a regulatory mechanism to prevent methylation prior to the initiation of packaging. Furthermore, it is necessary that only one or two pac sites in the concatemer are cleaved and that the remainder are protected from cleavage to ensure proper encapsidation. Presumably this occurs by the competitive binding to GATCs of some phage or host protein. Since the phage enzyme(s) responsible for cleavage (“pacase”) can bind to hemimethylated pac sites but cannot cleave, it may be that pacase itself prevents methylation [219].
Bacterial Virulence
dam mutants of S. enterica have attenuated virulence in the mouse model with the 50% lethal dose 10,000-fold or 1,000-fold higher than wildtype for the oral and intraperitoneal routes of administration respectively [220, 221]. Animals infected with the attenuated dam strain are resistant to super-infection by the wildtype offering the possibility of a vaccine [222–228]). There are several deficiencies associated with dam mutants that could contribute to attenuate virulence: decreased adhesion/invasion, reduced motility, envelope instability and sensitivity to bile (reviewed in [4, 229]). These deficiencies are probably due to altered gene expression as discussed above (Control of Gene Expression) except for bile sensitivity. The combination of these factors especially envelope instability are probably responsible for the high efficiency immune response in animals infected with dam bacteria. The bile sensitivity of S. enterica dam cells is due to killing by the dam-directed mismatch repair system probably by the formation of DNA double-strand breaks [230, 231]. At present it is not known what DNA modification produced by bile salts is recognized by the dam-directed mismatch repair system.
Overproduction of Dam also attenuates virulence of S. enterica [221]. This could be due to changing expression of genes that are normally unmethylated or by producing seqA phenocopies which change global transcription by altering the supercoiling of DNA [49] or by off-target methylation [29]. A S. enterica seqA mutant shows decreased virulence by the oral (but not intraperitoneal) route of administration and this is explained by the mutant’s increased sensitivity to bile salts [232].
Dam is an important component of virulence in certain pathogenic bacteria such as Salmonella enterica, Haemophilus influenzae, Actinobacillus actinomycetemcomitans, Klebsiella pneumoniae, enterohemorrhagic E. coli O157:H7, Campylobacter jejuni, Yersinia enterocolitica, Brucella abortus, Vibrio cholerae, Pasteurella multocida, Yersinia pseudotuberculosis, and Aeromonas hydrophila (reviewed in references [229, 233]). The effects of methylation on virulence have been studied by either constructing a dam mutant or overproducing Dam in these organisms. Although a detailed discussion of these studies is beyond the scope of this review, it is interesting that a Dam-mediated regulatory effect on the type III secretion system of Yersinia enterocolitica occurs through Clp-dependent proteolysis [234]. Type III secretion systems are widespread in pathogenic gram-negative bacteria.
Subsequent to the publications of the reviews ([229], [233]) discussing the bacteria listed above, Dam has been found to be involved in the pathogenesis of Edwardsiell tarda, a fish pathogen important for aquaculture [235], and in Streptococcus mutans, a dental pathogen [236] In both cases, a dam mutant was constructed and caused significant attenuation of overall virulence or altered expression of genes with cariogenic potential.
Type VI secretion systems (T6SS) of Gram-negative bacteria are responsible for bacterial killing and/or virulence toward different host cells. Both Fur (ferric iron uptake global transcriptional repressor) and Dam regulate the sci1 (T6SS) gene cluster as an epigenetic switch in enteroaggregative E. coli [237].
An additional virulence-associated phenotype influenced by Dam is conjugal transfer of the F and F-like plasmids including the pSLT virulence plasmid of S. enterica [96]. The latter plasmid bears the 7.8 kb spv region required for proliferation in the reticuloendothelial system and several other genes that may play roles in other stages of the infection process. The presence of the plasmid, which is self-transmissible, may impart an expanded host range on bacteria carrying it. Conjugal transfer in F and F-like plasmids is derepressed in dam mutants of both S. enterica and E. coli due to increased expression of the tra (transfer) operon. The increased transcription is mediated through effects on the regulatory genes, traJ and finP [238]. Dam methylation has opposite effects on these genes; transcription of traJ is increased in a dam mutant but transcription of finP, a small RNA that antagonizes traJ expression, is decreased. The interplay between these effects accounts for the level of tra operon expression [239]. The expression of the traJ gene is affected by Dam methylation like the pap gene and like the coupling of IS10 transposition to the replication fork [240]. Lrp binding to one of two GATCs in the promoter region determines the level of traJ expression and in the hemimethylated state binding to the non-coding strand is higher than the coding strand. This suggests that, like Tn10 transposition, expression of traJ is coupled to replication and is active on only one of the two hemimethylated configurations and only one of the daughter plasmids will be activated for transfer [239]. The effect of Dam methylation on finP transcription appears to be mediated through binding of the nucleoid protein H-NS, not at the promoter but as a global effect reminiscent of altered transcription in wild-type cells overexpressing Dam leading to a seqA phenocopy and altered global transcription [49].
Inactivation of the dam gene is not lethal in E. coli K-12 but dam mutants of enterohemorrhagic E. coli O157:H7 strain EDL933 cannot be isolated [241]. The reason for this is due to a lambda-like prophage, 933W, which encodes the genes for the production of Shiga toxin. Genetic studies showed that EDL933 dam mutants can be obtained only in cells cured of this prophage or in cells with inactive dam-directed mismatch repair. Given the demonstration that spontaneous induction of lambda prophage is increased in E. coli K-12 dam cells (Table 3) via increased SOS regulon expression [7], a similar model was proposed for EDL933 and prophage 933W [241]. The mechanism whereby prophage induction occurs in almost every cell inactivated for Dam remains to be determined. Increased excision of the defective prophage ST64B from a dam mutant of Salmonella enterica is also due to enhanced SOS regulon expression [242]. In this case, however, there was also a direct effect on transcription of genes putatively involved in phage induction due to the presence of dam sites in the regulatory region of these genes. The conclusion that loss of dam leads to inviability of EDL933 through prophage induction is a caution in studies where ability to delete a particular gene is often used to determine if it is essential or not to the viability of the organism. In this case, Dam does not perform an essential function but the cells die due to an indirect cause.
Post-transcriptional Regulation
A critical step during colonization and pathogenesis of enterohemorrhagic E. coli O157:H7 is the formation of “pedestals” that result from the accumulation of actin filaments beneath adherent bacteria elevating them above the surrounding cell surfaces [243]. Both intimate adhesion and actin pedestal formation result from the transfer of E. coli secreted proteins into the host cell where they interact with mammalian signaling molecules that control actin assembly. One of the secreted proteins is Tir (translocated intimin receptor) which is delivered to the cell membrane where it serves as a receptor for intimin, an adhesin on the outside of the bacterial cell. Wildtype E. coli O157:H7 show relatively poor pedestal formation on cultured mammalian cell lines.
Deletion of the dam gene of E. coli O157:H7 results in a dramatic increase in adherence and actin pedestal formation on cultured human cells compared to wild type [53]. Increases in adherence and pedestal formation in vitro correlated with elevated protein levels of intimin, Tir, and another secreted protein, EspFU. Consistent with its capacity for vigorous interaction with mammalian cells in vitro and in contrast to dam mutants of several other enteric pathogens, the dam mutant of E. coli O157:H7 was is capable of robust colonization of the intestines of infected animals [53].
The elevated protein levels of intimin and Tir did not result from an increase in mRNA levels as measured by microarrays and RT-PCR suggesting a post-transcriptional mechanism of regulation [53]. To further investigate the basis of this observation, an E. coli O157:H7 hfq mutant was constructed and pedestal formation was as robust as in a dam mutant (Brady, M., Leong, J.M., Marinus, M.G., unpublished data). The hfq mutant contains an elevated level of Tir (Marinus, M.G., unpublished data). One model to account for this observation is that translation of tir message is controlled, in part, by a small regulatory RNA that requires Hfq chaperone activity to bind the message. In an hfq mutant, no small RNA-mRNA binding is possible and translation of tir message is not impeded. In a dam mutant, however, transcription of the small RNA is decreased relative to wildtype leading to increased translation of tir message. This model is currently being tested.
The vsr gene is part of the “very small patch” (VSP) repair system discussed above (VSP Repair) and the efficiency of this system is reduced in dam bacteria [95]. The level of Vsr in wildtype is low in logarithmic phase cells and higher in stationary phase cells but in a dam mutant there is much less Vsr in stationary phase. Although the vsr mRNA level was not determined in wildtype and the dam mutant, the authors concluded that the reduction of Vsr was due to a post-transcriptional mechanism in the dam mutant [95]. Although the actual mechanism remains unknown, the involvement of a small regulatory RNA in vsr translation could explain the observations where its level is altered in the dam mutant by transcription and its action could be to alter either the rate of translation or message stability.
Dam overproduction in Yersinia enterocolitica imparts a hyper-invasive phenotype and results in many changes in cellular metabolism [244]. Among these is a change in the composition of lipopolysaccharide (LPS) O-antigen status where increased amounts of lipid A core, without O-antigen subunits, was observed. The O-antigen gene cluster consists of two transcriptional units but the transcript levels in the Dam overproducer, as measured by RT-PCR, of representative genes in each cluster (ddhA, gne, and rosA), was unchanged relative to wild type. The modulation of LPS structure is, therefore, due to an unknown posttranscriptional mechanism and undoubtedly contributes to the hyper-invasive phenotype.
DNA METHYLATION IN CAULOBACTER
The emphasis in this review has been on DNA methylation in E. coli and S. enterica, both members of the gamma-Proteobacteria. However, it is instructive to briefly review DNA methylation in Caulobacter crescentus, a member of the alpha-Proteobacteria which has defined morphological stages. The DNA methyltransferase in this organism is CcrM which methylates adenine in the sequence 5′-GANTC-3′ [56] in a distributive manner [245]. Unlike Dam, CcrM is essential for the life of the organism, is cell cycle regulated [246], and is not present at all stages of the life cycle. The life cycle consists of two cell types, stalked cells and swarmers. Chromosome replication occurs only in the stalked cell which has methylated DNA and involves the sequential action of three key unstable regulators: DnaA, GcrA and CtrA (Fig. 13[195]). The genes for these regulators are located sequentially on the chromosome with dnaA nearest the origin of replication (Cori) and ctrA the most distal. The action of these regulators, acting as a transcriptional cascade, is determined by the state of methylation of chromosomal DNA. DnaA initiates chromosome replication at the fully methylated Cori in a manner similar to that in E. coli. Since CcrM is not present at this stage, replication produces two hemimethylated daughter molecules during fork progression. As in E. coli, expression of the dnaA gene, which lies near Cori, is attenuated on hemimethylated DNA thereby reducing the possibility of premature initiation. DnaA also activates transcription of the gcrA gene, the product of which controls the transcription of replication genes encoding DNA polymerase III holoenzyme, DNA helicase and primase. GcrA in turn activates transcription of the ctrA gene the promoter of which contains two 5′-GANTC-3′ sequences in the upstream regulatory region and one is close to the -35 hexamer. This promoter also is active only when hemi-methylated and the expression of the gene is, therefore, co-coordinated with the cell cycle. CtrA is able to bind Cori to prevent premature initiation as well as activating transcription of the ftsZ and ccrM genes and repressing transcription from the gcrA gene. FtsZ is a key cell division protein and its production by CtrA enables a coupling between chromosome replication and cell division. Transcription of the ccrM gene occurs only in the hemi-methylated state and is activated by CtrA which binds to the upstream regulatory region of ccrM. This arrangement ensures that the concentration of CcrM increases toward the end of the replication cycle. The ccrM promoter also contains two 5′-GANTC-3′ sequences presumably ensuring auto-regulation of the gene. The production of CcrM is followed by methylation of the daughter chromosomes which silences the ctrA and ccrM genes and activates transcription of dnaA as well as preparing Cori for initiation by fully methylating it.
FIGURE 13.
Regulatory cascade in the Caulobacter cell cycle. The genes for dnaA, gcrA, ctrA and ccrM are shown together with their respective products. The genes are shown in the sequence they are replicated on the chromosome. Asterisks indicate CcrM recognition sites (GANTC). In addition to the genes shown in this figure, DnaA, GrcA and CtrA control about 40, 50 and 95 other genes respectively. Figure modified with permission from [195] and Copyright (2007) National Academy of Sciences, U.S.A.
After cell division the DNA of both cell types is fully methylated. In the swarmer cell, CtrA remains bound to Cori and the CcrM protein is degraded preventing further methylation thereby ensuring the origin is hemi-methylated and inert for further initiation. In the stalked cell, however, CtrA is destroyed by proteolysis allowing initiation to proceed on a fully methylated Cori.
PRACTICAL APPLICATIONS
Information about particular E. coli K-12 dam and dcm strains and suggestions about their handling and storage as well as plasmids expressing these genes can be found at http://users.umassmed.edu/martin.marinus/dstrains.html. A review dedicated to this subject that contains detailed protocols is also available [247].
Cleavage at dam and dcm Sites
The most frequent use of methylation-deficient strains is for the propagation of DNA molecules lacking either or both dam and dcm methylated sites [22]. This allows for the digestion by various endonucleases which have recognition sites overlapping methylation sequences. A list of such endonucleases can be found in most catalogs of restriction enzyme suppliers. A list of dam and dcm strains containing various genetic markers has been published [22] and is available from the E. coli Genetic Stock Center (http://cgsc.biology.yale.edu/).
An alternative use for Dam is the generation of specific cleavage sites. For example, the action of Dam on a specific DNA molecule will prevent the action of ClaI at overlapping dam sites (ATCGATC) but not at non-overlapping sites (ATCGATG/A/T). Hemimethylated GATC sites can be detected using restriction enzymes such as HphI, MboII and TaqI which cleave only one of the two replicated substrate sequences [26].
DNA Sequencing Procedures
DNA prepared in a dcm mutant avoids loss of bands corresponding to 5-meCyt by the Maxam-Gilbert chemical sequencing procedure. Methylation of adenine can be shown directly by automated dye terminator sequencing since the modification leads to an increase in the signal of the complementary thymine [248, 249].
Increased Transformation Efficiency
Shuttle-vector plasmid DNAs isolated from wild-type E. coli transform Streptomyces lividans at a very low frequency. Transformation efficiencies are increased 400- to 10,000-fold when the vector DNAs are prepared from a dam dcm mutant [250]. Similar results have been obtained with various Bacillus and Paracoccus species.
Propagation of Repeated Sequences
An array of 27 direct repeats consisting of 24-bp units in a plasmid was stably propagated in dam cells but not wild-type, recA, or mismatch repair-defective strains [251]. It is possible that SeqA binds to the 24 bp units (there are 2 GATCs per unit) and destabilizes them in wild-type but not in the dam cells where SeqA may not bind efficiently [251]. A 7 kb mutant version of the lac operon was constructed and its expansion into an amplified array was studied. Expansion occurred at an increased rate in a dam mutant [252].
Site-Directed Mutagenesis
dam-directed mismatch repair can reduce the yield of mutants in site-directed mutagenesis by removal of the desired mutated base. This can be avoided by preparing the template strand in a dam mutant, followed by annealing of the mutagenic primer, extension by polymerase and transformation of the appropriate dam strain [253]. Alternatively, if plasmid DNA is prepared from wildtype E. coli, the methylated template strand can be digested with DpnI after extension of the mutagenic primer by polymerase. DNA fragments produced by PCR should be unmethylated, and after transformation into a wild-type strain the mutant yield should not be affected [39].
Low-Level SOS-Inducing Agents
Many mutagenic and carcinogenic agents induce the SOS system of E. coli. Some mutagenic agents (e.g., 2-aminopurine) are too weak to elicit an SOS response in normal E. coli cells but do so in dam bacteria [254]. This method has been used to screen a large number of compounds [255].
Probing Chromosome Structure and Function
Expression of the cloned E. coil dam gene in organisms that do not have Dam methylation can be used as a probe for chromosome structure and function provided methylation is not lethal. The regions which are methylated can be identified by their susceptibility to restriction endonucleases which cleave methylated GATCs. For example, this technique has been used in yeast, where expressed genes tend to be Dam methylated to a greater extent than repressed genes in cells containing the dam plasmid [256].
The in vivo targets of chromatin proteins can be determined by tethering Dam to a specific protein and subsequently Dam methylation will occur at locations where the protein binds [257]. This technique was successfully applied in Drosophila cell cultures and whole flies using the GAL4 protein. Interestingly, even when about 50 % of total genomic GATCs are methylated, there seems to be little effect on fly development.
Another method using a fusion of the protein of interest with a mutant Dam (K9A) allows the protein to bind to target sites and methylate nearby GATCs. Immunoprecipitation is used to isolate DNA fragments that are methylated which can then be used for high-throughput sequencing, microarrays, etc. [258].
Salmonella dam Mutants as Live Vaccine Strains
Salmonellosis is a major problem in livestock management as is contamination of consumer meat products by these bacteria. The prophylactic and therapeutic use of antibiotics has been used for a number of years to control salmonellosis but the emergence of multi-drug resistant variants has necessitated other approaches. Vaccination is a proven prophylactic method to prevent disease and the use of Salmonella dam mutants for this purpose seems promising. Following on the original demonstration of significant protection by dam mutants from superinfection by wildtype strains in a murine model [220, 221], homologous and heterologous protection has also been demonstrated in avian [222, 224] and bovine models [223, 225, 226].
Dam Methyltransferase Inhibitors
An alternative strategy to control salmonellosis could be the use of a drug to inhibit Dam methyltransferase in Salmonella in the digestive tract of livestock thereby reducing virulence. Lead compounds that inhibit the enzyme in vitro have already been described [72, 73]. A potential problem, however, is that such agents could increase the virulence of related bacteria such as E. coli O157:H7 by stimulating the formation of Shiga toxin [53].
Acknowledgments
The research carried out in the M.G.M. laboratory described in this article was supported by the American Cancer Society, the National Science Foundation, and the National Institutes of Health. The research in the A.L.O laboratory was supported by the Danish Center for Microbiology, the Carlsberg Foundation, the Danish National Sciences Research Foundation, the Novo Nordisk Foundation, and the Danish Medical Research Council.
References
- 1.Marinus MG. Methylation of DNA. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2. Vol. 1. ASM Press; Washington, DC: 1996. pp. 782–791. [Google Scholar]
- 2.Løbner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low DA, Casadesus J. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr Opin Microbiol. 2008;11:106–112. doi: 10.1016/j.mib.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Broadbent SE, Balbontin R, Casadesus J, Marinus MG, van der WM. YhdJ, a nonessential CcrM-like DNA methyltransferase of Escherichia coli and Salmonella enterica. J Bacteriol. 2007;189:4325–4327. doi: 10.1128/JB.01854-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marinus MG, Morris NR. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974;85:309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- 8.Russell DW, Hirata RK. The detection of extremely rare DNA modifications. Methylation in dam− and hsd− Escherichia coli strains. J Biol Chem. 1989;264:10787–10794. [PubMed] [Google Scholar]
- 9.Marinus MG. Recombination is essential for viability of an Escherichia coli dam (DNA adenine methyltransferase) mutant. J Bacteriol. 2000;182:463–468. doi: 10.1128/jb.182.2.463-468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julio SM, Heithoff DM, Provenzano D, Klose KE, Sinsheimer RL, Low DA, Mahan MJ. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect Immun. 2001;69:7610–7615. doi: 10.1128/IAI.69.12.7610-7615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan ES, Waldor MK. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell. 2003;114:521–530. doi: 10.1016/s0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- 12.Barras F, Marinus MG. Arrangement of Dam methylation sites (GATC) in the Escherichia coli chromosome. Nucleic Acids Res. 1988;16:9821–9838. doi: 10.1093/nar/16.20.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Eichelmann MC, Ramirez-Santos J. Methylated cytosine at Dcm (CCATGG) sites in Escherichia coli: possible function and evolutionary implications. J Mol Evol. 1993;37:11–24. doi: 10.1007/BF00170457. [DOI] [PubMed] [Google Scholar]
- 14.Henaut A, Rouxel T, Gleizes A, Moszer I, Danchin A. Uneven distribution of GATC motifs in the Escherichia coli chromosome, its plasmids and its phages. J Mol Biol. 1996;257:574–585. doi: 10.1006/jmbi.1996.0186. [DOI] [PubMed] [Google Scholar]
- 15.Modrich P. Mechanisms and Biological Effects of Mismatch Repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 16.Russell DW, Zinder ND. Hemimethylation prevents DNA replication in E. coli. Cell. 1987;50:1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- 17.Vovis GF, Lacks S. Complementary action of restriction enzymes endo R-DpnI and Endo R-DpnII on bacteriophage f1 DNA. J Mol Biol. 1977;115:525–538. doi: 10.1016/0022-2836(77)90169-3. [DOI] [PubMed] [Google Scholar]
- 18.Ringquist S, Smith CL. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc Natl Acad Sci U S A. 1992;89:4539–4543. doi: 10.1073/pnas.89.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang MX, Church GM. A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature. 1992;360:606–610. doi: 10.1038/360606a0. [DOI] [PubMed] [Google Scholar]
- 20.Tavazoie S, Church GM. Quantitative whole-genome analysis of DNA-protein interactions by in vivo methylase protection in E. coli. Nat Biotechnol. 1998;16:566–571. doi: 10.1038/nbt0698-566. [DOI] [PubMed] [Google Scholar]
- 21.Hale WB, van der Woude MW, Low DA. Analysis of nonmethylated GATC sites in the Escherichia coli chromosome and identification of sites that are differentially methylated in response to environmental stimuli. J Bacteriol. 1994;176:3438–3441. doi: 10.1128/jb.176.11.3438-3441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer BR, Marinus MG. The dam and dcm strains of Escherichia coli--a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 23.Parniewski P, Kwinkowski M, Wilk A, Klysik J. Dam methyltransferase sites located within the loop region of the oligopurine-oligopyrimidine sequences capable of forming H-DNA are undermethylated in vivo. Nucleic Acids Res. 1990;18:605–611. doi: 10.1093/nar/18.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allers T, Leach DR. DNA palindromes adopt a methylation-resistant conformation that is consistent with DNA cruciform or hairpin formation in vivo. J Mol Biol. 1995;252:70–85. doi: 10.1006/jmbi.1994.0476. [DOI] [PubMed] [Google Scholar]
- 25.Collins M, Myers RM. Alterations in DNA helix stability due to base modifications can be evaluated using denaturing gradient gel electrophoresis. J Mol Biol. 1987;198:737–744. doi: 10.1016/0022-2836(87)90214-2. [DOI] [PubMed] [Google Scholar]
- 26.Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 27.Ogden GB, Pratt MJ, Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 28.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, Deikus G, Clark TA, Luong K, Murray IA, Davis BM, Keren-Paz A, Chess A, Roberts RJ, Korlach J, Turner SW, Kumar V, Waldor MK, Schadt EE. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. 2012 doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, Fomenkov A, Roberts RJ, Korlach J. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae SH, Cheong HK, Cheong C, Kang S, Hwang DS, Choi BS. Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition. J Biol Chem. 2003;278:45987–45993. doi: 10.1074/jbc.M306038200. [DOI] [PubMed] [Google Scholar]
- 31.Guo Q, Lu M, Kallenbach NR. Effect of hemimethylation and methylation of adenine on the structure and stability of model DNA duplexes. Biochemistry. 1995;34:16359–16364. doi: 10.1021/bi00050a016. [DOI] [PubMed] [Google Scholar]
- 32.Maynard-Smith MD, McKelvie JC, Wood RJ, Harmer JE, Ranasinghe RT, Williams CL, Coomber DM, Stares AF, Roach PL. Direct and continuous fluorescence-based measurements of Pyrococcus horikoshii DNA N-6 adenine methyltransferase activity. Anal Biochem. 2011;418:204–212. doi: 10.1016/j.ab.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 33.Bhagwat AS, McClelland M. DNA mismatch correction by Very Short Patch repair may have altered the abundance of oligonucleotides in the E. coli genome. Nucleic Acids Res. 1992;20:1663–1668. doi: 10.1093/nar/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merkl R, Kroger M, Rice P, Fritz HJ. Statistical evaluation and biological interpretation of non-random abundance in the E. coli K-12 genome of tetra- and pentanucleotide sequences related to VSP DNA mismatch repair. Nucleic Acids Res. 1992;20:1657–1662. doi: 10.1093/nar/20.7.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brooks JE, Blumenthal RM, Gingeras TR. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983;11:837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClelland M. Selection against dam methylation sites in the genomes of DNA of enterobacteriophages. J Mol Evol. 1984;21:317–322. doi: 10.1007/BF02115649. [DOI] [PubMed] [Google Scholar]
- 37.Blaisdell BE, Campbell AM, Karlin S. Similarities and dissimilarities of phage genomes. Proc Natl Acad Sci U S A. 1996;93:5854–5859. doi: 10.1073/pnas.93.12.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marinus MG, Morris NR. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975;28:15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- 39.Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983;104:571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ. The methylomes of six bacteria. Nucleic Acids Res. 2012;40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bale A, d’Alarcao M, Marinus MG. Characterization of DNA adenine methylation mutants of Escherichia coli K12. Mutat Res. 1979;59:157–165. doi: 10.1016/0027-5107(79)90153-2. [DOI] [PubMed] [Google Scholar]
- 42.Jonczyk P, Hines R, Smith DW. The Escherichia coli dam gene is expressed as a distal gene of a new operon. Mol Gen Genet. 1989;217:85–96. doi: 10.1007/BF00330946. [DOI] [PubMed] [Google Scholar]
- 43.Lobner-Olesen A, Boye E, Marinus MG. Expression of the Escherichia coli dam gene. Mol Microbiol. 1992;6:1841–1851. doi: 10.1111/j.1365-2958.1992.tb01356.x. [DOI] [PubMed] [Google Scholar]
- 44.Lyngstadaas A, Lobner-Olesen A, Boye E. Characterization of three genes in the dam-containing operon of Escherichia coli. Mol Gen Genet. 1995;247:546–554. doi: 10.1007/BF00290345. [DOI] [PubMed] [Google Scholar]
- 45.Rasmussen LJ, Marinus MG, Lobner-Olesen A. Novel growth rate control of dam gene expression in Escherichia coli. Mol Microbiol. 1994;12:631–638. doi: 10.1111/j.1365-2958.1994.tb01050.x. [DOI] [PubMed] [Google Scholar]
- 46.Rasmussen LJ, Lobner-Olesen A, Marinus MG. Growth-rate-dependent transcription initiation from the dam P2 promoter. Gene. 1995;157:213–215. doi: 10.1016/0378-1119(94)00619-4. [DOI] [PubMed] [Google Scholar]
- 47.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 48.Vaisvila R, Rasmussen LJ, Lobner-Olesen A, von Freiesleben U, Marinus MG. The LipB protein is a negative regulator of dam gene expression in Escherichia coli. Biochim Biophys Acta. 2000;1494:43–53. doi: 10.1016/s0167-4781(00)00209-8. [DOI] [PubMed] [Google Scholar]
- 49.Lobner-Olesen A, Marinus MG, Hansen FG. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc Natl Acad Sci U S A. 2003;100:4672–4677. doi: 10.1073/pnas.0538053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herman GE, Modrich P. Escherichia coli K-12 clones that overproduce dam methylase are hypermutable. J Bacteriol. 1981;145:644–646. doi: 10.1128/jb.145.1.644-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marinus MG, Poteete A, Arraj JA. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene. 1984;28:123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- 52.Radlinska M, Bujnicki JM. Cloning of enterohemorrhagic Escherichia coli phage VT-2 dam methyltransferase. Acta Microbiol Pol. 2001;50:161–167. [PubMed] [Google Scholar]
- 53.Campellone KG, Roe AJ, Lobner-Olesen A, Murphy KC, Magoun L, Brady MJ, Donohue-Rolfe A, Tzipori S, Gally DL, Leong JM, Marinus MG. Increased adherence and actin pedestal formation by dam-deficient enterohemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2007;63:1468–1481. doi: 10.1111/j.1365-2958.2007.05602.x. [DOI] [PubMed] [Google Scholar]
- 54.Bochow S, Elliman J, Owens L. Bacteriophage adenine methyltransferase: a life cycle regulator? Modelled using Vibrio harveyi myovirus like. J Appl Microbiol. 2012;113:1001–1013. doi: 10.1111/j.1365-2672.2012.05358.x. [DOI] [PubMed] [Google Scholar]
- 55.Dar ME, Bhagwat AS. Mechanism of expression of DNA repair gene vsr, an Escherichia coli gene that overlaps the DNA cytosine methylase gene, dcm. Mol Microbiol. 1993;9:823–833. doi: 10.1111/j.1365-2958.1993.tb01741.x. [DOI] [PubMed] [Google Scholar]
- 56.Marczynski GT, Shapiro L. Control of chromosome replication in Caulobacter crescentus. Annu Rev Microbiol. 2002;56:625–656. doi: 10.1146/annurev.micro.56.012302.161103. [DOI] [PubMed] [Google Scholar]
- 57.Liebert K, Hermann A, Schlickenrieder M, Jeltsch A. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J Mol Biol. 2004;341:443–454. doi: 10.1016/j.jmb.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 58.Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J Mol Biol. 2006;358:559–570. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stancheva I, Koller T, Sogo JM. Asymmetry of Dam remethylation on the leading and lagging arms of plasmid replicative intermediates. EMBO J. 1999;18:6542–6551. doi: 10.1093/emboj/18.22.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herman GE, Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982;257:2605–2612. [PubMed] [Google Scholar]
- 61.Bergerat A, Guschlbauer W, Fazakerley GV. Allosteric and catalytic binding of S-adenosylmethionine to Escherichia coli DNA adenine methyltransferase monitored by 3H NMR. Proc Natl Acad Sci U S A. 1991;88:6394–6397. doi: 10.1073/pnas.88.15.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson SN, Reich NO. GATC flanking sequences regulate Dam activity: evidence for how Dam specificity may influence pap expression. J Mol Biol. 2006;355:459–472. doi: 10.1016/j.jmb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 63.Bergerat A, Kriebardis A, Guschlbauer W. Preferential site-specific hemimethylation of GATC sites in pBR322 DNA by Dam methyltransferase from Escherichia coli. J Biol Chem. 1989;264:4064–4070. [PubMed] [Google Scholar]
- 64.Coffin SR, Reich NO. Modulation of Escherichia coli DNA methyltransferase activity by biologically derived GATC-flanking sequences. J Biol Chem. 2008;283:20106–20116. doi: 10.1074/jbc.M802502200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urig S, Gowher H, Hermann A, Beck C, Fatemi M, Humeny A, Jeltsch A. The Escherichia coli dam DNA methyltransferase modifies DNA in a highly processive reaction. J Mol Biol. 2002;319:1085–1096. doi: 10.1016/S0022-2836(02)00371-6. [DOI] [PubMed] [Google Scholar]
- 66.Pollak AJ, Reich NO. Proximal recognition sites facilitate intrasite hopping by DNA adenine methyltransferase: mechanistic exploration of epigenetic gene regulation. J Biol Chem. 2012;287:22873–22881. doi: 10.1074/jbc.M111.332502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boye E, Marinus MG, Lobner-Olesen A. Quantitation of Dam methyltransferase in Escherichia coli. J Bacteriol. 1992;174:1682–1685. doi: 10.1128/jb.174.5.1682-1685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calmann MA, Marinus MG. Regulated expression of the Escherichia coli dam gene. J Bacteriol. 2003;185:5012–5014. doi: 10.1128/JB.185.16.5012-5014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 70.Brezellec P, Hoebeke M, Hiet MS, Pasek S, Ferat JL. DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinformatics. 2006;22:1935–1941. doi: 10.1093/bioinformatics/btl336. [DOI] [PubMed] [Google Scholar]
- 71.Chahar S, Elsawy H, Ragozin S, Jeltsch A. Changing the DNA recognition specificity of the EcoDam DNA-(adenine-N6)-methyltransferase by directed evolution. J Mol Biol. 2010;395:79–88. doi: 10.1016/j.jmb.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 72.Mashhoon N, Pruss C, Carroll M, Johnson PH, Reich NO. Selective inhibitors of bacterial DNA adenine methyltransferases. J Biomol Screen. 2006;11:497–510. doi: 10.1177/1087057106287933. [DOI] [PubMed] [Google Scholar]
- 73.Naumann TA, Tavassoli A, Benkovic SJ. Genetic selection of cyclic peptide Dam methyltransferase inhibitors. Chembiochem. 2008;9:194–197. doi: 10.1002/cbic.200700561. [DOI] [PubMed] [Google Scholar]
- 74.Hobley G, McKelvie JC, Harmer JE, Howe J, Oyston PC, Roach PL. Development of rationally designed DNA N6 adenine methyltransferase inhibitors. Bioorg Med Chem Lett. 2012;22:3079–3082. doi: 10.1016/j.bmcl.2012.03.072. [DOI] [PubMed] [Google Scholar]
- 75.Posfai J, Bhagwat AS, Posfai G, Roberts RJ. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989;17:2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hardy LW, Finer-Moore JS, Montfort WR, Jones MO, Santi DV, Stroud RM. Atomic structure of thymidylate synthase: target for rational drug design. Science. 1987;235:448–455. doi: 10.1126/science.3099389. [DOI] [PubMed] [Google Scholar]
- 77.Hanck T, Schmidt S, Fritz HJ. Sequence-specific and mechanism-based crosslinking of Dcm DNA cytosine-C5 methyltransferase of E. coli K-12 to synthetic oligonucleotides containing 5-fluoro-2′-deoxycytidine. Nucleic Acids Res. 1993;21:303–309. doi: 10.1093/nar/21.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wyszynski MW, Gabbara S, Kubareva EA, Romanova EA, Oretskaya TS, Gromova ES, Shabarova ZA, Bhagwat AS. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 1993;21:295–301. doi: 10.1093/nar/21.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hattman S, Schlagman S, Cousens L. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J Bacteriol. 1973;115:1103–1107. doi: 10.1128/jb.115.3.1103-1107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marinus MG, Morris NR. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973;114:1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhagwat AS, Sohail A, Roberts RJ. Cloning and characterization of the dcm locus of Escherichia coli K-12. J Bacteriol. 1986;166:751–755. doi: 10.1128/jb.166.3.751-755.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 83.Lieb M. Specific mismatch correction in bacteriophage lambda crosses by very short patch repair. Mol Gen Genet. 1983;191:118–125. doi: 10.1007/BF00330898. [DOI] [PubMed] [Google Scholar]
- 84.Lieb M, Rehmat S. 5-Methylcytosine is not a mutation hot spot in nondividing Escherichia coli. Proc Natl Acad Sci U S A. 1997;94:940–945. doi: 10.1073/pnas.94.3.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lieb M, Bhagwat AS. Very short patch repair: reducing the cost of cytosine methylation. Mol Microbiol. 1996;20:467–473. doi: 10.1046/j.1365-2958.1996.5291066.x. [DOI] [PubMed] [Google Scholar]
- 86.Bhagwat AS, Lieb M. Cooperation and competition in mismatch repair: very short-patch repair and methyl-directed mismatch repair in Escherichia coli. Mol Microbiol. 2002;44:1421–1428. doi: 10.1046/j.1365-2958.2002.02989.x. [DOI] [PubMed] [Google Scholar]
- 87.Hennecke F, Kolmar H, Brundl K, Fritz HJ. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991;353:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- 88.Lieb M. Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J Bacteriol. 1987;169:5241–5246. doi: 10.1128/jb.169.11.5241-5246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feng G, Tsui HC, Winkler ME. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J Bacteriol. 1996;178:2388–2396. doi: 10.1128/jb.178.8.2388-2396.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macintyre G, Pitsikas P, Cupples CG. Growth phase-dependent regulation of Vsr endonuclease may contribute to 5-methylcytosine mutational hot spots in Escherichia coli. J Bacteriol. 1999;181:4435–4436. doi: 10.1128/jb.181.14.4435-4436.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mansour CA, Doiron KM, Cupples CG. Characterization of functional interactions among the Escherichia coli mismatch repair proteins using a bacterial two-hybrid assay. Mutat Res. 2001;485:331–338. doi: 10.1016/s0921-8777(01)00071-4. [DOI] [PubMed] [Google Scholar]
- 92.Monastiriakos SK, Doiron KM, Siponen MI, Cupples CG. Functional interactions between the MutL and Vsr proteins of Escherichia coli are dependent on the N-terminus of Vsr. DNA Repair (Amst) 2004;3:639–647. doi: 10.1016/j.dnarep.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 93.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 94.Tsutakawa SE, Muto T, Kawate T, Jingami H, Kunishima N, Ariyoshi M, Kohda D, Nakagawa M, Morikawa K. Crystallographic and functional studies of very short patch repair endonuclease. Mol Cell. 1999;3:621–628. doi: 10.1016/s1097-2765(00)80355-x. [DOI] [PubMed] [Google Scholar]
- 95.Bell DC, Cupples CG. Very-short-patch repair in Escherichia coli requires the dam adenine methylase. J Bacteriol. 2001;183:3631–3635. doi: 10.1128/JB.183.12.3631-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Torreblanca J, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium and a novel dam-regulated locus. Genetics. 1996;144:15–26. doi: 10.1093/genetics/144.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ritchie L, Podger DM, Hall RM. A mutation in the DNA adenine methylase gene (dam) of Salmonella typhimurium decreases susceptibility to 9-aminoacridine-induced frameshift mutagenesis. Mutat Res. 1988;194:131–141. doi: 10.1016/0167-8817(88)90015-6. [DOI] [PubMed] [Google Scholar]
- 98.Parker BO, Marinus MG. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci U S A. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McGraw BR, Marinus MG. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol Gen Genet. 1980;178:309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- 100.Glickman BW, Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980;77:1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Doutriaux MP, Wagner R, Radman M. Mismatch-stimulated killing. Proc Natl Acad Sci U S A. 1986;83:2576–2578. doi: 10.1073/pnas.83.8.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carraway M, Youderian P, Marinus MG. Spontaneous mutations occur near dam recognition sites in a dam− Escherichia coli host. Genetics. 1987;116:343–347. doi: 10.1093/genetics/116.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carraway M, Rewinski C, Wu TH, Marinus MG. Specificity of the Dam-directed mismatch repair system of Escherichia coli K-12. Gene. 1988;74:157–158. doi: 10.1016/0378-1119(88)90275-2. [DOI] [PubMed] [Google Scholar]
- 104.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 105.Wu TH, Clarke CH, Marinus MG. Specificity of Escherichia coli mutD and mutL mutator strains. Gene. 1990;87:1–5. doi: 10.1016/0378-1119(90)90488-d. [DOI] [PubMed] [Google Scholar]
- 106.Wang TC, Smith KC. Inviability of dam recA and dam recB cells of Escherichia coli is correlated with their inability to repair DNA double-strand breaks produced by mismatch repair. J Bacteriol. 1986;165:1023–1025. doi: 10.1128/jb.165.3.1023-1025.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nowosielska A, Marinus MG. Cisplatin induces DNA double-strand break formation in Escherichia coli dam mutants. DNA Repair (Amst) 2005;4:773–781. doi: 10.1016/j.dnarep.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 108.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Molec Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 109.Wyrzykowski J, Volkert MR. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J Bacteriol. 2003;185:1701–1704. doi: 10.1128/JB.185.5.1701-1704.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Robbins-Manke JL, Zdraveski ZZ, Marinus M, Essigmann JM. Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J Bacteriol. 2005;187:7027–7037. doi: 10.1128/JB.187.20.7027-7037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Microbiol. 2004;53:1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- 112.Peterson KR, Mount DW. Analysis of the genetic requirements for viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants. J Bacteriol. 1993;175:7505–7508. doi: 10.1128/jb.175.22.7505-7508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 114.Fujikawa N, Kurumizaka H, Nureki O, Tanaka Y, Yamazoe M, Hiraga S, Yokoyama S. Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein. Nucleic Acids Res. 2004;32:82–92. doi: 10.1093/nar/gkh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guarne A, Zhao Q, Ghirlando R, Yang W. Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex. Nat Struct Biol. 2002;9:839–843. doi: 10.1038/nsb857. [DOI] [PubMed] [Google Scholar]
- 116.Guarne A, Brendler T, Zhao Q, Ghirlando R, Austin S, Yang W. Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization. EMBO J. 2005;24:1502–1511. doi: 10.1038/sj.emboj.7600634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kang S, Han JS, Kim KP, Yang HY, Lee KY, Hong CB, Hwang DS. Dimeric configuration of SeqA protein bound to a pair of hemi-methylated GATC sequences. Nucleic Acids Res. 2005;33:1524–1531. doi: 10.1093/nar/gki289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brendler T, Austin S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 1999;18:2304–2310. doi: 10.1093/emboj/18.8.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Han JS, Kang S, Kim SH, Ko MJ, Hwang DS. Binding of SeqA protein to hemi-methylated GATC sequences enhances their interaction and aggregation properties. J Biol Chem. 2004;279:30236–30243. doi: 10.1074/jbc.M402612200. [DOI] [PubMed] [Google Scholar]
- 120.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 121.Odsbu I, Klungsoyr HK, Fossum S, Skarstad K. Specific N-terminal interactions of the Escherichia coli SeqA protein are required to form multimers that restrain negative supercoils and form foci. Genes Cells. 2005;10:1039–1049. doi: 10.1111/j.1365-2443.2005.00898.x. [DOI] [PubMed] [Google Scholar]
- 122.Kang S, Han JS, Kim SH, Park JH, Hwang DS. Aggregation of SeqA protein requires positively charged amino acids in the hinge region. Biochem Biophys Res Commun. 2007;360:63–69. doi: 10.1016/j.bbrc.2007.05.225. [DOI] [PubMed] [Google Scholar]
- 123.Bach T, Morigen, Skarstad K. The initiator protein DnaA contributes to keeping new origins inactivated by promoting the presence of hemimethylated DNA. J Mol Biol. 2008;384:1076–1085. doi: 10.1016/j.jmb.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 124.Hiraga S, Ichinose C, Niki H, Yamazoe M. Cell cycle-dependent duplication and bidirectional migration of SeqA- associated DNA-protein complexes in E. coli. Mol Cell. 1998;1:381–387. doi: 10.1016/s1097-2765(00)80038-6. [DOI] [PubMed] [Google Scholar]
- 125.Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells. 2000;5:327–341. doi: 10.1046/j.1365-2443.2000.00334.x. [DOI] [PubMed] [Google Scholar]
- 126.Brendler T, Sawitzke J, Sergueev K, Austin S. A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J. 2000;19:6249–6258. doi: 10.1093/emboj/19.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Molina F, Skarstad K. Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol Microbiol. 2004;52:1597–1612. doi: 10.1111/j.1365-2958.2004.04097.x. [DOI] [PubMed] [Google Scholar]
- 128.Skarstad K, Boye E, Steen HB. Timing of initiation of chromosome replication in individual E. coli cells. EMBO J. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yamaki H, Ohtsubo E, Nagai K, Maeda Y. The oriC unwinding by dam methylation in Escherichia coli. Nucleic Acids Res. 1988;16:5067–5073. doi: 10.1093/nar/16.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Løbner-Olesen A, Hansen FG, Rasmussen KV, Martin B, Kuempel PL. The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J. 1994;13:1856–1862. doi: 10.1002/j.1460-2075.1994.tb06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Skarstad K, Lueder G, Lurz R, Speck C, Messer W. The Escherichia coli SeqA protein binds specifically and co-operatively to two sites in hemimethylated and fully methylated oriC. Mol Microbiol. 2000;36:1319–1326. doi: 10.1046/j.1365-2958.2000.01943.x. [DOI] [PubMed] [Google Scholar]
- 132.Wold S, Boye E, Slater S, Kleckner N, Skarstad K. Effects of purified SeqA protein on oriC-dependent DNA replication in vitro. EMBO J. 1998;17:4158–4165. doi: 10.1093/emboj/17.14.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Torheim NK, Skarstad K. Escherichia coli SeqA protein affects DNA topology and inhibits open complex formation at oriC. EMBO J. 1999;18:4882–4888. doi: 10.1093/emboj/18.17.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boye E, Lobner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 135.Bach T, Skarstad K. Re-replication from non-sequesterable origins generates three-nucleoid cells which divide asymmetrically. Molecular Microbiology. 2004;51:1589–1600. doi: 10.1111/j.1365-2958.2003.03943.x. [DOI] [PubMed] [Google Scholar]
- 136.Olsson J, Dasgupta S, Berg OG, Nordstrom K. Eclipse period without sequestration in Escherichia coli. Mol Microbiol. 2002;44:1429–1440. doi: 10.1046/j.1365-2958.2002.02954.x. [DOI] [PubMed] [Google Scholar]
- 137.Morigen, Lobner-Olesen A, Skarstad K. Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol Microbiol. 2003;50:349–362. doi: 10.1046/j.1365-2958.2003.03695.x. [DOI] [PubMed] [Google Scholar]
- 138.Skarstad K, Lobner-Olesen A. Stable co-existence of separate replicons in Escherichia coli is dependent on once-per-cell-cycle initiation. EMBO J. 2003;22:140–150. doi: 10.1093/emboj/cdg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lobner-Olesen A, von Freiesleben U. Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J. 1996;15:5999–6008. [PMC free article] [PubMed] [Google Scholar]
- 140.Dasgupta S, Lobner-Olesen A. Host controlled plasmid replication: Escherichia coli minichromosomes. Plasmid. 2004;52:151–168. doi: 10.1016/j.plasmid.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 141.Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boye E, Lobner-Olesen A, Skarstad K. Limiting DNA replication to once and only once. EMBO Rep. 2000;1:479–483. doi: 10.1093/embo-reports/kvd116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nielsen O, Lobner-Olesen A. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep. 2008;9:151–156. doi: 10.1038/sj.embor.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Samitt CE, Hansen FG, Miller JF, Schaechter M. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989;8:989–993. doi: 10.1002/j.1460-2075.1989.tb03462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem. 2005;280:27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 147.Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ryan VT, Grimwade JE, Nievera CJ, Leonard AC. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol Microbiol. 2002;46:113–124. doi: 10.1046/j.1365-2958.2002.03129.x. [DOI] [PubMed] [Google Scholar]
- 149.Keyamura K, Fujikawa N, Ishida T, Ozaki S, Su’etsugu M, Fujimitsu K, Kagawa W, Yokoyama S, Kurumizaka H, Katayama T. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively Active Complexes of DnaA Protein and the Escherichia coli Chromosomal Origin Observed in the Electron Microscope. J Mol Biol. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 151.Nievera C, Torgue JJ, Grimwade JE, Leonard AC. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol Cell. 2006;24:581–592. doi: 10.1016/j.molcel.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Crooke E, Castuma CE, Kornberg A. The chromosome origin of Escherichia coli stabilizes DnaA protein during rejuvenation by phospholipids. J Biol Chem. 1992;267:16779–16782. [PubMed] [Google Scholar]
- 153.Yung BY, Crooke E, Kornberg A. Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J Biol Chem. 1990;265:1282–1285. [PubMed] [Google Scholar]
- 154.Grimwade JE, Torgue JJ, McGarry KC, Rozgaja T, Enloe ST, Leonard AC. Mutational analysis reveals Escherichia coli oriC interacts with both DnaA-ATP and DnaA-ADP during pre-RC assembly. Mol Microbiol. 2007;66:428–439. doi: 10.1111/j.1365-2958.2007.05930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin. J Biol Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 156.Riber L, Fujimitsu K, Katayama T, Lobner-Olesen A. Loss of Hda activity stimulates replication initiation from I-box, but not R4 mutant origins in Escherichia coli. Mol Microbiol. 2009;71:107–122. doi: 10.1111/j.1365-2958.2008.06516.x. [DOI] [PubMed] [Google Scholar]
- 157.Nishida S, Fujimitsu K, Sekimizu K, Ohmura T, Ueda T, Katayama T. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J Biol Chem. 2002;277:14986–14995. doi: 10.1074/jbc.M108303200. [DOI] [PubMed] [Google Scholar]
- 158.Riber L, Olsson JA, Jensen RB, Skovgaard O, Dasgupta S, Marinus MG, Lobner-Olesen A. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 2006;20:2121–2134. doi: 10.1101/gad.379506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 160.Riber L, Lobner-Olesen A. Coordinated replication and sequestration of oriC and dnaA is required for maintaining controlled once per cell cycle initiation in Escherichia coli. J Bacteriol. 2005 doi: 10.1128/JB.187.16.5605-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hansen FG, Christensen BB, Atlung T. The Initiator titration model: computer simulation of chromosome and minichromosome control. Res Microbiol. 1991;142:161–167. doi: 10.1016/0923-2508(91)90025-6. [DOI] [PubMed] [Google Scholar]
- 162.Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- 164.Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Su’etsugu M, Shimuta TR, Ishida T, Kawakami H, Katayama T. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J Biol Chem. 2005;280:6528–6536. doi: 10.1074/jbc.M412060200. [DOI] [PubMed] [Google Scholar]
- 166.Kasho K, Katayama T. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc Natl Acad Sci U S A. 2013;110:936–941. doi: 10.1073/pnas.1212070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 168.Grimwade JE, Ryan VT, Leonard AC. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol Microbiol. 2000;35:835–844. doi: 10.1046/j.1365-2958.2000.01755.x. [DOI] [PubMed] [Google Scholar]
- 169.Sanchez-Romero MA, Busby SJ, Dyer NP, Ott S, Millard AD, Grainger DC. Dynamic distribution of seqa protein across the chromosome of escherichia coli K-12. MBio. 2010;1 doi: 10.1128/mBio.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Waldminghaus T, Weigel C, Skarstad K. Replication fork movement and methylation govern SeqA binding to the Escherichia coli chromosome. Nucleic Acids Res. 2012;40:5465–5476. doi: 10.1093/nar/gks187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Waldminghaus T, Skarstad K. ChIP on Chip: surprising results are often artifacts. BMC Genomics. 2010;11:414. doi: 10.1186/1471-2164-11-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dame RT, Kalmykowa OJ, Grainger DC. Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in gram negative bacteria. PLoS Genet. 2011;7:e1002123. doi: 10.1371/journal.pgen.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Han JS, Kang S, Lee H, Kim HK, Hwang DS. Sequential binding of SeqA to paired hemi-methylated GATC sequences mediates formation of higher order complexes. J Biol Chem. 2003;278:34983–34989. doi: 10.1074/jbc.M304923200. [DOI] [PubMed] [Google Scholar]
- 174.Klungsoyr HK, Skarstad K. Positive supercoiling is generated in the presence of Escherichia coli SeqA protein. Mol Microbiol. 2004;54:123–131. doi: 10.1111/j.1365-2958.2004.04239.x. [DOI] [PubMed] [Google Scholar]
- 175.Weitao T, Nordström K, Dasgupta S. Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO Rep. 2000;1:494–499. doi: 10.1093/embo-reports/kvd106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.von Freiesleben U, Krekling MA, Hansen FG, Lobner-Olesen A. The eclipse period of Escherichia coli. EMBO J. 2000;19:6240–6248. doi: 10.1093/emboj/19.22.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bach T, Krekling MA, Skarstad K. Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. Embo Journal. 2003;22:315–323. doi: 10.1093/emboj/cdg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: Evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 179.Sawitzke J, Austin S. An analysis of the factory model for chromosome replication and segregation in bacteria. Mol Microbiol. 2001;40:786–794. doi: 10.1046/j.1365-2958.2001.02350.x. [DOI] [PubMed] [Google Scholar]
- 180.Hiraga S. Dynamic localization of bacterial and plasmid chromosomes. Annu Rev Genet. 2000;34:21–59. doi: 10.1146/annurev.genet.34.1.21. [DOI] [PubMed] [Google Scholar]
- 181.Sunako Y, Onogi T, Hiraga S. Sister chromosome cohesion of Escherichia coli. Mol Microbiol. 2001;42:1233–1241. doi: 10.1046/j.1365-2958.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- 182.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Han JS, Kang S, Lee H, Kim HK, Hwang DS. Sequential binding of SeqA to paired hemi-methylated GATC sequences mediates formation of higher order complexes. J Biol Chem. 2003;278:34983–34989. doi: 10.1074/jbc.M304923200. [DOI] [PubMed] [Google Scholar]
- 184.Adachi S, Fukushima T, Hiraga S. Dynamic events of sister chromosomes in the cell cycle of Escherichia coli. Genes Cells. 2008;13:181–197. doi: 10.1111/j.1365-2443.2007.01157.x. [DOI] [PubMed] [Google Scholar]
- 185.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci U S A. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fossum S, Crooke E, Skarstad K. Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli. EMBO J. 2007;26:4514–4522. doi: 10.1038/sj.emboj.7601871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Saint-Dic D, Kehrl J, Frushour B, Kahng LS. Excess SeqA leads to replication arrest and a cell division defect in Vibrio cholerae. J Bacteriol. 2008;190:5870–5878. doi: 10.1128/JB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Demarre G, Chattoraj DK. DNA adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet. 2010;6:e1000939. doi: 10.1371/journal.pgen.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Løbner-Olesen A, von Freiesleben U. Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J. 1996;15:5999–6008. [PMC free article] [PubMed] [Google Scholar]
- 190.Koch B, Ma X, Lobner-Olesen A. Replication of Vibrio cholerae chromosome I in Escherichia coli: dependence on dam methylation. J Bacteriol. 2010;192:3903–3914. doi: 10.1128/JB.00311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Val ME, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet. 2012;8:e1002472. doi: 10.1371/journal.pgen.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Reisenauer A, Kahng LS, McCollum S, Shapiro L. Bacterial DNA methylation: a cell cycle regulator? J Bacteriol. 1999;181:5135–5139. doi: 10.1128/jb.181.17.5135-5139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 194.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci U S A. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci U S A. 2007;104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Collier J. Regulation of chromosomal replication in Caulobacter crescentus. Plasmid. 2012;67:76–87. doi: 10.1016/j.plasmid.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 197.Casadesus J, Torreblanca J. In: Methylation-related epigenetic signals in bacterial DNA. Russo REA, Marteinssen RA, Riggs AD, editors. Cold Spring Harbor Press; Cold Spring harbor NY: 1996. pp. 141–153. [Google Scholar]
- 198.van der Woude MW. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol. 2011;14:205–211. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 199.Wallecha A, Munster V, Correnti J, Chan T, van der Woude M. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J Bacteriol. 2002;184:3338–3347. doi: 10.1128/JB.184.12.3338-3347.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wallecha A, Correnti J, Munster V, van der Woude M. Phase variation of Ag43 is independent of the oxidation state of OxyR. J Bacteriol. 2003;185:2203–2209. doi: 10.1128/JB.185.7.2203-2209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kaminska R, van der Woude MW. Establishing and maintaining sequestration of Dam target sites for phase variation of agn43 in Escherichia coli. J Bacteriol. 2010;192:1937–1945. doi: 10.1128/JB.01629-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Roberts D, Hoopes BC, McClure WR, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- 203.Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 204.Marinus MG. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- 205.Plumbridge J. The role of dam methylation in controlling gene expression. Biochimie. 1987;69:439–443. doi: 10.1016/0300-9084(87)90081-2. [DOI] [PubMed] [Google Scholar]
- 206.Barras F, Marinus MG. The great GATC: DNA methylation in E. coli. Trends Genet. 1989;5:139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 207.Barras F, Magnan M, Marinus MG. Evidence against a role for adenine methylation in the tryptophan biosynthetic pathway in Escherichia coli and for a growth phase-dependent induction of the trp promoter. Current Microbiology. 1991;23:21–25. [Google Scholar]
- 208.Oshima T, Wada C, Kawagoe Y, Ara T, Maeda M, Masuda Y, Hiraga S, Mori H. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol Microbiol. 2002;45:673–695. doi: 10.1046/j.1365-2958.2002.03037.x. [DOI] [PubMed] [Google Scholar]
- 209.Balbontin R, Rowley G, Pucciarelli MG, Lopez-Garrido J, Wormstone Y, Lucchini S, Garcia-del PF, Hinton JC, Casadesus J. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:8160–8168. doi: 10.1128/JB.00847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Riva A, Delorme MO, Chevalier T, Guilhot N, Henaut C, Henaut A. Characterization of the GATC regulatory network in E. coli. BMC Genomics. 2004;5:48. doi: 10.1186/1471-2164-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Braun RE, O’Day K, Wright A. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell. 1985;40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 212.Seshasayee AS. An assessment of the role of DNA adenine methyltransferase on gene expression regulation in E coli. PLoS ONE. 2007;2:e273. doi: 10.1371/journal.pone.0000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sun W, Hattman S. Escherichia coli OxyR protein represses the unmethylated bacteriophage Mu mom operon without blocking binding of the transcriptional activator C. Nucleic Acids Res. 1996;24:4042–4049. doi: 10.1093/nar/24.20.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hattman S. Unusual modification of bacteriophage Mu DNA. J Virol. 1979;32:468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci U S A. 1983;80:7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976;70:17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- 217.Hattman S, Sun W. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind hemimethylated Pmom promoter DNA. Nucleic Acids Res. 1997;25:4385–4388. doi: 10.1093/nar/25.21.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sternberg N, Sauer B, Hoess R, Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986;187:197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- 219.Sternberg N, Coulby J. Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proc Natl Acad Sci U S A. 1990;87:8070–8074. doi: 10.1073/pnas.87.20.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Garcia-del Portillo F, Pucciarelli MG, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc Natl Acad Sci U S A. 1999;96:11578–11583. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 222.Dueger EL, House JK, Heithoff DM, Mahan MJ. Salmonella DNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect Immun. 2001;69:7950–7954. doi: 10.1128/IAI.69.12.7950-7954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dueger EL, House JK, Heithoff DM, Mahan MJ. Salmonella DNA adenine methylase mutants elicit early and late onset protective immune responses in calves. Vaccine. 2003;21:3249–3258. doi: 10.1016/s0264-410x(03)00252-4. [DOI] [PubMed] [Google Scholar]
- 224.Dueger EL, House JK, Heithoff DM, Mahan MJ. Salmonella DNA adenine methylase mutants prevent colonization of newly hatched chickens by homologous and heterologous serovars. Int J Food Microbiol. 2003;80:153–159. doi: 10.1016/s0168-1605(02)00152-6. [DOI] [PubMed] [Google Scholar]
- 225.Mohler VL, Heithoff DM, Mahan MJ, Walker KH, Hornitzky MA, McConnell CS, Shum LW, House JK. Cross-protective immunity in calves conferred by a DNA adenine methylase deficient Salmonellaenterica serovar Typhimurium vaccine. Vaccine. 2006;24:1339–1345. doi: 10.1016/j.vaccine.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 226.Mohler VL, Heithoff DM, Mahan MJ, Walker KH, Hornitzky MA, Shum LW, Makin KJ, House JK. Cross-protective immunity conferred by a DNA adenine methylase deficient Salmonella enterica serovar Typhimurium vaccine in calves challenged with Salmonella serovar Newport. Vaccine. 2008;26:1751–1758. doi: 10.1016/j.vaccine.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 227.Sarnacki SH, Marolda CL, Noto LM, Giacomodonato MN, Valvano MA, Cerquetti MC. Dam methylation controls O-antigen chain length in Salmonella enterica serovar enteritidis by regulating the expression of Wzz protein. J Bacteriol. 2009;191:6694–6700. doi: 10.1128/JB.00839-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Giacomodonato MN, Sarnacki SH, Llana MN, Garcia Cattaneo AS, Uzzau S, Rubino S, Cerquetti MC. Impaired synthesis and secretion of SopA in Salmonella Typhimurium dam mutants. FEMS Microbiol Lett. 2009;292:71–77. doi: 10.1111/j.1574-6968.2008.01473.x. [DOI] [PubMed] [Google Scholar]
- 229.Heusipp G, Falker S, Schmidt MA. DNA adenine methylation and bacterial pathogenesis. Int J Med Microbiol. 2007;297:1–7. doi: 10.1016/j.ijmm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 230.Prieto AI, Ramos-Morales F, Casadesus J. Bile-induced DNA damage in Salmonella enterica. Genetics. 2004;168:1787–1794. doi: 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Prieto AI, Ramos-Morales F, Casadesus J. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics. 2006;174:575–584. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Prieto AI, Jakomin M, Segura I, Pucciarelli MG, Ramos-Morales F, Garcia-del PF, Casadesus J. The GATC-binding protein SeqA is required for bile resistance and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:8496–8502. doi: 10.1128/JB.01156-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Falker S, Schmidt MA, Heusipp G. Altered Ca(2+) regulation of Yop secretion in Yersinia enterocolitica after DNA adenine methyltransferase overproduction is mediated by Clp-dependent degradation of LcrG. J Bacteriol. 2006;188:7072–7081. doi: 10.1128/JB.00583-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sun K, Jiao XD, Zhang M, Sun L. DNA adenine methylase is involved in the pathogenesis of Edwardsiella tarda. Vet Microbiol. 2010;141:149–154. doi: 10.1016/j.vetmic.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 236.Banas JA, Biswas S, Zhu M. Effects of DNA methylation on expression of virulence genes in Streptococcus mutans. Appl Environ Microbiol. 2011;77:7236–7242. doi: 10.1128/AEM.00543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Brunet YR, Bernard CS, Gavioli M, Lloubes R, Cascales E. An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 2011;7:e1002205. doi: 10.1371/journal.pgen.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Camacho EM, Casadesus J. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol Microbiol. 2002;44:1589–1598. doi: 10.1046/j.1365-2958.2002.02981.x. [DOI] [PubMed] [Google Scholar]
- 239.Camacho EM, Serna A, Madrid C, Marques S, Fernandez R, de la CF, Juarez A, Casadesus J. Regulation of finP transcription by DNA adenine methylation in the virulence plasmid of Salmonella enterica. J Bacteriol. 2005;187:5691–5699. doi: 10.1128/JB.187.16.5691-5699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Camacho EM, Casadesus J. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol Microbiol. 2005;57:1700–1718. doi: 10.1111/j.1365-2958.2005.04788.x. [DOI] [PubMed] [Google Scholar]
- 241.Murphy KC, Ritchie JM, Waldor MK, Lobner-Olesen A, Marinus MG. Dam methyltransferase is required for stable lysogeny of the Shiga toxin (Stx2)-encoding bacteriophage 933W of enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2008;190:438–441. doi: 10.1128/JB.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Alonso A, Pucciarelli MG, Figueroa-Bossi N, Garcia-del Portillo F. Increased excision of the Salmonella prophage ST64B caused by a deficiency in Dam methylase. J Bacteriol. 2005;187:7901–7911. doi: 10.1128/JB.187.23.7901-7911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hayward RD, Leong JM, Koronakis V, Campellone KG. Exploiting pathogenic Escherichia coli to model transmembrane receptor signalling. Nat Rev Microbiol. 2006;4:358–370. doi: 10.1038/nrmicro1391. [DOI] [PubMed] [Google Scholar]
- 244.Falker S, Schilling J, Schmidt MA, Heusipp G. Overproduction of DNA adenine methyltransferase alters motility, invasion, and the lipopolysaccharide O-antigen composition of Yersinia enterocolitica. Infect Immun. 2007;75:4990–4997. doi: 10.1128/IAI.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Albu RF, Jurkowski TP, Jeltsch A. The Caulobacter crescentus DNA-(adenine-N6)-methyltransferase CcrM methylates DNA in a distributive manner. Nucleic Acids Res. 2012;40:1708–1716. doi: 10.1093/nar/gkr768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Collier J. Epigenetic regulation of the bacterial cell cycle. Curr Opin Microbiol. 2009;12:722–729. doi: 10.1016/j.mib.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 247.Rasmussen LJ, Marinus MG. Use of DNA methylation deficient strains in molecular genetics. In: Adolph KW, editor. Microbial Gene Techniques. Academic Press; San Diego: 1995. pp. 267–279. [Google Scholar]
- 248.Rao BS, Buckler-White A. Direct visualization of site-specific and strand-specific DNA methylation patterns in automated DNA sequencing data. Nucleic Acids Res. 1998;26:2505–2507. doi: 10.1093/nar/26.10.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bart A, van Passel MW, van AK, van der EA. Direct detection of methylation in genomic DNA. Nucleic Acids Res. 2005;33:e124. doi: 10.1093/nar/gni121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.MacNeil DJ. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J Bacteriol. 1988;170:5607–5612. doi: 10.1128/jb.170.12.5607-5612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Troester H, Bub S, Hunziker A, Trendelenburg MF. Stability of DNA repeats in Escherichia coli dam mutant strains indicates a Dam methylation-dependent DNA deletion process. Gene. 2000;258:95–108. doi: 10.1016/s0378-1119(00)00420-0. [DOI] [PubMed] [Google Scholar]
- 252.Poteete AR. Expansion of a chromosomal repeat in Escherichia coli: roles of replication, repair, and recombination functions. BMC Mol Biol. 2009;10:14. doi: 10.1186/1471-2199-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zoller MJ, Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982;10:6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Craig RJ, Arraj JA, Marinus MG. Induction of damage inducible (SOS) repair in dam mutants of Escherichia coli exposed to 2-aminopurine. Mol Gen Genet. 1984;194:539–540. doi: 10.1007/BF00425572. [DOI] [PubMed] [Google Scholar]
- 255.Quillardet P, Hofnung M. Induction of the SOS system in a dam-3 mutant: a diagnostic strain for chemicals causing DNA mismatches. Mutat Res. 1987;177:17–26. doi: 10.1016/0027-5107(87)90017-0. [DOI] [PubMed] [Google Scholar]
- 256.Singh J, Klar AJ. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 257.van SB, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 258.Xiao R, Roman-Sanchez R, Moore DD. DamIP: a novel method to identify DNA binding sites in vivo. Nucl Recept Signal. 2010;8:e003. doi: 10.1621/nrs.08003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Marinus MG, Konrad EB. Hyper-recombination in dam mutants of Escherichia coli K-12. Mol Gen Genet. 1976;149:273–277. doi: 10.1007/BF00268528. [DOI] [PubMed] [Google Scholar]
- 260.Marinus MG, Carraway M, Frey AZ, Brown L, Arraj JA. Insertion mutations in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;192:288–289. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- 261.Parker B, Marinus MG. A simple and rapid method to obtain substitution mutations in Escherichia coli: isolation of a dam deletion/insertion mutation. Gene. 1988;73:531–535. doi: 10.1016/0378-1119(88)90517-3. [DOI] [PubMed] [Google Scholar]
- 262.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.O’Reilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol. 2004;186:7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Efimova EP, Delver EP, Belogurov AA. Alleviation of type I restriction in adenine methylase (dam) mutants of Escherichia coli. Mol Gen Genet. 1988;214:313–316. doi: 10.1007/BF00337727. [DOI] [PubMed] [Google Scholar]
- 265.Karran P, Marinus MG. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 266.Yallaly P, Eisenstark A. Influence of DNA adenine methylase on the sensitivity of Escherichia coli to near-ultraviolet radiation and hydrogen peroxide. Biochem Biophys Res Commun. 1990;169:64–69. doi: 10.1016/0006-291x(90)91433-s. [DOI] [PubMed] [Google Scholar]
- 267.Sutera VA, Jr, Lovett ST. The role of replication initiation control in promoting survival of replication fork damage. Mol Microbiol. 2006;60:229–239. doi: 10.1111/j.1365-2958.2006.05093.x. [DOI] [PubMed] [Google Scholar]
- 268.Peterson KR, Wertman KF, Mount DW, Marinus MG. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol Gen Genet. 1985;201:14–19. doi: 10.1007/BF00397979. [DOI] [PubMed] [Google Scholar]
- 269.Lundblad V, Kleckner N. Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. Genetics. 1985;109:3–19. doi: 10.1093/genetics/109.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Messer W, Bellekes U, Lother H. Effect of dam methylation on the activity of the E. coli replication origin, oriC. EMBO J. 1985;4:1327–1332. doi: 10.1002/j.1460-2075.1985.tb03780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Smith DW, Garland AM, Herman G, Enns RE, Baker TA, Zyskind JW. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J. 1985;4:1319–1326. doi: 10.1002/j.1460-2075.1985.tb03779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Abeles A, Brendler T, Austin S. Evidence of two levels of control of P1 oriR and host oriC replication origins by DNA adenine methylation. J Bacteriol. 1993;175:7801–7807. doi: 10.1128/jb.175.24.7801-7807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Gammie AE, Crosa JH. Roles of DNA adenine methylation in controlling replication of the REPI replicon of plasmid pColV-K30. Mol Microbiol. 1991;5:495–503. doi: 10.1111/j.1365-2958.1991.tb02133.x. [DOI] [PubMed] [Google Scholar]
- 274.Onogi T, Yamazoe M, Ichinose C, Niki H, Hiraga S. Null mutation of the dam or seqA gene suppresses temperature-sensitive lethality but not hypersensitivity to novobiocin of muk null mutants. J Bacteriol. 2000;182:5898–5901. doi: 10.1128/jb.182.20.5898-5901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hernday AD, Braaten BA, Low DA. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol Cell. 2003;12:947–957. doi: 10.1016/s1097-2765(03)00383-6. [DOI] [PubMed] [Google Scholar]
- 276.Correnti J, Munster V, Chan T, van der Woude M. Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol Microbiol. 2002;44:521–532. doi: 10.1046/j.1365-2958.2002.02918.x. [DOI] [PubMed] [Google Scholar]