Abstract
The northern region of the Indian subcontinent is a vast landscape interlaced by diverse ecologies, for example, the Gangetic Plain and the Himalayas. A great number of ethnic groups are found there, displaying a multitude of languages and cultures. The Tharu is one of the largest and most linguistically diverse of such groups, scattered across the Tarai region of Nepal and bordering Indian states. Their origins are uncertain. Hypotheses have been advanced postulating shared ancestry with Austroasiatic, or Tibeto-Burman-speaking populations as well as aboriginal roots in the Tarai. Several Tharu groups speak a variety of Indo-Aryan languages, but have traditionally been described by ethnographers as representing East Asian phenotype. Their ancestry and intra-population diversity has previously been tested only for haploid (mitochondrial DNA and Y-chromosome) markers in a small portion of the population. This study presents the first systematic genetic survey of the Tharu from both Nepal and two Indian states of Uttarakhand and Uttar Pradesh, using genome-wide SNPs and haploid markers. We show that the Tharu have dual genetic ancestry as up to one-half of their gene pool is of East Asian origin. Within the South Asian proportion of the Tharu genetic ancestry, we see vestiges of their common origin in the north of the South Asian Subcontinent manifested by mitochondrial DNA haplogroup M43.
INTRODUCTION
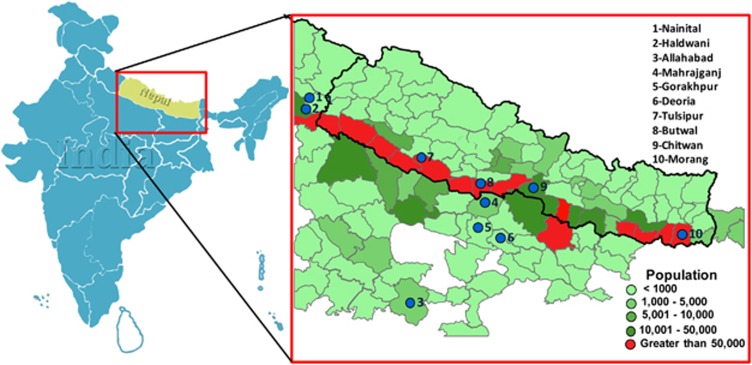
The Tarai (or Terai) is a region stretching between the northernmost reaches of the Gangetic Plain and the southern foothills of the Greater Himalayas, an elongated area that straddles southern Nepal and northern India. This region is ecologically diverse, hosts a variety of cultures and languages, and has historically been fragmented into numerous local polities.1 Tharu is an ‘umbrella-ethnonym', uniting a variety of historically endogamous groups that speak different dialects and are also distinct in many socio-cultural aspects. The Tharu live in the Tarai and surrounding areas of Nepal and are also found in Indian states of Uttar Pradesh and Uttarakhand (Figure 1), where they are recorded as scheduled tribes.
Figure 1.
The geographical distribution of the Tharu in South Asia. The sampling locations are shown by circles and districtwise Tharu population coverage is colour coded.
The Tharu were described by ethnographers in the nineteenth century as representing either an East Asian phenotype or a somatological amalgam of ‘Indo-Chinese' or ‘Mongoloid racial stock' and a ‘negrito' aboriginal substrate.2, 3 The Tharu speak a number of related Indo-Aryan dialects collectively known as Thāruvānī.4 The Linguistic Survey of India has described the peculiarities that distinguish Tharu speech from that of their Indo-Aryan neighbours as ‘a few ignorant corruptions'.5 After this observation, made by Grierson at the beginning of the twentieth century, no systematic linguistic investigation of any of the Tharu dialects has been undertaken thus far. In fact, the Tharu forms of speech are going extinct without being documented and the linguistic reality is far more complex. There is no single Thāruvānī language. Rather, the Tharu speak their own local varieties of the Indo-Aryan languages that are widespread in the Tarai belt, that is, Awadhi, Bhojpuri and Maithili, and are distinct from the mainstream form of each of these languages. Moreover, most Tharu live in a state of diglossia, with one form of the language being used in public domains – such as the marketplace and in interactions with their Indo-Aryan neighbours, and another form of the language confined to their own hearth and the family circle. The more intimate varieties of Tharu speech are not only grammatically distinct, but also retain non-Indo-Aryan lexical items, some of which appear to be of Tibeto-Burman origin, for example, intimate household Maithili Tharu tẽ ‘eye' vs general marketplace Maithili Tharu yākhī ‘eye'.6 These links of Thāruvānī with Tibeto-Burman languages complement the findings of genetic signals prevalent among contemporary Tibeto-Burman speakers among the Tharu.7, 8, 9
‘eye' vs general marketplace Maithili Tharu yākhī ‘eye'.6 These links of Thāruvānī with Tibeto-Burman languages complement the findings of genetic signals prevalent among contemporary Tibeto-Burman speakers among the Tharu.7, 8, 9
Despite their linguistic heterogeneity, the Tharu are strongly associated with the ecological region of the Tarai and claim a shared pan-Tharu ethnic identity10 – a view supported by the results of some genetic studies.8, 11 Resistance to malaria is widespread among the Tharu.11, 12 This may support an origin for the Tharu within the Tarai jungles as prolonged exposure to swampy, that is, malaria-rich environments which could have led to natural selection favouring resistance.12, 13 Classical and mitochondrial DNA (mtDNA) RFLP markers have corroborated the presence of an East Asian genetic component in the Tharu.7, 8, 9 Yet a recent study of mtDNA and Y-chromosomal markers found that the genetic diversity of the Tharu, sampled from a couple of districts in Nepal has been shaped also by Indian and West Eurasian sources.14 Furthermore, the East/Southeast Asian component was shown to exhibit diverse maternal ancestry and mtDNA haplogroup (hg) sharing with Malaysians, Japanese and Andaman Islanders.
On the basis of the available cultural and genetic information on the Tharu, we tested two major models that would explain the elevated genetic diversity patterns of this group: (1) admixture of population(s) with East/Southeast Asian ancestry and a subsequent expansion of the Tharu from the north of the subcontinent (possibly from the Tarai region), followed by extensive admixture with local populations (ie, pan-Tharu ancestry),1, 8, 10, 11, 15 and (2) assimilation of different tribal groups with notable genetic ancestry in East/Southeast Asia, into a single ethnolinguistic entity called Tharu – somewhat similar to the mixed ancestry observed in the Harijans.1, 16, 17 As both models predict high level of genetic sharing between the Tharu groups and their local neighbours and a substantial East/Southeast Asian component in the gene pool of the Tharu, the main question to be addressed is – Do we see any exclusively common South Asian-specific genetic patterns uniting the different Tharu groups?
Haploid genetic systems have four times smaller effective population size than autosomal markers and are thus particularly prone to genetic drift. Therefore, to test the two proposed models, this study provides a broader framework for reconstructing the genetic history of the Tharu with autosomal coupled with high-resolution mtDNA and Y-chromosomal markers. We estimate the proportion of different ancestry components in the Tharu in context of other Eurasian populations. To assess the intra and inter-group relationships of various Tharu groups with their neighbours, we analysed two Tharu groups from Uttar Pradesh and Uttarakhand states in northern India for thousands of autosomal markers across the whole genome published elsewhere18, 19 and ten groups of the Tharu from Nepal and northern India for mtDNA and Y-chromosomal markers. Additionally, we have genotyped hair thickness and mammary gland morphology associated c.1540T>C polymorphism in the EDAR gene (EDARV370A), which has been estimated to have arisen in central China ∼30 000 years ago.20 In India, it occurs specifically in populations of East or Southeast Asian ancestry.21
MATERIALS AND METHODS
Sampling
About 2–5 ml blood samples obtained with informed written consent were collected from 240 Tharu individuals from 8 different districts of India and Nepal (Figure 1). During the data collection donors were selected through interviews to carefully choose blood unrelated individuals (at least for the past three generations). This project was approved by the Institutional Ethical Committee of the Estonian Biocentre (Tartu, Estonia) and CSIR-Centre for Cellular and Molecular Biology (Hyderabad, India).
Genotyping
The hypervariable segment I (HVS-I) of mtDNA and haplotype determination were scored against the Reconstructed Sapiens Reference Sequence (RSRS).22 Hgs were assigned based on HVS-I variations and they were further confirmed by genotyping the coding regions mutations (Supplementary Table 8), published till date in PhyloTree (www.phylotree.org.).
Over 35 Y-chromosomal biallelic markers were used in this study for assigning the hg to each individual. In total, 86 M134, M17 and M82 derived samples were typed and analysed for 17 loci Y-STRs by using the AmpFℓSTR Y-filer PCR amplification Kit (Applied Biosystems, Grand Island, NY, USA) following the manufacturer protocol. The alleles were noted for all except changes in the two loci where ‘DYS389I' was used as ‘DYS389cd' and ‘DY389ab'=(DYS389II-DYS389I). Out of 17 loci obtained, two DYS385 loci were excluded from the current analyses because they could not be distinguished using the typing method employed. Thus, all the analysis linked with Y-STR data were carried out with 15 loci.
mtDNA and Y-chromosome data analysis
Principal component analysis (PCA) was run using popstr (http://harpending.humanevo.utah.edu/popstr/) to infer population relationships based on hg frequencies (Supplementary Tables 5 and 6). We limited the geographical coverage to the regions surrounding the Tharu and removed outliers for a zoomed genetic landscape. Comparative data sets were taken from the literature (Supplementary Tables 5 and 6). We completed mtDNA sequences of a novel branch of M43 hg to reconstruct the hg phylogeny. Coalescence time for M43 was calculated by the rho statistic according to the mutation-rate estimation of Soares et al.23 Hg M43 isofrequency maps were generated by using Surfer 8 of Golden Software (Golden Software Inc., Golden, CO, USA), following the Kriging procedure.
A median-joining network, resolved with the MP algorithm, was constructed for three major hgs (H1a1a-M82, O3a2c1-M134 and R1a-M17) using the Network package (version 4.6) (www.fluxus-engineering.com); one Steiner tree is shown in Supplementary Figure 6. Age was estimated from microsatellite variation within each hg using the method described by Zhivotovsky et al24 and updated in Sengupta et al.25
Autosomal data analysis
The study was performed using control samples collected, genotyped and published for various population studies (Supplementary Table 1). We used different numbers of populations and data sets for various analyses to obtain the maximum resolution: for PCA and ADMIXTURE, we used two different types of data sets containing 4507 and 98 189 common SNPs (Data 1–3). The wider coverage of Pan Asian data set was required to differentiate Tibeto-Burman, Austronesian and Austroasiatic components, which was possible only with the 4507 number of common SNPs by including both the Tharu groups (Supplementary Table 1). A check for closely related individuals was carried out within each population study by calculating average identity by state (IBS) scores for all pairs of individuals.26
We used PLINK 1.0726 to filter our data set to include only SNPs on the 22 autosomal chromosomes with minor allele frequency >1% and genotyping success >99%. Because background linkage disequilibrium (LD) can affect both PCA27 and structure-like analyses,28 we thinned the data set by removing one SNP of any pair in strong LD r2>0.4 in a window of 200 SNPs (sliding the window by 25 SNPs at a time).
We carried out PC analysis using smartpca programme (with default settings) of the EIGENSOFT package27 to capture genetic variability described by the first five PCs. The fraction of the total variation described by a PC is the ratio of its eigenvalue to the sum of all eigenvalues. In the final settings, we ran ADMIXTURE with a random seed number generator on the LD pruned data set 25 times at K=2 to K=12. Because the top values of the resulting log-likelihood scores were stable (virtually identical) within the runs of each K from K=2 to K=9, we can with some confidence argue, that convergence at global maximum was reached. Thus, we omitted runs at K=10 to K=12 from further analysis.
Mean pairwise Fst differences between different population groups were calculated using the methods of Cockerham and Weir29 and method explained in Bhatia et al.30 Both the methods yielded same result for all the populations. The average heterozygosity values were calculated on the basis of ∼4500 SNPs for different population groups. We estimated the amount of East Asian ancestry among the Tharu and their neighbour Indo-European populations using the f4 ancestry ratio test.18 We used Japan and CEU as an outgroup and calculated the Han ancestry among the Tharu and others; East Asian ancestry=f4 (Japan,Kurumba;Tharu,CEU)/f4 (Japan,Kurumba;Han,CEU). The EDAR c.1540T>C, a non-synonymous SNP in exon 12 was genotyped among different Tharu groups by PCR-direct sequencing protocol described elsewhere.21 The allele frequency of EDAR was estimated by gene counting.
For haplotype-based analysis (fineSTRUCTURE),31 we merged two data sets,18, 19 which yielded 11 695 common SNPs (Supplementary Table 1 – Data 4).18, 19 Samples were phased with Beagle 3.3.2.32 A coancestry matrix was constructed using ChromoPainter31 with the default settings. FineSTRUCTURE was used to perform an MCMC iteration on the coancestry matrix generated by ChromoPainter using 10 000 000 burning and runtime and 10 000 MCMC samples. A tree was built using fineSTRUCTURE with default settings. All these information are plotted in fineSTRUCTURE Gui supplied by the developer. Additionally, we have also calculated mean chunk sharing of each of the Indian groups.
RESULTS AND DISCUSSION
Genetic analysis of genome-wide variation in the Tharu
For the autosomal analyses, we combined published data on the Tharu sampled from two Indian states (Uttar Pradesh and Uttarakhand).18, 33 We used different combinations of data sets and analysed the Tharu in the context of a wide range of Eurasian populations (Supplementary Table 1). The comparison of mean pairwise population Fst values suggested that the Uttar Pradesh Tharu and Uttarakhand Tharu are distinct from each other and cluster more closely with their immediate geographic neighbours. To put these results into a perspective, the Fst differentiation between Uttar Pradesh Tharu with Uttarakhand Tharu (0.027) was, for example, twice as high as that between Pakistani populations and Indian Dravidians (0.013) (Figure 2a; Supplementary Table 2). Therefore, for all group-level analyses we combined the Tharu samples as Tharu 1 for Uttar Pradesh Tharu and Tharu 2 for Uttarakhand Tharu. Both the Tharu groups show similar levels of average genome-wide heterozygosity (Supplementary Figure 1).
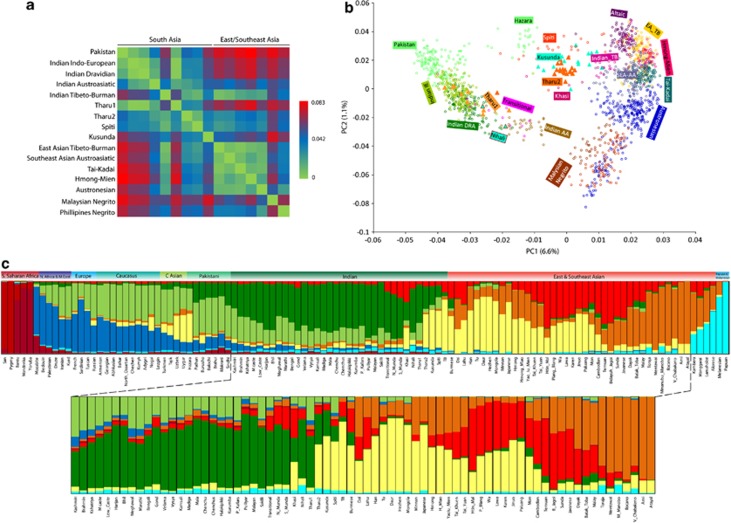
Figure 2.
(a) Mean pairwise Fst comparison of the Tharu from various South and East/Southeast Asian language and ethnic groups obtained from the autosomal SNP data. (b) Principal component analysis (PCA) of the combined autosomal genotypic data of individuals from South and East/Southeast Asia; (c) Results of the populationwise ADMIXTURE analysis (K=9) of world population projecting South and East/Southeast Asian populations. AA, Austroasiatic; C Asian, Central Asian; DRA, Dravidian; IE, Indo-European; TB, Tibeto-Burman.
The first principal component in the PCA27 (Figure 2b) differentiates populations on the East to West axis with Southeast and East Asians (from Altaic to Malaysian Negrito) being at one side and most, but not all South Asians (from Indo-European to Dravidian and to Austroasiatic – SA1) at the opposite. The second PC captures the genetic variation on the North to South axis. A distinctive group of South Asian populations (Hazara, Spiti, Kusunda, Khasi, Indian Tibeto-Burman – SA2) stands intermediate on the East-West axis of PC1 and follow the general North to South cline along PC2. This third grouping becomes critical for interpreting the diversity of the Tharu populations, since one of the groups analysed here (Tharu 1) clearly overlaps within SA1, while the other (Tharu 2) forms its own small cluster at the heart of the second South Asian population cluster (SA2).
To reveal detailed individual-level genetic structure in our sample, we inferred each sample's genetic ancestry proportion in constructed genetic clusters often referred to as ‘constructed ancestral populations'. We used the model-based population structure analysis tool implemented in the ADMIXTURE program.28 In agreement with previous studies,18, 33 the populationwise ADMIXTURE plot showed two major components among the Indian populations (Figure 2b). The relative proportions of these components among the Tharu vary and are similar to their neighbouring populations (Figure 2b; Supplementary Figures 2 and 3). Strikingly, the two Tharu groups differ substantially in the membership to the genetic ancestry component prevalent in East Asia. In fact, this East Asian component is the dominant one among the Uttarakhand Tharu (Tharu 2) while among the Tharu from Uttar Pradesh (Tharu 1) membership to this cluster remains at the background level, quite like among a wide range of other Indian populations. Sharing of East and Southeast Asian components in South Asians can be attributed to a recent admixture (eg, Austroasiatic and Tibeto-Burman expansions) or to shared ancestry predating the split of ancestral East and South Asian populations. Such a deep sharing should be rather uniform between and within extant populations in the region. Thus, clear outliers like Tharu 2, Kusunda, Spiti, Khasi and Indian Munda speakers likely represent more recent admixture.
To quantify the East Asian admixture, we performed the f4 ancestry ratio statistic test18 among both the Tharu groups and their neighbouring Indo-European populations. The results showed that the two Tharu groups differ significantly in the percentage of East Asian admixture (Table 1). The estimates of East Asian admixture were significant only in case of Tharu 2 and suggested that approximately half of their genome is derived from a different source than Tharu 1, who showed low but insignificant presence of the East Asian ancestry (Z score <2). Thus, irrespective of the potential existence of a common genetic origin or ‘homeland' for different Tharu groups in South Asia, it is clear that for at least some of them substantial proportions of their genomes share ancestry with populations of East and Southeast Asia.
Table 1. The proportion of East Asian ancestry among the Tharu and their neighbours.
| Population | East Asian ancestry (%) | Z score |
|---|---|---|
| Tharu 1 | 3.73 | 1.45684 |
| Tharu 2 | 52.9 | 14.6152 |
| Uttar Pradesh Scheduled caste | −2.06 | −0.71172 |
| Vaish | −1.06 | −0.45358 |
To identify whether the two Tharu groups have any uniquely shared ancestry components, we used ChromoPainter and performed fineSTRUCTURE analysis.31 In the present analysis, all the individuals form a total of 25 clusters in the tree (Supplementary Table 3). Individuals from Tharu 1 were scattered in four clusters that they shared with Indian Indo-European, Dravidian and Indian Austroasiatic speakers. Tharu 2 individuals cluster predominantly in their own single cluster, with the closest population being Spiti in the fineSTRUCTURE tree (Supplementary Figure 4). Importantly, in higher level clustering Tharu 2 and the Spiti falls within the East and Southeast Asian cluster together with, among others, the Tibeto-Burman speakers of northeastern India. The patterns of mean chunk sharing of various South vs East and Southeast Asian ancestry components for Indian Indo-European, Dravidian, Austroasiatic and Tibeto-Burmans are consistent with the PCA and ADMIXTURE analysis (Figures 2b, c and 3a).
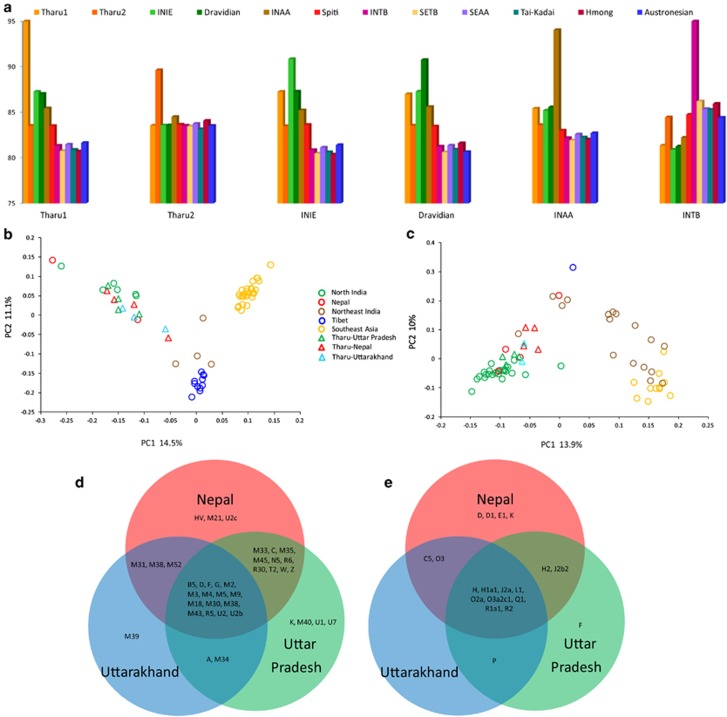
Figure 3.
(a) The mean sharing of various chunk lengths among different ethnolinguistic South and East/Southeast Asian groups. Principal component analysis (PCA) based on mtDNA (b) and Y-chromosome (c) relative frequencies of haplogroups within populations. The mtDNA (d) and Y-chromosomal (e) haplogroup sharing among the different Tharu groups. INAA, Indian Austroasiatic; INIE, Indian Indo-European; INTB, Indian Tibeto-Burman; SEAA, Southeast Asian Austroasiatic; SETB, Southeast Asian Tibeto-Burman.
The mean chunk sharing of Tharu 1 and Tharu 2 showed that the haplotype chunks donated from the other groups of populations differ greatly (Figure 3a). Tharu 1 contains largely similar chunks as their neighbouring Indo-European and Dravidic speakers, whereas Tharu 2 receives longer chunks from East and Southeast Asian populations. These results are suggestive of relatively massive and recent admixture of Tharu 2 with population(s) of the East Asian gene pool. Here, the clustering of Tharu 2 with the Spiti population in sharing common haplotype tracks in fineSTRUCTURE analysis is a suggestive smoking gun for mediators of such an admixture. Furthermore, it appears from these results that the two Tharu groups do not share with each other more chunks than they share with their own geographic neighbours. Together, these results support the second hypothesis postulated above, suggesting that at least the two different Tharu groups analysed here may share little, if any, exclusively common genetic ancestry and they may show geographically very different origins. However, care must be taken in extending these results to the Tharu as such. Only two Tharu groups could be included to the current analysis of genetic diversity on the autosomal level. Furthermore, one of these (Tharu 1) was sampled from the Allahabad district, which was the westernmost sampling point of the Tarai Tharu groups and consequently seems to represent one extreme in the distribution of the East vs South Asian ancestry proportions in the Tharu. However, relative to their Uttar Pradesh Indo-Aryan-speaking neighbours, three samples within Tharu 1 showed elevated (8–18%) membership to the East Asian-specific genetic clusters (Supplementary Table 4). Thus, further studies need to address this issue using a more comprehensive set of the Tharu groups.
Patterns of mtDNA and Y-chromosome structure in the Tharu
A PCA of mtDNA and Y-chromosomal markers segregates South Asia from Southeast Asia along the first PC (Figures 3b and c). The northeast Indian groups and Tibetans are positioned largely in between the two, while the differentiation of the Tibetans from all other groups defines the second PC in both mtDNA and Y-chromosome PCA plots. The Tharu groups are generally positioned close to the north Indian populations while being shifted slightly towards Southeast Asian and Tibetan groups. The latter is consistent with previous observations showing that to a large extent the East Asian-specific genetic component among the Nepali and the Tharu groups has entered the region from the north over the Himalayas.14, 34, 35
Consistent with the autosomal analysis, we found that the Tharu groups differ substantially in the overall proportions of East and South Asian-specific hgs in both haploid loci. However, it is clear that notable part (15–50%) of the maternal and paternal lineages among the Tharu trace back to East Asian populations (Table 2).
Table 2. mtDNA and Y-chromosomal haplogroup profiles in various geographical and ethnic groups.
| mtDNA | Y-chromosome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | South Asian | E/SE Asian | West Eurasian | Unresolved | n | South Asian | E/SE Asian | West Eurasian | Unresolved | |
| Nepal others | 270 | 53.43 | 33.21 | 9.75 | 3.61 | 188 | 23.96 | 38.83 | 10.61 | 26.6 |
| Uttarakhand others | 123 | 51.39 | 32.82 | 11.46 | 4.33 | 127 | 39.32 | 5.57 | 15.79 | 39.32 |
| Uttar Pradesh others | 144 | 74.31 | 0 | 3.47 | 22.22 | 561 | 57.39 | 2.32 | 10.7 | 29.59 |
| Tharu-Nepal | 204 | 44.12 | 51.47 | 2.45 | 1.96 | 202 | 34.65 | 40.59 | 13.37 | 11.39 |
| Tharu-UT | 45 | 48.89 | 42.22 | 0 | 8.89 | 45 | 37.78 | 26.67 | 8.89 | 26.66 |
| Tharu-UP | 164 | 65.85 | 25 | 6.71 | 2.44 | 164 | 42.68 | 14.63 | 15.86 | 26.83 |
Abbreviations: n, number of samples; UP, Uttar Pradesh; UT, Uttarakhand. South Asian haplogroups:- mtDNA: M2-6, M33-65, R5-8, R31-32; Y-chromosome: C5, F, H, L and R2; E/SE-Asian haplogroups:- mtDNA: A-G, M7-12, R22, N9; Y-chromosome: C2, C3, D and M-O; unresolved haplogroups: M*, R*, N* including other lineages, eg, M31 and West Eurasian specific; Y-chromosome: C*, G, I-K*, P*, Q and R1.
To investigate further whether the Tharu groups have a common genetic heritage, we specifically looked for inter-population and intra-population hg sharing between the Tharu and their local neighbours (Figures 3d and e; Table 2; Supplementary Tables 5 and 6). In contrast to the autosomal results, which were based on a limited number of samples and the Tharu groups, we find a high level of common hg sharing between different Tharu groups (Figures 3d and e). Notably, the hg shared by different Tharu groups is found among both East and Southeast Asian-specific hgs (mtDNA: B5, D, F, G and M9; Y-chromosome: O3a2c1) shows a cline from east to west only among the Tharu groups and is virtually absent among local Uttar Pradesh populations (Table 2; Supplementary Tables 5 and 6), thus reflecting at least some degree of common ancestry among the Tharu (Tables 2 and 3; Supplementary Tables 5 and 6). The diversity of East Asian-specific hgs among the Tharu and other Nepali populations was lower than that of South Asian hgs (Supplementary Table 7). This is indeed the case considering both haploid loci and can be explained by extensive admixture of the Tharu with local populations, which would increase the diversity of South Asian but not East Asian-specific lineages.35
Table 3. The mtDNA haplogroup frequency among different Tharu groups.
| Country | State | District | n | A | B5a | C | D1 | D2 | D3 | D4 | F1 | G | G2 | J | H | HV | K | M | M18 | M2 | M21 | M30 | M31 | M33 | M3 | M34 | M35 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nepal | Nepal | Butwal | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nepal | Nepal | Tulsipur | 21 | 0 | 4.76 | 9.52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.76 | 0 | 0 | 4.76 | 4.76 | 0 | 4.76 | 0 | 0 | 4.76 | 0 | 0 |
| Nepal | Nepal | Chitwan | 133 | 0 | 5.26 | 3.01 | 0.75 | 0.75 | 0.75 | 6.77 | 6.02 | 9.02 | 14.29 | 0 | 0.75 | 0 | 0 | 0 | 0.75 | 0 | 1.5 | 0.75 | 1.5 | 3.76 | 0 | 0 | 0.75 |
| Nepal | Nepal | Morgan | 40 | 0 | 2.5 | 2.5 | 0 | 0 | 0 | 10 | 5 | 2.5 | 10 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 7.5 | 2.5 | 0 | 5 |
| India | Uttarakhand | Haldwani | 22 | 4.55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13.64 | 0 | 0 | 0 | 0 | 0 | 13.64 | 4.55 | 4.55 | 0 | 4.55 | 0 | 0 | 0 | 4.55 | 0 |
| India | Uttarakhand | Nainital | 23 | 0 | 4.35 | 0 | 0 | 0 | 0 | 17.39 | 4.35 | 13.04 | 17.39 | 0 | 0 | 0 | 0 | 0 | 0 | 4.35 | 0 | 0 | 4.35 | 0 | 0 | 0 | 0 |
| India | Uttar Pradesh | Gorakhpur | 38 | 0 | 5.26 | 2.63 | 0 | 0 | 0 | 2.63 | 2.63 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.89 | 0 | 0 | 5.26 | 0 | 0 |
| India | Uttar Pradesh | Allahabad | 37 | 0 | 8.11 | 2.7 | 0 | 0 | 0 | 2.7 | 0 | 8.11 | 8.11 | 0 | 0 | 0 | 2.7 | 0 | 2.7 | 8.11 | 0 | 0 | 0 | 0 | 2.7 | 0 | 2.7 |
| India | Uttar Pradesh | Deoria | 47 | 0 | 2.13 | 4.26 | 0 | 0 | 0 | 2.13 | 0 | 4.26 | 4.26 | 0 | 0 | 0 | 2.13 | 2.13 | 2.13 | 4.26 | 0 | 0 | 0 | 0 | 6.38 | 2.13 | 0 |
| India | Uttar Pradesh | Mahrajganj | 42 | 2.38 | 0 | 2.38 | 0 | 0 | 0 | 7.14 | 0 | 0 | 7.14 | 2.38 | 0 | 0 | 0 | 2.38 | 2.38 | 2.38 | 0 | 2.38 | 0 | 2.38 | 9.52 | 0 | 0 |
| Country | State | District | n | M38 | M39 | M4 | M4a | M40 | M43 | M45 | M5 | M52 | M9a1 | N5 | R | R30 | R5 | R6 | T2b | U1a | U2a | U2b | U2c | U2 | U7 | W | Z |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nepal | Nepal | Butwal | 10 | 0 | 0 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 20 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 |
| Nepal | Nepal | Tulsipur | 21 | 0 | 0 | 0 | 0 | 0 | 4.76 | 9.52 | 9.52 | 0 | 0 | 4.76 | 0 | 0 | 14.29 | 0 | 0 | 0 | 0 | 9.52 | 0 | 0 | 0 | 9.52 | 0 |
| Nepal | Nepal | Chitwan | 133 | 0.75 | 0 | 1.5 | 0 | 0 | 9.77 | 0 | 8.27 | 0 | 15.04 | 0 | 0 | 1.5 | 0 | 0 | 0.75 | 0 | 0 | 3.01 | 0 | 0 | 0 | 0 | 3.01 |
| Nepal | Nepal | Morgan | 40 | 20 | 0 | 0 | 0 | 0 | 2.5 | 0 | 5 | 2.5 | 0 | 0 | 0 | 5 | 0 | 2.5 | 0 | 0 | 0 | 7.5 | 2.5 | 0 | 0 | 0 | 0 |
| India | Uttarakhand | Haldwani | 22 | 9.09 | 4.55 | 4.55 | 0 | 0 | 4.55 | 0 | 9.09 | 4.55 | 0 | 0 | 0 | 0 | 4.55 | 0 | 0 | 0 | 0 | 4.55 | 0 | 4.55 | 0 | 0 | 0 |
| India | Uttarakhand | Nainital | 23 | 0 | 0 | 4.35 | 0 | 0 | 8.7 | 0 | 4.35 | 0 | 8.7 | 0 | 0 | 0 | 8.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| India | Uttar Pradesh | Gorakhpur | 38 | 21.05 | 0 | 5.26 | 0 | 0 | 2.63 | 5.26 | 5.26 | 0 | 0 | 2.63 | 2.63 | 10.53 | 2.63 | 0 | 0 | 2.63 | 5.26 | 5.26 | 0 | 0 | 0 | 2.63 | 0 |
| India | Uttar Pradesh | Allahabad | 37 | 0 | 0 | 2.7 | 0 | 2.7 | 16.22 | 2.7 | 5.41 | 0 | 2.7 | 0 | 2.7 | 0 | 2.7 | 0 | 0 | 0 | 0 | 0 | 0 | 2.7 | 0 | 2.7 | 8.11 |
| India | Uttar Pradesh | Deoria | 47 | 2.13 | 0 | 2.13 | 4.26 | 0 | 8.51 | 2.13 | 8.51 | 0 | 4.26 | 0 | 0 | 2.13 | 4.26 | 4.26 | 4.26 | 0 | 2.13 | 0 | 0 | 6.38 | 0 | 2.13 | 6.38 |
| India | Uttar Pradesh | Mahrajganj | 42 | 9.52 | 0 | 0 | 2.38 | 4.76 | 4.76 | 7.14 | 7.14 | 0 | 0 | 2.38 | 0 | 2.38 | 0 | 0 | 2.38 | 0 | 4.76 | 4.76 | 0 | 2.38 | 2.38 | 0 | 0 |
Because hgs specific to South Asia are found in many different ethnic groups other than the Tharu, we focused our search further on mtDNA lineages presenting all the Tharu groups (Supplementary Table 8). The rare hg M43 is notably found among all the Tharu groups (Table 3). To gain a better temporal and spatial view of the spread of this hg, we examined all the M43 haplotypes in our existing Indian data (Supplementary Figure 5). We found that the M43 haplotypes, specific to the Tharu, are restricted geographically and found only among the Tharu, Nepali, northern and northeastern Indian populations (Supplementary Figure 5; Supplementary Table 5). The phylogenetic tree of this hg reveals a deep rooting coalescent time comparable to that of other South Asian M subclades.36 The spatial dispersal of this rare hg is extremely limited, and it is absent among East/Southeast Asian and South Indian populations (Supplementary Figure 5; Supplementary Table 5). High variation of hg M43 among Nepali-north Indian populations likely suggests its origin in the north of the subcontinent. Therefore, it appears that the distribution of M43 highlights the minor but nevertheless present layer of common maternal ancestry between all the Tharu groups studied here.
Individuals screened for mtDNA were also assayed for Y-chromosome diversity (Supplementary Tables 6 and 9). Both South Asian (hgs H, H1a1a, L1 and R2) and West Eurasian (hg J2a) lineages are shared among all the groups. However, the O3a2c1-M134 hg, indicative of an East Asian origin, is also reflected in the autosomal analysis, could be directly co-associated with the Tharu dispersal from the Tarai to Uttarakhand and Uttar Pradesh (Figures 3d and e). The ancestral form of this lineage occurs widely throughout East and Southeast Asia,37 consistent with dispersal from Tibet and Southeast Asia to northern India via Nepal.34, 35, 38 Gene flow in the opposite direction is equally likely for some of the lineages (eg, hgs C, H, J, L and R). An infrequent hg O2a-M95 is also shared among all the groups, although it is uncertain whether this marker arrived in northern India by way of the Tharu or through the Austroasiatic expansion, or both.21
We analysed the Y-STR data from three major Y-chromosomal hgs H1a1a-M82, O3a2c1-M134 and R1a-M17 (Supplementary Table 10). The lack of clear clustering in the STR-based phylogenetic networks (Supplementary Figure 6) suggests that the scenario of a simple founder effect is highly unlikely, and argues for the involvement of more than one founding lineage in the migration. We have also observed that the relative ages are inconsistent with a simple dispersal from the Tarai to other areas (Table 4). Instead, the relative ages would fit better with broad population movements during the Holocene.
Table 4. The variance and age of three Y-chromosomal major haplogroups of the Tharu, among different regional and ethnic groups.
| Haplogroup | Group | n | Variance | Age (Kya) |
|---|---|---|---|---|
| O3a2c1-M134 | Tibet | 46 | 0.3 | 11.2±3 |
| Nepal | 120 | 0.34 | 13±3.8 | |
| NE India | 45 | 0.38 | 15.9±2 | |
| Tharu-All | 23 | 0.25 | 12.2±0.8 | |
| Tharu-Nepal | 8 | 0.23 | 9.7±1.2 | |
| Tharu-UT | 7 | 0.2 | 10±1.6 | |
| Tharu-UP | 8 | 0.19 | 8.5±1.2 | |
| R1a-M17 | Nepal | 44 | 0.3 | 13.7±2.9 |
| India-North | 39 | 0.34 | 14.6±2.9 | |
| Tharu-All | 49 | 0.31 | 14.5±0.8 | |
| Tharu-Nepal | 5 | 0.16 | 7.3±1.7 | |
| Tharu-UT | 8 | 0.22 | 8.2±1.9 | |
| Tharu-UP | 36 | 0.32 | 15±1 | |
| H1a1a-M82 | India-North | 35 | 0.49 | 20.8±4.4 |
| Tharu-All | 19 | 0.24 | 11.3±1.5 | |
| Tharu-Nepal | 4 | 0.27 | 8.5±1.7 | |
| Tharu-UT | 4 | 0.22 | 7.3±1 | |
| Tharu-UP | 11 | 0.19 | 9.4±1.7 |
Abbreviations: NE India, Northeast India; UP, Uttar Pradesh; UT, Uttarakhand. The age was estimated from the ρ statistic (the mean number of mutations from the assumed root of each and every founder), using a 25-year generation time and the TD statistic, assuming a mutation rate of 6.9 × 10−4 (Zhivotovsky et al24), based on variation at 15 common Y-STR loci.
In our previous study,21 we observed that the c.1540T>C polymorphism in the EDAR gene was highly informative for assessing East and Southeast Asian ancestries in Indian populations. Genotyping of this SNP among different Tharu groups revealed a decreasing gradient of the frequency of the c.1540T>C allele from Nepal-Tulsipur to Uttar Pradesh-Allahabad sampling points of the Tharu (Supplementary Table 11). Taken together, the occurrence of considerable East Asian mtDNA, Y-chromosomal and EDAR polymorphisms among all the Tharu subgroups suggests that the expansion of the Tharu from Tarai occurred after an influx of East Asian lineages into this region (Tables 2 and 3; Supplementary Tables 8, 9 and 11).
CONCLUSION
Our aim here was to search for any exclusively common genetic signatures uniting the different Tharu groups that would testify for at least some degree of shared genetic ancestry for (all) the Tharu. Regardless of the genetic system explored, the overall proportion of East Asian genetic ancestry in the Tharu groups gave way to South Asian ancestry on a general East to West axis. The analyses of autosomal data revealed a high level of heterogeneity among the two studied Tharu groups and thus supported the model of cultural rather than demic unity of the Tharu. However, sharing of specific mtDNA and Y-chromosomal hgs between all the studied Tharu groups and lack of them among other Uttar Pradesh populations can be seen as a vestige of a shared ancestry for the Tharu groups. These haploid lineages shared among the Tharu include both autochthonous (mtDNA M43) and East Asian-specific hgs (like Y-chromosomal hg O3a2c1-M13439). The latter suggests that the East Asian layer in the genetic ancestry of the Tharu is older than the expansion of the Tharu within the subcontinent (potentially from the Tarai region). However, nearly exclusive sharing of mtDNA hg M43 by the Tharu indicates some degree of shared genetic ancestry originating in the north of the subcontinent. The expansion of the Tharu was followed by extensive admixture and assimilation with the local populations, which is testified by higher diversity of the autochthonous South Asian mtDNA and Y-chromosomal lineages among the Tharu.39 Finally, beyond the particular history of the Tharu, our study has identified the first deeply rooted mtDNA hg originating in the north of the subcontinent, providing the first insights into the earliest human populations in this region.
Acknowledgments
We thank the Tharu community for the kind participation to this study. Chandana Basu Mallick for useful discussion and proof read. This study was supported by the European Union European Regional Development Fund through the Centre of Excellence in Genomics to Estonian Biocentre and University of Tartu to RV, MM and GC, by Tartu University grant (PBGMR06901) to TK and other supporting funding sources were ERC Starting Investigator grant (FP7 – 261213) to TK; Estonian Institutional Research grant IUT24-1 to RV; European Commission grant (ECOGENE 205419) to RV; Estonian Science Foundation grant (8973) to MM and UK-India Education Research Initiative (RG47772) grant to FC, MML, KT and TK. KT was supported by the Council of Scientific and Industrial Research, Government of India (GENESIS: BSC0121) and (BSC 0208). LS was supported by Bhatnagar (CSIR) and JC Bose (DST) Fellowships, Government of India. AKP and RT were supported by the European Social Funds Doctoral Studies and Internationalisation Programme DoRa. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was carried out in part in the High Performance Computing Center of University of Tartu.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Guneratne A. Many Tongues, One People: The Making of Tharu Identity in Nepal. Ithaca, NY, USA: Cornell University Press; 2002. [Google Scholar]
- Buchanan-Hamilton F.History, Antiquities, Topography and Statistics of Eastern IndiaMartin M (ed). Wm. H. Allen & Co.: London; 18382705–706. [Google Scholar]
- Crooke W. The Tribes and Castes of the North-Western Provinces and Oudh. Office of the Superintendent of Government Printing: India; 1896. [Google Scholar]
- Grierson GA.(ed.):: Linguistic Survey of India (Vol V. Indo-Aryan Family, Eastern Group, Part II: Specimens of the Bihari and Oriya Languages)Calcutta, India: Superintendent of Government Printing, 1903
- Grierson GA. Linguistic Survey of India: Including Khassi and Tai. Mo-Khme and Siamese-Chinese Families. Office of the Superintendent of Government Printing: India; 1904. [Google Scholar]
- van Driem G.(ed.):: Languages of the HimalayasLeiden2001
- Passarino G, Semino O, Modiano G, Santachiara-Benerecetti AS. COII/tRNA(Lys) intergenic 9-bp deletion and other mtDNA markers clearly reveal that the Tharus (southern Nepal) have Oriental affinities. Am J Hum Genet. 1993;53:609–618. [PMC free article] [PubMed] [Google Scholar]
- Passarino G, Semino O, Pepe G, et al. MtDNA polymorphisms among Tharus of eastern Terai (Nepal) Gene Geogr. 1992;6:139–147. [PubMed] [Google Scholar]
- Koirala S, Shah S, Baral P.A comparative anthropometric study of mongoloid and Tharu ethnic races in Eastern Nepal.: WebmedCentral Anatomy 20123WMC003512
- Chhetri RB. The plight of the Tharu Kamaiyas in Nepal: a review of the social, economic and political facets. Occas Papers Sociol Anthropol. 2005;9:22–46. [Google Scholar]
- Modiano G, Morpurgo G, Terrenato L, et al. Protection against malaria morbidity: near-fixation of the α-thalassemia gene in a Nepalese population. Am J Hum Genet. 1991;48:390–397. [PMC free article] [PubMed] [Google Scholar]
- Terrenato L, Shrestha S, Dixit K, et al. Decreased malaria morbidity in the Tharu people compared to sympatric populations in Nepal. Ann Trop Med Parasitol. 1988;82:1–11. doi: 10.1080/00034983.1988.11812202. [DOI] [PubMed] [Google Scholar]
- Skar H.Nepal, indigenous issues and civil rights: the Plight of the Rana Tharuin Barnes R, Gray A, Kingsbury B (eds):: Indigenous Peoples of AsiaAnn Arbor, MI, USA: Association for Asian Studies, 1993173–194.
- Fornarino S, Pala M, Battaglia V, et al. Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation. BMC Evol Biol. 2009;9:154. doi: 10.1186/1471-2148-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopff G.An ‘Indigenous Minority' in a border area: Tharu Ethnic Associations, NGOs, and the Nepalese Statein Gellner D (ed.):: Resistance and The 79 State: Nepalese ExperiencesNewDelhi: Social Science Press2007199–243.
- Jeffery R, Jeffery P. Population, Gender and Politics: Demographic Change in Rural North India. Cambridge University Press; 1997. [Google Scholar]
- Ghurye GS. Caste and Race in India. Popular Prakashan; 1969. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGO Pan-Asian SNP Consortium. Abdulla MA, Ahmed I, et al. Mapping human genetic diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- Kamberov YG, Wang S, Tan J, et al. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, Metspalu M, Choi Y, et al. Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol Biol Evol. 2011;28:1013–1024. doi: 10.1093/molbev/msq288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, van Oven M, Rosset S, et al. A ‘copernican' reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioglu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham CC, Weir BS. Covariances of relatives stemming from a population undergoing mixed self and random mating. Biometrics. 1984;40:157–164. [PubMed] [Google Scholar]
- Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu M, Romero IG, Yunusbayev B, et al. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Hum Genet. 2011;89:731–744. doi: 10.1016/j.ajhg.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayden T, Cadenas AM, Regueiro M, et al. The Himalayas as a directional barrier to gene flow. Am J Hum Genet. 2007;80:884–894. doi: 10.1086/516757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Li YC, Sun F, et al. Revisiting the role of the Himalayas in peopling Nepal: insights from mitochondrial genomes. J Hum Genet. 2012;57:228–234. doi: 10.1038/jhg.2012.8. [DOI] [PubMed] [Google Scholar]
- Petraglia M, Clarkson C, Boivin N, et al. Population increase and environmental deterioration correspond with microlithic innovations in South Asia ca. 35,000 years ago. Proc Natl Acad Sci USA. 2009;106:12261–12266. doi: 10.1073/pnas.0810842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JD, Peng MS, Quang HH, et al. Patrilineal perspective on the Austronesian diffusion in Mainland Southeast Asia. PLoS ONE. 2012;7:e36437. doi: 10.1371/journal.pone.0036437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Chaubey G, Kivisild T, et al. Maternal footprints of Southeast Asians in North India. Hum Hered. 2008;66:1–9. doi: 10.1159/000114160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driem G. The ethnolinguistic identity of the domesticators of Asian rice. Comptes Rendus Palevol. 2012;11:117–132. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.