Key Points
Stimulation of the B-cell receptor of chronic lymphocytic leukemia cells results in activation of an unfolded protein response.
Unfolded protein response activation following surface immunoglobulin M stimulation in vitro is dependent on the activity of BTK and SYK.
Abstract
B-cell receptor (BCR) signaling plays a key role in the behavior of chronic lymphocytic leukemia (CLL). However, cellular consequences of signaling are incompletely defined. Here we explored possible links between BCR signaling and the unfolded protein response (UPR), a stress response pathway that can promote survival of normal and malignant cells. Compared with normal B cells, circulating CLL cells expressed increased, but variable, levels of UPR components. Higher expression of CHOP and XBP1 RNAs was associated with more aggressive disease. UPR activation appeared due to prior tissue-based antigenic stimulation because elevated expression of UPR components was detected within lymph node proliferation centers. Basal UPR activation also correlated closely with surface immunoglobulin M (sIgM) signaling capacity in vitro in both IGHV unmutated CLL and within mutated CLL. sIgM signaling increased UPR activation in vitro with responders showing increased expression of CHOP and XBP1 RNAs, and PERK and BIP proteins, but not XBP1 splicing. Inhibitors of BCR-associated kinases effectively prevented sIgM-induced UPR activation. Overall, this study demonstrates that sIgM signaling results in activation of some components the UPR in CLL cells. Modulation of the UPR may contribute to variable clinical behavior, and its inhibition may contribute to clinical responses to BCR-associated kinase inhibitors.
Introduction
Chronic lymphocytic leukemia (CLL) provides a unique opportunity to understand how antigen can influence the behavior of malignant lymphocytes. It also acts as a model for the development of novel therapies targeted toward B-cell receptor (BCR) signaling pathways.1-4 CLL comprises 2 major subsets with differing levels of somatic hypermutation of tumor IGV genes. CLL with unmutated IGV (U-CLL) derives from naïve CD5+CD27− B cells of the normal natural antibody repertoire, whereas CLL with mutated IGV genes (M-CLL) may derive from postgerminal center CD5+CD27+ cells.5,6 Importantly, these subsets have distinct clinical behavior, and U-CLL has a more aggressive clinical course. Antigen signaling is thought to be ongoing in both subsets and, rather than the presence or absence of signaling, it is the balance between distinct types of responses that appears to determine clinical behavior.1 Anergy, a state of cellular lethargy that is induced following antigen engagement in the absence of T-cell help,7 is observed in all CLL but is particularly prominent in M-CLL.1 By contrast, positive antigen signaling leading to proliferation and survival appears more evident in U-CLL. The importance of antigen signaling for CLL is emphasized by recent results that have demonstrated the clinical effectiveness of inhibitors of BCR-associated kinases.8
Antigen engagement in vivo is thought to occur within proliferation centers (PCs) found predominantly in the lymph nodes (LNs) of CLL patients. Following stimulation, CLL cells enter the circulation and therefore carry a temporary “imprint” of their prior tissue-based stimulation.9,10 Thus, markers of anergy,7 including strong down-modulation of surface immunoglobulin M (sIgM) expression and signaling capacity, raised extracellular signal-regulated kinase (ERK)1/2 phosphorylation, and nuclear factor of activated T cells expression can be detected in blood CLL cells, most prominently in M-CLL.11-13 In contrast to M-CLL, blood cells from patients with U-CLL tend to retain sIgM expression and signaling responsiveness and express higher levels of markers of positive BCR signaling, including the proliferation and survival-promoting proteins MYC and MCL1.14,15 Positive signaling can be mimicked in vitro by treating CLL cells with anti-IgM antibodies, which increases expression of these markers in samples that retain sIgM responsiveness.16,17 Although the overall behavior of U-CLL and M-CLL is distinct, there is heterogeneity within these subsets, especially within M-CLL.11 For example, high levels of sIgM expression and signaling in M-CLL may highlight a subset at higher risk of progression. Indeed, our previous study demonstrated that anti-IgM–induced BIM phosphorylation was associated with requirement for treatment, including within the M-CLL subset.18
Despite recent advances, the consequences of BCR stimulation in CLL remain incompletely understood. In this work, we investigated the effects of sIgM stimulation on the unfolded protein response (UPR). The UPR has been most widely studied as a stress response pathway that responds to accumulation of unfolded/misfolded proteins and/or elevated secretory protein synthesis within the endoplasmic reticulum lumen.19,20 See supplemental Figure 1, available on the Blood Web site for a summary of UPR molecules and pathways.
In B cells, the UPR plays key roles in differentiation because production of secreted immunoglobulin by plasma cells requires a compensatory increase in protein production capacity mediated by UPR induction.21 Thus, XBP1 and IRE1 are essential for plasma cell development.22-24 The UPR is also essential for the survival of multiple myeloma cells and is an established therapeutic target in this disease.25-27 However, the UPR plays other roles in B cells, independent of its requirement to support increased secretory immunoglobulin synthesis per se, including for differentiation beyond the pro-B-cell stage.24 In mature B cells, differentiation-promoting factors, such as interleukin (IL)4 or lipopolysaccharide, rapidly activate a subset of UPR components prior to increased immunoglobulin synthesis, and the UPR is activated normally in cells that lack the ability to secrete IgM.23,28-30 BCR stimulation has also been shown to increase some UPR components, although this stimulation alone is not sufficient to promote differentiation.31 Thus, UPR activation is not simply a consequence of stress but can be a signal-regulated pathway that induces a partial anticipatory response that prepares B cells for subsequent antibody production. In contrast to these physiological prosurvival responses, prolonged, high-level UPR activation in response to pharmacological agents (such as proteasome inhibitors that cause accumulation of mis-folded proteins) induces a cell death-promoting UPR response.19,20
Previous studies have shown that CLL cells express some UPR components and that pharmacological inducers of the UPR promote apoptosis of CLL cells in vitro.32-36 However, the potential regulation of the UPR following BCR stimulation of CLL cells has not been studied. In this paper, we demonstrate for the first time that sIgM stimulation results in a partial activation of the UPR, with selective activation of specific downstream UPR effector pathways. Higher levels of UPR activation correlated with more aggressive disease and BCR-targeted kinase inhibitors decreased UPR activation, suggesting that this response may contribute to disease progression and that its inhibition may be important for clinical activity of drugs such as ibrutinib.
Materials and methods
Patients and cell samples
Patients were recruited after written informed consent was provided in accordance with Ethics Committee approvals and the Declaration of Helsinki. Blood was obtained from patients with IgM+IgD+ CLL with a diagnostic phenotype who attended Hematology outpatient clinics at the Leicester Royal Infirmary, Portsmouth Hospital, Southampton General Hospital, the Royal Wolverhampton Hospitals National Health Service Trust, or the Royal Berkshire Hospital, Reading (all in the United Kingdom [UK]). Clinical details for the patients studied are given in supplemental Table 1. The majority of samples were obtained at or shortly after diagnosis and mainly prior to any therapy for CLL. Where treatment of CLL had taken place, this was ≥6 months prior to sample collection. Disease was considered to be more aggressive if there were signs of clinical progression and/or the patient was treated for CLL at any point following diagnosis.
Blood samples were processed as previously described.11 Cell viability determined by trypan blue exclusion was ≥90%. The proportion of CD5+CD19+ CLL cells was >80% in all cases. IGHV mutation status, expression of cell surface CD5, CD19, and CD38, and ZAP70 were determined as previously described.11,37 IgM signaling capacity was determined by measuring the percentage of cells with increased intracellular calcium following stimulation with soluble goat F(ab′)2 anti-IgM and using a cutoff value of ≥5% responding cells to define samples as sIgM responsive.11 Normal B cells were isolated from peripheral blood or buffy coats from healthy donors using the B cell Isolation Kit II with the addition of anti-CD138 Microbeads (both from Miltenyi Biotec, Bisley, UK) to ensure effective depletion of plasma cells.
Additional methods are provided as supplemental Materials.
Results
Basal activation of UPR-associated pathways in CLL and normal B cells
We first analyzed basal activation of the UPR (ie, in unstimulated cells) in CLL samples isolated from the blood of 40 patients using quantitative polymerase chain reaction (qPCR) to quantify expression of XBP1 and CHOP RNAs. The samples comprised 20 U-CLL, which, as previously described,11 generally retained sIgM signaling responsiveness. We also analyzed 20 M-CLL samples. These samples were selected to contain a substantial proportion of sIgM signal-competent samples to allow us to probe potential correlations between UPR activation and sIgM signaling within this subset. Circulating B cells from healthy individuals were analyzed as controls. To validate the qPCR assays, CLL samples were treated with the pharmacological UPR inducer thapsigargin. As expected, thapsigargin substantially increased XBP1 and CHOP RNA expression in CLL samples (supplemental Figure 2A).
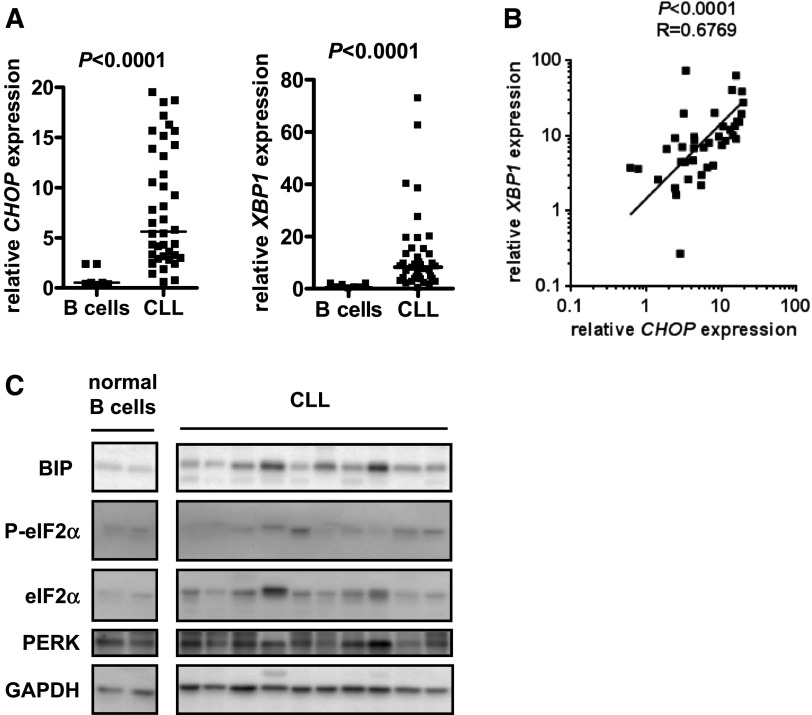
Although basal expression of CHOP and XBP1 RNAs were variable between individual CLL samples, median CHOP and XBP1 RNA expression levels were significantly higher than normal B cells (Figure 1A). CHOP and XBP1 RNA expression levels were closely correlated, demonstrating that these RNAs are generally coexpressed in individual CLL samples (Figure 1B).
Figure 1.
Expression of UPR components in unstimulated CLL samples and normal B cells. (A) CHOP and XBP1 RNA expression was quantified by qPCR in CLL samples (n = 40) and normal B cells (n = 7). Expression values were normalized so that the average value in normal B cells was set to 1.0. Graphs show median and individual data points, and the statistical significance of differences between CLL samples and normal B cells (Mann-Whitney test). (B) Correlation between CHOP and XBP1 RNA expression in CLL samples. The line shows results of linear regression, and the statistical significance of the correlation is shown (Spearman correlation). (C) Immunoblot analysis of BIP, total and phospho-eIF2α, PERK, and glyceraldehyde-3-phosphate dehydrogenase (loading control) in normal B cells (2 preparations shown) and CLL samples. Results shown are representative of >30 samples studied across a series of separate immunoblots.
We extended these results by examining other features of UPR activation in unstimulated CLL cells including BIP, PERK, and the PERK substrate eIF2α. We were unable to identify antibodies suitable for reliable analysis of XBP1 and CHOP protein expression in CLL cells. As expected, thapsigargin increased BIP protein expression and phosphorylation of PERK (detected by reduced migration) and eIF2α (detected using a phospho-specific antibody) (supplemental Figure 2B). Immunoblotting demonstrated that basal expression of BIP protein was elevated in some CLL samples compared with normal B cells (Figure 1C). We also detected moderately increased PERK expression in some CLL samples compared with normal B cells but not a clear decrease in PERK mobility as observed in thapsigargin-treated CLL cells. Consistent with weak PERK activation in CLL cells, we detected only very modest levels eIF2α phosphorylation in some samples.
Although we detected raised XBP1 RNA in unstimulated CLL samples, there was little evidence for accumulation of XBP1S; very low levels of basal expression of XBP1S RNA were detected in only 2 of 18 untreated CLL cell samples (data not shown). XBP1S expression was detected in thapsigargin-treated cells, confirming the validity of the assay. However, even in thapsigargin-treated cells, XBP1S RNA levels were relatively low level (supplemental Figure 3).
Overall, these results demonstrate substantial but variable basal activation of some UPR components in CLL blood cells.
Correlations between basal UPR activation and sIgM signaling capacity in vitro
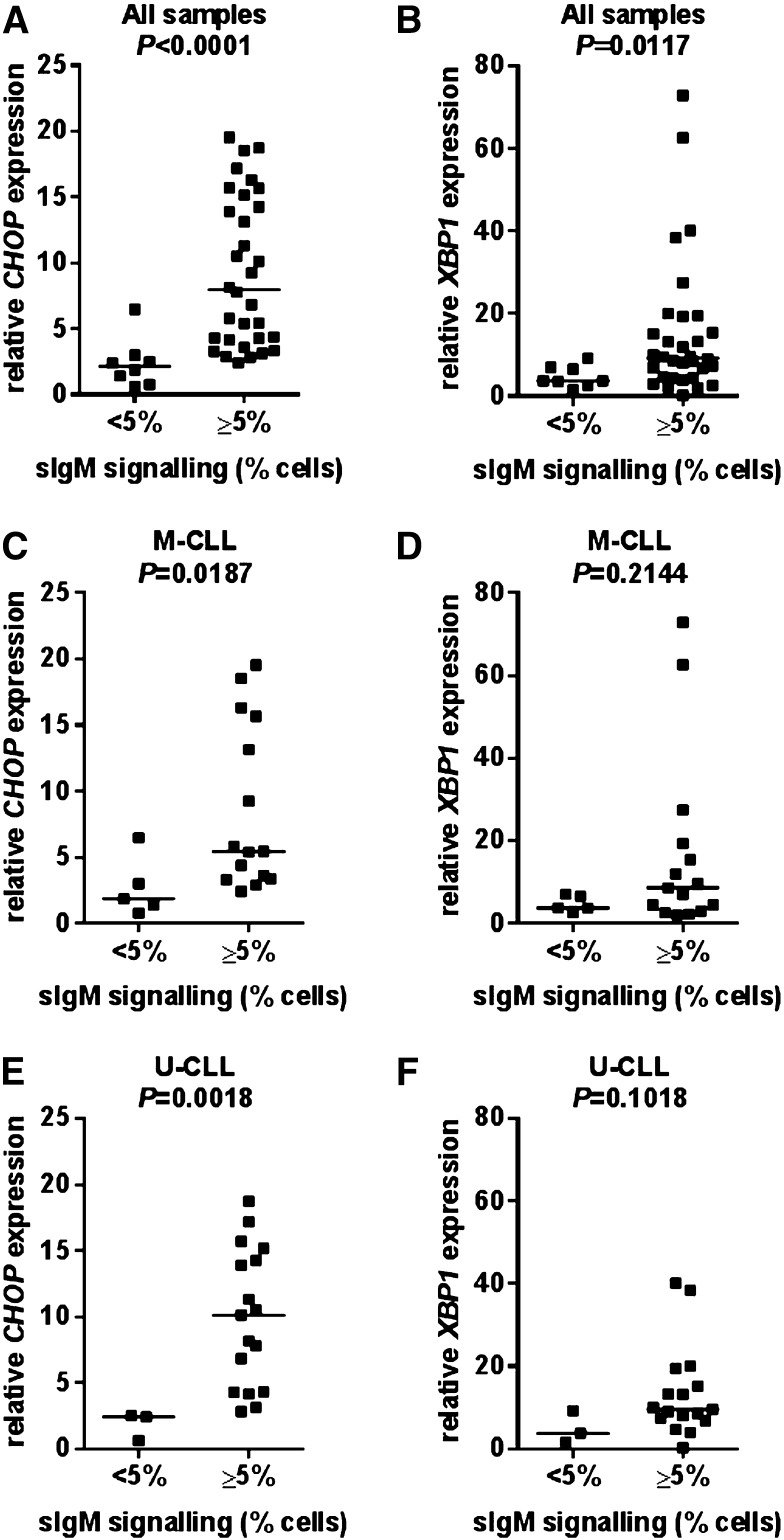
We next investigated potential correlations between basal UPR activation and sIgM signaling capacity measured using anti-IgM–induced intracellular Ca2+ mobilization. When considering the total cohort, there were significant correlations between sIgM signaling capacity in vitro and CHOP and XBP1 RNA expression levels with higher basal level expression of these RNAs associated with retained sIgM signaling capacity (Figure 2A-B). Similar to the complete cohort, there was a positive correlation between signaling capacity and CHOP RNA levels when U-CLL and M-CLL samples were considered separately (Figure 2C,E). There was a similar trend for XBP1 RNA, but this did not reach statistical significance (Figure 2D,F). Thus, basal UPR activation correlates with sIgM signaling capacity in vitro in both the M-CLL and U-CLL subsets. Consistent with the correlation between UPR activation and retained signal capacity, there were trends toward increased CHOP/XBP1 RNA expression in U-CLL (supplemental Figure 4). However, it is important to emphasize, that these differences did not reach statistical significance, most likely due to the enrichment for M-CLL signal competent samples in the current cohort.
Figure 2.
Correlations between CHOP and XBP1 RNA expression and sIgM signaling capacity. Correlations between basal CHOP and XBP1 RNA expression and anti-IgM signaling responsiveness in (A-B) all samples, (C-D) M-CLL, and (E-F) U-CLL. The statistical significance of differences was analyzed using the Mann-Whitney test.
Correlation between basal UPR activation and clinical behavior
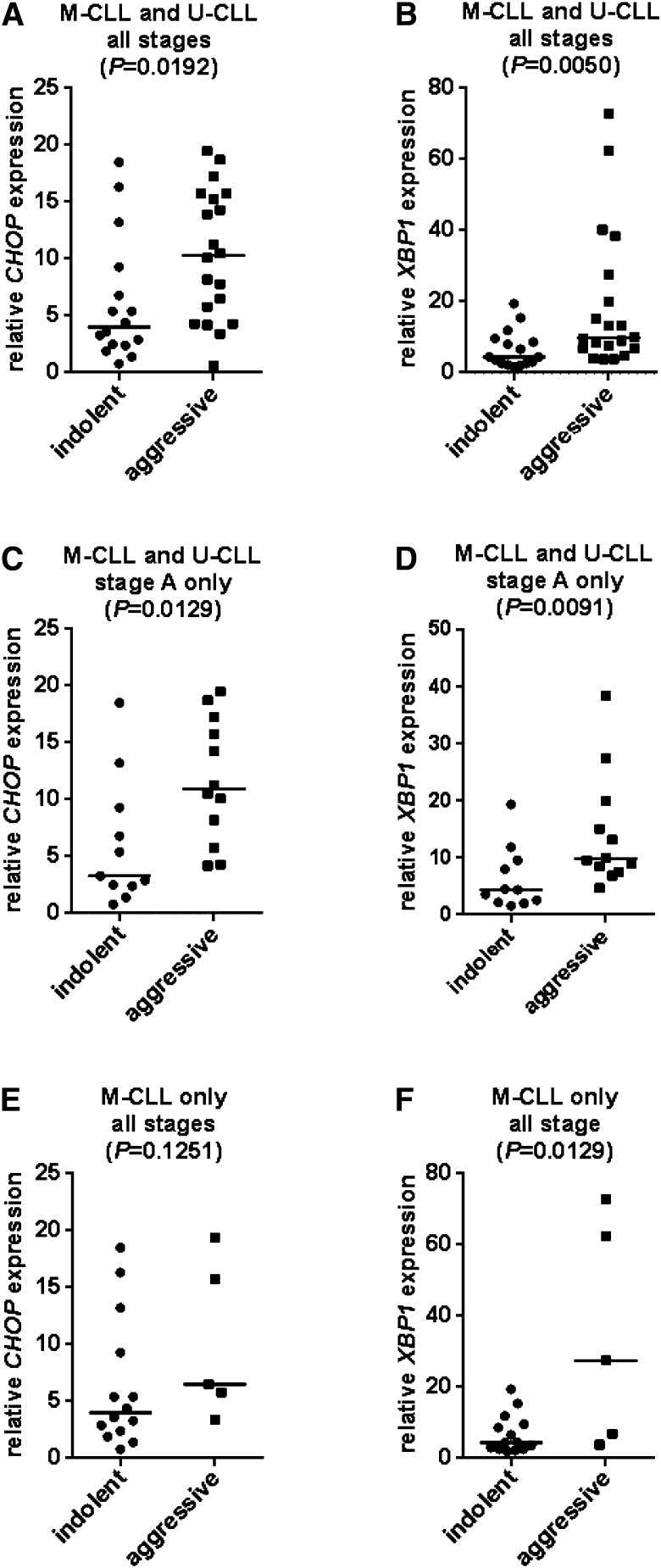
To begin to probe the potential clinical significance of UPR activation, we also investigated whether variable basal UPR activation correlated with clinical behavior depending on whether the patient had indolent or more aggressive disease (Materials and methods). Higher basal CHOP or XBP1 RNA levels were associated with more aggressive disease in the total cohort (Figure 3A-B). Similar correlations were detected when only Binet stage A disease (U-CLL and M-CLL combined) was analyzed (n = 23) (Figure 3C-D). There was also consistently higher expression of CHOP or XBP1 RNAs in more aggressive disease compared with indolent disease specifically within the M-CLL subset (all stages), although this was only significant for XBP1 (Figure 3E-F). There were only 2 cases of indolent disease among the 17 U-CLL samples analyzed, where outcome data were available precluding meaningful analysis of this subset. These observations provide further support for the idea that high basal UPR activation is associated with retained sIgM signaling and that these features may be associated with relatively aggressive disease, possibly even within M-CLL.
Figure 3.
Correlations between CHOP and XBP1 RNA expression and clinical behavior. Correlations between basal CHOP and XBP1 RNA expression and indolent/aggressive disease for (A-B) all CLL/all stages (n = 31), (C-D) stage A (M-CLL and U-CLL combined; n = 23), and (E-F) M-CLL (all stages; (n = 19). The statistical significance of differences was analyzed using Mann-Whitney test.
Effect of sIgM engagement on UPR activation
The correlation between basal UPR activation and retained sIgM signaling capacity suggested that UPR activation was directly linked to the capacity to respond to antigen stimulation in vivo. Activation of sIgM in vitro using anti-IgM antibodies mimics positive BCR signaling in CLL. Therefore, to determine directly whether sIgM stimulation activated the UPR in CLL cells, we investigated the effects of anti-IgM on XBP1/CHOP RNA expression. Normal B cells were analyzed as controls.
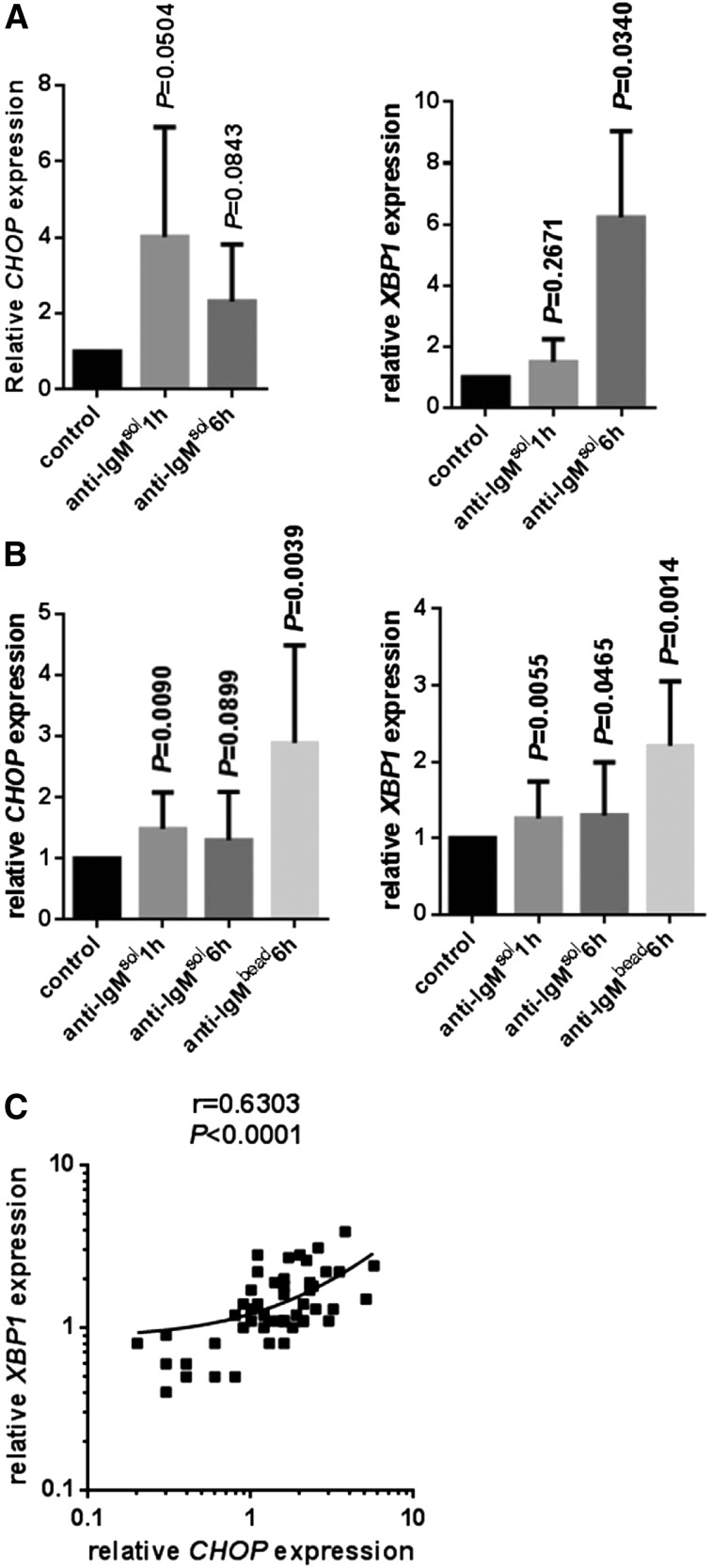
In normal B cells, soluble anti-IgM increased expression of CHOP RNA most strongly at 1 hour and less so at 6 hours after stimulation (Figure 4A). Induction of XBP1 RNA was greatest at 6 hours after stimulation. Similar experiments were performed using CLL samples, all of which were classed as sIgM signal responsive. There were significant increases in CHOP and XBP1 RNA expression following treatment with soluble anti-IgM compared with control cells (Figure 4B). However, similar to other sIgM signaling responses,38 increases in CHOP/XBP1 RNAs were much weaker than in normal B cells.
Figure 4.
Regulation of CHOP and XBP1 RNA expression by anti-IgM. (A) Normal B cells and (B) CLL samples were stimulated with soluble (sol) or bead-bound (bead) anti-IgM for 1 or 6 hours, and expression of CHOP and XBP1 RNAs was analyzed by qPCR. Expression values were normalized so that the average value in control normal and CLL cells was set to 1.0. Graphs show mean values ± standard deviation for data obtained with 6 or 4 preparations of normal B cells (for CHOP and XBP1 analysis, respectively). For experiments with CLL, 15 samples were used to compare responses between soluble anti-IgM at 1 and 6 hours, and 10 further samples were used to compare responses to soluble and bead-bound anti-IgM. The statistical significance of differences between treated and control cells are shown for each condition (Student t test). (C) Correlation between fold induction of XBP1 and CHOP RNAs in soluble/bead-bound anti-IgM–treated CLL samples (1- and 6-hour data combined; linear regression and Spearman correlation shown).
The weak induction of CHOP/XBP1 RNAs in CLL samples may reflect the low level of sIgM expression in these cells, a consequence of anergy-promoting interactions in vivo.7,11 Because BCR signal strength in CLL cells can be enhanced by treating cells with immobilized anti-IgM,16 we also stimulated sIgM signal responsive CLL samples with anti-IgM bound to Dynabeads (Figure 4B). Cells were analyzed at 6 hours after stimulation because the onset of signaling is delayed in cells treated with bead-bound9 compared with soluble antibodies,17 presumably due to potentially slower engagement of sIgM. Compared with soluble antibodies, bead-bound anti-IgM triggered larger increases in CHOP/XBP1 RNA expression (Figure 4B). Considering all data for anti-IgM–treated cells, there was a strong positive correlation between induction of XBP1 and CHOP RNAs (Figure 4C). Consistent with the stronger signal, increases in phosphorylation of ERK1/2 and AKT was greater and longer lasting in cells treated with bead-bound compared with soluble anti-IgM (supplemental Figure 5). There was considerable variation in the extent of anti-IgM–induced CHOP/XBP1 RNA expression (Figure 4B). However, the fold increase in CHOP or XBP1 RNA expression did not differ between M-CLL and U-CLL in this cohort of signaling-competent samples and did not correlate with ZAP-70 expression or sIgM expression (data not shown).
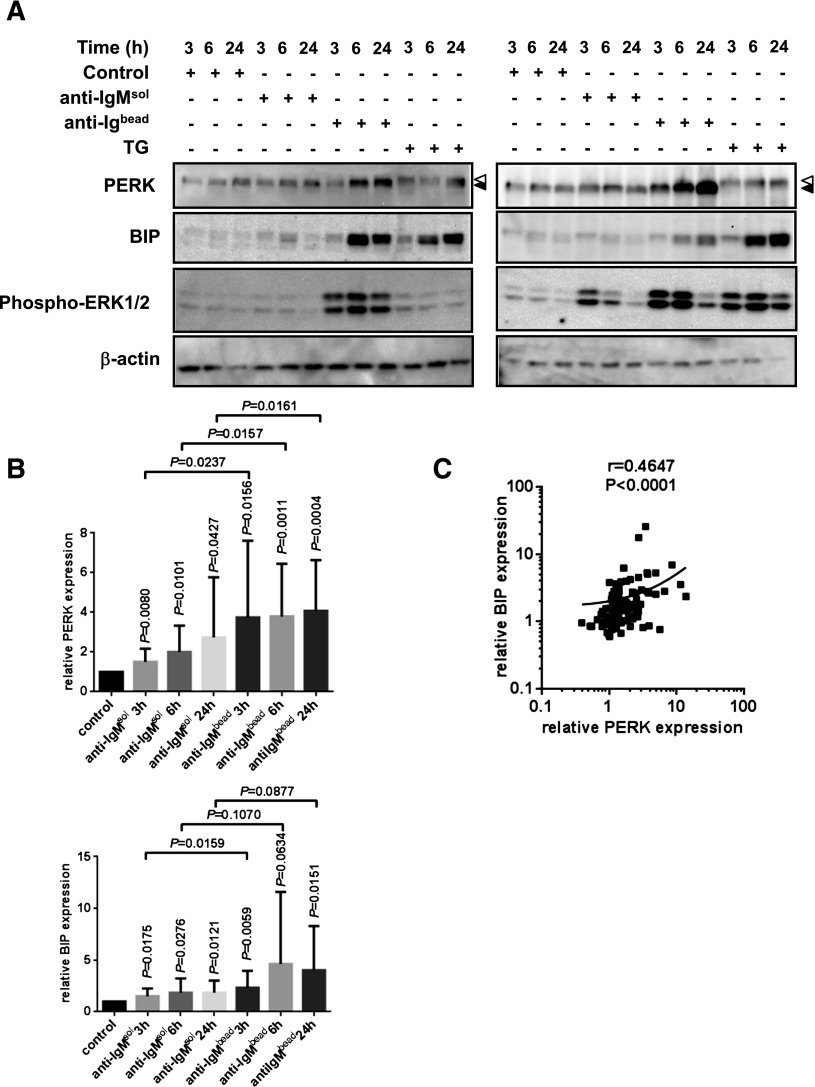
We performed similar experiments to determine whether anti-IgM also induced protein markers of the UPR in CLL using 15 signaling responsive samples. Treatment with anti-IgM increased expression of both PERK and BIP (Figure 5A-B). The induction by soluble anti-IgM was significant for some time points; however, levels of induction were greater for bead-bound anti-IgM. Similar to CHOP/XBP1 RNAs, there was considerable variation in the extent of anti-IgM–induced PERK/BIP expression (Figure 5A-B), although these parameters correlated closely within individual samples, indicating coregulation (Figure 5C). As expected, anti-IgM also induced ERK1/2 phosphorylation (Figure 5A). Thapsigargin also induced ERK1/2 phosphorylation in a subset of samples.
Figure 5.
Regulation of BIP and PERK expression by anti-IgM. CLL samples (n = 15) were stimulated with soluble (sol) or bead-bound (bead) anti-IgM or thapsigargin as a control (TG; 15 μM) for up to 24 hours, and expression of PERK, BIP, and phosphorylated ERK1/2 was analyzed by immunoblotting. (A) Representative results from 2 CLL samples. Phosphorylated and nonphosphorylated forms of PERK are indicated by white and black triangles, respectively. (B) Quantitation of results. Expression values were normalized so that the average value in control cells at each time point was set to 1.0, and graphs show mean values (±standard deviation). The statistical significance of differences is shown. Vertical values show the P values for difference between that condition and control cells, whereas horizontal values show P values for the differences between soluble and bead-bound anti-IgM–treated samples at each time point (paired Student t test). (C) Correlation between fold induction of PERK and BIP in soluble/bead-bound anti-IgM–treated CLL samples (3-, 6-, and 24-hour data combined; linear regression and Spearman correlation shown).
Variable induction of PERK/BIP was not clearly different between M-CLL and U-CLL samples and did not correlate with ZAP-70 expression or sIgM expression. However, variation in the extent of PERK/BIP induction did appear to be related to the strength of sIgM-induced signaling analyzed using other readouts. First, there was no evidence for induction of PERK or BIP expression in 4 nonresponsive samples (supplemental Figure 6). Second, there was a modest, but significant, correlation between anti-IgM–induced ERK1/2 phosphorylation and BIP/PERK induction in a subset of signal-responsive samples (supplemental Figure 7). sIgM stimulation also increased BIP and PERK expression in normal B cells, although analysis was technically difficult due to the small number of B cells obtained for immunoblot analysis (supplemental Figure 8). Similar to unstimulated cells, we did not detect increased XBP1S expression in CLL cells treated with anti-IgM (data not shown).
Expression of UPR associated components in vivo
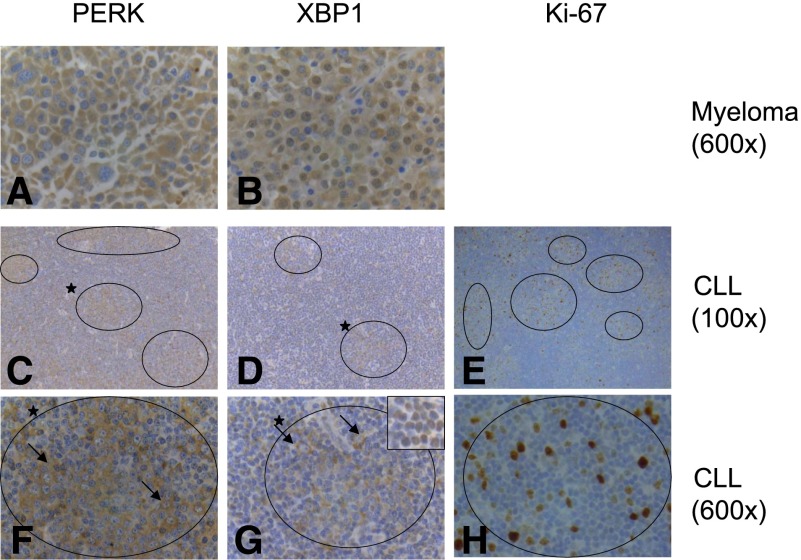
We performed immunohistochemistry to investigate UPR activation in the LNs of patients with CLL/small lymphocytic lymphoma (SLL) (Figure 6; supplemental Table 2). Because of the absence of suitable antibodies, analysis was restricted to PERK and XBP1. Comparisons were made to multiple myeloma, known to be associated with UPR activation.39
Figure 6.
UPR activation in CLL/SLL LNs. Immunohistochemical analysis of expression of (A,C,F) PERK, (B,D,G) XBP1, and (E,H) Ki-67 in (A-B) multiple myeloma and (C-H) CLL/SLL LNs. Original magnification of images are shown. Results are representative of a total of 11 biopsies analyzed. CLL PCs are circled, and higher-magnification images of the PC marked (*) in C and D are shown in F and G. The inset in G shows the nuclear expression of XBP1 in the small CLL cells in contrast to the cytoplasmic expression in the large blasts in the PCs. Arrows highlight large blasts in F and G. Images for C and F (PERK) and D and G (XBP1) are from samples 8 and 11, respectively. Inset shown in G is from sample 1.
Overall, PERK and XBP1 were widely detected in LN samples; 11 of 11 and 10 of 11 samples were positive for PERK and XBP1 expression, respectively. Similar to myeloma samples, PERK immunostaining was largely extranuclear, consistent with endoplasmic reticulum localization. In CLL, PERK was more strongly expressed in cells within PCs compared with surrounding cells in 6 of 11 samples. In 1 additional sample, expression was only detected in malignant cells within PCs. In the other samples, PERK expression was not different between cells within and outside of PCs. There was also variability in the distribution of XBP1 between individual samples, but 2 broad patterns of expression were observed. In 5 of 10 positive samples, XBP1 was predominantly detected in the nucleus (similar to the localization in myeloma samples) in cells outside of PCs. In the other positive samples, XBP1 expression was predominantly localized outside of the nucleus, and in these samples, expression was mainly detected in leukemic blasts within PCs. Overall, the analysis demonstrates that UPR-associated proteins were expressed within malignant LNs. Although there was substantial intrasample variation, features were frequently more prominent in cells within PCs consistent with the idea that UPR activation in CLL cells is a consequence of antigen engagement in vivo. Clinical data and/or matched blood samples were not available for these samples, so we were unable to correlate this variation to outcome or variable sIgM signaling capacity.
To explore further potential regulation of the UPR in vivo, we analyzed expression of BIP, CHOP, and XBP1 RNAs using Gene Expression Assay data from a study comparing CLL cells derived from blood and LNs.40 The 3 RNAs were more highly expressed in LN samples compared with blood. Differences were significant (paired Student t tests) for BIP and CHOP (P < .0001 and P = .0014, respectively) but not for XBP1 (P = .8995).
Effect of BCR signaling inhibitors on UPR regulation
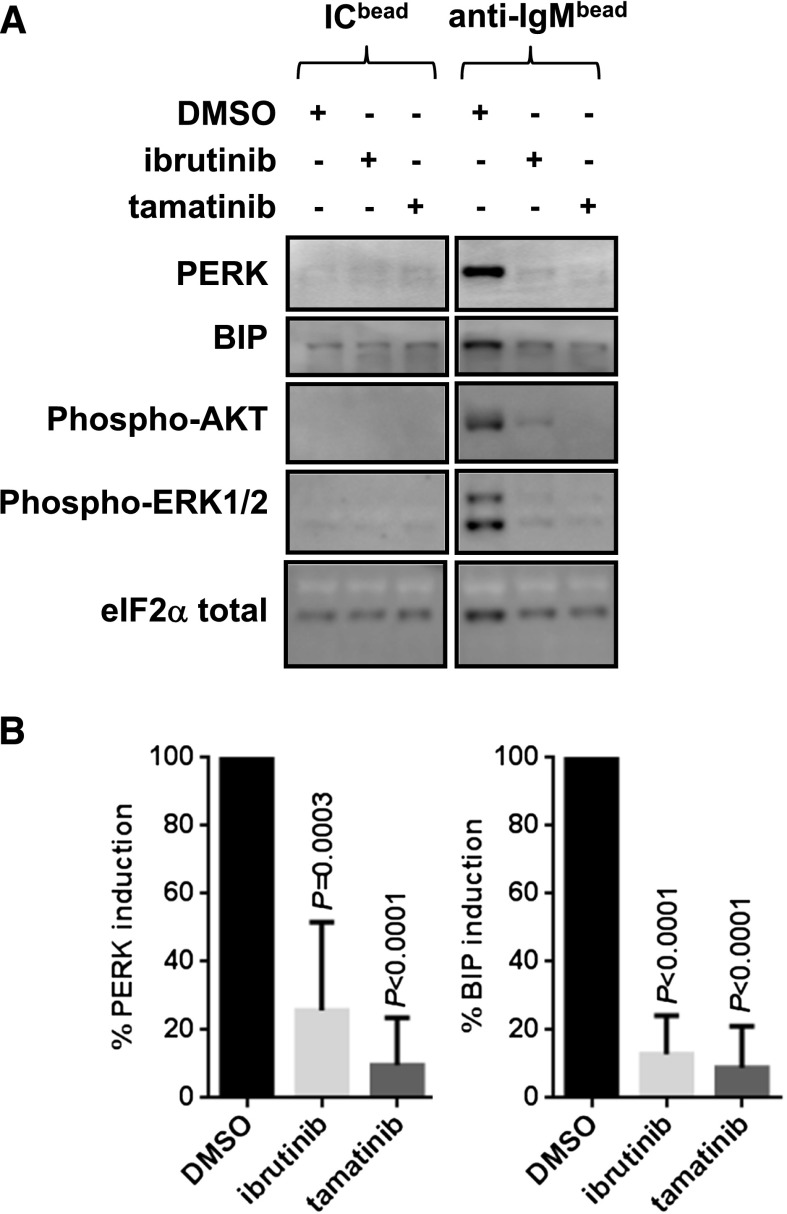
To investigate whether UPR induction was a direct consequence of activation of signaling pathways, CLL cells were pretreated with inhibitors of BCR-associated kinases prior to stimulation with bead-bound anti-IgM. The inhibitors tested were the clinical BTK and SYK inhibitors ibrutinib and tamatinib (the active form of fostamatinib). Both compounds significantly reduced anti-IgM–induced BIP and PERK expression (Figure 7). As expected, both inhibitors also effectively blocked induction of phosphorylation of both AKT and ERK1/2 (Figure 7). Thus, sIgM-induced UPR activation appears to mediate via kinase-dependent signaling pathways, and its inhibition may contribute to the therapeutic activity of agents such as ibrutinib.
Figure 7.
Effect of signaling inhibitors on anti-IgM–induced UPR activation. Cells were pretreated with dimethylsulfoxide, ibrutinib, or tamatinib for 30 minutes before being stimulated with bead-bound anti-IgM or control antibodies. Expression of PERK, BIP, phosphorylated ERK1/2, and phosphorylated AKT was analyzed at 24 hours. (A) Representative immunoblots. (B) Quantitation of results for all samples (n = 6 for ibrutinib and tamatinib). Graphs show inhibition of PERK/BIP expression with anti-IgM/dimethylsulfoxide–treated cells set to 100%. The statistical significance of differences between control and compound-treated control cells is shown (Student t test).
Discussion
BCR signaling has emerged as a key determinant of the clinical behavior of CLL and as an effective target for therapeutic attack. It is important, therefore, to define the functional consequences of sIg stimulation. In this work we investigated potential links between the BCR and the UPR, a multifunctional response pathway that can promote cell survival or death, dependent on the extent and duration of the activating signal. Several studies have shown that pharmacological inducers of the UPR promote apoptosis of CLL cells in vitro.33,34,36 However, the potential regulation of the UPR following BCR stimulation of CLL cells has not been studied previously.
Our results demonstrate that sIgM stimulation results in activation of a partial UPR. This conclusion is based on 3 lines of evidence. First, variable levels of basal activation of the UPR in unstimulated, circulating CLL cells correlated closely with sIgM signal capacity and were associated with more aggressive disease. Second, stimulation of sIgM in vitro increased expression of UPR components, and this was effectively blocked by BCR-targeted kinase inhibitors, including ibrutinib. Third, immunohistochemistry and GEA analysis demonstrated relatively high levels of UPR components in LNs in vivo. Interestingly, activation and therapeutic targeting of the UPR has also been reported during leukemogenesis in the Eμ-TCL1 mouse model of CLL,35,41 although the relevance of antigen signaling in vivo in this model remains unclear.
Our analysis demonstrated that sIgM stimulation, especially using bead-bound anti-IgM, triggered UPR induction using signal-responsive samples from both the M-CLL and U-CLL subsets. By contrast, anti-IgM did not significantly induce UPR activation in nonsignaling samples, indicating that the competency for UPR induction broadly correlates with sIgM signaling responsiveness measured using canonical readouts. There was variation in the extent of UPR induction within the signal-responsive samples. Although this did not obviously correlate with IGHV mutation status, ZAP-70 expression, or sIgM expression, there did appear to be a correlation between variable BIP/PERK induction and the strength of sIgM signaling (measured by parallel analysis of ERK1/2 phosphorylation) among signal-responsive samples. Further studies are required to probe relationships between sIgM-induced UPR activation and other signaling responses; however, UPR induction is likely to be part of a constellation of responses, coregulated downstream of sIgM in signal-responsive samples.4 Consistent with this, pretreatment of samples with ibrutinib or tamatinib effectively inhibited anti-IgM–induced UPR activation, providing functional evidence for linkage between kinase activation and UPR activation. We did not address consequences of sIgD stimulation in this study but have shown previously that, although competent for triggering initial calcium responses, anti-IgD fails to effectively engage downstream responses.17 Consistent with this, a recent GEA study showed that BIP RNA was induced in CLL samples following stimulation of sIgM but not sIgD.42
An important finding of the study was that UPR activation in CLL cells was partial. There was clear evidence for increased expression of CHOP and XBP1 RNAs and BIP protein. However, the PERK arm appeared to be only weakly activated because PERK expression was increased, but without substantial phosphorylation, and there were only modest levels of phosphorylation of its substrate eIF2α. Despite the induction of full-length XBP1 RNA, there was little evidence for IRE1-dependent processing to XBP1S, consistent with a previous study demonstrating only low expression of XBP1S protein in CLL/SLL LNs.43 Treatment of CLL cells with thapsigargin resulted in activation of all arms of the UPR. Thus, failure to activate some specific parts of the UPR likely represents the consequences of selective regulation rather than inherent defects that prevent induction of these specific arms. However, it was noticeable that XBP1S splicing was low, even in thapsigargin-treated cells, consistent with the idea that activation of this pathway may be relatively weak in CLL.33
Direct analysis of the functional consequences of UPR activation was not explored in this work; this would require knockdown of multiple proteins, which is technically difficult in any cell system, especially in CLL where RNA interference is extremely demanding. However, the molecular hallmarks of the partial UPR activation in CLL cells is very reminiscent of the anticipatory UPR that has been described in normal B cells. In this situation, selective activation of some UPR components is thought to prepare the cells for subsequent immunoglobulin secretion. For example, CHOP and XBP1 RNAs are induced within 1 to 2 hours following treatment of mouse B cells with IL4, whereas XBP1S splicing is detected much later at 48 hours after treatment and is dependent on enhanced immunoglobulin production.44
As in normal B cells, the partial UPR activation in CLL cells is likely to have a prosurvival function. First, anti-IgM–induced UPR activation in CLL cells lacks components typically associated with proapoptotic responses. IRE1 is the principle mediator of UPR-associated apoptosis, via downstream activation of proapoptotic kinases such as ASK1 and JNK (supplemental Figure 1). However, the absence of substantial XBP1S splicing, which is catalyzed by IRE1’s endonuclease activity, indicates that IRE1 is not effectively activated in CLL cells. Moreover, anti-IgM stimulation only very weakly induces JNK phosphorylation in CLL cells.16 Although CHOP is commonly considered as a proapoptotic factor, analysis of Chop-deficient mouse B cells has clearly demonstrated that CHOP does not play a proapoptotic role in B cells.31,45 Second, UPR activation in CLL cells is associated with increased expression of BIP, a chaperone with prosurvival functions.46 For example, in diffuse large B-cell lymphoma, a high level of BIP expression is associated with poor prognosis, and its overexpression confers resistance to apoptosis in vitro.47 BIP is induced in normal murine T cells following stimulation in vitro, and its ablation using RNAi promotes apoptosis in mouse EL4 T-lymphoma cells.48 The conclusion that partial UPR activation in CLL cells promotes survival is consistent with the previous observation that RNA interference-mediated knockdown of BIP promotes CLL cell apoptosis in vitro.32 However, it is possible that UPR activation has additional functional consequences. For example, Xbp1 is required for optimal signaling via sIgM and CXCR4, although the functional basis for these effects are unknown.28,41
The close correlation between basal UPR activation and retained sIgM signal capacity supports the idea that UPR activation is not simply an artifact of stimulation in vitro, but can also be a consequence of antigen engagement in vivo. Although antigen engagement is thought to be ongoing in all CLL, distinct biological responses appear to determine clinical behavior.1 Antigen-induced anergy is associated with strong down-modulation of sIgM signaling and is most prominent in M-CLL.11 By contrast, positive signaling is generally more evident in U-CLL and is associated with retained signaling capacity.11 Although the overall behavior of U-CLL and M-CLL is distinct, there is heterogeneity within these subsets, especially within M-CLL,11 and high levels of retained signaling in M-CLL may highlight cases at higher risk of progression.18 Overall, UPR activation appears to be one of several markers detected in circulating cells that reveal prior positive signaling within tissues (supplemental Figure 9). By contrast, strong down-modulation of sIgM signaling responses in vitro (including reduced capacity to enhance UPR activation) and lower levels of basal UPR activation are associated with anergy. Activation of an anticipatory UPR is linked to differentiation, and this linkage between anergy and reduced sIgM-induced UPR activation is consistent with the observation that differentiation responses are reduced in anergic cells in nonmalignant model systems.7 Moreover, very recent data demonstrate that IL21-induced differentiation responses are suppressed in anergic CLL cells.49 Further studies will be required to more accurately define the relationship between UPR activation, sIgM signal capacity, and disease behavior in larger, unselected cohorts. However, the expression of UPR components, along with other markers such as MYC and MCL1 that are also induced following sIgM-stimulation in vitro, may have utility as prognostic or predictive markers, including for new BCR-targeted kinase inhibitors, possibly including within the M-CLL subset.
In summary, our studies led to the novel observation that sIgM stimulation in CLL cells results in partial activation of the UPR. UPR activation appears to contribute to the growth promoting effects of BCR stimulation and is associated with more aggressive disease. Inhibition of UPR activation may contribute to the therapeutic effects of novel drugs targeted toward BCR-associated signaling kinases, including BTK and SYK.
Acknowledgments
The authors thank the patients involved in this study for the kind gift of samples. The authors also thank Drs Simon Wagner, Robert Corser, Abraham Jacob, and Henri Grech and their associated clinical teams for support and Isla Henderson and Ian Tracy for technical support.
This work was supported by the Kay Kendall Leukaemia Fund, Leukaemia and Lymphoma Research, Worldwide Cancer Research, Cancer Research UK, the Southampton Experimental Cancer Medicine Centre, and the University of Southampton. M.S.H. gratefully acknowledges support from a Postgraduate National Program of José Castillejo fellowship and financial sponsorship from the Spanish Ministry of Education.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.K., V.C., M.S.H., A.L., and M.C. performed research and analyzed data; S.K., A.J.S., M.A.-K., F.K.S., and G.P. designed the research and analyzed data; K.N.P., B.K., A.S.D., and F.F. provided patient samples and analyzed clinical data; S.K. and G.P. wrote the initial draft of the manuscript; and all authors contributed to the modification of the draft and approved the final submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.K. is Barts Cancer Institute, Queen Mary, University of London, John Vane Science Centre, Charterhouse Square, London EC1M 6BQ, UK.
The current affiliation for V.C. is Hematology Department, University College of London–Cancer Institute, 72 Huntley St, London WC1E 6BT, UK.
The current affiliation for M.S.H. is Department of Pharmacology, University of Seville, No. 2, 41012 Seville, Spain.
Correspondence: Sergey Krysov, Barts Cancer Institute, Queen Mary, University of London, John Vane Science Centre, Charterhouse Square, London, EC1M 6BQ, UK; e-mail: s.krysov@qmul.ac.uk.
References
- 1.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118(16):4313–4320. doi: 10.1182/blood-2011-06-338855. [DOI] [PubMed] [Google Scholar]
- 2.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120(6):1175–1184. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang S, Kipps TJ. The pathogenesis of chronic lymphocytic leukemia. Annu Rev Pathol. 2014;9:103–118. doi: 10.1146/annurev-pathol-020712-163955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Packham G, Krysov S, Allen A, et al. The outcome of B-cell receptor signaling in chronic lymphocytic leukemia: proliferation or anergy. Haematologica. 2014;99(7):1138–1148. doi: 10.3324/haematol.2013.098384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forconi F, Potter KN, Wheatley I, et al. The normal IGHV1-69-derived B-cell repertoire contains stereotypic patterns characteristic of unmutated CLL. Blood. 2010;115(1):71–77. doi: 10.1182/blood-2009-06-225813. [DOI] [PubMed] [Google Scholar]
- 6.Seifert M, Sellmann L, Bloehdorn J, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med. 2012;209(12):2183–2198. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cambier JC, Gauld SB, Merrell KT, Vilen BJ. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nat Rev Immunol. 2007;7(8):633–643. doi: 10.1038/nri2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coelho V, Krysov S, Steele A, et al. Identification in CLL of circulating intraclonal subgroups with varying B-cell receptor expression and function. Blood. 2013;122(15):2664–2672. doi: 10.1182/blood-2013-02-485425. [DOI] [PubMed] [Google Scholar]
- 10.Calissano C, Damle RN, Marsilio S, et al. Intraclonal complexity in chronic lymphocytic leukemia: fractions enriched in recently born/divided and older/quiescent cells. Mol Med. 2011;17(11-12):1374–1382. doi: 10.2119/molmed.2011.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mockridge CI, Potter KN, Wheatley I, Neville LA, Packham G, Stevenson FK. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood. 2007;109(10):4424–4431. doi: 10.1182/blood-2006-11-056648. [DOI] [PubMed] [Google Scholar]
- 12.Muzio M, Apollonio B, Scielzo C, et al. Constitutive activation of distinct BCR-signaling pathways in a subset of CLL patients: a molecular signature of anergy. Blood. 2008;112(1):188–195. doi: 10.1182/blood-2007-09-111344. [DOI] [PubMed] [Google Scholar]
- 13.Apollonio B, Scielzo C, Bertilaccio MT, et al. Targeting B-cell anergy in chronic lymphocytic leukemia. Blood. 2013;121(19):3879–3888, S1-S8. doi: 10.1182/blood-2012-12-474718. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Kater AP, Widhopf GF, II, et al. B-cell activating factor and v-Myc myelocytomatosis viral oncogene homolog (c-Myc) influence progression of chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2010;107(44):18956–18960. doi: 10.1073/pnas.1013420107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pepper C, Lin TT, Pratt G, et al. Mcl-1 expression has in vitro and in vivo significance in chronic lymphocytic leukemia and is associated with other poor prognostic markers. Blood. 2008;112(9):3807–3817. doi: 10.1182/blood-2008-05-157131. [DOI] [PubMed] [Google Scholar]
- 16.Petlickovski A, Laurenti L, Li X, et al. Sustained signaling through the B-cell receptor induces Mcl-1 and promotes survival of chronic lymphocytic leukemia B cells. Blood. 2005;105(12):4820–4827. doi: 10.1182/blood-2004-07-2669. [DOI] [PubMed] [Google Scholar]
- 17.Krysov S, Dias S, Paterson A, et al. Surface IgM stimulation induces MEK1/2-dependent MYC expression in chronic lymphocytic leukemia cells. Blood. 2012;119(1):170–179. doi: 10.1182/blood-2011-07-370403. [DOI] [PubMed] [Google Scholar]
- 18.Paterson A, Mockridge CI, Adams JE, et al. Mechanisms and clinical significance of BIM phosphorylation in chronic lymphocytic leukemia. Blood. 2012;119(7):1726–1736. doi: 10.1182/blood-2011-07-367417. [DOI] [PubMed] [Google Scholar]
- 19.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 20.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 21.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8(9):663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 22.Reimold AM, Iwakoshi NN, Manis J, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 23.Iwakoshi NN, Lee AH, Glimcher LH. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev. 2003;194:29–38. doi: 10.1034/j.1600-065x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115(2):268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA. 2003;100(17):9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrasco DR, Sukhdeo K, Protopopova M, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11(4):349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aronson LI, Davies FE. DangER: protein ovERload. Targeting protein degradation to treat myeloma. Haematologica. 2012;97(8):1119–1130. doi: 10.3324/haematol.2012.064923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu CC, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28(11):1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277(50):49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- 30.van Anken E, Romijn EP, Maggioni C, et al. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18(2):243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 31.Skalet AH, Isler JA, King LB, Harding HP, Ron D, Monroe JG. Rapid B cell receptor-induced unfolded protein response in nonsecretory B cells correlates with pro- versus antiapoptotic cell fate. J Biol Chem. 2005;280(48):39762–39771. doi: 10.1074/jbc.M502640200. [DOI] [PubMed] [Google Scholar]
- 32.Rosati E, Sabatini R, Rampino G, et al. Novel targets for endoplasmic reticulum stress-induced apoptosis in B-CLL. Blood. 2010;116(15):2713–2723. doi: 10.1182/blood-2010-03-275628. [DOI] [PubMed] [Google Scholar]
- 33.Lust S, Vanhoecke B, Van Gele M, et al. Xanthohumol activates the proapoptotic arm of the unfolded protein response in chronic lymphocytic leukemia. Anticancer Res. 2009;29(10):3797–3805. [PMC free article] [PubMed] [Google Scholar]
- 34.Carew JS, Nawrocki ST, Krupnik YV, et al. Targeting endoplasmic reticulum protein transport: a novel strategy to kill malignant B cells and overcome fludarabine resistance in CLL. Blood. 2006;107(1):222–231. doi: 10.1182/blood-2005-05-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriss CL, Pinilla-Ibarz JA, Mailloux AW, et al. Overexpression of TCL1 activates the endoplasmic reticulum stress response: a novel mechanism of leukemic progression in mice. Blood. 2012;120(5):1027–1038. doi: 10.1182/blood-2011-11-394346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahoney E, Maddocks K, Flynn J, et al. Identification of endoplasmic reticulum stress-inducing agents by antagonizing autophagy: a new potential strategy for identification of anti-cancer therapeutics in B-cell malignancies. Leuk Lymphoma. 2013;54(12):2685–2692. doi: 10.3109/10428194.2013.781168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanham S, Hamblin T, Oscier D, Ibbotson R, Stevenson F, Packham G. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood. 2003;101(3):1087–1093. doi: 10.1182/blood-2002-06-1822. [DOI] [PubMed] [Google Scholar]
- 38.Efremov DG, Gobessi S, Longo PG. Signaling pathways activated by antigen-receptor engagement in chronic lymphocytic leukemia B-cells. Autoimmun Rev. 2007;7(2):102–108. doi: 10.1016/j.autrev.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Vincenz L, Jäger R, O’Dwyer M, Samali A. Endoplasmic reticulum stress and the unfolded protein response: targeting the Achilles heel of multiple myeloma. Mol Cancer Ther. 2013;12(6):831–843. doi: 10.1158/1535-7163.MCT-12-0782. [DOI] [PubMed] [Google Scholar]
- 40.Herishanu Y, Pérez-Galán P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117(2):563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang CH, Ranatunga S, Kriss CL, et al. Inhibition of ER stress-associated IRE-1/XBP-1 pathway reduces leukemic cell survival. J Clin Invest. 2014;124(6):2585–2598. doi: 10.1172/JCI73448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavolaro S, Peragine N, Chiaretti S, et al. IgD cross-linking induces gene expression profiling changes and enhances apoptosis in chronic lymphocytic leukemia cells. Leuk Res. 2013;37(4):455–462. doi: 10.1016/j.leukres.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 43.Maestre L, Tooze R, Cañamero M, et al. Expression pattern of XBP1(S) in human B-cell lymphomas. Haematologica. 2009;94(3):419–422. doi: 10.3324/haematol.2008.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4(4):321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 45.Masciarelli S, Fra AM, Pengo N, et al. CHOP-independent apoptosis and pathway-selective induction of the UPR in developing plasma cells. Mol Immunol. 2010;47(6):1356–1365. doi: 10.1016/j.molimm.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Lee AS. Stress induction of GRP78/BiP and its role in cancer. Curr Mol Med. 2006;6(1):45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- 47.Mozos A, Roué G, López-Guillermo A, et al. The expression of the endoplasmic reticulum stress sensor BiP/GRP78 predicts response to chemotherapy and determines the efficacy of proteasome inhibitors in diffuse large b-cell lymphoma. Am J Pathol. 2011;179(5):2601–2610. doi: 10.1016/j.ajpath.2011.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takano S, Ando T, Hiramatsu N, et al. T cell receptor-mediated signaling induces GRP78 expression in T cells: the implications in maintaining T cell viability. Biochem Biophys Res Commun. 2008;371(4):762–766. doi: 10.1016/j.bbrc.2008.04.132. [DOI] [PubMed] [Google Scholar]
- 49.Duckworth A, Glenn M, Slupsky JR, Packham G, Kalakonda N. Variable induction of PRDM1 and differentiation in chronic lymphocytic leukemia is associated with anergy. Blood. 2014;123(21):3277–3285. doi: 10.1182/blood-2013-11-539049. [DOI] [PubMed] [Google Scholar]