Key Points
The majority of mutations are found in genes that have low or no detectable biological expression.
Mutated genes often show differential allelic expression in multiple myeloma patient samples.
Abstract
Recent work has delineated mutational profiles in multiple myeloma and reported a median of 52 mutations per patient, as well as a set of commonly mutated genes across multiple patients. In this study, we have used deep sequencing of RNA from a subset of these patients to evaluate the proportion of expressed mutations. We find that the majority of previously identified mutations occur within genes with very low or no detectable expression. On average, 27% (range, 11% to 47%) of mutated alleles are found to be expressed, and among mutated genes that are expressed, there often is allele-specific expression where either the mutant or wild-type allele is suppressed. Even in the absence of an overall change in gene expression, the presence of differential allelic expression within malignant cells highlights the important contribution of RNA-sequencing in identifying clinically significant mutational changes relevant to our understanding of myeloma biology and also for therapeutic applications.
Introduction
Multiple myeloma (MM) is an incurable neoplastic disease involving the proliferation of monoclonal antibody producing plasma cells.1 MM is a heterogeneous disease but the hallmark genetic changes include several genomic rearrangements, such as translocations involving the IgH locus or hyperdiploidy.2 The pathogenic molecular changes and the processes that drive genomic instability during the development and evolution of the disease are complex and incompletely understood. Elucidating the precise genetic changes that drive malignant transformation, affect the phenotypic behavior of the disease, and alter treatment response is an essential component of our drive toward individualized therapy.3 Recent studies have focused on attempts to identify individual driver mutations that might provide both prognostic information and unique therapeutic targets. Whole genome and whole exome sequencing of increasingly large numbers of patient samples have identified a number of commonly mutated genes in MM patients, such as CCND1, NRAS, KRAS, BRAF, TP53, and FAM46C.4-8 However, none of these mutations are found in more than one quarter of patients, and most are found in less than 10% of samples sequenced.
We recently reported a large cohort of MM exome sequences involving 84 samples from 67 patients. Of these patients, 15 contributed samples from multiple time points during disease evolution.9 We again identified a diverse set of gene mutations with significant heterogeneity across our cohort, with a median of 52 (range, 21-488) mutations per sample. Computational approaches can be used to prioritize mutations that are expected to alter protein structure and function, or to lie in likely driver genes. It is more challenging to determine which mutations are likely to be clinically meaningful or even expressed in MM patients.
Although a number of studies have interrogated the role of mutational changes in cancer and in myeloma, the phenotypic impact has not been evaluated for the majority of these changes.10 The ultimate significance of these genetic changes will depend on whether the mutated allele is expressed, and whether the mutation affects expression, splicing, or function of the gene product. Deep sequencing of RNA (RNA-seq), in combination with whole exome sequencing, provides an opportunity for direct quantitation of allele-specific gene expression, as well as a tool to answer these clinically relevant questions.
In this study, we perform RNA-seq on 14 samples from 10 patients, for which we have previously evaluated exome sequence, some of whom have samples from 2 time points during disease evolution. For the first time, we report allele-specific expression and correlate it to the DNA mutant allele frequency in MM patient samples. We find that the majority of identified DNA mutations are not expressed at detectable levels, and that unbalanced allelic expression of mutant alleles is a relatively common occurrence in MM patients.
Materials and methods
Clinical samples
We have performed whole exome sequencing on purified myeloma cells. Their mutational spectrum was previously described.9 Fourteen of these samples with adequate quantity of available RNA were chosen to evaluate allelic-specific expression. Samples were collected after written informed consent was obtained. Samples and data were obtained and managed in accordance with the Declaration of Helsinki under protocol 08/H0308/303: somatic molecular genetics of human cancers, melanoma, and myeloma (Dana-Farber Cancer Institute). The same protocol was approved by the National Research Ethics Service Committee East of England—Cambridge Central.
RNA collection, sequencing, and read mapping
RNA purification and preparation was performed as previously described.9 Total RNA was first put through quality control (QC). RNA quantity was determined on the Qubit using the Qubit RNA Assay Kit (Life Technologies, Carlsbad, CA) and RNA quality was determined on the Bioanalyzer using the RNA Pico Kit (Agilent, Santa Clara, CA). Using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA), 100 ng of total RNA was converted into a DNA library following the manufacturer’s protocol, with no modifications. Following library construction, DNA libraries were then put through QC. Library quantity was determined using the Qubit High Sensitivity DNA Kit (Life Technologies) and library size was determined using the Bioanalyzer High Sensitivity Chip Kit (Agilent). Finally, libraries were put through quantitative polymerase chain reaction (PCR) using the Universal Library Quantification Kit for Illumina (Kapa Biosystems, Wilmington, MA) and run on the 7900HT Fast quantitative PCR machine (ABI, Grand Island, NY). Libraries passing QC were diluted to 2 nM using sterile water, and then sequenced on the HiSequation 2000 (Illumina, San Diego, CA) at a final concentration of 12 pM, following all manufacturer’s protocols. Two samples per lane of data were sequenced via duplex sequencing, yielding a theoretical maximum of approximately 100 million 50-nucleotide paired-end sequenced reads per sample (200 million reads per lane). The actual number of sequenced reads per sample, along with other information, is given in supplemental Data Set NM 1 on the Blood Web site.
We mapped RNA-seq reads to the human genome (build hg19) using TopHat 2.0.10 with default parameters,11 and a gene annotation file from the Illumina iGenomes Web site corresponding to Ensembl GRCh37. We quantified gene abundances from mapped reads using the “htseq-count” function from HTSequation 0.6.1,12 also utilizing this annotation file, and the options “-m intersection nonempty -t exon -i gene_id.” We transformed gene-level counts to fragments per kilobase per million (FPKM) fragments mapped values, dividing each count by the total length of nonoverlapping exons of the gene. To determine the percentage of reads originating from the immunoglobulin heavy (IGH) locus for each patient, we summed gene-level counts from HTSeq and divided the total counts from IGH-related genes by this quantity. For comparison purposes, we also computed estimated gene-level counts values using RSEM software,13 which uses an internal mapping procedure independent of TopHat to quantify gene level abundances, adjusting counts for multiply mapping reads and biases in the data. We used default parameters, except for the “–estimate-rspd–no-bam-output–paired-end–calc-ci” options. We found that this alternative approach caused little change in the percentage of gene-level counts originating from IGH-related genes (supplemental Data Set 1).
Mutant allele abundance, QC, quantification, and testing
For our RNA-seq data, we took several QC measures to ensure accurate quantification of relative allele abundances at each mutation location identified in the previous exome sequencing study. We first filtered and mapped RNA-seq reads for each sample, to keep only uniquely mapped reads, using SAMtools 0.1.17,14 and then further removed potential PCR duplicates using the MarkDuplicates function in Picard 1.7, with default parameters. These QC steps have been shown to reduce the likelihood of false positives and biases in observed mutant allele frequencies during mutation calling using RNA-seq data.15 We calculated allelic counts at each single-nucleotide variant position in each sample using the SAMtools mpileup function with default parameters, except for the “-A” option and the “-l” option to specify the list of mutation coordinates, which we took, for each sample, from the list of validated mutations in our prior study.9 We recorded the number of overlapping sequences containing the mutant and wild-type (WT) allele, as identified in the previous study for each position and sample. We defined the mutant allele frequency as the number of covering RNA-seq reads containing the mutant allele at that position, divided by the total number of RNA-seq reads overlapping that position. We compared these frequencies to those calculated analogously at the DNA level in the previous exome sequencing study.
For a given mutation observed in a patient, we tested for significant departures in RNA and DNA mutational burden through a Bayesian hypothesis-testing framework,16 accounting for uncertainty in allelic abundance due to variability in total read depth at the mutation locus in each data type (supplemental Methods). Briefly, for each mutation and patient, we computed probabilities that the observed mutant allele frequencies in both the RNA and DNA would occur, assuming the true frequencies are either the same (null hypothesis) or distinct (alternative hypothesis). The ratio of these probabilities yields a Bayes Factor, where values >10 indicate strong evidence favoring a difference in mutant allele frequencies. We use these Bayes Factors to measure evidence in favor or against differences in the DNA and RNA mutant allele frequencies. We also extend this test to 2 time points, looking for joint changes in both RNA and DNA mutant allele frequency over time points.
Results
Limited detectable expression of mutant allele in MM
In 14 samples from 10 patients with exome sequencing data and sufficient RNA available, we performed RNA-seq (supplemental Data Set 1) using the standardized pipeline described in “Materials and methods.” Of these 10 patients, 4 had RNA samples collected at 2 time points (labeled “early” and “late”) matching those used for our prior evolutionary analysis.9 The remaining 6 patients had RNA and DNA sequencing data from only a single time point (supplemental Table 1). A total of 981 validated DNA mutations were previously identified across these 14 samples (supplemental Data Set 2). The number of validated mutations per sample varied significantly among patients (supplemental Figure 1), reflecting the heterogeneity in mutational profiles observed between patients. Overall, mapping rates for our samples were high (65% to 89%) and the number of reads mapping to the IGH locus did not dominate our RNA-seq data (supplemental Data Set 1).
We first evaluated whether our previously identified mutations were found in expressed transcripts from our RNA-seq data after QC with the goal of determining the relative frequency of the variant sequence of a given mutation in expressed transcripts. We define “presence” as having at least one covering RNA-seq read containing the mutant allele. To show the specificity of our approach, we also demonstrate that our QC steps reduce the probability of identifying the mutant allele in nonmutated genes in other samples (supplemental Figure 2). We consider the presence of the mutant allele in nonmutated genes from other samples to be a false positive due to technical artifacts. From here onwards, the term “RNA-seq read coverage” refers to the number of reads that remain after all QC steps.
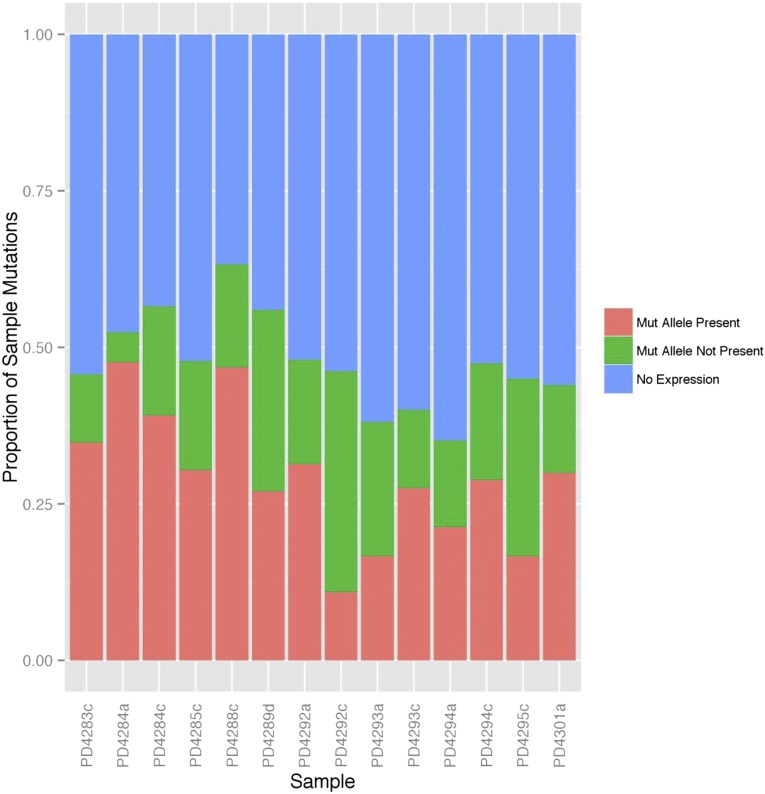
In all, only 47% (462/981; range, 35% to 65% across patients) of the total number of mutant genes from all samples had non-zero RNA-seq read coverage, and only 27% (261/981; range, 11% to 48% of mutations) had at least one covering RNA-seq read containing the mutant allele (mutant allele present). This percentage again varied considerably across samples (plotted in Figure 1) but is in line with previously published studies.17,18 The majority of mutations showing “no expression” in Figure 1 originated from genes with low or no detectable expression, defined as having FPKM <3 (supplemental Figure 1, blue and purple boxes). However, among those mutations that have at least 10 supporting RNA-seq reads, the percentage expressing the mutant allele is 69.9% (144/206 single-nucleotide variants; supplemental Figure 3). Local RNA-seq read coverage is related to the level of gene expression (supplemental Figure 4), therefore, higher biological expression allows for easier detection of the mutant allele.
Figure 1.
The majority of mutations are not expressed in MM patient samples. Mutations identified in the exome sequencing data were classified into 3 groups: mutations with the mutant allele found in the RNA-seq (salmon), those with the mutant allele not found in the RNA-seq despite RNA-seq read coverage of the mutation region (green), and those with no covering RNA-seq reads (blue, “No Expression”). The percentage of each type of mutation within each sample is shown.
The majority of mutations (64%) were found in genes with low or no detectable expression. We define low gene-level expression as FPKM <3. This value corresponds to the 60th percentile of expression across all samples. If we use a threshold FPKM of 1, this percentage drops to 53 and other results do not change significantly (supplemental Figures 5 and 6).
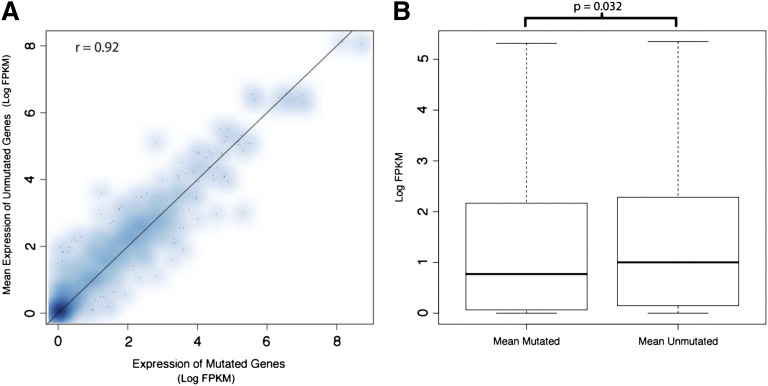
We next asked whether the presence of a mutation itself altered the expression of the mutant genes. In Figure 2A, the expression level of each mutated gene was plotted against the mean expression of that same gene from samples not containing the mutation. We found that mutant and WT gene expression across patients, for each gene, was similar (Spearman correlation of 0.92). Because of the limited number of samples, and the limited number of genes mutated in multiple samples, our study does not provide sufficient cases to formally investigate a link between the presence of an individual mutation and gene expression in our study. However, when we compared the overall expression of mutated genes to the expression of nonmutated versions of the same genes in other samples, we observed lower expression among the mutated genes relative to the nonmutated genes (permutation P value for difference in log median expression = .032; Figure 2B).
Figure 2.
The average expression of genes carrying a mutation is similar to those in samples without the mutation, but aggregated expression of mutated genes is lower. (A) The expression level (as measured by log FPKM) of genes in the mutant sample was plotted against the average expression of the same gene in samples not harboring the mutation. The Spearman correlation is 0.92. Due to the limited sample size and limited number of samples carrying a mutated copy of a given gene, we do not have strong evidence to demonstrate lower expression of mutated genes on an individual gene basis. (B) The distribution of expression (log FPKM) for all mutated genes aggregated together is plotted with the expression of the unmutated versions of these genes in other samples. We find slightly lower expression within the mutant gene group (permutation P = .032). Such aggregation of the data helps to overcome the per-gene sample size limitation mentioned previously.
Expression pattern of frequently mutated genes in MM
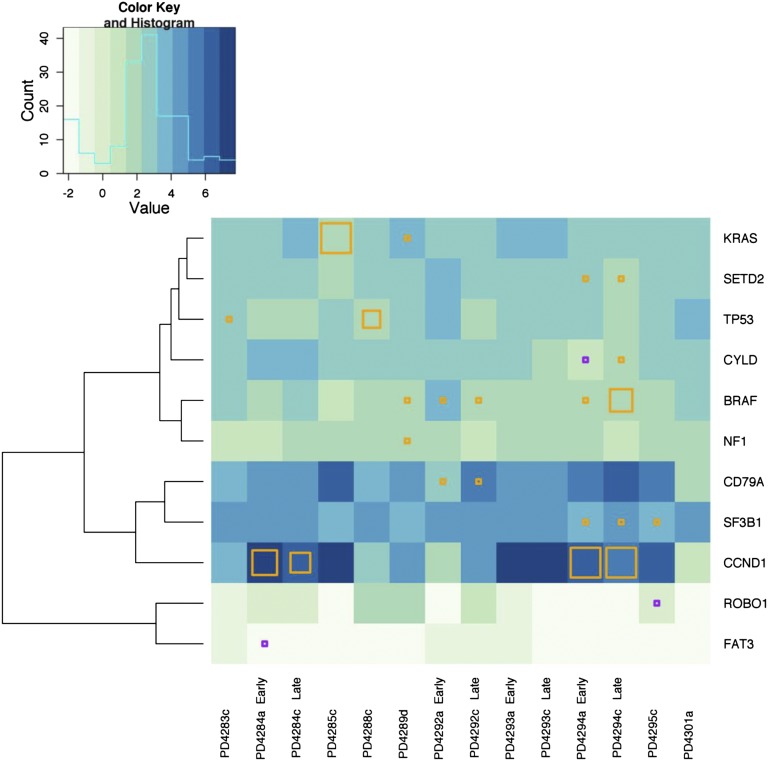
Our prior exome sequencing study identified a subset of genes mutated at increased frequency across the patient panel and these were identified as possible driver mutations. We evaluated whether these frequently mutated genes were expressed in our subset of MM patients. We found that the expression of these genes varied across samples. In Figure 3, we clustered the samples based upon the relative expression level of these frequently mutated genes, marking the mutational status in each of our 14 samples. We found that the majority of the mutated genes were expressed at a level above 3 FPKM (Figure 3 and supplemental Data Set 2). Exceptions were FAT3 (FPKM = 0.9), ROBO1 (FPKM = 1.2), and CYLD (FPKM = 2.4). Although there was heterogeneity in terms of gene expression between patients, early and late samples from the same patient tended to be similar (Figure 3). Interestingly, the exception to this was patient PD4292, who previously demonstrated a pattern of marked clonal change between time points.9
Figure 3.
Frequently mutated genes are expressed in MM patient samples. Hierarchical clustering across all 14 samples based upon gene expression of our previously identified, frequently mutated genes was performed. Only those genes with at least 1 mutation in our RNA sequenced subset of 14 samples are shown. The presence of a mutation in a gene with FPKM >3 is denoted by an orange square and genes with an FPKM <3 is denoted with a purple square. The size of the square reflects the frequency of the mutant allele as compared with the WT within our RNA-seq data. A histogram of log FPKM is displayed in the upper left corner to illustrate the distribution of the logarithm of gene expression levels across patients and the genes displayed in the figure.
Mutant and WT alleles may be differentially expressed
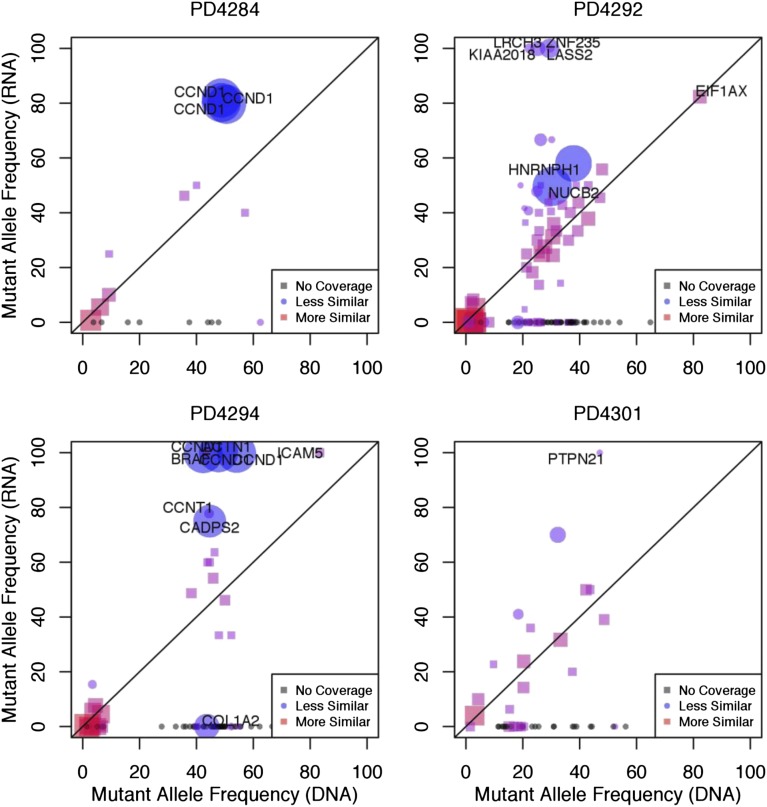
The frequency with which a specific mutation is found within expressed RNA transcripts will depend on the clonal frequency of the mutation at the DNA-level, in addition to the relative expression level of RNA transcripts containing the mutant allele vs those containing the WT allele. To determine whether mutant alleles were being differentially expressed as compared with their WT counterpart, we compared the frequency of each mutant sequence in the exome sequencing data to its corresponding frequency in the RNA-seq data. If each allele was used equally during transcription, we would expect that the prevalence of the mutant allele in the RNA would be consistent with its prevalence in DNA.17 We plotted the frequency with which each mutation is found within the RNA-seq data against DNA mutant allele frequency (Figure 4 and supplemental Figure 7). We observed a number of mutations for which expression of the mutant allele was absent in the RNA, and conversely, many others where the expression of the WT allele was absent. Because the accuracy of this approach is dependent upon the number of reads covering the mutant sequence, we computed the relative probabilities of the frequencies being equal vs different using a Bayesian hypothesis test (supplemental Methods), accounting for variable read coverage in both DNA and RNA, as well as possible technical variability in the underlying mutant allele frequency from other sources.
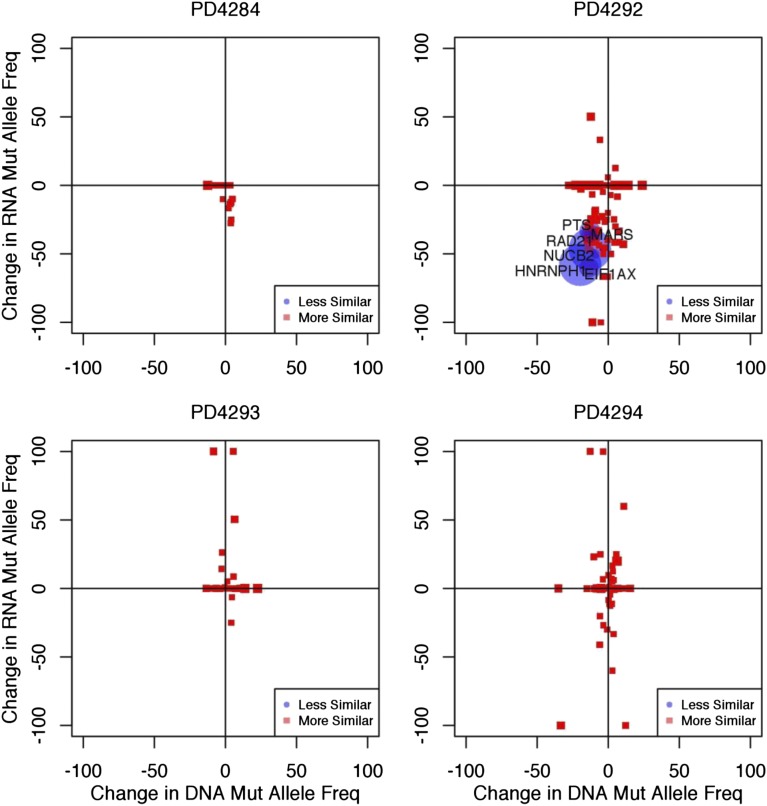
Figure 4.
Mutant alleles are differentially expressed. The mutant allele frequency of each mutation observed in the RNA-seq data (y-axis) was plotted against the mutant allele frequency observed in the exome sequencing data (x-axis). We define the mutational frequency as the number of sequencing reads covering a mutation that contain the mutant allele divided by the total number of covering sequencing reads. The degree of similarity between the exome and RNA-seq data are represented by red squares (more similar) and blue circles (less similar). Plots are shown for 4 representative samples. The size of the point on the plot (circle or square) represents the level of statistical certainty, as measured by a Bayesian hypothesis test assessing the dissimilarity in the mutant allele frequencies, given the coverage of a mutation in the RNA-seq and exome sequencing data. Some genes, such as CCND1 contain multiple mutations, and are therefore represented by more than 1 point on the graph. Gray points on the plot correspond to mutations that have zero RNA-seq read coverage and are placed on the plot to show their mutant allele-frequency in DNA.
For example, the mutant allele of the driver gene CCND1 in patient PD4294 was expressed almost exclusively, compared with the WT allele in both the early and late time points (Bayes Factor >10), indicating significant differential expression at this locus (Figure 4 and supplemental Table 2). In patient PD4284, the CCND1 mutant allele was similarly expressed at a higher level than would be predicted based on DNA frequencies (Figure 4 and supplemental Table 2). Another gene showing a similar pattern in patient PD4288 was PARP4 (supplemental Data Set 2). Conversely, in patient PD4292, the mutant allele frequency of EIF1AX was lower than would be expected, despite the EIF1AX gene’s overall expression (FPKM = 18.9) in that particular sample. These results indicate that mutations found in DNA may be variably transcribed or not transcribed at all.
Clonal change in RNA, mirrors that seen in the DNA
In our prior study, we found distinct patterns of clonal evolution in MM over time, defining evolutionary patterns such as linear evolution, branching evolution, and differential clonal response.9 We next plotted the change in mutant allele frequency within the RNA between time points against the corresponding change within the DNA (Figure 5). The small number of mutations covered adequately at both time points limited our analysis of clonal change in the RNA. We found that concomitant changes in both DNA and RNA mutant allele frequency were only observed in the samples that were previously observed to show mutational evolution over time (PD4292). We observed no significant changes in both DNA and RNA in samples showing no change over time (Figure 5 and supplemental Figure 8). In PD4292, such changes in DNA and RNA were largely decreases, indicating a loss of mutant allele frequency, possibly related to treatment between the time points, leading to loss of subclones expressing some mutations.
Figure 5.
Change in RNA expression of mutant alleles over time correlates with change in mutant allele frequency. The change in the mutant allele frequency in the RNA between time points (y-axis) was plotted against the change in the mutant allele frequency in DNA (x-axis) from the early time point to the late time point for each patient. The size of the point on the plot represents the degree of statistical certainty as measured by a Bayesian hypothesis test assessing simultaneous change in both DNA and RNA mutant allele frequency between time points. The degree of the similarity is represented by red (more similar) and blue (less similar), and the significance of the difference is represented by the size of point (larger point indicates greater significance).
Discussion
MM is a disease with heterogeneous molecular and genetic changes.2 We previously identified a number of gene mutations in MM patient samples, some common across multiple patients.9 Although this and other mutational analyses identify recurrent mutations, the relevance of these mutations for myeloma cell growth, survival, or its biological behavior and response or resistance to therapy remains unclear. In this study, we have for the first time in MM, used RNA-seq to examine the relationship between mutational status of a gene and its allelic expression as an indicator of its ability to affect cellular behavior. The majority of the mutated genes identified in the previous study (64%) have low or no detectable biological expression, suggesting that many, if not most, mutations may be biologically silent bystander events. We predict that these mutations are therefore unlikely to be functionally relevant. Our target sequencing depth of 100-million paired-end reads provide an adequate number of reads to determine the expression status of the majority of mutant genes, as those genes not covered are unlikely to be expressed at a biologically significant level. The ability to detect mutant transcripts is associated with the level of gene expression (supplemental Figures 1 and 3-6); therefore, we believe that increasing sequencing depth would be unlikely to significantly increase our yield. Moreover, we assume mutations that would be detected via deeper sequencing would likely have lower functional impact, as they would be found in lowly expressed transcripts that were not detected at our original levels of sequencing.
Our finding that the mutant allele is present in the RNA-seq data for only 27% of our initial set of mutations (on average, across patients) is in line with other cancer genome and transcriptome-sequencing reports.17-20 There is no strong evidence that the majority of mutations directly alter the overall expression levels of their associated genes (Figure 2A), but we do see that mutated genes have slightly lower expression than nonmutated genes overall (Figure 2B). Previous studies have shown that highly mutated gene regions tend to have lower levels of gene expression21; however, because our mutation data comes from exome sequencing, we are unable to assess whether mutations found within enhancer or promoter regions affect gene expression. We would also like to emphasize that the main conclusions of our study stem from a patient-by-patient analysis, and that none of the mutation-level analyses and tests were performed on the patient samples as a whole. Our main goal in this study is to show that patient-specific mutations found in DNA are not always found in RNA, or have limited or differential expression. For other generalized conclusions, a significantly larger study will be required. We also note that we ran several QC measures to ensure that the mutant alleles being detected were of high quality, in order to reduce the probability that our conclusions may result from experimental artifacts.
Many of the single nucleotide changes identified in our exome sequencing were predicted to produce nonsense mutations. One might expect these to be absent from the sequenced transcriptome due to nonsense-mediated RNA decay. Although this may be the case for a small subset of genes, we think it is unlikely to explain the majority of our findings, as we would expect it to lead to a significant overall decrease in the expression of mutant genes, which is something that we did not observe. In contrast to the mutated gene pool at large, we previously identified a subset of commonly mutated genes within our MM patient panel. The majority of these genes were expressed at a moderate or high level in MM cells. Of the 28 genes in our study, only 11 exhibited mutations in the 14 samples we examined, and we were able to detect mutant transcripts in 9 of these 11 genes. These genes could play an important role in disease pathogenesis, especially considering that many of them are known to be involved in other cancer types. Among these genes, mutations found in the majority of the malignant cells (clonal) and also showed expression of the mutant transcript, would be candidate functional driver mutations worthy of future evaluation. However, we found that only a small number of these mutated genes showed detectable expression of the mutated allele, consistent with the allele fraction of the mutation as shown by whole exome sequencing. The strongest example for this was in the case of CCND1, whose WT allele was completely absent in patient PD4294, and was silenced to a large extent in patient PD4284. Copy number array data9 indicated no significant changes at this locus, suggesting that overexpression of the mutant allele may occur through other means, such as structural rearrangements or translocations. Indeed, PD4294 and PD4284 harbor the t(11:14) translocation, which is known to include the CCND1 gene. Our data cannot determine whether the translocation included the mutant or WT allele.2,9
We previously discussed concerns that using BRAF inhibitors in patients with coexistent RAS mutations might promote secondary tumors, as the inhibitors may have paradoxical extracellular signal-regulated kinase activating effects.22,23 However, there would be less concern for this paradoxical activation if the mutated genes were not actually expressed. Direct examination of mutant allele expression in those patients carrying mutations in both genes will be needed to further evaluate this phenomenon and might provide one basis for clinical testing using RNA-seq prior to initiating targeted therapies. This emphasizes the need for continued study beyond mutational analysis at the DNA level and may explain why therapies targeted at mutant oncogenes can be ineffective or produce paradoxical effects.24
In conclusion, we have correlated allele-specific RNA expression with exome mutant allele frequency in MM patient samples. Although a large number of mutations have been described in MM, only a small fraction of the mutant alleles have detectable expression in our data, and are found in genes with low or no detectable expression. This suggests that the majority of observed mutations may actually be bystanders with limited, if any, functional implications, and highlights the importance of using RNA-seq to evaluate allele-specific expression. These results are intriguing as they provide a possible explanation as to why it is that multiple prior expression studies have failed to identify any overlapping set of genes that correlates with disease state. It is possible that the pathogenic changes in MM lead to alteration of allele-specific expression or alternative splicing that are not reflected in the overall RNA level expressed from a particular locus. Future study with a larger number of samples and correlation with in vitro studies will be needed to more fully evaluate this hypothesis.
Acknowledgments
This study was supported by National Institutes of Health, National Cancer Institute grants PO1-155258 (N.C.M., P.J.C., K.C.A., G.P., H.A.L., and S.M.), RO1-124929 (N.C.M.), P50-100007 (K.C.A. and N.C.M.), PO1-78378 (K.C.A. and N.C.M.), RCA125711C (M.A.S.), and a Wellcome Trust grant 077012/Z/05/Z. P.V.L. is a postdoctoral researcher of the Research Foundation–Flanders. P.J.C. is personally funded through a Wellcome Trust Senior Clinical Research Fellowship. K.C.A. is an American Cancer Society Clinical Research Professor. N.B. is a European Hematology Association fellow and was supported by a grant from the Lady Tata Memorial Trust. The authors thank Yaoyou Wang and Renee Rubio at the Center for Cancer Computational Biology for assistance with sequencing and analysis.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: N.U.R. performed research, contributed vital new analytical tools, analyzed data, and wrote the paper; A.S.S. designed research, performed research, analyzed data, and wrote the paper; N.B. performed research and analyzed data; D.C.W. and P.V.L. analyzed data; Y.T.T. provided vital new reagents; M.A.S., M.F., M.K.S., P.G.R., F.M., S.M., P.A.F., K.C.A., H.A.L., and P.J.C. analyzed data; G.P. analyzed data and wrote the paper; and N.C.M. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nikhil C. Munshi, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana B106, Boston, MA; e-mail: nikhil_munshi@dfci.harvard.edu; and Peter J. Campbell, Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton CB10 1SA, United Kingdom; e-mail: pc8@sanger.ac.uk.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Bergsagel PL, Drach J, et al. International Myeloma Working Group. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girnius S, Munshi NC. Individualized therapy in multiple myeloma: are we there? Semin Oncol. 2013;40(5):567–576. doi: 10.1053/j.seminoncol.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer [published correction appears in Nature. 2013;502(7470):258]. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egan JB, Shi CX, Tembe W, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120(5):1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker BA, Wardell CP, Melchor L, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120(5):1077–1086. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 8.Walker BA, Wardell CP, Melchor L, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28(2):384–390. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997–3010. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14(10):703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anders S. HTSeq: analysing high-throughput sequencing data with Python. http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html. 2010. Accessed June 5th, 2014. [DOI] [PMC free article] [PubMed]
- 13.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323–337. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinn EM, Cormican P, Kenny EM, et al. Development of strategies for SNP detection in RNA-seq data: application to lymphoblastoid cell lines and evaluation using 1000 Genomes data. PLoS ONE. 2013;8(3):e58815. doi: 10.1371/journal.pone.0058815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen S, Park JW, Huang J, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40(8):e61. doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callahan MK, Rampal R, Harding JJ, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med. 2012;367(24):2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert C, Arnault JP, Mateus C. RAF inhibition and induction of cutaneous squamous cell carcinoma. Curr Opin Oncol. 2011;23(2):177–182. doi: 10.1097/CCO.0b013e3283436e8c. [DOI] [PubMed] [Google Scholar]
- 24.Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K. Latest advances and current challenges in the treatment of multiple myeloma. Nat Rev Clin Oncol. 2012;9(3):135–143. doi: 10.1038/nrclinonc.2012.15. [DOI] [PubMed] [Google Scholar]