Abstract
Whole genome sequencing was completed on 1,325 individuals from 602 families, identifying 27 million autosomal variants. Genetic association tests were conducted for those individuals who had been assessed for one or more of 17 endophenotypes (N range = 802–1,185). No significant associations were found. These 27 million variants were then imputed into the full sample of individuals with psychophysiological data (N range = 3,088–4,469) and again tested for associations with the 17 endophenotypes. No association was significant. Using a gene-based variable threshold burden test of nonsynonymous variants, we obtained five significant associations. These findings are preliminary and call for additional analysis of this rich sample. We argue that larger samples, alternative study designs, and additional bioinformatics approaches will be necessary to discover associations between these endophenotypes and genomic variation.
Descriptors: Endophenotype, Psychophysiology, Whole genome sequencing, Rare variant, P300, Startle, Antisaccade, EEG
Five of the companion articles in this special issue describe genome-wide association studies (GWAS) from a fixed genotyping array with a prespecified set of 527,829 variants. Such genotyping arrays are designed primarily to capture common variants, those with a minor allele frequency, or MAF, greater than .05. The other original research article in this issue (Vrieze et al., 2014) describes an association study between the 17 putative endophenotypes and rare nonsynonymous exonic variants specifically, which are variants in coding regions that affect protein structure. In the current study, we extended these analyses by employing whole genome sequencing in an attempt to discover nearly all single nucleotide polymorphisms (SNPs) present in any given individual, including those on the GWAS and exome arrays as well as tens of millions of additional variants. Because all variants are directly measured and genotyped, this results in increased power for common variants and the ability to test rare variants throughout the entire genome on a far larger scale than the other articles in this special issue.
Whole genome sequencing interrogates the entire genome to discover and accurately genotype variants from across the allelic spectrum, from private mutations possessed by a single person (or family), to common variants genotyped on typical microarrays. The past few years have seen significant advances in population genetics and characterization of rare genomic variation, which were only possible with genome sequencing technology. The 1000 Genomes Project, for example, combined exome and whole genome sequencing to discover 38 million SNPs in 1,092 individuals from 14 ancestral populations (1000 Genomes Project Consortium, 2012). The Exome Sequencing Project (Fu et al., 2013) and analogous exome sequencing projects (Nelson et al., 2012) have extensively interrogated exonic regions of the genome and characterized a wide diversity of rare coding variants. In the present study, we found 27.1 million autosomal SNPs, 21.3 million of which have minor allele frequency less than 5%. Almost none of these 21 million variants were tested in the other articles of this special issue.
1000 Genomes, Exome Sequencing Project, ENCODE, and many related projects represent breathtaking technological and analytical achievements, delivering insight into molecular biology, genomics, evolutionary history, migratory patterns, and disease biology, to name a few (Lander, 2011). Genome sequencing has been less widely used in the study of human behavior, with notable exceptions including advances in the genetics of autism (Neale et al., 2012; O’Roak et al., 2011) and schizophrenia (Fromer et al., 2014; Purcell et al., 2014). These studies employed exome sequencing, which interrogates only the exons for each of ~20,000 protein coding genes throughout the genome. The exome is an important, but small, section of the genome, comprising less than 2% of all sequence in the genome. The remainder of the genome, colorfully referred to in the past as “junk DNA,” is everything but that. Work by the ENCODE consortium (ENCODE Project Consortium, 2012) and others have verified that noncoding DNA harbors genetic variation critical to genomic function. While coding variants can affect protein structure, which is undoubtedly important, noncoding DNA can affect which, when, and how frequently genes are expressed, termed “gene regulation” more broadly. Indeed, early work suggests that a majority of disease-associated variants are in noncoding regions, with regulatory regions likely enriched for genome-wide significant variants (Maurano et al., 2012; Pickrell, 2014). Exhaustively interrogating genetic variation in coding and noncoding regions requires whole genome sequencing.
In the accompanying papers in this special issue, we described a variety of genetic association studies in a sample of 4,905 individuals using different genotyping technologies to identify variants associated with 17 psychophysiological phenotypes (for an overview, see Iacono, Malone, Vaidyanathan, & Vrieze, 2014). These endophenotypes include P300 amplitude, antisaccade direction errors, startle eye blink magnitude and modulation by affective stimuli, skin conductance level and responses in a habituation paradigm, and measures of resting EEG.
Although some of these endophenotypes are robust candidates, and despite the hope that endophenotypes would provide increased power to detect associated genes, the investigations described in the companion articles of this special issue yielded few significant findings. In analyses of common variants, only antisaccade error was significantly associated with an individual SNP (Vaidyanathan, Malone, Donnelly et al., 2014). Tests of rare exonic variants also produced one significant association, between a nonsynonymous SNP in PARD3 and electroencephalogram (EEG) theta power (Vrieze et al., 2014). Gene-based tests of common variants, which aggregate the effect of all SNPs within a given gene into a single score, yielded several significant associations. P3 amplitude was associated with MYEF2 (Malone, Vaidyanathan et al., 2014), EEG delta power was associated with three genes (DEFA4, DEFA6, and GABRA1; Malone, Burwell et al., 2014), antisaccade performance was associated with two genes on Chromosome 2—B3GNT7 and NCL—and the aversive difference startle modulation score was associated with the PARP14 gene on Chromosome 3. Gene-based tests of rare exonic variants yielded one significant association with the pleasant difference startle modulation score and PNPLA7 (Vrieze et al., 2014), which was not readily interpretable.
The present article appears last in this special issue because it is our most comprehensive and most powerful attempt to discover novel genetic loci associated with these endophenotypes. In this article, we describe three primary analyses. First, we test for association between 27 million autosomal SNPs and each of the 17 endophenotypes in 1,706 individuals with whole genome sequences. Second, we conduct gene-based tests of nonsynonymous variants in these same 1,706 individuals. Third, we use the combination of genotype arrays and sequences to impute all 27 million variants into the full Minnesota Center for Twin and Family Research (MCTFR) sample with psychophysiological endophenotypes (N = 4,905) and conduct the same single variant and gene-based burden tests in this larger sample.
Methods
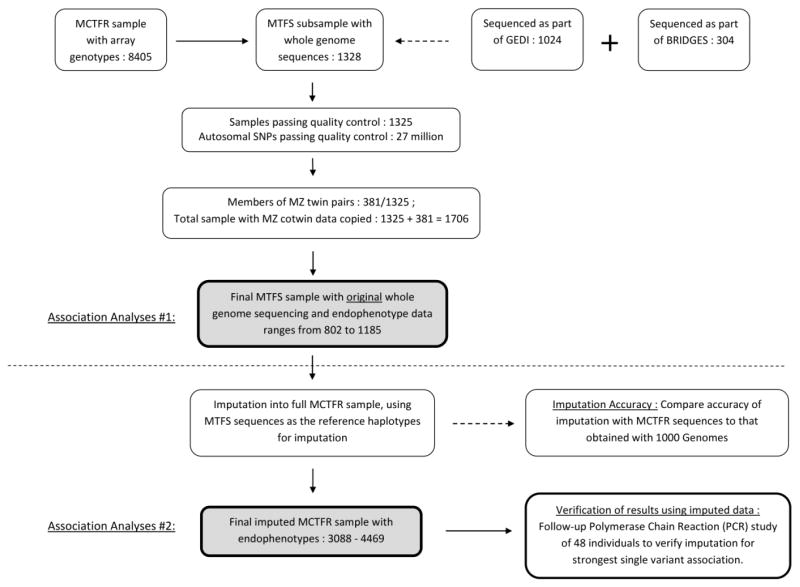
A schematic overview of the methodological sequence is displayed in Figure 1.
Figure 1.
Schematic analysis overview. For additional details see Iacono et al., 2014.
Participants
Individuals were selected from the Minnesota Twin Family Study (MTFS) for moderate-depth whole genome sequencing. While we originally collected molecular genetic data from 7,845 individuals in the MCTFR (Iacono et al., 2014), we undertook whole genome sequencing for a subsample of 1,328 individuals due to prohibitive costs. Of these, 1,325 individuals from 602 families passed quality control checks and were available for association study. First, 1,038 individuals were sequenced as part of the NIDA Genes, Environment, and Development Initiative (GEDI; McGue et al., 2013). Next, 304 individuals were sequenced as part of a study of bipolar disorder (operating under the acronym BRIDGES and led by M. Boehnke at University of Michigan and R. Myers at HudsonAlpha) that is not yet completed and for which there are no currently available published sources. Sample selection within the MCTFR began by considering only individuals with self-reported European ancestry confirmed by genome-wide principal components, as described previously (Miller et al., 2012). The GEDI sample was selected in an attempt to maximize power to detect associations with measures of alcohol use and externalizing psychopathology (see details in online supporting information). BRIDGES samples had been selected from the MCTFR cohorts to serve as additional controls in a case-control genetic association study of bipolar disorder. As such, these samples happened to have been screened for major forms of psychopathology to be included in the BRIDGES study. BRIDGES selection criteria were at least 23 years of age; no history of mania, major depression, or alcohol dependence; and no first-degree relative with a history of bipolar disorder or major depressive disorder. In addition, the samples were matched for northern European ancestry to other BRIDGES bipolar cases using genetic principal components. Not all of these GEDI- and BRIDGES-selected individuals participated in the psychophysiology lab.
Of these 1,325 individuals, 381 were one member of a monozygotic (MZ) twin pair. Zygosity has been validated in this sample through questionnaire, in-person review of the appearance of the twins by experts, anthropomorphic measurements, DNA concordances for all fraternal twins, and DNA concordances for many MZ twin pairs, resulting in a zygosity diagnosis error rate well under 1%. To maximize the yield of phenotypic data, co-twins of MZ twins who were sequenced were added to the sample. Genotypes were simply copied from the sequenced MZ twin to his/her unsequenced co-twin, under the assumption that the sequence was identical between MZ twins. This is a reasonable assumption here, especially for common and low-frequency SNPs, because differences between members of an MZ twin pair would be indistinguishable from sequencing errors at the sequencing depth obtained here (10×). For rare variants, specifically singletons, copying genotypes from one MZ twin to the other will introduce errors due to somatic mutations (Poduri, Evrony, Cai, & Walsh, 2013), which are present only in one MZ twin and not the other. If we assume that each person carries 50 somatic SNP mutations in their genome (Neale et al., 2012), and 3.5 million SNPs (1000 Genomes Project Consortium, 2012), somatic mutations will represent a genotype error rate of approximately 50/3,500,000 = .001% in the MZ twins only. In the context of the present study, we suggest that the increase in power obtained by adding 381 individuals outweighs the increased genotype error expected from somatic mutations. Furthermore, we do not evaluate singletons in our single variant tests. Therefore, somatic mutations in the present study will only affect gene-based tests in the exome, where we expect less than one somatic exonic mutation per individual.
After copying genotypes for MZ co-twins, there were 1,706 total sequenced individuals for association analysis. As noted, these individuals were selected to be sequenced based on phenotypic characteristics that were independent of their having been assessed in the psychophysiology lab. Because not all MCTFR participants completed a psychophysiological assessment, overlap between this sample and the sample for the psychophysiology GWAS used here is incomplete. The actual number with both sequences and psychophysiological measurements ranged from 802 to 1,185, depending on the phenotype. See Table 1 for more complete information on the study sample.
Table 1.
Description of Sequenced and Imputed Individuals with Psychophysiology Measures
| Phenotype | Abbreviation | Sequenced samples | Imputed samples | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N sequenced | % Female | % Fathers | % Mothers | % Offspring | # Sequenced SNPs with MAC ≥ 3 | N Imputed* | # Imputed SNPs with MAC ≥ 3 | ||
| P3 amplitude reduction | P3 | 1,149 | 46.1 | 21.3 | 4.4 | 74.2 | 14,879,798 | 4,166 | 21,426,324 |
| P3 genetic factor | gP3 | 802 | 43.9 | 28.3 | 6.1 | 65.6 | 12,816,986 | 3,088 | 19,284,812 |
| Antisaccade | SAC | 1,185 | 47.0 | 21.5 | 4.8 | 73.7 | 15,124,572 | 4,469 | 21,812,431 |
| Skin conductance level | SCL | 1,181 | 46.5 | 21.6 | 4.8 | 73.6 | 15,131,818 | 3,791 | 21,085,834 |
| Electrodermal activity factor | EDA | 1,185 | 46.5 | 21.6 | 4.8 | 73.6 | 15,146,067 | 4,424 | 21,774,103 |
| Overall startle | STRTL | 840 | 54.9 | 11.8 | 5.2 | 83.0 | 13,186,144 | 3,323 | 19,722,535 |
| Aversive difference startle | aSTRTL | 840 | 54.9 | 11.8 | 5.2 | 83.0 | 13,186,144 | 3,321 | 19,719,458 |
| Pleasant difference startle | pSTRTL | 840 | 54.9 | 11.8 | 5.2 | 83.0 | 13,186,144 | 3,322 | 19,720,993 |
| Skin conductance response frequency | fSCR | 1,141 | 46.5 | 20.9 | 4.9 | 74.1 | 14,902,096 | 4,299 | 21,572,833 |
| Skin conductance response amplitude | aSCR | 1,108 | 46.7 | 20.5 | 4.5 | 75.0 | 14,732,802 | 4,102 | 21,289,320 |
| EEG power (α, β, θ, δ and total power) at CZ | [α/β/θ/δ/tot]Power | 1,045 | 46.1 | 20.7 | 4.4 | 74.9 | 14,330,024 | 3,948 | 20,980,108 |
| Alpha EEG power O1O2 | αPowerO1O2 | 1,056 | 46.1 | 20.6 | 4.5 | 74.9 | 14,416,605 | 3,966 | 21,032,956 |
| Alpha EEG frequency O1O2 | αFreqO1O2 | 1,056 | 46.1 | 20.6 | 4.5 | 74.9 | 14,416,605 | 3,966 | 21,032,956 |
These Ns are the same as reported in the other empirical papers comprising this special issue, and are characterized in detail in the accompanying methods article (Iacono et al., 2014). Please also see that publication for the breakdown of family structure.
Endophenotypes
The endophenotypes examined here are listed below. An overview of all measures is provided in Appendix 1 in Iacono et al. (2014), while the accompanying five GWAS articles in this special issue provide additional details about each measure, including laboratory procedures and evidence supporting each measure as a candidate endophenotype.
-
P300 amplitude (Malone, Vaidyanathan et al., 2014):
-
1
P3 event-related potential (P3)
-
2
P3 genetic factor (gP3)
-
1
-
Antisaccade eye tracking error rate (Vaidyanathan, Malone, Donnelly et al., 2014)
-
3
Antisaccade tracking error rate (SAC)
-
3
-
Electrodermal activity (Vaidyanathan, Isen et al., 2014)
-
4
Skin conductance level (SCL)
-
5
Skin conductance response frequency (fSCR)
-
6
Skin conductance response amplitude (aSCR)
-
7
Electrodermal activity factor (EDA), a general factor derived from SCL, fSCR, and aSCR
-
4
-
Startle blink reflex and affective startle modulation (Vaidyanathan, Malone, Miller, McGue, & Iacono, 2014)
-
8
Overall startle (STRTL)
-
9
Aversive difference startle modulation (aSTRTL)
-
10
Pleasant difference startle modulation (pSTRTL)
-
8
-
Resting state EEG (Malone, Burwell et al., 2014)
-
11
Total EEG power (totPower)
-
12
Alpha EEG power (αPower)
-
13
Beta EEG power (βPower)
-
14
Theta EEG power (θPower)
-
15
Delta EEG power (δPower)
-
16
Alpha EEG power O1O2 (αPowerO1O2)
-
17
Alpha EEG frequency O1O2 (αFreqO1O2)
-
11
Whole Genome Sequences and Concordance with Array Genotypes
All DNA samples used in this sequencing study were whole blood, and were obtained from participants of the MTFS, which is included in the MCTFR. All sequencing was done on Illumina HiSeq technology with 100 or 150 base pair paired-end reads. An introductory overview of sequencing methodology is provide in Appendix 1, with a more complete description in the online supporting information. Sequencing was conducted at two separate institutions, the University of Michigan Sequencing Core and the HudsonAlpha Institute of Technology. After mapping, duplicate read removal, and clipping of overlapping paired-end reads, average depth was 10.47. The variant-calling pipeline, Genomes on the Cloud (GotCloud; Jun, Wing, Abecasis, & Kang, 2014), discovered 27,103,144 autosomal biallelic SNPs in the sample as a whole (see supporting information Table S1 for additional summary information about the genotype calls).
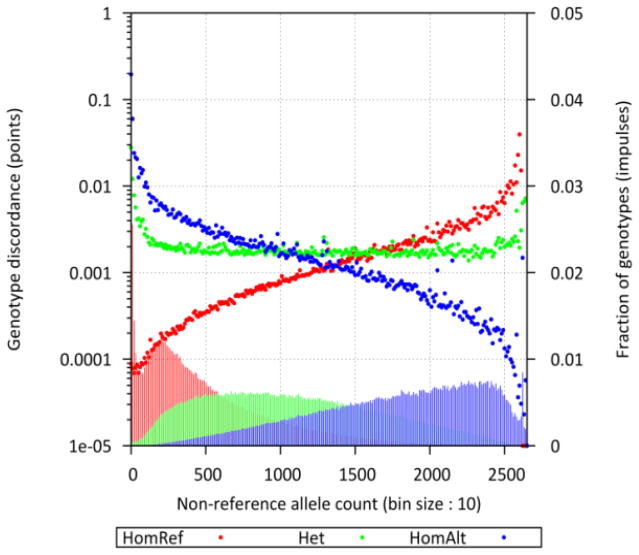
Genotype concordance was tested between the sequence-based genotypes and the array-based genotypes obtained by integrating the 660W-Quad and HumanExome arrays, as described in detail in a companion article to this special issue (Vrieze et al., 2014). All 1,325 directly sequenced individuals had array genotype data, and autosomal genotypes showed 99.91% concordance between the sequenced and array-based genetic variants. Array-based genotyping technology is highly accurate, and this result indicates that the sequence genotypes were also highly accurate. We expect accuracy to decline for rarer genotypes, which can be seen in Figure 2, where we display genotype concordance between the sequence genotypes and array genotypes across the minor allele frequency spectrum, from rare variants to common variants.
Figure 2.
Discordance rates between the integrated array genotypes and sequence genotypes. This plot provides a description of the accuracy of the genotype calls from whole genome sequencing. The bar chart along the bottom gives the fraction of genotypes that were homozygous reference (HomRef), heterozygous (Het), and homozygous alternate (HomAlt), for the full range of possible nonreference allele counts. For example, if an individual in the study was called homozygous reference on the array (i.e., homozygous for the same allele that exists on the reference genome GRCh37), then the red dots give the rate at which that individual was called something other than homozygous reference in the sequence data. For SNPs with a nonreference allele count of 1–10, the sequence error rate was approximately 1 in 10,000. For SNPs with nonreference allele count of 1,500 (MAF ~50%), the sequence error rate was approximately 1 in 1,000. Similarly, if an individual was called heterozygous for some SNP on the array, then the green dots give the rate at which individuals were called heterozygous in the sequences. For a site with nonreference allele count of 1–10, the sequence error rate was about 20%. (Note that this 20% is based on only 141 genotypes—individuals homozygous for an alternate allele are rare themselves.) For sites with nonreference allele counts of 1,500, the rate was a little over 1 in 1,000. In general, sequencing was highly accurate, with accuracy falling off for the rarest variants.
We also examined the power of our 10× sequencing to discover rare genotypes on the integrated array. The 10× sequencing in this sample discovered 7,567 of 10,328 singletons, or SNPs where the rare allele is observed only once (73%), 7,322 of 8,745 doubletons (84%), 8,367 of 8,809 tripletons (95%), and 3,886 of 3,966 quadrupletons (98%) on the integrated array. Of monomorphic sites on the integrated array, sequencing erred in a small fraction of instances, calling 2,515 of 150,329 monomorphic sites on this array as polymorphic (1.7%). While errors in variant discovery are likely due to sequencing errors, it is also possible that these errors are due to incorrect genotype calls on the integrated array, which can be more challenging for rare variants. In summary, 10× whole genome sequencing does reasonably well in genotyping singletons and other rare variants.
Genotype Imputation
As noted, not everyone in the sequenced sample of 1,706 individuals had psychophysiological data. The sequenced individuals who had psychophysiological data for any given endophenotype were a subset of the full 4,905 individuals with psychophysiology data included in the other articles of this special issue (see Figure 1 in Iacono et al., 2014). In addition to conducting association analyses in the sequenced individuals, we also attempted to utilize all available psychophysiological data from as many participants as possible. To do this, we imputed the 27 million sequenced variants into the full array genotyped sample. In imputation, one uses the more complete sequence information to fill in the many millions of variants that were not included on a particular genotype array. Imputation is well known to increase power for association and improve resolution of fine-mapping efforts to pinpoint likely causal variants (Howie, Fuchsberger, Stephens, Marchini, & Abecasis, 2012; Li, Willer, Ding, Scheet, & Abecasis, 2010).
The imputation reference haplotype panel (e.g., 1000 Genomes or, in this case, our sequences) is a critical component of imputation. This is especially true for imputation of less common variants, which is highly sensitive to the degree of ancestry matching between the genotyped sample (to be imputed in) and the reference haplotype panel (e.g., 1000 Genomes), as well as the number of individuals in the reference haplotype panel. The number of individuals in the panel is important because imputation accuracy is in part a function of how many copies of a variant exist in the haplotype panel. If only one copy exists (i.e., a singleton), that variant will likely be difficult to impute accurately. One simple way to increase the number of copies of a variant is to increase the number of individuals in the haplotype panel. Since the number of individuals of European ancestry sequenced in the MTFS was over three times larger than the number of individuals of European ancestry in 1000 Genomes, we expected the use of our MTFS haplotype reference panel to provide significantly greater imputation accuracy over 1000 Genomes.
Through imputation, we were able to take advantage of the moderate-depth sequences we generated, and the wealth of genetic information contained in them: over 27 million autosomal variants, many of which are rare. The first step was to phase the array genotypes for the full sample of 7,278 genotyped individuals in the MCTFR (excluding ungenotyped MZ co-twins). We used the full MCTFR sample in order to improve phasing accuracy with SHAPEIT (Delaneau, Zagury, & Marchini, 2013) by using as much family information as possible. By using the full sample for this purpose, we retained all available family information, thereby maximizing phasing accuracy.
For all single variant association analyses, we used the full set of 27 million imputed SNPs. For gene-based burden tests, we restricted the set of imputed variants to those imputed with sufficient accuracy, as judged by an imputation minimac RSQ > .3 (Li et al., 2010).
Evaluation of Imputation Accuracy
We evaluated imputation accuracy in several ways. First, we compared minimac RSQ values between imputation results with MTFS sequences and 1000 Genomes sequences. Minimac RSQ is an estimate of the true squared correlation between the imputed allele dosage and the true allele dosage, based on the hidden Markov model in minimac (Li et al., 2010). RSQ > .3 is a conventional cutoff to conclude a variant has been imputed with sufficient accuracy. For instance, variants with RSQ ≤ .3 are commonly excluded in association studies.
In order to provide a more direct comparison of imputation accuracy between the MTFS and 1000 Genomes reference sequences, we compared imputation accuracy using dosage R2, the squared Pearson correlation between the minor allele genotype count from rare variants on the exome array and the imputed dosage. In this comparison, we restricted phasing and imputation to Chromosome 20 from the 660W-Quad only. We purposefully excluded all variants from the exome array during phasing and imputation, imputed all the exome chip variants that were discovered through sequencing, and then evaluated the accuracy of that imputation. We restricted this subsample to individuals with European ancestry, just as we have throughout this special issue in all association analyses. Finally, we included only one member of each MZ twin pair, because the other member is entirely redundant for purely genetic analyses such as imputation (recall the genotypes are copied from the genotyped twin to his/her co-twin). This resulted in a subsample of 6,610 individuals of European ancestry for 1,369 SNPs, for the sole purpose of evaluating imputation accuracy for rare variants.
Follow-Up Genotyping
Several imputed variants in an intron of the ALK gene were significantly associated with EEG alpha frequency. We genotyped these variants in 48 DNA samples from individuals in 12 families, including 13 putative carriers of the rare allele in at least one site, 10 of whom had endophenotype data. For each sample, 1 ul DNA at 20 ng/ul was genotyped in a 25 ul reaction along with 12.5 ul TaqMan Universal PCR Master Mix (Life Technologies, Grand Island, NY), 0.625 ul of 40X primer/probe mix, and 10.875 ul water. We used a manufacturer-recommended PCR program: an initial step of 95°C for 10 min, then a 15-s step at 92°, and a 1.5-min step at 60°. Steps 2 and 3 were cycled through 50 times. Each of the four polymorphisms was amplified separately, and the allelic variants for each polymorphism were labeled with VIC and FAM 5′-fluorescent labels. We used an Applied Biosystems PRISM sequence detection 7500 Real Time PCR System to discriminate between alleles, which uses an optical reading of fluorescent markers following a 1-min period of activation at 60°C to assess the amount of probe sequence product.
Results
Association Analysis in Individuals with Endophenotype Data
We conducted single variant tests on inverse normalized phenotypes in EPACTS using EMMAX (Kang et al., 2010), which produces a genetic kinship matrix that is used to correct for population stratification and familial structure. Depending on the endophenotype, there were approximately 14 million variants with a minor allele count greater than three that were tested for association with an endophenotype, resulting in an approximate Bonferroni correction of 4 × 10−9 to obtain genome-wide significance. At this threshold, no single variant was significant. Q-Q plots and Manhattan plots for each endophenotype are available in the supporting information.
In order to evaluate whether the aggregate effect of rare nonsynonymous variants in genes produced associations with the endophenotypes, we conducted burden tests using a variable threshold count-based method (Price et al., 2010) to group nonsynonymous variants within genes. Variants were annotated using EPACTS (Kang, 2014) against GENCODE v11. All missense and nonsense nonsynonymous SNPs, including essential splice SNPs, were included in all gene-based tests. Depending on the phenotype, we tested from 15,816 (P3 genetic factor) to 16,394 (antisaccade) genes that had at least two nonsynonymous variants and a burden allele count of at least three, considering only variants with MAF < .05, resulting in Bonferroni corrections of ~3.2 × 10−6. No gene was associated with any endophenotype at these levels of significance.
Genotype Imputation with Sequences into Full Sample
We then imputed into these 7,278 phased haplotypes with the MCTFR sequences and again separately with the 1000 Genomes as the haplotype reference panels, using minimac with 200 states and 5 rounds (Howie et al., 2012). Imputation accuracy statistics for both reference panels are provided in Table 2. While it may appear that the MTFS sequences perform better than 1000 Genomes because higher minimac RSQ values were obtained, a direct comparison using the summaries in Table 2 is not entirely appropriate. The 1,325 sequenced individuals are included in these estimates and bias them upward—that is, for 1,325 individuals we used their own sequences to conduct imputation into their array genotypes.
Table 2.
Imputation Accuracy into All Available Individuals of European Ancestry
| Imputation with MTFS sequences | Imputation with 1000 Genomes Aug 2011 release | |||||
|---|---|---|---|---|---|---|
| N | Mean RSQ | RSQ > .3 | N | Mean RSQ | RSQ > .3 | |
| < .001 | 11,874,879 | .479 | 78.8% | 19,028,251 | .278 | 35.5% |
| MAF [.001–.005) | 5,137,513 | .639 | 95.8% | 6,641,963 | .456 | 64.9% |
| MAF [.005–.01) | 1,325,092 | .764 | 98.5% | 1,690,255 | .568 | 79.7% |
| MAF [.01–.05) | 2,673,865 | .862 | 99.0% | 3,022,145 | .726 | 90.1% |
| MAF ≥ .05 | 5,872,931 | .965 | 99.5% | 6,604,512 | .911 | 97.3% |
Note. SNPs were selected using the 1000 Genomes imputed minor allele frequency. RSQ = minimac-estimated quality metric based on the imputation hidden Markov model; RSQ > .3 = percentage of variants with minimac RSQ greater than .3, a conventional threshold used to discard poorly imputed variants.
Evaluation of Imputation Accuracy with Dosage R2
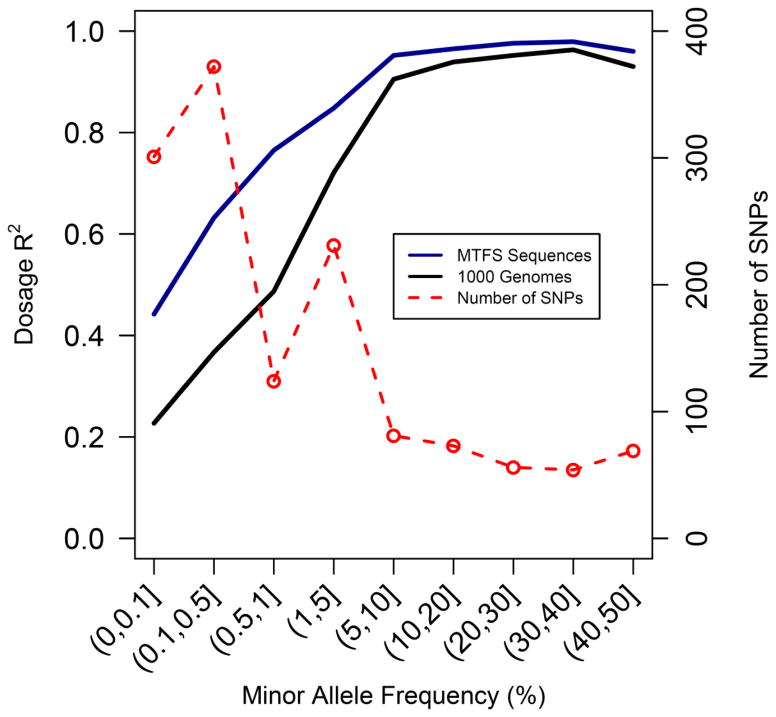
For 333 common variants (MAF > 5%), imputation accuracy was comparable between 1000 Genomes and MCTFR sequences (dosage R2 = .905 and .952, respectively). For less common variants, with MAF ≤ 5%, imputation using the MCTFR sequences performed noticeably better, resulting in increases of .2–.3 in the dosage R2 value for SNPs with MAF < 1%. The results are displayed in Figure 3, where one notices that MCTFR outperforms 1000 Genomes for each minor allele frequency bin.
Figure 3.
Imputation accuracy comparison between MTFS sequence and 1000 Genomes reference panels. This plot provides a comparison of the imputation accuracy on Chromosome 20 obtained with 1000 Genomes compared to MCTFR sequence. Plotted in solid lines is the squared Pearson correlation between the imputed dosage and the genotyped minor allele counts for a range of allele frequencies, using MCTFR sequences (black) and 1000 Genomes sequences (blue) as imputation reference panels. To make a direct comparison, the SNPs are restricted imputation to sites on the genome-wide 660W-Quad SNP array, and then tested accuracy on rare variants on the exome chip. The number of SNPs contributing to each window are given in red, and each window was centered on each dot in the red line. The plots show that the MTFS reference panel provides imputation results for all MAFs, and especially better results for less common alleles.
Association Results with Imputed Genotypes
We conducted single variant and burden association tests using the imputed genotypes, in the full sample of individuals with psychophysiological phenotypes reported on in the companion articles in this special issue (N ranging from 3,088 to 4,469). Q-Q plots and Manhattan plots are displayed in the supporting information. Depending on the phenotype, the number of SNPs with an imputed dosage allele count of at least three ranged from 19,284,812 (P3 genetic factor) to 21,812,431 (antisaccade error), resulting in Bonferroni corrections ranging from 2.6e−9 to 2.3e−9. At these levels of significance, there were two variants (chr2:29994680 and chr2:29978404) significantly associated with alpha EEG frequency at O1O2. However, follow-up genotyping found an imputation error in marker chr2:29994680, which affected dosage counts for both variants. After accounting for this error, the p values for these SNPs dropped to nonsignificant levels. The results are displayed in Table 3, which includes all p values less than the conventional genome-wide significance threshold of 5e−8, even though p < 5e−8 is not genome-wide significant in this context.
Table 3.
Single Variant Results for Imputed Variants with p < 5 × 10−8 and Imputation RSQ > .3
| Phenotype | Chr | Base Pair | Ref | Alt | N | Dosage AC | RSQ | Genotype counts (REF/HET/ALT)* | MAF | Beta | Bonferroni threshold | p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aSCR | 14 | 78792316 | C | T | 4,102 | 1552.36 | .984 | 2713/1222/167 | .1892 | −0.17 | 2.3e-9 | 3.7e-8 |
| SAC | 2 | 232296082 | C | T | 4,469 | 2751.04 | .989 | 2138/1910/421 | .3078 | 0.14 | 2.3e-9 | 3.8e-8 |
| STRTL | 2 | 81204732 | A | C | 3,323 | 53.25 | .645 | 3278/45/0 | .0080 | −1.06 | 2.5e-9 | 3.0e-9 |
| aSTRTL | 3 | 117162298 | C | T | 3,321 | 11.9 | .747 | 3310/11/0 | .0018 | −1.86 | 2.5e-9 | 3.2e-8 |
| aSTRTL | 3 | 122371714 | G | A | 3,321 | 383.07 | .994 | 2956/346/19 | .0477 | −0.29 | 2.5e-9 | 4.0e-8 |
| pSTRTL | 2 | 218395145 | T | C | 3,322 | 1402.99 | .971 | 2060/1123/139 | .2112 | 0.17 | 2.5e-9 | 4.1e-8 |
| βPower | 18 | 19453259 | G | C | 3,948 | 5.31 | .439 | 3946/2/0 | .0007 | −4.10 | 2.4e-9 | 1.4e-8 |
| βPower | 7 | 115086918 | T | G | 3,948 | 3.51 | .726 | 3944/4/0 | .0004 | −3.56 | 2.4e-9 | 3.1e-8 |
| βPower | 12 | 128908244 | G | A | 3,948 | 530.5 | .838 | 3435/501/12 | .0672 | 0.31 | 2.4e-9 | 4.4e-8 |
| θPower | 4 | 6020367 | G | T | 3,948 | 4258.6 | .945 | 832/1988/1128 | .4607 | 0.14 | 2.4e-9 | 4.9e-8 |
| αFreqO1O2 | 2 | 29943375 | G | A | 3,966 | 10.35 | .722 | 3957/9/0 | .0013 | 2.00 | 2.4e-9 | 1.0e-8 |
| αFreqO1O2 | 2 | 29946190 | A | T | 3,966 | 10.37 | .720 | 3957/9/0 | .0013 | 2.00 | 2.4e-9 | 1.0e-8 |
| αFreqO1O2 | 2 | 29978404 | G | T | 3,966 | 10.57 | .694 | 3957/9/0 | .0013 | 2.00 | 2.4e-9 | 8.8e-9 |
| αFreqO1O2 | 2 | 29994680 | A | G | 3,966 | 10.80 | .688 | 3956/9/0 | .0014 | 2.00 | 2.4e-9 | 8.3e-9 |
| αFreqO1O2 | 17 | 40076444 | G | A | 3,966 | 42.78 | .736 | 3922/44/0 | .0054 | 1.00 | 2.4e-9 | 4.0e-8 |
Note. p values reported for αFreqO1O2 are computed taking into account follow-up genotyping described in the Method. Ref = reference allele reported on GRCh37 reference genome; Alt = alternate allele, or nonreference allele; N = samples size with genotypes and phentoypes; Dosage AC = the number of expected minor alleles determined from imputation, which helps account for statistical uncertainty in the imputation procedure; RSQ = minimac imputation RSQ, a measure of imputation accuracy. A conventional RSQ threshold to exclude clearly bad variants is RSC < .3. MAF = minor allele frequency. Beta = standardized effect size.
Genotype counts are organized as homozygote REFerence/HETerozygote/homozygote ALTernate genotypes.
We conducted variable threshold gene-based burden tests for all nonsynonymous variants imputed with MTFS sequences with minimac RSQ > .3, a conventional cutoff. After considering only genes that had at least two called SNPs with a burden allele count of three or greater, the number of genes tested ranged from 16,070 to 16,263, with corresponding Bonferroni cutoffs of ~2.8e-6. Four genes were significantly associated with an endophenotype and are displayed in Table 4. The variable threshold collapsing and multivariante count (VTCMC) test identified two genes, annexin A3 (ANXA3) associated with antisaccade, and solute carrier family 27 (fatty acid transporter) member 6 (SLC27A6) associated with aversive difference startle modulation. The SKAT test also identified two genes, GBX2 and KIF18A, as significantly associated with EEG beta power and pleasant difference startle modulation, respectively.
Table 4.
Burden Test Results and Results for Individual SNPs Within Each Gene in the Imputed Genotypes with Minimac RSQ > .3
| Phenotype | Test | Gene | Gene-based p value | N | CHR | Position | Ref | Alt | Dosage AC | RSQ | REF/HET/ALT | MAF | Beta (SE) | p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| βPower | SKAT | GBX2 | 2.9e-6 | 3,948 | 2 | 237074980 | G | T | 1.89 | 0.461 | 3947/1/0 | .0002 | −3.23 (.86) | 1.8e-4 |
| 237076595 | G | A | 2.31 | 0.368 | 3947/1/0 | .0003 | −2.93 (.86) | 6.9e-4 | ||||||
| pSTRTL | SKAT | KIF18A | 1.1e-6 | 3,322 | 11 | 28045380 | G | A | 2.35 | 0.533 | 3320/2/0 | .0004 | −0.06 (.73) | .94 |
| 28080499 | A | C | 5.83 | 0.688 | 3318/4/0 | .0009 | −0.03 (.50) | .95 | ||||||
| 28080557 | C | T | 26.52 | 0.664 | 3303/19/0 | .0040 | 0.06 (.24) | .79 | ||||||
| 28090850 | C | T | 1.44 | 0.478 | 3321/1/0 | .0002 | −2.25 (1.0) | .02 | ||||||
| 28106253 | G | A | 128.26 | 0.996* | 3194/128/0 | .0193 | −0.38 (.09) | 2.8e-5 | ||||||
| 28110151 | T | C | 280.07 | 1.000* | 3047/270/5 | .0422 | 0.13 (.06) | .03 | ||||||
| 28116342 | C | A | 2.01 | 0.992* | 3320/2/0 | .0003 | 0.29 (.71) | .69 | ||||||
| 28116352 | G | A | 217.10 | 0.919 | 3105/212/5 | .0368 | 0.23 (.07) | 1.0e-3. | ||||||
| 28119309 | T | G | 85.38 | 0.990* | 3238/82/2 | .0129 | −0.04 (.11) | .70 | ||||||
| SAC | VTCMC | ANXA3 | 2.3e-6 | 4,469 | 4 | 79492179 | T | A | 2.45 | 0.231 | 4467/2/0 | .0003 | −1.62 (1.4) | .22 |
| 79500235 | G | A | 2.92 | 0.670* | 4465/4/0 | .0003 | 0.04 (.80) | .96 | ||||||
| 79507418 | C | T | 111.14 | 0.982* | 4359/109/1 | .0124 | −0.47 (.10) | 6.9e-6 | ||||||
| 79522685 | C | T | 13.06 | 0.994* | 4456/13/0 | .0015 | −0.34 (.31) | .27 | ||||||
| 79531211 | C | T | 3.00 | 0.996* | 4466/3/0 | .0003 | −1.79 (.57) | 1.5e-3 | ||||||
| aSTRTL | VTCMC | SLC27A6 | 2.5e-6 | 3,321 | 5 | 128301391 | G | A | 2.02 | 0.621 | 3320/1/0 | .0003 | 2.19 (.97) | .02 |
| 128301916 | A | G | 23.00 | 0.998* | 3298/23/0 | .0035 | −0.12 (.22) | .58 | ||||||
| 128302185 | G | A | 1.00 | 1.000* | 3320/1/0 | .0002 | 0.44 (1.0) | .66 | ||||||
| 128302288 | C | T | 8.96 | 0.993* | 3312/9/0 | .0014 | −0.40 (.34) | .25 | ||||||
| 128320963 | C | T | 1.00 | 0.994* | 3320/1/0 | .0002 | 2.47 (1.0) | .01 | ||||||
| 128351629 | C | T | 0.08 | 0.613 | 3321/0/0 | .00001 | 37.7 (25.0) | .13 | ||||||
| 128359377 | A | G | 6.93 | 0.457 | 3317/4/0 | .0010 | 1.33 (.66) | .04 | ||||||
| 128365323 | T | G | 1.51 | 0.393 | 3320/1/0 | .0002 | 2.31 (1.4) | .09 | ||||||
| 128365384 | G | A | 1.14 | 0.880* | 3320/1/0 | .0002 | −0.65 (.99) | .51 | ||||||
| 128368848 | T | G | 3.80 | 0.641 | 3318/3/0 | .0006 | 1.93 (.61) | 1.6e-3 |
Note. Variants in bold are those included in the variable threshold burden test. The minor allele threshold determined from the VTCMC method was 4 for SLC27A6 and 110 for ANXA3. Variants that fell below this threshold, and were therefore included in the VTCMC test, are in bold. All SNPs were nonsynonymous. No SNPs were stop or start gain or loss. The Bonferroni correction for gene-based p values was 2.9e-6. SKAT = sequence kernel association test; VTCMC = variable threshold collapsing and multivariante count. All other abbreviations are defined in Table 3.
Variants were directly genotyped on either the 660W-Quad or HumanExome array.
Discussion
We reported results for association tests between psychophysiological endophenotypes and SNPs discovered through whole genome sequencing. Association results identified no genome-wide significant variants, after accounting for the many millions of tests conducted here. Gene-based burden tests identified four potential signals, in ANXA3, GBX2, KIF18A, and SLC27A6 genes, associated with antisaccade error, EEG beta power, pleasant difference startle modulation, and aversive difference startle modulation, respectively. ANXA3 is part of a signal transduction pathway and the regulation of cell growth, and has not been associated previously with endophenotypes or phenotypes relevant to antisaccade performance. GBX2, gastrulation brain homeobox 2, is involved in brain development in the mid/hindbrain region, and controls the proper expression of other genes during embryogenesis. The GBX2 association was marginally significant, based on only two relatively poorly imputed singletons, and should be interpreted with additional skepticism until replication. SLC27A6 is a solute carrier not expressed in the brain and KIF18A is involved in chromosome congression during mitosis and meiosis, limiting interpretative speculation about their roles in modulated startle.
Whole genome sequences provide an immense amount of information about genomic variation that is only beginning to be tapped in the present article, and we consider the present results preliminary. Nevertheless, the results suggest that hunting for genes associated with complex phenotypes, including complex endophenotypes, will require alternative approaches to those considered here. The present article is the largest and most comprehensive test of genetic association for psychophysiological endophenotypes undertaken to date. The study sample is richly phenotyped and genotyped, but clearly naïve single variant analyses and gene-based tests with nonsynonymous annotation will not be sufficient to discover strong genetic signals in a sample of this size. The results should bring pause to arguments about the utility of endophenotypes, or intermediate phenotypes, to dramatically increase power to detect individual variants or genes associated with them, or with their relevant clinical phenotypes. Of course, it is possible and perhaps likely that some endophenotypes will serve this purpose in samples of this size, but those are not among our 17.
The endophenotypes described in this special section were initially conceived and selected two decades ago, around the time the Human Genome Project began. Nevertheless, many of our endophenotypes were as much measures of basic brain function then as they are now. We therefore contend that the present findings are highly relevant to current endophenotypic research including exciting new efforts to identify neural systems involved in behavior and psychiatric disorder such as the RDoC (Insel et al., 2010), insofar as investigators attempt to understand the genetic architecture of those systems.
In aggregate, there were fewer significant and biologically plausible associations than might be expected, if one assumes that endophenotypes will provide greater power to identify genes relevant for psychiatric disease or behavioral traits. Given the results reported in this special issue, this assumption may be unrealistic for these psychophysiological endophenotypes, although one might contend that they remain relatively complex and genetically distal measures of basic brain function and task-related brain activity. At the very least, one may conclude the endophenotypic increase in power to detect genes is not great enough to be observed in a sample of the size reported here (N < 4,500).
There was some reason to expect that these endophenotypes would provide sufficient power to detect at least a handful of variants in our sample of between 3,088 and 4,469 individuals. More direct measures of biological function, for example, have shown greater power in detecting associated loci in prior work. As we noted in the accompanying method paper (Iacono et al., 2014), genetic association studies of bone mineral density, cholesterol levels, and QT interval all identified genetic associations in samples of fewer than 5,000 individuals (e.g., see Figure 2b in Visscher, Brown, McCarthy, & Yang, 2012). In contrast, investigations of more distal phenotypes, such as height and body mass index, required closer to 20,000 individuals before any significant and replicable loci were discovered.
Limitations and future directions
The present article represents a first and preliminary step. We have conducted here a variety of obvious initial analyses but much remains to be done with the data available here, by us and by external investigators interested in working with these data. The present sequencing study is ongoing. We are expanding our sample: increasing the number of sequencing reads for a large minority of individuals, refining genotype calls, and extending variant calling to indels, structural variants, and the sex chromosomes. Structural variants and indels have been implicated in autism (Glessner et al., 2009) and schizophrenia (Rees et al., 2014), and represent an additional source of genetic variation of potential value. Plans are also underway to evaluate naturally occurring knockouts (e.g., stop-gains) in great detail in individuals who are homozygous, heterozygous, or compound heterozygous. Indeed, even in the present study with 1,325 sequences there were 54 stop-gain variants (almost entirely rare) in 195 autosomal candidate genes identified by the NIDA Center for Genetic Studies discussed elsewhere in this special issue (https://zork5.wustl.edu/nida/neurosnp.html). Finally, we will leverage the whole genome sequences to conduct enrichment tests of regulatory regions using publicly available epigenomic data. Each of these endeavors is a natural extension of the results reported here.
One major, but necessary, obstacle encountered in the current article is the strong multiple test correction. It is tempting to bypass this restriction by conducting targeted analyses of candidate genes, correcting only for the number of tests conducted on those genes. Indeed, targeted sequencing has produced important results in prior work (Bevilacqua et al., 2010), but it remains to be seen whether new sequencing technologies will overcome the known limitations of the candidate gene approach (Hirschhorn, Lohmueller, Byrne, & Hirschhorn, 2002; Sullivan, 2007). Without strong a priori evidence for a candidate gene–phenotype association, and clear genomic function of candidate variants within that gene, we caution that targeted approaches with relaxed statistical or experimental control should be interpreted with circumspection until consistently replicated. If statistical stringency or gene candidacy criteria were relaxed, we would expect a bevy of false-positive association results, and the use of valuable resources to falsify those erroneous findings. The problem is compounded in whole genome sequencing by the large number of protein-coding genes in the human genome and the many different ways to annotate variants within and around these genes. Despite these concerns, we believe that analysis of whole genome sequence data is not complete until a variety of sensible analyses are attempted and subjected to replication, including detailed study of strong candidate genes as we have argued elsewhere (Vrieze, Iacono, & McGue, 2012).
Conclusion
We tested 27 million SNPs for association with 17 endophenotypes in a moderately sized study sample. No single variant was significant. Gene-based tests identified four associated genes, and replication is required. The findings suggest that these endophenotypes may not provide sufficient power to discover individual variants or genes relevant to clinical phenomena, at least in a sample of this size using the brute force analytical methods presented here. However, additional analyses are underway and remain to be done, leveraging additional genomic variation, structure, function, and experimental design to test the utility of this sample to detect genetic variants relevant for these endophenotypes.
Supplementary Material
Acknowledgments
This research was supported by NIH grants DA024417, DA05147, AA09367, DA13240, DA036216, DA034606, HG007022, and HL117626. Dr. Vrieze is now at the Department of Psychology and Neuroscience, and the Institute for Behavioral Genetics, at the University of Colorado Boulder.
Footnotes
Additional supporting information may be found in the online version of this article:
Figures S1-S34: Q-Q plots and Manhattan plots are provided for single variant tests for each of the 17 phenotypes.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua L, Doly S, Kaprio J, Yuan Q, Tikkanen R, Paunio T, Goldman D. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature. 2010;468:1061–1066. doi: 10.1038/nature09629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nature Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, O’Donovan MC. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WQ, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM NHLBI Exome Sequencing Project. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/Nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genetics in Medicine. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genetics. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Psychophysiology. 2014. Genome-wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, Cuthbert BN, Garvey M, Heinssen R, Pine DS, Quinn K, Wang P. Research Domain Criteria (RDoC): Toward a new classification framework for research on mental disorders. American Journal of Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Jun G, Wing MK, Abecasis GR, Kang HM. An efficient and scalable analysis framework for variant extraction and refinement from population scale DNA sequence data. 2014 doi: 10.1101/gr.176552.114. Manuscript in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM. Efficient and parallelizable association container toolbox (EPACTS) 2014 Retrieved from http://genome.sph.umich.edu/wiki/EPACTS.
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–354. doi: 10.1038/Ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiology. 2010;34:816–834. doi: 10.1002/Gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Burwell SJ, Vaidyanathan U, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of resting EEG activity: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12344. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Vaidyanathan U, Basu S, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of P3 event-related brain potential amplitude: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12345. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, Iacono WG. A genome-wide association study of behavioral disinhibition. Behavior Genetics. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, McGue M. The Minnesota Center for Twin and Family Research genome-wide association study. Twin Research and Human Genetics. 2012;15:767–774. doi: 10.1017/thg.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Daly MJ. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Mooser V. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics. 2011;43:585–589. doi: 10.1038/Ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. American Journal of Human Genetics. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. American Journal of Human Genetics. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Walters JT, Chambert KD, O’Dushlaine C, Szatkiewicz J, Richards AL, Kirov G. CNV analysis in a large schizophrenia sample implicates deletions at 16p12.1 and SLC1A1 and duplications at 1p36.33 and CGNL1. Human Molecular Genetics. 2014;23:1669–1676. doi: 10.1093/hmg/ddt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF. Spurious genetic associations. Biological Psychiatry. 2007;61:1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan U, Isen JD, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of electrodermal activity: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12346. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Donnelly JM, Hammer MA, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of antisaccade eye tracking error rate: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12347. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of acoustic startle eye blink and affectively modulated startle response: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12348. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. American Journal of Human Genetics. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Iacono WG, McGue M. Confluence of genes, environment, development, and behavior in a post genome-wide association study world. Development and Psychopathology. 2012;24:1195–1214. doi: 10.1017/S0954579412000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Pankratz N, Vaidyanathan U, Miller MB, Kang HM, Iacono WG. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12349. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.