Abstract
We review and summarize seven molecular genetic studies of 17 psychophysiological endophenotypes that comprise this special issue of Psychophysiology, address criticisms raised in accompanying Perspective and Commentary pieces, and offer suggestions for future research. Endophenotypes are polygenic, and possibly influenced by rare genetic variants. Because they are not simpler genetically than clinical phenotypes, they are unlikely to assist gene discovery for psychiatric disorder. Once genetic variants for clinical phenotypes are identified, associated endophenotypes are likely to provide valuable insights into the psychological and neural mechanisms important to disorder pathology. This special issue provides a foundation for informed future steps in endophenotype genetics, including the formation of large sample consortia capable of fleshing out the many genetic variants contributing to individual differences in psychophysiological measures.
Keywords: Endophenotype, Molecular genetics, Genome-wide association study, Sequencing association analysis
“... as we know, there are known knowns; there are things that we know that we know. We also know there are known unknowns; that is to say we know there are some things we do not know. But there are also unknown unknowns, the ones we don't know we don't know.”
— Donald Rumsfeld on the nature of the evidence for weapons of mass destruction in Iraq, February 12, 2002
This special issue represents an unprecedented attempt to lay bare the genetic bases of 17 measures representing extensively studied fundamental psychophysiological processes. The genetic bases of these processes are of considerable interest because of their potential to serve as endophenotypes for psychiatric disorder. Although previous work has examined the heritability of these measures and produced a smattering of candidate gene findings related to many of them, few published genome-wide studies of any putative endophenotypes exist. Our aim was to provide a comprehensive, multifaceted, uniformly applied approach to the molecular-genetic investigation of a wide range of psychophysiological measures varying in their psychological complexity, heritability, and status as endophenotypes. For each endophenotype, we conducted biometric heritability analyses using twin and family data, molecular-genetic heritability analyses examining the degree to which individuals with similar endophenotype characteristics showed corresponding similarity in their genotypes, a genome-wide association study (GWAS) of the strength of association of common SNPs (single nucleotide polymorphisms) with the endophenotype, a genome-wide study of the strength of association between autosomal genes and each endophenotype, candidate SNP and gene studies undertaken to corroborate hypothesized associations derived from previous findings reported in the literature, association analysis based on rare genetic variants in the human exome, and an association analysis using variants identified through whole-genome sequencing.
Like Mr. Rumsfeld, who failed to find what he was looking for, our search for specific genetic effects on endophenotypes came up short. Nonetheless, our work supports the notion that such effects exist. Here, we discuss the strengths of the studies described in this special issue, the limitations noted by the distinguished group of commentators, and some directions for future searches.
Strengths of the MTFS Endophenotype Genetic Analyses
Our approach has a number of strengths, which we enumerate here.
Large Sample Size
We created the largest single sample ever employed for the genetic analysis of putative endophenotypes, using laboratory procedures intended to achieve continuity in variable operationalization over a two-decade time span.
Genetically Informative Sample
The sample consisted of parent and twin offspring (N ~4,900) who were part of the Minnesota Twin Family Study (MTFS). Our use of nuclear twin families made possible the investigation of the molecular-genetic basis of the endophenotypes in the same sample from which the biometric heritability of each could be estimated. There was very wide variation in the biometric estimates of heritability, from very high to essentially zero, which provided important context to evaluate the molecular-genetic results for each endophenotype individually and for the entire set considered together.
Due to the nature of our twin-family sample, the fact that we obtained few significant results cannot easily be dismissed as due to poor measurement. Almost all of our measures showed strong evidence of heritability as well as similarity within monozygotic (MZ) twin pairs. To the extent that MZ twins are parallel forms of the same person, high MZ twin similarity provides direct evidence of measurement reliability.
Broad Approach to Endophenotype Evaluation
Our endophenotypes represent broadly the domain of psychophysiological measures that have been used to inform our understanding of psychopathology. Some of the endophenotypes we studied tap basic processes, like orienting and habituation (electrodermal reactivity), autonomic and central nervous system arousal (electrodermal activity and resting state electroencephalogram [EEG] power), and eye blink startle. Others constitute more complex measures related to information processing and emotion, like oddball P3 amplitude, inhibitory control over a prepotent response in the antisaccade task, and affectively modulated startle. Although we do not argue that these measures provide a representative sampling of psychophysiological endophenotypes and we acknowledge that there may exist some endophenotypes that are genetically more informative, our results are more broadly applicable than they would be if our conclusions were dependent on results from a single selectively analyzed and reported variable.
Evaluation Using Both Atheoretical and Hypothesis-Driven Approaches
We used both agnostic, discovery-based and candidate-gene guided approaches. A unique feature of our work was the examination of both common and rare variants. We used p-value significance thresholds that ranged from very conservative to somewhat less so, depending on the nature of the analysis and the number of statistical tests conducted. Should other investigators wish to use a different p-value threshold, our results are reported at a level of detail that allows them to use our tabled data in the articles. We expect the publication and archiving of our findings to be of particular value in years to come to other investigators examining the molecular-genetic basis of these endophenotypes and related measures. These scientists will be able to use our archived data to examine the strength of association we found for our psychophysiological measures to genetic variants they identify in their work. Finally, our data is to be archived in government repositories like dbGaP (database of Genotypes and Phenotypes), and made accessible to qualified investigators who wish to apply different analytic methods and strategies.
Known Unknowns: Key Findings
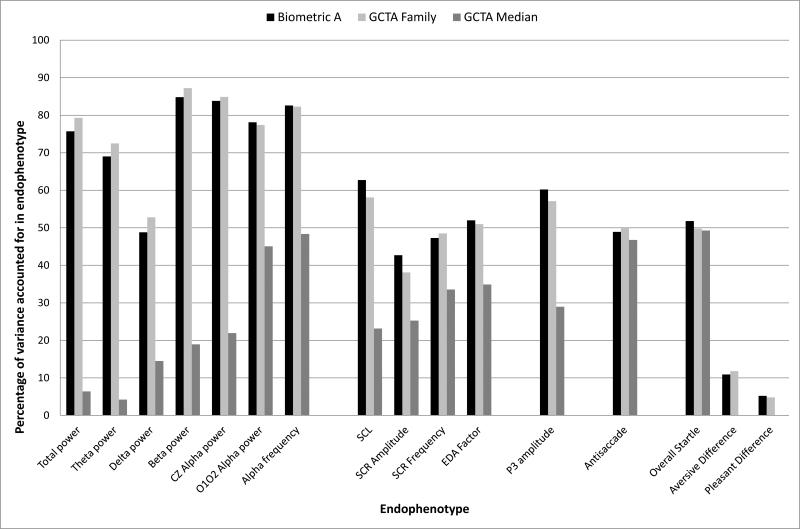
Figure 1 and Table 1 summarize results from various methods that we used across the seven empirical articles to study our 17 endophenotypes. Figure 1 shows that biometric heritability estimates derived from the family data and from the SNP-based genome-wide complex trait analysis (GCTA)-family method, which statistically controls shared environmental influences, generally match each other, and show considerable range. The EEG measures show especially high heritability, with several estimates exceeding 80%. Most of the other endophenotypes, plus one of the EEG measures (delta power), show moderate heritability, falling in the 40‒60% range. Affective modulated startle, by contrast and unlike overall startle, shows little evidence of heritability. The GCTA values derived from unrelated individuals (GCTA-Median in the figure) that provide an estimate of SNP heritability fall considerably below the other two estimates for EEG, electrodermal, and P3 measures. This pattern, consistent with what is called “missing heritability” in the literature, is often seen with other phenotypes, and is interpreted to indicate that rare variants and genetic mechanisms besides additive polygenicity are contributing to heritability. For antisaccade eye tracking error and overall startle amplitude, there is little evidence of missing heritability. While this could indicate that the genetic architecture of these endophenotypes is different from the others, the standard errors of these GCTA estimates across measures are large and overlapping, indicating that replication is desired to rule out the possibility that chance fluctuations based on sample characteristics account for the SNP heritability differences across endophenotypes. The SNP heritability of the two modulated startle measures is essentially zero, supporting the conclusion that startle difference scores are not heritable and likely poor endophenotype candidates despite the substantial literature linking them to several psychiatric disorders.
Figure 1.
Summary of heritability results from biometric analyses and genome-wide complex trait analyses (GCTA) for each endophenotype. The x axis indicates the psychophysiological variable, while the y axis shows the percentage of variance accounted for. “Biometric A” (black bars) denotes the estimate of the additive genetic component obtained in conventional family-based biometric models of each endophenotype. “GCTA Family” (light gray bars) represents GCTA estimates of endophenotypic variance accounted for by the combined effect of all 527,829 single nucleotide polymorphisms (SNPs) used in GWAS analyses with all MTFS participants included. This GCTA model accounts for the effect of all causal variants, including nonadditive genetic effects, but the estimate provided removes the effect contributed by shared environmental influence. “GCTA Median” (dark gray bars) represents the average GCTA value for each endophenotype based on the GCTA estimates for unrelated people. It indicates the additive heritability conferred by all the SNPs on the genotyping array using two different GCTA models (based on unweighted SNPs or weighting SNPs by local linkage disequilibrium patterns) and three different subsamples of unrelated participants (using genetic relatedness matrix values of .025, .05, and .10). The P3 genetic factor score is not included in the figure because it represents only genetic variance, i.e., has a biometric heritability of 100%.
Table 1.
Summary of Statistically Significant Results for SNP- and Gene-Based Tests for All 17 Endophenotypes from the Seven Empirical Studies in this Special Issue
| Endophenotype | Common variants | Rare variants | Whole genome | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP test | Gene test | ||||||||
| Genome-wide (N = 527,829) | Endophenotype-general (N = 1,180) | Endophenotype-specific | Genome-wide (N = 17,601) | Endophenotype-general (N = 204) | COGS (N = 92) | Endophenotype-specific | SNP/gene | SNP/gene | |
| Total power | - | - | - | - | - | - | - | - | - |
| Theta power | - | - | - | - | - | - | - | rs139550735 on Chr 10 in PARD3 | - |
| Delta power | - | - | - | DEFA4/DEFA6 | GABRA1 | - | - | gene- | - |
| Beta power | - | - | - | - | [GABRA2]1 | - | - | - | GBX22 |
| CZ alpha power | - | - | - | - | - | - | - | - | - |
| O1O2 | - | - | - | - | - | - | - | - | - |
| Alpha power | |||||||||
| Alpha frequency | - | - | - | - | - | - | - | - | - |
| SCL | - | - | - | - | - | - | - | - | - |
| SCR amplitude | - | - | - | - | - | - | - | - | - |
| SCR frequency | - | - | - | - | - | - | - | - | - |
| EDA factor | - | - | - | - | - | - | - | - | - |
| P3 amplitude | - | - | - | MYEF2 | - | - | - | - | - |
| Genetic factor score | - | - | - | MYEF2 | - | - | - | - | - |
| Antisaccade | Chr 2 hotspot around rs4973397 | - | - | B3GNT7, NCL | - | - | - | - | ANXA33 |
| Overall startle | - | - | - | - | GRIK3 | GRIK3 | - | - | - |
| Aversive difference scores | Chr 3 hotspot around rs790110 | - | - | PARP14 | - | - | - | - | SLC27A63 |
| Pleasant difference scores | - | - | - | - | - | - | - | PNPLA72 | KIF18A2 |
Note. SNP = single nucleotide polymorphism; Chr = chromosome; Hotspot = region of chromosome with several SNPs within close proximity of each other that each came close to statistical significance value of 5 × 10−8 in GWAS analyses, but no single SNP was significant in predicting endophenotype; COGS = Consortium on the Genetics of Schizophrenia; SCL = skin conductance level; SCR = skin conductance response; EDA = electrodermal activity.
p = .014, which did not exceed Bonferroni threshold of .003.
Statistically significant for gene-based sequence kernel association test after Bonferroni correction.
Statistically significant for gene-based variable threshold collapsing and multivariate count test after Bonferroni correction.
Table 1 provides a summary of molecular-genetic findings. What is striking is that most (89%) of the 153 cells are empty. The discovery-based analyses involving genome-wide testing of the approximately 527,000 SNPs, 17,000 genes, 85,000 exome chip rare variants, and 27 million sequenced variants produced only a few findings. By current convention, none can be considered valid discoveries in the absence of replication. Although one advantage of the discovery-based approach is the opportunity to capitalize on novel etiological insights that might arise from unexpected effects (see how Ford in this issue interpreted the delta power DEFA4/DEFA6 finding in light of the hypothesized role of inflammation to the pathophysiology of schizophrenia), unreplicated “discoveries” appear more plausible if they can be linked to the endophenotype through a known biological mechanism. Of the 11 genes in Table 1 identified through genome-wide studies, four appear likely to affect brain function. MYEF2, myelin expression factor 2, stands out because of the importance of myelin sheathing to nerve conduction. PARD3, PNPLA7, and GBX2 concern brain function. The latter three represent tentative discoveries based on rare variants, and all are in need of replication.
Findings for candidate SNP and gene analyses were even scanter, especially given the relaxed p-value thresholds relative to those adopted for the genome- and exome-wide tests. We examined 1,180 endophenotype-general candidate SNPs selected as likely to be relevant to psychiatric disorders related to the endophenotypes based on the best leads available in the literature. We also examined SNPs reported in the literature based on their prior association with a particular endophenotype. None of these SNP analyses produced a significant result.
Three sets of candidate gene analyses were also undertaken: 204 endophenotype-general candidate genes selected from the neuroSNP data base (Saccone et al., 2009) because they are involved in neural systems or substance-metabolizing pathways that might reasonably be expected to affect one or more of the endophenotypes; 92 genes from the Consortium on the Genetics of Schizophrenia (COGS) that have received considerable support as relevant to schizophrenia endophenotypes (Greenwood et al., 2013), some of which are in the set of endophenotypes we examined as well; and endophenotype-specific genes that were different for each endophenotype. Only two findings emerged as significant, GABRA1 for delta power and GRIK3 for overall startle. GABRA1 encodes for a GABA receptor and has been associated with alcohol misuse (Dick et al., 2006). However, it has not been previously linked to delta power and, in MTFS analyses, it was not associated with alcohol use or misuse (Irons et al., 2014; McGue et al., 2013; Vrieze et al., 2013). GRIK3, one of the COGS genes selected for its relevance to schizophrenia, is involved in the glutamate system. It has not previously been associated with startle. In the exome chip paper, the endophenotype-general candidate genes were also evaluated for the influence of rare variant effects if they possessed sufficient nonsynonymous variation. None of the gene-based tests was significant.
Although GABRA2 is listed in the table, this entry is bracketed because the result did not exceed the Bonferroni threshold for significance (α = 3.12 × 10−3). We list it, however, because, among the endophenotype-specific candidate genes we examined for EEG measures, the relationship between GABRA2 and EEG beta power has, compared to the other endophenotype-specific effects examined, perhaps the strongest track record as a replicated effect. Given that, had we examined only this one possible association, we would have been delighted to report our finding, with a p value of .014, as corroboration of prior reports. But in the context of the many tests we carried out and the assumption implicit in Bonferroni correction that all hypotheses are equally plausible, it is not significant. This indicates the dilemma every scientist faces when deciding how to separate wheat from chaff in molecular-genetic research.
To summarize, almost all of our endophenotypes showed moderate to strong biometric heritability determined at least in part by the combined effect of hundreds of thousands of SNPs. However, using a wide array of molecular-genetic analytic approaches, no solid leads were identified when examining individual genes or SNPs; these remain very much unknown. The genetics of our endophenotypes are thus like that of other complex traits, including psychiatric disorders. They are not simple and therefore not likely to lead to the identification of important risk alleles for psychiatric disorders.
Unknown Unknowns: Strategies for Exploration
What are the potential reasons for our lack of significant findings? The Perspective and Commentary pieces raise a number of important issues regarding challenges and limitations confronted in the execution of these studies, and potential strategies to finding the molecular-genetic bases of endophenotypes. Here, we respond to key points that were raised.
Statistical Power
Several commentaries drew attention to ways in which power to detect many more SNPs or genes could have been limited by characteristics of the MTFS sample. The comments are instructive, and provide useful examples of ways in which analytical strategy and psychophysiological theory can lead to possibly improved study design for the detection of genetic associations with endophenotypes.
Underrepresentation of pathological extremes
Ours is a general population sample, so extreme pathology (like schizophrenia or autism) is not represented to any significant extent. Hence, as Cuthbert (2014, this issue) points out, it is possible that we would have more promising results had we overselected for cases at the extremes. We agree, but it is nonetheless the case that common mental disorders are amply represented in the MTFS, with rates suggesting that a thousand or more individuals in these endophenotype studies are affected with disorders like depression and alcoholism. Examining lifetime prevalence of selected disorders in MTFS older cohort twins based on in-person structured psychiatric interviews, Hamdi and Iacono (2014) reported the following rates: antisocial personality disorder (7.7%), cannabis dependence (9.9%), alcohol dependence (21.2%), major depression (27.0%), and nicotine dependence (32.8%). The age-11 prevalence of ADHD among younger-cohort and ES twins is 7% (Keyes et al., 2009). The parents of the twins also show high lifetime rates (20‒22%) of alcohol dependence and illicit drug abuse or dependence (Holdcraft & Iacono, 2002, 2004). Other MTFS reports also document significant rates of offspring and parent internalizing and externalizing disorders (Hicks, DiRago, Iacono, & McGue, 2009; Marmorstein, Iacono, & McGue, 2009; Wilson, Vaidyanathan, Miller, McGue, & Iacono, in press). While it is possible that more extreme cases would have helped boost our yield of hits, our results cannot be attributed to our having studied only mentally healthy individuals.
Sample too small
One presumed advantage of neurobiologically informed endophenotypes is that their associated genetic effect sizes should be larger than they are for their more complex and distal but related clinical phenotypes. We had 80% power to detect effects accounting for 1.4% of the variance in P3 amplitude, as one example. When we began this molecular-genetic project in 2007, we were thus optimistic that our large sample would prove adequate for the purpose. Our results indicate that a much larger sample would be needed. While it would of course be desirable to substantially increase sample size, it is important to consider how large would be large enough given the cost, time, and effort to collect this type of laboratory data. For P3 amplitude, which has strong support as an endophenotype for substance use and related child and adult externalizing disorders (which, as noted above, are well represented in the MTFS), our largest effect accounted for .33% of the variance. We would need 20,000 individuals to have sufficient power to detect an effect of this magnitude. Even with such a large sample, that would likely be the only effect we would have the power to detect because most effects can be expected to be considerably smaller in magnitude. If samples of tens of thousands are required to unravel endophenotype genetics and considering, as Baker (2014, this issue) points out, that their relevance to the genetics of psychopathology would still need to be determined, they are not likely to serve well their intended purpose of facilitating psychiatric disorder gene finding.
Sample developmentally heterogeneous
Our sample is ethnically homogenous, but composed of distinctly different age groups comprising middle-aged parents and age-17 offspring. Although we adjusted for age, generation, and birth year to account for possible age-related effects, as Baker noted (2014, this issue), it is possible that the genes contributing to variation in our endophenotypes are different at various points in development, a factor that could weaken our ability to detect an effect. Multivariate, longitudinal approaches can assess genetic contributions to “innovations,” or new sources of variance in a longitudinal context, as well as genetic contributions that are relatively constant. They can also be sensitive to gene-environment interactions (Kaprio, 2012). Indeed, in other work in the MTFS sample, we have shown that genetic variants associated with height and smoking in adulthood show smaller effects for those respective measures during adolescence, and we look forward to future work evaluating similar hypotheses for endophenotypes. For the moment, however, we caution that we are not aware of studies that show developmentally heterogeneous genetic effects for our chosen endophenotypes at the ages we examined. For the one endophenotype that we have studied developmentally, P3 amplitude, the evidence suggests that the same genetic influences span adolescence to young adulthood (from age 17 to 24; Carlson & Iacono, 2006).
Choice of p-Value Thresholds for Significance
It is possible that our p-value cutoffs are either too stringent or two liberal. There is no easily achieved consensus regarding how to set these thresholds. Across all the empirical papers, we carried out in excess of 500 million statistical tests, expecting over 25 million to be significant at p < .05. Had we not published these papers as a set following prescribed procedures standardizing the analytic approach across them, readers would not easily recognize the predicament created by advancing a handful of “significant” findings in the context of so many tests. Faced with this reality, we believe we had little choice but to adopt conventional p-value cutoffs to control the familywise error rate on a per-phenotype basis, and to be cautious in the interpretation of our results. However, the thresholds we adopted in this special issue still come with the obvious cost incurred by the many false negatives buried in our data. Schumann (2014, this issue) and Patrick (2014, this issue) both made valuable suggestions regarding how to move beyond the impasse created by the burden of correcting for multiple tests, and we agree with the majority of our commentators that multi-investigator consortia built from pooled, harmonized data collected across many laboratories is one way to overcome this problem.
Possible Contribution of Nonadditive Effects
We are not measuring potentially important nonadditive effects, such as dominance and recessive single-locus effects. However, research on complex traits in humans has provided scant evidence that such effects are overwhelmingly important or detectable even with massive sample sizes. Had we adjusted our models to test for them, we would have added even more to the already high multiple testing burden. In response to the concern that we might be missing dominance effects, as a follow-up to our GCTA analyses, we reanalyzed the endophenotypes using a variant of the GCTA model recommended for family data (Yang, Lee, Goddard, & Visscher, 2013) in which we modeled, and thus statistically controlled, dominance effects (as well as shared environmental influences) from family relationships while estimating the magnitude of shared genetic influences. For the 17 endophenotypes, dominance accounted for 0 to 13% of the variance (median .05), producing 95% confidence intervals that did not overlap zero (and thus indicated significant dominance effects) for eight of the measures (antisaccade error [7%], skin conductance level [10%], and response frequency [7%], and several EEG measures: occipital-parietal alpha power [5%], beta [4%], theta [10%], delta [13%], and total power [5%]). These results, which were largely nonsignificant or produced small effects, indicate that our molecular-genetic findings were unlikely to be substantially affected by our focus on the additive effects of SNPs.
As Goldman (2014, this issue) notes, epistasis, which is key to the type of emergenic traits and network interactions that Miller et al. (Miller, Clayson, & Yee, 2014, this issue) and Schumann (2014, this issue) discussed, is another possible contributing factor we did not evaluate. In our experience, epistasis is only evaluated once a number of known genetic loci have been identified, and then pairwise tests of epistasis are conducted on those known loci. This is due to the overwhelming multiple testing burden incurred by naively testing all pairwise SNP combinations. For instance, if we confined ourselves to examine just pairwise interactions for the SNPs we evaluated in our GWAS analyses, we would need to adopt a p-value cutoff of 5 × 10−16. If we included the 27 million variants from the sequencing study, we would need to calculate hundreds of trillions of statistical tests! Methods are being developed to prioritize SNPs for exploring interaction effects using statistical learning algorithms (Lubke et al., 2013). Nevertheless, the computational burden is significant, and accommodating the family structure of our sample may not always be simple. Although some nonadditive effects may be important, additive effects comprise the most obvious initial candidate to explore, which we have done comprehensively.
Refine the Endophenotype
Some commentaries raised concerns about how the endophenotypes were operationally defined, measured, and analyzed. As Baker (2014, this issue) noted, our endophenotypes could be profitably recast, using multivariate modeling, in a way that facilitates identifying shared genetic influences across measures and phenotypes, possibly including DSM clinical phenotypes. Bivariate analyses of endophenotype and associated trait or disorder provide an opportunity to converge on the genetic overlap between the two, which is certain to be small in magnitude. Focusing on P3 amplitude, Ford (2014, this issue) presented a wealth of creative ideas intended to improve the genetic yield by enhancing reliability and heritability as well as parsing P3 amplitude into components that may prove more genetically tractable (providing “endophenotypes for endophenotypes”; see Miller & Rockstroh, 2013). These are worthy goals, and their implementation could lead to the identification of some associated SNPs, perhaps uncovering important genetic clues that were missed.
However, we are doubtful that implementation of these ideas would dramatically change the genetic landscape for P3 or our conclusions about its genetic architecture. High heritability is not a requirement, nor does it portend success, for finding SNPs or rare variants. The multivariate P3 genetic factor score we examined has a heritability of 1.0 yet produced results that varied indistinguishably from those associated with P3 amplitude. Moreover, heritability varied across the set of endophenotypes from exceptionally high (~.85) to zero, yet the likelihood of obtaining significant molecular-genetic results clearly did not depend on endophenotype heritability; approximately 30% of the filled cells in Table 1 are for affective modulated startle indices, which are the least heritable of our measures. Nor did the likelihood of obtaining a significant “hit” depend on the apparent complexity of the endophenotype; those with the simplest neural circuitry, such as electrodermal orienting and eye blink startle, were no more likely to produce an association than the more complex ones.
Our analysis of P3 endophenotype-specific candidate genes, which produced null results for P3 recorded at parietal leads, included genes identified in prior GWAS using an oddball event-related potential protocol (Kang et al., 2012; Zlojutro et al., 2011). However, the identified genes were associated with a time-frequency component of frontal theta power associated with the P3 event-related potential. Had we included this same measure, perhaps we would have affirmed these results. However, using MTFS samples, we found that the time-frequency constituent components of P3 are strongly correlated with the time-domain P3 amplitude measure we used (Gilmore, Malone, Bernat, & Iacono, 2010; Gilmore, Malone, & Iacono, 2010; Yoon, Malone, Burwell, Bernat, & Iacono, 2013). Moreover, in MTFS twins, P3 and its time-frequency components show stronger association with the clinical phenotypes in the externalizing spectrum when measured at parietal as opposed to frontal sites (Yoon et al., 2013). Thus, considering the consistency in our findings across many measures, it becomes difficult to resist the notion that, to borrow from Gertrude Stein, there is not much “there there,” and thus little reason to expect further refinement of the endophenotype to lead to valid genetic associations.
To summarize, our commentators have made a number of valuable suggestions that represent possible reasons for our lack of significant findings and strategies to implement in the future that may yield discoveries. It is worth noting that our knowledge of psychiatric genetics remains very much in its infancy—very much a world of “unknown unknowns.” Thus, as Patrick (2014, this issue) points out, publication of both positive and negative findings (i.e., considering all available information) is required before conclusions of any sort are warranted.
Looking to the Future
In addition to the suggestions made by the commentators, such as increasing sample size through meta-analysis, several steps can be taken to further genetic association studies of endophenotypes.
Prioritize Sets of Genetic Variants of Known/Predicted Function that Are Enriched for Association
Our analytic strategy encompassed two naïve extremes, one represented by tests of each individual SNP in isolation and the other by a single GCTA test of all in aggregate. The genome-wide scan might be improved upon by differentially weighting groups of SNPs (Roeder, Devlin, & Wasserman, 2007), such as those expressed in brain versus those not, or those implicated by prior research. Examining multiple markers, rather than one SNP at a time, is another possibility, which can provide increased power (Pan, 2008) and which also permits pathway or network analysis. Such methods incorporate prior biological knowledge or information about the topological relationship among genes in a network. This might be valuable for assessing matrix-based endophenotypes (Schumann, 2014, this issue). These methods also provide a means to evaluate gene-gene interactions while constraining the search space. It is now possible to focus on those genetic variants known to influence gene expression (so-called expression quantitative trait loci, or eQTLs), rather than examine all SNPs. Massive publicly available datasets now provide comprehensive maps of enhancers, insulators, promoters, and eQTLs, all part of the genetic regulatory system that controls gene expression levels. Chromatin state information can be used to prioritize SNPs with genomic function (Pickrell, 2014). Whole-genome sequences are especially valuable for these tests, as they provide exhaustive directly genotyped variants within functional regions instead of relying on tag SNPs and common variants. In fact, we are currently pursuing such analyses ourselves. When a significantly associated variant is eventually found and replicated, these functional categories can prove valuable in understanding how that variant influences the genome to affect the trait of interest.
Expand Genetic Diversity
Individuals of European ancestry, such as those making up our sample, possess only a small fraction of the total genetic variation in the human population (1000 Genomes Project Consortium, 2012); Africa alone has more genetic diversity than the rest of the world combined. There are myriad examples of genetic effects that are largely limited to specific ancestral or founder populations, and the absence of a significant finding in a European sample does not preclude the discovery of a large-effect variant identified in a different ancestral group. This is especially true for rare variants, where ancestral divergence is the rule.
Extend Sequencing Analysis
One major hurdle in rare variant association analysis is statistical power—rare variants may have very large effects but still account for tiny fractions of population disease burden. For example, recent research has identified rare variants affecting macular degeneration with odds ratios of 20:1 (Raychaudhuri et al., 2011) and similarly large effects on other diseases (Cohen & Hobbs, 2013; Sigma Type 2 Diabetes Consortium et al., 2014; TG and HDL Working Group of the Exome Sequencing Project, 2014). However, even a completely penetrant rare variant can only account for a tiny fraction of disease burden, because it affects so few people. Cardiovascular disease causes over half a million deaths annually, but a rare variant present in only 1 in 10,000 individuals can only be responsible for 50 of those deaths. For analogous reasons, any individual rare variant cannot account for substantial heritability in any given population, even with a huge effect size (yet another way that heritabilities are unreliable guides in the discovery of gene associations).
Achieving sufficient power to detect rare variant effects (even large ones) is an important consideration in the design of future studies, of endophenotypes or otherwise. Rare variants of known function, such as a protein-truncating stop-gain variant, or an insertion-deletion that displaces a motif within a strong enhancer, can provide directional and biologically plausible tests of association. As Goldman (2014, this issue) suggests, detailed and systematic study of functional rare variants, with follow-up of promising signals in carrier family members, is a way to efficiently obtain many copies of an otherwise rare variant (e.g., see Bevilacqua et al., 2010). Indeed, the longitudinal MTFS cohorts are ideally suited to recruit additional family members in order to replicate promising findings concerning rare variants. The sequencing analyses reported in this special issue only scratched the surface of possibility, and we are already actively expanding that analysis in many ways.
Conclusions: Limits of the Knowns and Unknowns
These MTFS papers represent the first comprehensive effort to examine the possible etiologic relevance of common and rare genetic polymorphisms for a wide range of psychophysiological endophenotypes using genome-wide and candidate-targeted methods applied to the largest sample employed for such a purpose to date. We make no claim that it is appropriate to generalize our conclusions to every possible variable that could be considered an endophenotype. However, given our results and the current state of the literature on psychiatric endophenotypes, in the absence of contrary evidence, we believe it is fair to apply our conclusions broadly. It is also worth noting that the variables we tested ranged in neurobiological complexity from being generated by networks of brain structures (e.g., EEG power spectra and P3) to those presumed to have simple underlying circuitry (e.g., startle). Regardless of the variable tested, our conclusions were largely the same. Considering our results across the seven empirical papers in this special issue, we offer the following conclusions.
Our Findings Warrant Further Investigation
As Table 1 highlights, we do have significant findings. While many could be false positives, they nevertheless emerged against the backdrop of a carefully considered and conservative data analytic approach, thus warranting attention in future investigations. To our knowledge, this work provides the first examination of the possible contribution of rare variants to endophenotype genetics using exome chip and whole-genome sequencing methods. The results suggest that these approaches, like GWAS based on common variants, are also worth pursuing with psychophysiological measures. As Wilhelmsen (2014, this issue) noted, findings based on small effects may have important biological significance and rare variants can readily implicate genomic regions and etiological mechanisms, with especially profound consequences for the families that carry them. As can be seen from our tabled data in the articles and supporting information, we have many effects that, despite not attaining statistical significance, are associated with quite small p values. Almost certainly, some of these are true associations. It will be up to future investigators to coax the signal out of the data by showing that their effects can be corroborated by ours.
Endophenotypes Are Massively Polygenic
Our findings suggest that, like other complex traits, endophenotypes are polygenic, reflecting the contribution of a very large number of genetic variants each contributing very small effects.
Endophenotypes Likely Reflect the Influence of Rare Variants
Polygenic inheritance does not preclude the possibility, perhaps strong, of rare variants with large effects. Such variants can only account for a tiny fraction of heritability in a population for the simple reason that they affect only a very small number of individuals. While we tested rare variants in the present work and found a few potential signals, this work was conducted with approximately one third the whole sample. Falling costs will permit studying larger samples, and additional studies in carefully selected samples will be required to further evaluate the contribution of rare variants in these endophenotypes.
Endophenotypes Will Not Simplify Gene-Finding for Psychiatric Disorder
The promise of endophenotypes has been oversold. Even if endophenotypes are conceptually simpler than DSM (Diagnostic and Statistical Manual of Mental Disorders) diagnoses and closer to underlying biology, endophenotype genetics are not sufficiently simpler genetically to aid in gene discovery in a sample the size of MTFS. Consequently, the same challenges that make psychiatric genetics difficult are likely to make endophenotype genetics difficult.
Endophenotype Genetics Might Contribute Important Biologic Insights for Psychiatric Disorders
As de Geus (2010; 2014, this issue) and Munafò and Flint (2014, this issue) have noted, the value of endophenotypes may best be realized after genetic variants for a disorder, trait, or biological process are identified through other means such as large-sample meta-analyses of relevant traits and work with model organisms. Once genetic variants for clinical phenotypes are identified, we can determine their relevance to specific endophenotypes, in turn generating insights into neural and psychological processes important to clinical pathology. A related benefit of this approach will be better understanding the neurobiology of endophenotypes and the workings of endophenotype-relevant brain systems.
Next Steps
A theme echoed repeatedly throughout the special issue articles and commentaries is the advantage of forming consortia to enhance sample size such that the small effects of the genetic signals buried in genetic noise can be identified without ballooning the rate of false positives that comes by relaxing p values. For laboratory measures, amassing large samples is challenging even when pooling across laboratories. However, consortia like ENIGMA (Enhancing Neuro Imaging through Meta-Analysis; http://enigma.ini.usc.edu/), which includes an EEG offshoot, indicate that success is possible. In addition, it should be possible to develop methods that facilitate large-scale data collection outside the laboratory using inexpensive monitors, such as physiological recording devices already in popular use by fitness enthusiasts. Smartphones permit recording photoplethysmographic data, while mobile EEG recording devices are proliferating. Brain-computer interfaces and other technologies to assist individuals with disabilities may provide useful tools for “crowd-sourcing” data collection as well.
Our results suggest that the focus of endophenotypic theory and research should change, moving away from gene finding to using the results of gene finding to understand psychophysiological mechanisms of etiological relevance to psychopathology. As Braff (2014, this issue) notes in his commentary, the value of these MTFS endophenotype papers may best be realized in what comes next with projects that combine the study of molecular genetics, endophenotypes, and psychiatric disorder, such as COGS. As we and others continue to expand our work, we must actively seek to share data readily, rapidly, and unconditionally with other researchers, preferably without embargo. Open science and open consents may not always be practical or appropriate but should be pursued to the fullest extent possible. It is our hope that this special issue provides a foundation for such cooperation, informing future steps in endophenotype genetics that enable a more satisfying answer than Mr. Rumsfeld's about what is known and what is not.
Acknowledgments
The research was supported by NIH grants: DA 05147, DA 13240, DA 024417, DA 036216, AA09367. DA 034606, HG 007022, and HL 117626.
References
- 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LA. Do our “big data” in genetic analysis need to get bigger? Psychophysiology. 2014 doi: 10.1111/psyp.12351. (in press) [DOI] [PubMed] [Google Scholar]
- Bevilacqua L, Doly S, Kaprio J, Yuan Q, Tikkanen R, Paunio T, Goldman D. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature. 2010;468:1061–1066. doi: 10.1038/nature09629. doi: 10.1038/nature09629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL. Genomic substrates of neurophysiological endophenotypes: Where we've been and where we're going. Psychophysiology. 2014 doi: 10.1111/psyp.12352. (in press) [DOI] [PubMed] [Google Scholar]
- Carlson SR, Iacono WG. Heritability of P300 amplitude development from adolescence to adulthood. Psychophysiology. 2006;43:470–480. doi: 10.1111/j.1469-8986.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Hobbs HH. Simple genetics for a complex disease. Science. 2013;340:689–690. doi: 10.1126/science.1239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert BN. Translating intermediate phenotypes to psychopathology: The NIMH research domain criteria. Psychophysiology. 2014 doi: 10.1111/psyp.12342. (in press) [DOI] [PubMed] [Google Scholar]
- de Geus EJ. From genotype to EEG endophenotype: A route for post-genomic understanding of complex psychiatric disease? Genome Medicine. 2010;2:1–4. doi: 10.1186/gm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Geus EJ. Molecular genetic psychophysiology: A perspective on the Minnesota contribution. Psychophysiology. 2014 doi: 10.1111/psyp.12341. (in press) [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, Xuei XL, Goate A, Hesselbrock V, Foroud T. Association between GABRA1 and drinking behaviors in the collaborative study on the genetics of alcoholism sample. Alcoholism: Clinical and Experimental Research. 2006;30:1101–1110. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Ford JM. Decomposing P300 to identify its genetic basis. Psychophysiology. 2014 doi: 10.1111/psyp.12353. (in press) [DOI] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Bernat EM, Iacono WG. Relationship between the P3 event-related potential, its associated time-frequency components, and externalizing psychopathology. Psychophysiology. 2010;47:123–132. doi: 10.1111/j.1469-8986.2009.00876.x. doi: 10.1111/j.1469-8986.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Iacono WG. Brain electrophysiological endophenotypes for externalizing psychopathology: A multivariate approach. Behavior Genetics. 2010;40:186–200. doi: 10.1007/s10519-010-9343-3. doi: 10.1007/s10519-010-9343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D. The missing heritability of behavior: The search continues. Psychophysiology. 2014 doi: 10.1111/psyp.12362. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Swerdlow NR, Gur RE, Cadenhead KS, Calkins ME, Dobie DJ, Lazzeroni LC. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the consortium on the genetics of schizophrenia. American Journal of Psychiatry. 2013;170:521–532. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdi NR, Iacono WG. Lifetime prevalence and co-morbidity of externalizing disorders and depression in prospective assessment. Psychological Medicine. 2014;44:315–324. doi: 10.1017/S0033291713000627. doi: 10.1017/S0033291713000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks BM, DiRago AC, Iacono WG, McGue M. Gene-environment interplay in internalizing disorders: Consistent findings across six environmental risk factors. Journal of Child Psychology and Psychiatry and Allied Disciplines. 2009;50:1309–1317. doi: 10.1111/j.1469-7610.2009.02100.x. doi: 10.1111/j.1469-7610.2009.02100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdcraft LC, Iacono WG. Cohort effects on gender differences in alcohol dependence. Addiction. 2002;97:1025–1036. doi: 10.1046/j.1360-0443.2002.00142.x. [DOI] [PubMed] [Google Scholar]
- Holdcraft LC, Iacono WG. Cross-generational effects on gender differences in psychoactive drug abuse and dependence. Drug and Alcohol Dependence. 2004;74:147–158. doi: 10.1016/j.drugalcdep.2003.11.016. doi: 10.1016/j.drugalcdep.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Irons DE, Iacono WG, Oetting WS, Kirkpatrick RM, Vrieze SI, Miller MB, McGue M. Gamma-aminobutyric acid system genes—No evidence for a role in alcohol use and abuse in a community-based sample. Alcoholism: Clinical and Experimental Research. 2014;38:938–947. doi: 10.1111/acer.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang J-C, Wetherill L, Hinrichs T, Dick D. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes, Brain and Behavior. 2012;11:712–719. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprio J. Twins and the mystery of missing heritability: The contribution of gene-environment interactions. Journal of Internal Medicine. 2012;272:440–448. doi: 10.1111/j.1365-2796.2012.02587.x. doi: 10.1111/j.1365-2796.2012.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes MA, Malone SM, Elkins IJ, Legrand LN, McGue M, Iacono WG. The enrichment study of the Minnesota Twin Family Study: Increasing the yield of twin families at high risk for externalizing psychopathology. Twin Research and Human Genetics. 2009;12:489–501. doi: 10.1375/twin.12.5.489. doi: 10.1375/twin.12.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubke G, Laurin C, Walters R, Eriksson N, Hysi P, Spector T, Boomsma D. Gradient boosting as a SNP filter: An evaluation using simulated and hair morphology data. Journal of Data Mining Genomics Proteomics. 2013;4:143. doi: 10.4172/2153-0602.1000143. doi: 10.4172/2153-0602.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein NR, Iacono WG, McGue M. Alcohol and illicit drug dependence among parents: Associations with offspring externalizing disorders. Psychological Medicine. 2009;39:149–155. doi: 10.1017/S0033291708003085. doi: 10.1017/S0033291708003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, Iacono WG. A genome-wide association study of behavioral disinhibition. Behavior Genetics. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GA, Clayson PE, Yee CM. Hunting genes, hunting endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12354. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GA, Rockstroh B. Endophenotypes in psychopathology research: Where do we stand? Annual Review of Clinical Psychology. 2013;9:177–213. doi: 10.1146/annurev-clinpsy-050212-185540. doi: 10.1146/annurevclinpsy-050212-185540. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Flint J. The genetic architecture of psychophysiological phenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12355. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. Network-based model weighting to detect multiple loci influencing complex diseases. Human Genetics. 2008;124:225–234. doi: 10.1007/s00439-008-0545-1. doi: 10.1007/s00439-008-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick CJ. Genetics, neuroscience, and psychopathology: Clothing the emperor. Psychophysiology. 2014 doi: 10.1111/psyp.12356. (in press) [DOI] [PubMed] [Google Scholar]
- Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. American Journal of Human Genetics. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Iartchouk O, Chin K, Tan PL, Tai AK, Ripke S, Yu Y. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nature Genetics. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, Devlin B, Wasserman L. Improving power in genome-wide association studies: Weights tip the scale. Genetic Epidemiology. 2007;31:741–747. doi: 10.1002/gepi.20237. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Bierut LJ, Chesler EJ, Kalivas PW, Lerman C, Saccone NL, Rutter JL. Supplementing high-density SNP microarrays for additional coverage of disease-related genes: Addiction as a paradigm. PloS One. 2009;4:e5225. doi: 10.1371/journal.pone.0005225. doi: 10.1371/journal.pone.0005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G. Are we doing enough to extract genomic information from our data? Psychophysiology. 2014 doi: 10.1111/psyp.12357. (in press) [DOI] [PubMed] [Google Scholar]
- Sigma Type 2 Diabetes Consortium. Estrada K, Aukrust I, Bjørkhaug L, Burtt NP, Mercader JM, Flannick J. Association of a low-frequency variant in HNF1A with Type 2 diabetes in a Latino population. Journal of the American Medical Association. 2014;311:2305–2314. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TG and HDL Working Group of the Exome Sequencing Project Loss-of-function mutations in APOC3, triglycerides, and coronary disease. New England Journal of Medicine. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Feng S, Miller MB, Hicks BM, Pankratz N, Abecasis GR, McGue M. Non-synonymous exonic variants in addiction and behavioral disinhibition. Biological Psychiatry. 2013;75:783–789. doi: 10.1016/j.biopsych.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen KC. The feasibility of genetic dissection of endophenotype. Psychophysiology. 2014 doi: 10.1111/psyp.12366. (in press) [DOI] [PubMed] [Google Scholar]
- Wilson S, Vaidyanathan U, Miller MB, McGue M, Iacono WG. Premorbid risk factors for major depressive disorder: Are they associated with early onset and recurrent course? Development and Psychopathology. doi: 10.1017/S0954579414001151. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide complex trait analysis (GCTA): Methods, data analyses, and interpretations. In: Gondro C, van der Werf J, Hayes B, editors. Genome-wide association studies and genomic prediction. Vol. 1019. Humana Press; New York, NY: 2013. pp. 215–236. 2013/06/13 ed. [DOI] [PubMed] [Google Scholar]
- Yoon HH, Malone SM, Burwell SJ, Bernat EM, Iacono WG. Association between P3 event-related potential amplitude and externalizing disorders: A time-domain and time-frequency investigation of 29-year-old adults. Psychophysiology. 2013;50:595–609. doi: 10.1111/psyp.12045. doi: 10.1111/psyp.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Kuperman S. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156:44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]