Abstract
This article provides an introductory overview of the investigative strategy employed to evaluate the genetic basis of 17 endophenotypes examined as part of a 20-year data collection effort from the Minnesota Center for Twin and Family Research. Included are characterization of the study samples, descriptive statistics for key properties of the psychophysiological measures, and rationale behind the steps taken in the molecular genetic study design. The statistical approach included (a) biometric analysis of twin and family data, (b) heritability analysis using 527,829 single nucleotide polymorphisms (SNPs), (c) genome-wide association analysis of these SNPs and 17,601 autosomal genes, (d) follow-up analyses of candidate SNPs and genes hypothesized to have an association with each endophenotype, (e) rare variant analysis of nonsynonymous SNPs in the exome, and (f) whole genome sequencing association analysis using 27 million genetic variants. These methods were used in the accompanying empirical articles comprising this special issue, Genome-Wide Scans of Genetic Variants for Psychophysiological Endophenotypes.
Descriptors: Biometric modeling, Genome-wide complex trait analysis, Genome-wide association study, Exome chip, Whole genome sequencing, Endophenotype
Over the course of its 25-year history, the Minnesota Center for Twin and Family Research (MCTFR) has been among the leading contributors to research in developmental psychopathology, taking advantage of large, genetically informative, prospectively studied parent-offspring samples to generate insights into the nature of the genetic liability that underlies risk for the development of mental disorders. One of the central aims of the MCTFR has been the evaluation of psychophysiological measures for their potential as psychiatric endophenotypes. MCTFR studies have examined the heritability of psychophysiological variables, their degree of association with psychopathology, and the extent to which they identify those at risk for the development of mental disorders. In 2007, funding was obtained to procure DNA samples from MCTFR participants to enable studies of the molecular genetic basis of psychiatric disorder-relevant traits, including MCTFR candidate endophenotypes, which has led to the development of this special issue. This article lays the foundation for the seven accompanying empirical papers by detailing the analytic methods used and providing descriptive findings that characterize the candidate endophenotypes.
Endophenotypes are laboratory-based quantitative measures indexing genetic risk for a psychiatric disorder. Because they are presumed to be more proximal to gene effects and thus more indicative of the genetic pathways that underlie complex psychiatric disorders, it has been suggested that endophenotypes might facilitate finding genes relevant to the associated disorder (cf. Gottesman & Gould, 2003; Iacono & Malone, 2011). Indeed, substantial interest in endophenotypes has emerged in recent years in psychophysiological, psychiatric (Braff et al., 2008; Flint & Munafò, 2007), and molecular genetic (Wood & Neale, 2010) research. However, the degree to which endophenotypes may assist gene finding remains an open question (e.g., for contrasting perspectives, see Flint & Munafò, 2007; Jonas & Markon, in press)—one that the current relatively broad-based approach using a large sample such as the MCTFR is better designed to address than past attempts using small samples focused on candidate genes. Moreover, independent of their status as endophenotypes, psychophysiological measures tap into neurobiological and psychological constructs (e.g., arousal mechanisms, attention, working memory, emotion regulation), the genetic basis of which are of interest in their own right (e.g., see Anokhin, 2014, who makes a strong case for the value of understanding how genetic factors influence psychophysiological measures).
Each of the first five empirical papers in this special issue, which deal with common genetic variants, is based on a different measure (P3 amplitude, antisaccade error rate) or set of measures (electroencephalogram [EEG] characteristics, multiple measures of electrodermal activity, and modulation of the startle eye blink) that, to varying degrees can be considered endophenotypes for different disorders. Each paper reviews evidence for considering a particular measure as an endophenotype, taking into account (when possible) criteria we have recently enumerated and that emphasize a developmental perspective (Iacono & Malone, 2011). Key criteria involve heritability, association with one or more clinical phenotypes believed to share a common genetic liability, presence in unaffected individuals at high genetic risk, and the ability to predict prospectively the development of disorder.
Overview of the MCTFR Endophenotypes
At the time the MCTFR was begun 25 years ago, endophenotype research was in a relatively nascent state, and it was not clear what psychophysiological measures might best tap into genetic liability for psychopathology in a general population sample of prospectively studied twin children. Ultimately, the measures chosen were selected from those reported in the literature to show strong evidence of heritability and association with psychopathology, including those derived from established laboratory paradigms that showed that first-degree relatives of affected individuals scored outside “normal” range. This led to the selection of measures that, at the time, had demonstrated potential as endophenotypes for alcoholism, mood disorders, and schizophrenia (Iacono, 1985, 1998; Iacono, Lykken, & McGue, 1996). P3 event-related potential amplitude, resting EEG spectral characteristics, eye tracking performance, and electrodermal habituation met these criteria and became part of the standard psychophysiological assessment battery used for all participants. The acoustic startle reflex and its affective modulation were added to the battery later, and therefore were not assessed on all participants. At the time these startle measures were added, there was scant evidence supporting startle as an endophenotype. Their addition was instead motivated primarily by research supporting the potential of the startle paradigm to provide insights into the neurobiology of psychiatric disorder (Vaidyanathan, Patrick, & Cuthbert, 2009), a feature that has resulted in its prominence in the National Institute of Mental Health’s Research Domain Criteria (RDoC, Insel et al., 2010).
The psychophysiological measures and evidence supporting them as endophenotypes are described in detail in each of the five empirical papers examining their association with common molecular genetic variants (using genome-wide association study [GWAS] methods). All 17 are considered together in the two papers dealing with rare genetic variants (using an exome chip array and sequencing methods). Appendix 1 provides a brief description of the five psychophysiological paradigms and the measures derived from each.
Overview of the MCTFR and the Samples Used in Endophenotype Studies
Characterization of MCTFR samples
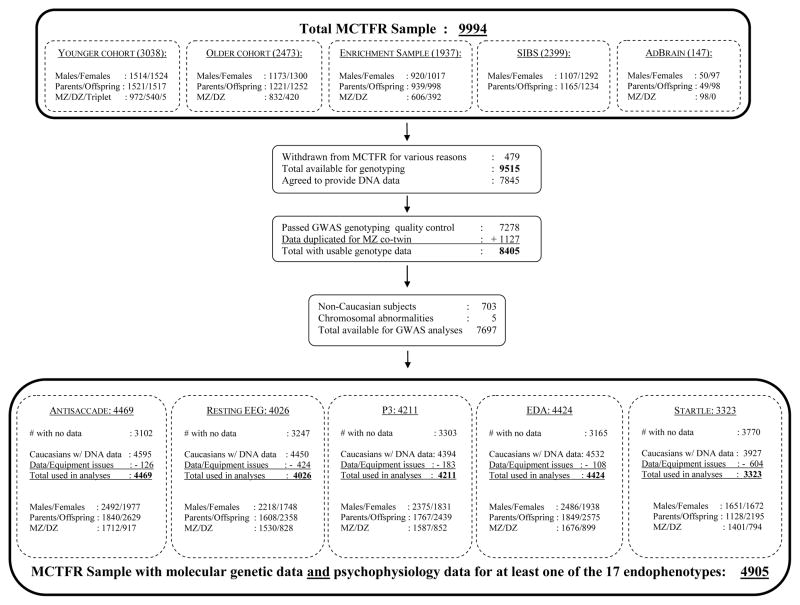
The MCTFR oversees a set of longitudinal investigations focused on families with twin and adoptive children. Initiated by David Lykken in 1990, the MCTFR has enrolled and assessed five parent-offspring samples totaling 9,994 participants (see Figure 1). All these community-based samples were recruited using epidemiological procedures intended to maximize inclusiveness and minimize sampling bias. MCTFR research is ongoing, and continues to track offspring development into adulthood, with those first enrolled in the project now being reassessed 24 years after their initial recruitment.
Figure 1.
Sample flow chart providing an overview of the MCTFR participant samples used in the molecular genetic studies of endophenotypes derived from five different psychophysiological protocols. See text for key to abbreviations.
The MCTFR embraces three twin samples comprising the Minnesota Twin Family Study (MTFS). The MTFS began as a cross-sequential study of preadolescent (younger cohort) and late adolescent (older cohort) monozygotic (MZ) and same-sex dizygotic (DZ) twins and their parents (for details, see Iacono, Carlson, Taylor, Elkins, & McGue, 1999; Iacono & McGue, 2002). Younger cohort families were recruited during the year the twins turned age 11, and the older cohort was recruited when the twins turned 17 (see Figure 1). Twin families constituted a statewide sample identified using Minnesota state birth records. Families with children whose cognitive ability was insufficient to provide informed assent or consent were not recruited. No medical, psychological, or psychiatric exclusionary criteria were used to screen out participants. These families were broadly representative of Minnesota families with children living at home according to the 2000 U.S. Census for Minnesota (Holdcraft & Iacono, 2004). Data collection for a third MTFS twin sample (the Enrichment Sample, ES) was launched in 2000 (for details, see Iacono, McGue, & Krueger, 2006; Keyes et al., 2009). ES focused on families with 11-year-old twins, half of whom were selected to be at high risk for developing substance use disorders, and the other half of whom were selected using the same methods as were followed in the MTFS. Only participants from these three MTFS studies (younger cohort, older cohort, and ES) were tested in the psychophysiology laboratory, and thus only these MTFS families were included in the evaluation of the molecular genetic basis of the endophenotypes.
MCTFR participants from two other studies, the Sibling Interaction and Behavior Study (SIBS) and the Adolescent Brain (AdBrain) Development Study, also provided DNA samples and were genotyped using the same procedures followed for the MTFS samples (for details, see McGue et al., 2013; Miller et al., 2012). However, SIBs participants were never evaluated in the psychophysiology lab, and the AdBrain twins did not undergo the MTFS psychophysiological testing protocol, so neither of these samples is included in the molecular genetic analyses of the endophenotypes. Their data was valuable, however, for optimizing the quality control procedures used to process the genotyped data in all the papers. In addition, these samples were used to enhance the accuracy of the imputation of genetic marker variants in this sequencing study (Vrieze, Malone, Vaidyanathan et al., 2014). Given their peripheral role in this series of papers, we refer readers to other publications for additional detail regarding these two studies (Malone, Luciana et al., 2014; McGue et al., 2007).
How participants came to have both psychophysiological and molecular genetic data
The cohort-sequential nature of the MTFS design is such that the MTFS younger cohort and ES twins were reassessed approximately 6 years after study intake, and thus these samples, like the older cohort twins, were seen at age 17. For this series of molecular genetic investigations, we targeted the age-17 assessment of all twins for the collection of endophenotype data. This resulted in the largest possible twin sample and represents a stage of development when adolescents are on the cusp of adulthood. Most parents completed an identical laboratory assessment; those who did were included. Fathers completed the laboratory assessment during the initial family intake visit to the university. Because mothers were asked to provide comprehensive information about other family members at the intake assessment, there was no time available to accommodate the psychophysiological assessment. For younger cohort and ES mothers, psychophysiological assessment took place when they accompanied the twins for their age-14 follow-up visit. Because the older cohort twins were legal adults when they returned for their first follow-up at age 20, there was no need for their mothers to accompany them, so their mothers were never asked to complete a psychophysiological lab session. In addition, as a result of adjustments to the design precipitated by a cut in funding to one of the supporting grants, the majority of mothers of younger cohort female twins were not asked to complete the laboratory assessment. Finally, as noted previously, the startle paradigm was not part of the original MTFS psychophysiological battery, so some twins (those in the ES study) were evaluated at other than age 17, and many twins and parents were never evaluated with startle (for additional details, see Vaidyanathan, Malone, Miller, McGue, & Iacono, 2014). Because these aspects of the design account for the great majority of missing data, these data can reasonably be treated as missing at random (Little & Rubin, 2002), which can be accommodated in our approaches to statistical analysis and permits unbiased statistical estimation. All participants in MCTFR studies gave written informed consent or assent, if under the age of 18, to participate in the initial study, to provide a DNA sample, and to allow their phenotype and genotype data plus a sample of their DNA to be placed in a public repository to be shared with other researchers.
As Figure 1 illustrates, there were 7,697 participants who provided DNA and passed our genotyping quality control screening. Only Caucasian participants were included because ethnic differences in allele frequencies can create spurious associations in genetic association studies. Of these 7,697 subjects, 4,905 had data for at least one endophenotype, with the vast majority of the remainder not included because they were from the SIBs or AdBrain samples, and some because they did not have data even though they were in the MTFS (e.g., younger cohort twins who did not return at age 17). There were 1,715 families, 64.1% of them MZ twin families, with the number of individuals in a family (including stepparents) ranging from 1 to 5. Families with 3 members were most common (48%), and approximately 91% comprised 2–4 members. As can be seen in Table 1, for each endophenotype, somewhat different subsets of these individuals provided valid data for the genetic analyses. Most of the participants were offspring, and as would be expected given that mothers (unlike fathers) were not always asked to complete a psychophysiological assessment, about twice as many fathers contributed data as did mothers.
Table 1.
Sample Sizes and Family Composition
| Psychophysiological measure | N | Sex and family composition | ||||
|---|---|---|---|---|---|---|
| % Female | % Fathers | % Mothers | % Offspring | % Stepparents | ||
| Antisaccade | 4,469 | 44.2 | 26.2 | 13.3 | 58.8 | 1.7 |
| EEG power (α, β, θ, and δ) at CZ | 3,948 | 44.3 | 25.3 | 13.6 | 59.5 | 1.6 |
| Alpha EEG (power, frequency) at O1O2 | 3,966 | 44.1 | 25.4 | 13.5 | 59.5 | 1.6 |
| P3 amplitude | 4,166 | 43.4 | 26.7 | 13.4 | 58.1 | 1.7 |
| P3 genetic factor | 3,088 | 43.2 | 33.5 | 17.1 | 49.4 | 0.0 |
| Skin conductance level | 3,791 | 43.0 | 30.8 | 15.9 | 51.3 | 1.9 |
| Skin conductance response amplitude | 4,102 | 43.6 | 25.5 | 12.6 | 60.3 | 1.6 |
| Skin conductance response frequency | 4,299 | 43.9 | 26.0 | 13.8 | 58.7 | 1.6 |
| Electrodermal activity factor | 4,424 | 43.8 | 26.5 | 13.7 | 58.2 | 1.6 |
| Overall startle | 3,323 | 50.3 | 17.9 | 14.8 | 66.1 | 1.2 |
| Aversive difference eye blink startle | 3,321 | 50.3 | 17.9 | 14.8 | 66.1 | 1.2 |
| Pleasant difference eye blink startle | 3,322 | 50.3 | 17.9 | 14.8 | 66.0 | 1.2 |
Note. EEG = electroencephalogram; P3 = P300 wave of the event-related potential.
Figure 1 documents that there is some variability across measures in the total number of individuals with data for each, ranging from a high of 4,469 for the antisaccade task to a low of 3,323 for startle. It took over 20 years to collect the psychophysiological data from all of these MTFS participants. It was not always possible to obtain data for everyone on each measure (as noted previously, this was especially true for startle, which was added late to the assessment protocol). In addition, there was also obvious variability across measures in the likelihood that collected data could be used in analyses. Our procedure required that data be collected on individuals who ordinarily would have been excluded for an assessment based on preexisting status (e.g., having a bad cough on the day of assessment, taking medications or psychoactive substances that might interfere with psychophysiological recording), or having a physical problem or condition likely to affect the validity of the psychophysiological measurement or neurophysiological state at the time of testing (e.g., serious head injury, neurological disorder). Adolescents taking medication for attention deficit hyperactivity disorder (ADHD), such as methylphenidate, were asked to refrain from doing so the day of their assessment. If any reported taking these medications, they were excluded from analysis. This nonexclusionary approach to laboratory assessment was necessitated by privacy concerns (e.g., family members came together and could easily determine if one member was excluded from a procedure, raising questions regarding why), the desire to optimize future participation in our longitudinal research (e.g., by not inadvertently creating the impression that some participant data would not have value), and the need to keep participants occupied and with staff for the entirety of a day-long assessment. In addition, someone who might be seen as inappropriate for one procedure might nevertheless be seen as appropriate for another, making it awkward to explain why they were qualified for certain procedures but not others. Finally, participants were also excluded due to psychophysiological recording problems, which ranged from excessive artifact and recording equipment malfunction to the failure of disk storage.
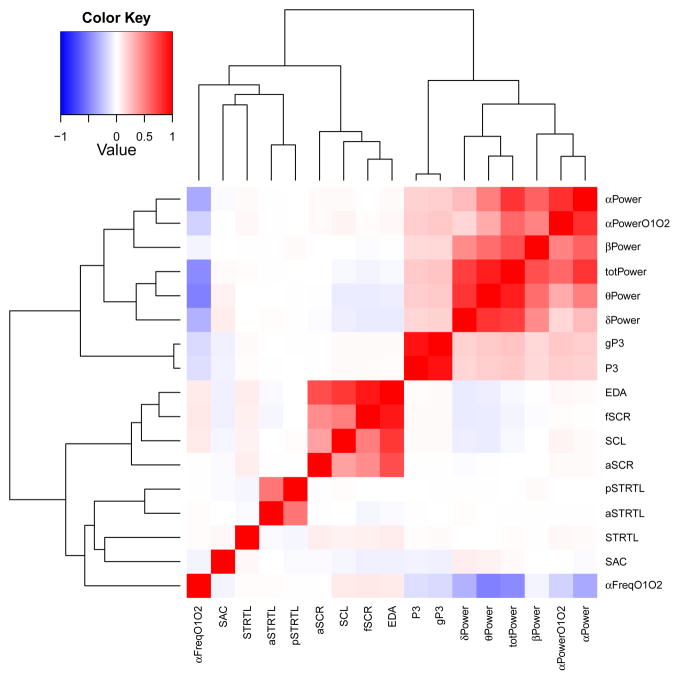
Tables 1 and 2 describe the participant samples and data used in the five common variant studies as well as the exome chip rare variant article. Figure 2 provides a heat map representation of the correlations among the 17 endophenotypes. The heat map shows that, with a few exceptions, each of the five psychophysiological protocols yielded variables that were much more strongly correlated with each other than they were with variables from other protocols. One exception arose with startle where the amplitude of the overall startle response showed little correlation with aversive and pleasant startle difference measures, which were derived from z scores. Another involved the EEG and P3 measures wherein P3 amplitude and the genetic factor score were on average correlated .19 with the EEG power measures. Alpha frequency showed a strong negative correlation (averaging −.38) with EEG power in all spectral bands except beta (−.05).
Table 2.
Means and Standard Deviations for Each Endophenotype, Stratified by Family Member
| Psychophysiological measure | Male twins | Female twins | Fathers | Mothers | ||||
|---|---|---|---|---|---|---|---|---|
| M | SD | M | SD | M | SD | M | SD | |
| Antisaccade % error | 0.31 | (0.22) | 0.27 | (0.19) | 0.34 | (0.23) | 0.36 | (0.23) |
| Alpha EEG power | 5.54 | (0.80) | 5.83 | (0.83) | 5.57 | (0.87) | 5.56 | (0.57) |
| Beta EEG power | 2.89 | (0.57) | 3.22 | (0.57) | 3.25 | (0.66) | 3.31 | (0.69) |
| Theta EEG power | 5.83 | (0.54) | 6.03 | (0.52) | 5.39 | (0.63) | 5.40 | (0.64) |
| Delta EEG power | 6.03 | (0.43) | 6.12 | (0.38) | 5.56 | (0.51) | 5.62 | (0.55) |
| Total EEG power | 7.00 | (0.47) | 7.18 | (0.48) | 6.72 | (0.57) | 6.76 | (0.56) |
| Alpha power (O1O2) | 4.95 | (1.04) | 5.16 | (1.02) | 4.45 | (0.99) | 4.33 | (1.00) |
| Alpha frequency (O1O2) (Hz) | 9.71 | (0.54) | 9.78 | (0.57) | 9.78 | (0.56) | 9.79 | (0.53) |
| P3 amplitude (μ) | 23.29 | (7.76) | 25.98 | (8.61) | 14.43 | (6.58) | 15.10 | (7.19) |
| P3 genetic factor | −0.05 | (0.82) | 0.04 | (0.93) | 0.04 | (0.67) | −0.07 | (0.72) |
| Skin conductance level (mS1/2) | 1.93 | (0.48) | 1.74 | (0.48) | 1.52 | (0.45) | 1.20 | (0.43) |
| Skin conductance response amplitude (mS1/2) | 0.48 | (0.24) | 0.56 | (0.27) | 0.33 | (0.15) | 0.38 | (0.17) |
| Skin conductance Response frequency | 9.84 | (4.77) | 9.59 | (5.07) | 8.46 | (5.35) | 5.85 | (5.31) |
| Electrodermal activity factor | 0.30 | (0.76) | 0.26 | (0.83) | −0.22 | (0.76) | −0.62 | (0.78) |
| Overall startle magnitude (μ) | 38.94 | (35.88) | 52.11 | (44.86) | 26.30 | (26.15) | 37.82 | (38.94) |
| Aversive difference eye blink startle (z score) | 0.16 | (0.66) | 0.18 | (0.65) | 0.17 | (0.62) | 0.24 | (0.62) |
| Pleasant difference eye blink startle (z score) | −0.09 | (0.64) | 0.05 | (0.60) | 0.05 | (0.60) | 0.02 | (0.60) |
Note. For ease of understanding, statistics are given here for the raw (unresidualized) variables. EEG power measures are log-transformed. All are from Cz, at the vertex, unless otherwise indicated. O1O2 indicates the average of two bipolar recordings: O1-P7 and O2-P8. Skin conductance amplitude and level were square-root transformed. EEG = electroencephalogram; P3 = P300 wave of the event-related potential.
Figure 2.
Heat map representation of phenotype correlations among the 17 phenotypes. All measures were covariate adjusted (see text). The dendrogram shows measure clustering based on the correlations. totPower = total EEG power at electrode Cz; μPower = power in the alpha, beta, theta, and delta bands at Cz, respectively; αPowerO1O2 = alpha power at electrodes 01–02; αFreqO1O2 = alpha frequency at electrodes 01–02; P3 = P300 amplitude; gP3 = genetic factor score for P300 amplitude; EDA = electrodermal factor score; SCL = skin conductance level; fSCR = skin conductance response frequency; aSCR = skin conductance response amplitude; aSTRTL = aversive difference startle score; pSTRTL = pleasant difference startle score; STRTL = overall startle amplitude; SAC = antisaccade tracking error rate.
Genotyping and Quality Control
Across all five MCTFR samples (see top of Figure 1), 9,515 participants were eligible to provide DNA because they were still living and had not withdrawn from the MCTFR study in which they were enrolled. From this group, 7,845 provided DNA, with 7,278 (93%) providing blood and 567 providing saliva samples. However, when the MZ co-twins of the MZ twins in this sample of 7,845 are added to the total, more than 88% (N = 8,405) agreed to participate. Of the 12% who did not, the majority could not be contacted within the time allocated for obtaining consent or had concerns about providing a DNA sample. Samples were stored at the Rutgers University Cell and DNA Repository, which followed standard procedures to extract DNA. All genotyping, including the Illumina 660W-Quad, Illumina HumanExome, and whole genome sequencing, was conducted on these DNA samples. The Illumina 660W-Quad (Illumina, 2008–2013) contained 657,366 variants, 561,490 of which were single nucleotide polymorphisms (SNPs; see Appendix 2 for a glossary of commonly used terminology), which are the focus of the first five articles in this special issue (the remaining 95,876 markers were for copy number variants that were not analyzed here). The plates used for genotyping contained 96 wells, and DNA samples were distributed randomly across plates with two exceptions: each plate included samples from two members of a three-member family from the Centre d’Etude du Polymorphisme Humaine (CEPH), the genotypes of whom are known, with the specific individuals rotated across plates, as well as a randomly selected MCTFR duplicate sample. These two types of samples allowed us to assess the accuracy and quality of genotyping.
Genotyping produces measures of intensity for each allele (which we will refer to as A and a), which reflect the degree to which DNA binds to specific allele probes. When plotted against one another, the pairs of intensity values ideally yield three distinct clusters, one cluster corresponding to AA homozygotes, another to aa homozygotes, and the third to Aa heterozygotes. A subset of 1,508 SNPs out of the total of 561,490 could not be called because the clustering of intensity values was not sufficiently distinct to permit identifying the three genotypes reliably. The remaining markers (559,982) were subjected to a series of quality control filters and were excluded for any of the following, if (a) Illumina scientists identified the marker as untrustworthy; (b) duplicate samples did not yield identical results more than once; (c) the call rate was less than 99%, indicating that the algorithm used to estimate the probability that a genotype at an individual SNP is aa, AA, or Aa failed for a nontrivial number of DNA samples; (d) the minor allele frequency (MAF) was less than 1 in 100 subjects; (e) there were more than two Mendelian inconsistencies within families, indicating a mismatch of alleles between parents and offspring; (f) allele frequencies were inconsistent with Hardy-Weinberg equilibrium, an indicator of stability in a population and a necessary precondition for genetic analysis (p < 10−7 in the Caucasian subsample); (g) if the marker was associated with the particular plate used for processing; or (h) the marker was associated with sex (also at p < 10−7), which would indicate a source of systematic error. This resulted in the elimination of 32,153 markers (5.7%), leaving 527,829 SNPs for analysis. The majority of SNPs dropped had a MAF less than .01 (19,999, or 3.6% of the total).
The quality of each individual’s DNA sample was assessed using five criteria, and it was excluded if (1) more than 5,000 SNPs could not be called, suggesting poor quality of the sample; (2) GenCall scores produced by Illumina’s BeadStudio software, indexing confidence in each call, were below an empirically derived threshold (Cunningham et al., 2008); (3) samples had apparently been mixed; (4) a sample was characterized by excessive homozygosity or heterozygosity; or (5) known genetic relationships could not be confirmed. This process, which took advantage of the family relationships to identify errors in labeling samples, eliminated 160 samples. The final sample of 7,278 that passed all quality-control filters included 1,127 samples from individuals whose monozygotic twin had not been genotyped, in which case the genotypes of genotyped twins were assigned to the nongenotyped identical twin, which resulted in a final sample of 8,405. Five individuals with X chromosome anomalies, such as Turner syndrome, were subsequently eliminated due to concerns about potential cognitive correlates.
The Illumina HumanExome BeadChip array was genotyped in a similar fashion as the 660W-Quad, with additional steps and filters to deal with the very rare variants genotyped on the exome array. Details are provided in the relevant article (Vrieze, Malone, Pankratz et al., 2014). For whole genome sequencing, we took advantage of the results of our 660W-Quad genotyping to select samples that had quality DNA, genome-wide genotypes (useful in evaluating sequencing accuracy), and were of European ancestry. See Vrieze, Malone, Vaidyanathan et al. (2014) for a full description of the whole genome sequencing experiment.
Confounding by Ethnicity
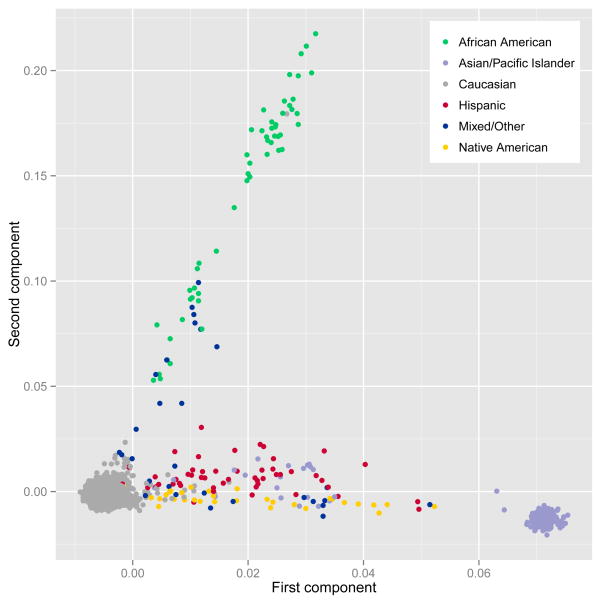
Ethnic differences in allele frequencies are common. They can be confounded with ethnic differences in mean levels of a phenotype or rates of a disorder, in which case a spurious association between genetic variants and phenotype can exist. Ethnic differences in genotype can be assessed by means of multivariate techniques such as principal component analysis (PCA), which captures the major sources of genetic variation in a reduced subspace. Figure 3 depicts the first two components from an analysis of the entire MCTFR sample using the program EIGENSTRAT (http://genepath.med.harvard.edu/~reich/Software.htm) (Price et al., 2006), which detects and corrects for population stratification in genome-wide association studies. The PCA method explicitly models ancestry differences along continuous axes of variation. Close pairwise relationships (e.g., parent-child, siblings) were avoided when determining the major dimensions of variation, but the genotypes of those individuals excluded were then projected onto the components extracted from the unrelated subsample (for additional details, see Miller et al., 2012). The first principal dimension in Figure 3 consists of a component anchored by individuals who reported European ancestry at one end and by individuals whose self-reported ancestry is East Asian (Korean adoptees from the SIBS project) at the other. The second principal dimension differentiated individuals of European ancestry from those who reported African-American ancestry. Because the majority of the MCTFR sample is Caucasian, broadly representative of the racial composition of the state of Minnesota during the birth years from which the different samples were drawn, we restricted genetic analyses to Caucasian individuals of European ancestry (e.g., Caucasians of Middle Eastern ancestry were not included). PCA was conducted separately on these subjects in EIGENSTRAT to identify the major dimensions of genetic variation in this otherwise ethnically homogeneous sample. As is common practice, the 10 components (PCs) accounting for the most variance were included as covariates in our genetic association analyses (cf. Price et al., 2006) to account for subtle genotypic variation that might create spurious associations.
Figure 3.
First two components from a principal components analysis of ancestry differences in 4,756 unrelated subjects in the MCTFR genotyped sample using the program EIGENSTRAT. Each dot represents an individual. Dots are color coded by self-reported ethnicity. The principal axis of variation is anchored at one end by those of self-reported European ancestry and at the other by East Asians. The second principal dimension differentiates European ancestry from those who reported African-American ancestry.
Genetic Analyses
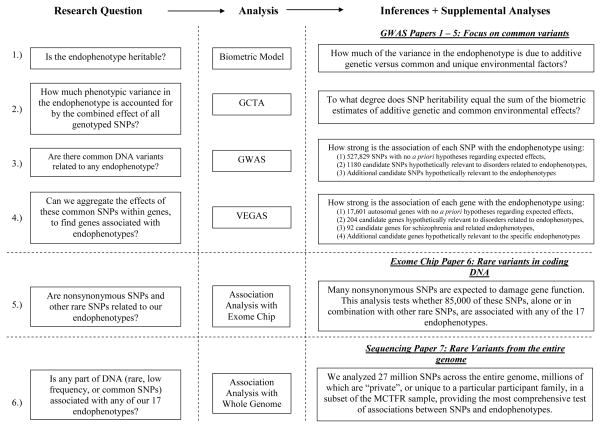
The analytic approach for the seven empirical articles in this special issue is illustrated in the flowchart in Figure 4. We describe the approach first for the five GWAS articles examining common variants, then for the exome chip article examining effects of rare nonsynonymous variants in coding regions, and finally for the sequencing paper covering rare polymorphic SNPs throughout the genome.
Figure 4.
Flow chart highlighting the research questions posed and the analytic methods used to address them in the seven accompanying empirical articles. GCTA = genome-wide complex trait analysis; GWAS = genome-wide association study; VEGAS = versatile gene-based association study; SNP = single nucleotide polymorphism; MCTFR = Minnesota Center for Twin & Family Research.
Associations between common variants and endophenotypes: GWAS-based analyses
Our approach to assessing the influence of common variants in our GWAS studies comprised four prongs: biometric, genome-wide complex trait (GCTA), GWAS, and versatile gene-based association (VEGAS) studies. Figure 4 depicts these analyses in a simplified form; in what follows, we provide a more detailed explication of the methods and assumptions behind these analyses.
1. Biometric models
First, we conducted biometric model-fitting analyses for the purpose of estimating the magnitude of heritable differences in each measure, using standard biometric approaches to modeling twin-family data (M. C. Neale, Boker, Xie, & Maes, 2003). Analyses were conducted using the OpenMx package (Boker et al., 2011) for the R statistical computing environment (R Development Core Team, 2010). Such approaches consist of estimating the parameters in latent variable models, which treat the observed values of a phenotype as due to (caused by) the influence of four latent variables: additive genetic influence (A); nonadditive dominance genetic influence (D), which reflects interactive effects between alleles at the same locus; common or shared environmental influence (C); and unique or unshared environmental influence (E). The observed correlations (or covariances, more commonly) are compared to the correlations implied by the model, which, given standard biometric assumptions, are determined by the known genetic and environmental correlations among family members with respect to the latent factors. Parents and offspring share half their genes by descent, whereas DZ twins share half their segregating genes, on average; the genetic correlation in these pairs is therefore 0.5. MZ pairs share all genes, yielding a genetic correlation of 1 for both A and D. By contrast, the probability that DZ pairs will share both alleles at a locus, which is necessary for dominance effects, is 1/4; this is the dominance genetic correlation (0.25). All family members by definition share the common environment, whereas E reflects environmental factors that are unique to each individual; it does not contribute to within-family correlations. Our models did not allow for assortative mating, the tendency for people with similar characteristics to marry (“like marries like”), as likely influences on the endophenotypes. That this is a reasonable assumption is supported by the mother-father correlation in Table 3, which is very close to 0 across the 17 endophenotypes we examined in this special issue. Similarly, we assumed no environmentally mediated “vertical” transmission of psychophysiological features from parents to offspring. As we indicate in the Adjusting for Covariates section below, all measures were adjusted for any effects on mean levels of particularly relevant covariates, including the first 10 PCs from EIGENSTRAT. Although the latter might seem to overcorrect familial resemblance, it simply adjusts for any effects on mean levels of unknown stratification factors captured by the 10 PCs. However, adjusting for covariate effects on mean levels cannot account for any effects of covariates on phenotypic variances, which we observed most commonly in relation to gender and age cohort. Biometric models therefore allowed for gender and age-cohort effects on variances. For the majority of measures, there were significant differences between cohorts in the phenotypic variance. Moderator effects were included in our biometric models even if they were not significant, however, in order to maintain consistency across measures, thereby facilitating comparison.
Table 3.
Median Within-Family Correlations for the 17 Endophenotypes
| Relationship | Median r |
|---|---|
| Mother-father | .01 |
| Mother-offspring | .23 |
| Father-offspring | .18 |
| MZ twins | .64 |
| DZ twins | .34 |
Note. Correlations were produced by rapid feasible generalized least squares (RFGLS), our analytic method for GWAS that is described in the text, which models within-family correlations. All measures were adjusted for the same set of covariates: chronological age, gender, generation (adolescent or adult), any task-specific factors that might affect mean levels, and scores on 10 principal components reflecting residual population stratification in our Caucasian sample.
In order to adopt a common framework for all five articles, we began by examining the pattern of within-family correlations. The aforementioned Table 3 presents the median correlation across the 17 measures for each family relationship. The MZ twin correlation was large and approximately twice the magnitude of the DZ twin correlation. Because the genetic correlation in MZ pairs is twice the correlation in DZ pairs, this pattern is consistent with additive genetic influence. Because shared environment is shared equally by MZ and DZ twins (the equal environments assumption), a DZ twin correlation exceeding half the MZ twin correlation is consistent with a shared environmental influence. The DZ correlation is only slightly greater than half the MZ correlation, suggesting at best a weak shared environmental effect. However, the mother-father correlation, which can be due to shared environment or assortative mating, was effectively 0, suggesting neither is influencing the endophenotypes. Taking the pattern of twin and parent correlations into account provides little support for a shared environmental effect.
Under the additive ACE model, we expect parent-offspring and DZ twin correlations to be equal, but they are not. In fact, the DZ twin correlation is larger than the parent-offspring correlation. This is what one might expect if there were dominance effects. Because dominance effects are 25% shared by DZ twins, but completely unshared by parents and children, they would lead to a DZ twin (or sibling) correlation that is higher than the parent-offspring correlation. However, the parent-offspring correlation can also be deflated for other reasons, including the special twin environment (which each DZ twin has but the parent does not (Maes, Neale, & Eaves, 1997), and gene-environment interaction (also discussed as cryptic genetic variation, Paaby & Rockman, 2014), which causes different genetic effects to be expressed at different developmental stages or in different cohorts (Eaves, Last, Young, & Martin, 1978).
Dominance effects are also suggested when the DZ correlation is much less than half the MZ correlation, but that pattern is not evident in Table 3. Evidence of dominance effects is thus weak and inconsistent in the family correlations, and our molecular genetic heritability analyses (i.e., GCTA, described next) can only accommodate additive genetic effects. We therefore opted to focus on ACE models in the accompanying articles. However, this may cause us to somewhat overestimate the narrow-sense heritability of our measures (heritability due solely to additive genes). We therefore also fit ADE models and report the results of these if they suggest that dominance effects are an important influence on a particular measure. We also examined ACE and ADE models fit to the twins only, in part due to concerns about gene-environment effects in the parental generation, as described above, and in part to facilitate comparison with the published findings of other researchers working with twin samples. Our goal was not careful explication of the structure of these measures from a biometric perspective, but rather to establish that they are heritable, and broadly to characterize the magnitude of heritable differences, which provides an estimate of the genetic target in genome-wide analyses of individual genetic variants. Because our sample is genetically informative, we are able to do both in the same sample, which provides a relatively unique opportunity to establish the magnitude of heritability and identify relevant genetic variants at the same time.
2. SNP heritability
As an adjunct to biometric analyses, we also conducted genome-wide complex trait analysis (GCTA; Yang, Lee, Goddard, & Visscher, 2011), which, for each endophenotype, assesses the additive effect of all SNPs in linkage disequilibrium (LD) with the 527,829 SNPs on the Illumina genotyping array. LD creates a correlation among SNPs that are commonly inherited together in a given chromosomal region. An apparent association between SNPs on the genotyping array and a phenotype may be due not to the genotyped SNPs, but rather to variants in LD with those SNPs. GCTA thus estimates the variance in the phenotype explained by all SNPs in aggregate, rather than estimating associations between each individual SNP and endophenotype, as is done in GWAS. This is accomplished by treating each SNP as a random effect in a linear mixed model. Fixed effects in the model in our analyses, which are normally of interest in regression analyses, consisted of the covariates described in the Adjusting for Covariates section that follows. Each genotype is a standardized count of the number of minor alleles. A simple reparameterization expresses the random effects in terms of a matrix of pairwise genetic relationships among all participants (the genetic relatedness matrix). Restricted maximum likelihood is used to estimate the random effect variance, which is the total variance in the phenotype accounted for by SNPs on the genotyping array or in LD with them.
In samples comprising families, estimates of the additive genetic variance in a phenotype are driven by the phenotypic correlations among family members, which can be influenced by common environmental effects and nonadditive genetic effects, thus leading to biased estimates. Estimates of additive genetic variance can also be influenced by other classes of genetic variation (e.g., rare variants). Thus, the GCTA estimates derived from families will reflect all causal variants, including rare variants not well tagged by SNPs on the genotyping array, rather than only those on the array itself or in LD with SNPs on the array. GCTA results based on families thus have a very different interpretation than results obtained in unrelated individuals. Yang and colleagues therefore recommend filtering family samples using several values of genetic relatedness to exclude close relatives, which ideally yields stable estimates across different thresholds. Because the choice of a cutoff is arbitrary, we used values of .025, .05, and .10, the most stringent of which (.025) corresponds to approximately third to fourth cousins. Because SNP heritability estimates can be inflated by SNPs in LD with “causal” variants (Speed, Hemani, Johnson, & Balding, 2012), in addition to the commonly employed GCTA procedure of Yang et al. (2011), we also used the program LDAK to derive LD-adjusted kinship coefficients that weight SNPs by local LD patterns (http://dougspeed.com/ldak). Hence, each article provides GCTA estimates based on the use of three different cutoffs, using both the GCTA approach most commonly employed in the literature, and a less commonly employed version that has the advantage of taking into account LD patterns. Our goal was thus not to provide a single GCTA point estimate for each endophenotype, but rather to examine how the estimates vary using different procedures, providing us with the opportunity to examine the extent to which our results depend on the assumptions inherent to each. Of particular interest was the degree to which convergence was evident in point estimate across the six analyses.
In addition to analyses based on subsamples of unrelated individuals, we conducted two different GCTA analyses using the whole sample. One used the procedure of Yang et al. (2011) without filtering subjects on the basis of genetic relatedness. GCTA estimates produced by this approach are driven by phenotypic relationships, including effects of shared environment (the C latent variable in biometric models). We also carried out a second analysis of the whole sample using a method recommended by Yang and colleagues (Yang, Lee, Goddard, & Visscher, 2013) for simultaneously modeling the genetic and environmental influences shared by family members. This produces an estimate unconfounded by the contribution of C. It also provides an estimate of C effects, which offers an opportunity to corroborate or disconfirm biometric model-fitting results. (We do not report these estimates, however, because they can be inferred from the magnitude of the difference in GCTA estimates from the two family-based models.) To summarize, we examined SNP heritability using three different versions of GCTA, and where possible, different cutoffs for relatedness in our sample. The online supporting information for this paper provides further details regarding exactly how the different GCTA models were applied for the analyses carried out in each of the empirical papers.
GCTA is essentially descriptive; its purpose is not to identify specific SNPs that influence a trait. As such, it complements the biometric model-fitting analyses by focusing on the molecular-genetic basis of phenotypic similarity rather than fitting models based on phenotypic covariances. It tells us to what extent biometric heritability can be accounted for by a truly additive model of one class of genetic variants, common SNPs (and everything in LD with these SNPs). This in turn gives us some insight into the utility of additional investigations into other types of genetic variants, such as copy number variants, variable nucleotide repeats, insertions and deletions, rare SNPs, epistasis, or dominance, none of which is accounted for by GCTA despite possibly contributing to biometric heritability.
3. Analysis of individual SNPs in GWAS papers of common variants
The third prong in our analytic strategy was to conduct genome-wide association studies (GWAS). In the GWAS, we conducted regression analyses of effects on each psychophysiological measure of each of the 527,829 SNPs on the Illumina 660W-Quad genotyping array that survived quality control filters. Whereas GCTA considers all SNPs together, GWAS considers each SNP alone. The focus of GWAS is often on relatively common SNPs—typically those with a MAF of at least 5% (those with a MAF less than 1% were discarded as part of our quality control procedure). A focus on common SNPs is consistent with the “common disease–common variant” model (for a review of the development of this concept, see Visscher, Brown, McCarthy, & Yang, 2012). For this model, the underlying genetic architecture differs between common and rare disorders, with common disorders (here, more appropriately, an endophenotype for a common disorder) thought to be heavily influenced by variants that are relatively common in the population, although this does not exclude the possibility that rare variants may also be involved. This model was influenced by discoveries of susceptibility variants for common diseases that have large MAFs, such as alleles in the apolipoprotein E gene (APOE) that confer risk for Alzheimer’s disease and alleles in the PPARG gene that confer risk for type II diabetes (Bush & Moore, 2012). Importantly, these procedures were not those used in the exome chip and sequencing papers to appropriately analyze large numbers of rare variants (see the next section for details and Vrieze, Malone, Pankratz et al., 2014; Vrieze, Malone, Vaidyanathan et al., 2014).
GWAS using MCTFR data is complicated by the nested structure of our sample, which induces a correlation among family members. To account for the lack of independence in family data, we used rapid feasible generalized least squares (RFGLS; X. Li, Basu, Miller, Iacono, & McGue, 2011). RFGLS is a computationally efficient form of generalized least squares (GLS). GLS can be appropriate when residuals are correlated (or heteroskedastic). GLS assumes that the residual covariance structure (e.g., within higher-order units, in our case, families) is known. If it is not, the observed variances and covariances can be used as an estimate of the unknown covariance structure, an approach known as feasible GLS (FGLS). In the present case, data were clustered in families comprising one to four members, with three family types: MZ and DZ families and stepparents. RFGLS estimates the residual covariance matrix separately for each type. FGLS would require us to estimate the residual covariance structure conditional on model covariates and a given SNP for each of the 527,829 SNPs, which is computationally inefficient. RFGLS estimates the residual covariance matrix once, conditional only on model covariates, based on the assumption that SNP effects on the residual covariances will be negligible. This produces significant savings in computational time and minimal bias or loss of power (X. Li et al., 2011). Constraints are imposed on several elements of the residual covariance matrix in order to reduce the number of parameters to be estimated, thereby avoiding problems with algorithm convergence. The mother-offspring and father-offspring correlations are constrained equal, as are variances for the two members of a twin pair. In all, four correlations (MZ or DZ twin pair, mother-offspring, father-offspring, mother-father) and four variances (twin, mother, father, stepparent) were estimated in the investigations described here. The independent variable in each analysis was a count of the number of minor alleles (0, 1, or 2) for each SNP, with the variables described below as covariates. The causal model implicit in using a count of minor alleles is that SNP effects are additive. Each SNP association was assessed via a test with 1 df.
We used the conventional p-value threshold of 5 × 10−8, a genome-wide significance criterion used in GWAS that is considered robust to false positives because it tightly controls the familywise error rate arising from the testing of hundreds of thousands of SNPs. This is based on the notion of genome-wide significance, which corrects for the total number of effective independent regions in the genome, based on LD patterns in a particular population. Although the threshold adopted is stringent, we are applying it on a per phenotype and per experiment basis instead of correcting for all the possible different phenotypes we are evaluating across all the different papers. We believe there is an advantage to adopting this approach when all the tests we are conducting appear as part of a collection of papers presented together (as opposed to publishing each independently in different sources spread out over an extended period of time) because it allows the reader to make an informed opinion about the evidence for association in the context of a transparent overall approach. Nevertheless, for each tested endophenotype, there undoubtedly will be SNPs that are related to the endophenotypes that do not cross this stringent but necessary significance threshold. Therefore, we point out “suggestive” associations with each measure, although we do not interpret them. In this vein, each GWAS paper is accompanied by a supplement, which includes a list of SNPs associated with that paper’s endophenotype(s) at a significance level of p < 10−4. Although many of the SNPs with p values this small will represent false positives, a small subset is likely to constitute a valid signal in the genetic pathway mediating the development of a particular endophenotype. It is here that future molecular genetic investigators interested in a psychophysiological measure might look for evidence that their small p-value findings overlap with and are in effect replicated by ours.
In addition to this genome-wide scan, we used GWAS results to explicitly assess associations for two sets of candidate SNPs. The first set comprised 1,180 SNPs related to disorders or traits that are likely a priori to be associated with the different endophenotypes. These were identified through MEDLINE and included meta- and mega-analyses of alcohol (Wang et al., 2011) and drug (C. Y. Li et al., 2011) dependence, cocaine abuse (Clarke et al., 2013), smoking and nicotine dependence (Belsky et al., 2013; Bierut et al., 2008; Furberg et al., 2010; Liu, Tozzi et al., 2010; Thorgeirsson et al., 2010), ADHD (B. M. Neale et al., 2010), schizophrenia, bipolar disorder, and major depression (Greenwood et al., 2011; Hek et al., 2013; Ripke et al., 2012; Smoller et al., 2013; Sullivan, Daly, & O’Donovan, 2012), or related phenotypes, such as heavy drinking (Heath et al., 2011) and the maximum number of drinks consumed at one time (Kapoor et al., 2013; Pan et al., 2013), and the personality characteristic of excitement seeking (Terracciano et al., 2011).
The second candidate SNP set was different for each investigation, consisting of SNPs that have been reported in previous research to be associated with the specific endophenotypes investigated. SNPs in either of these two sets that were not on the Illumina array were imputed, using the program Minimac (Howie, Fuchsberger, Stephens, Marchini, & Abecasis, 2012), after genotypes had first been phased using Beagle (Browning & Browning, 2009), which uses known familial structure to improve phasing accuracy. Genotypes were imputed with 1000 Genomes reference haplotypes (1000 Genomes Project Consortium, 2012). Imputation produces an allele dosage for each variant site in 1000 Genomes, which is a weighted count of the minor allele; each genotype (AA, Aa, and aa, represented as 0, 1, and 2, respectively) is weighted by the posterior probability of that genotype as estimated by the imputation algorithm. Analyses of imputed SNPs used the allele dosage as the independent variable. We only used SNPs that had been imputed accurately, with an imputation r2 of at least .30 (http://www.ncbi.nlm.nih.gov/pubmed/21058334). A Bonferroni-corrected significance threshold was adopted for both candidate sets, which corresponded to α = 4.24 × 10−5 for the set of 1,180 SNPs and a different value for the second set of candidate SNPs that varied from one endophenotype to another.
4. Aggregating SNPs within a gene
The fourth aspect of our analysis strategy consisted of testing associations between individual genes and the endophenotypes, using VEGAS, which stands for a “versatile gene-based association study” (Liu, Mcrae et al., 2010). VEGAS combines into a single score evidence of association between all SNPs in a gene and a phenotype. This approach can be particularly powerful when several SNPs located in a gene are causally related to the phenotype, in which case the p value associated with any of them may not be small enough to be distinguishable from noise. VEGAS assigns SNPs to a gene by reference to the UCSC Genome Browser assembly, including all SNPs within 50 kilobases of the 3′ and 5′ untranslated region of a given gene in order to capture regulatory SNPs and SNPs in LD with those in the gene itself. Individual p values for each SNP are converted into chi-squared statistics with 1 df and summed. Although other gene-based approaches exist, VEGAS easily accommodates the clustered nature of our sample because the p values it uses were produced by RFGLS and accurately reflect the nested structure of our data. LD between SNPs causes the SNPs and their p values to be correlated. Therefore, the null distribution of the gene score in the presence of LD must be determined. VEGAS uses Monte Carlo methods and the LD structure of a reference sample from the International HapMap Project (International HapMap Consortium, 2005). We selected the CEPH sample of Utah residents of European ancestry in HapMap (CEU) for this purpose.
We used VEGAS to conduct gene-based tests of association in a manner parallel to our analyses of individual SNPs. We tested the association between each of 17,601 autosomal genes and our endophenotypes in a genome-wide scan comparable to our GWAS of SNPs. The VEGAS algorithm we used did not consider allosomes. A threshold of p = 2.84 × 10−6 was used for determining statistical significance, which corrects for the number of different genes. In addition, we evaluated three sets of candidate genes. The first set comprised 204 genes selected because they are likely a priori to be related to the endophenotypes by virtue of particular characteristics: they belong to one of the major neurotransmitter or neuromodulatory systems (dopamine, noradrenaline, acetylcholine, GABA, glutamate, and serotonin), they belong to the endogenous opioid or cannabinoid systems, or they are implicated in metabolizing alcohol and nicotine. Relevant genes were identified through the NeuroSNP database (https://zork5.wustl.edu/nida/neurosnp.html). A threshold of 2.45 × 10−5 was used for determining the significance of any genes in this set. The second set consisted of 92 autosomal candidate genes identified by the Consortium on the Genetics of Schizophrenia (Greenwood et al., 2011), which reported evidence of association between these genes and some candidate endophenotypes broadly similar to those studied here. The third set was unique to each article, consisting of any candidate genes that have been found in previous research to be associated with the particular endophenotype examined.
Adjusting for Covariates
The measures we examined in these seven articles are potentially influenced by several demographic-related characteristics. For instance, gender differences in mean levels are sometimes observed for these measures considered as a group. Moreover, the sample comprises two age cohorts: adolescent twins and their parents, who are primarily middle aged. The actual ages vary within each cohort, and all of our measures are likely to change somewhat in mean level over the course of the life span, including during the late-adolescent period spanned by twins in this sample. We therefore adjusted all measures for these covariates in order to remove them as potential sources of confounding in our analyses. The covariate set, which was common to the five investigations of common variants, also included the 10 genetic PCs derived from EIGENSTRAT to adjust for any effects of unknown population stratification factors, in addition to age cohort, gender, and chronological age. Because data for these investigations were collected over a span of approximately 20 years, there were sometimes changes in protocol or recording system. The covariate set for each investigation therefore included dummy variables as necessary to accommodate variation in procedure or differences between protocols that might be specific to an experimental task. With the exception of the 10 genetic PCs, the same covariates were used in the two papers examining rare variants.
Associations Between Rare Variants and Endophenotypes: Exome Chip Analysis
Whereas the first five papers in this special issue examine the role of common variants in accounting for variance in the different putative endophenotypes, the sixth empirical report focuses on rare variants. In general, results from the common variant papers indicated that estimates of SNP heritability from GCTA were less than estimates of phenotypic heritability from fitting biometric models. This pattern suggests that not all of the genetic influence on these endophenotypes is accounted for by the common variants on the Illumina genotyping array. This finding is common in medical and psychiatric genetics, and it has led many to consider the role of rare variants (Zuk et al., 2014). In the investigation described in this paper, we examined associations between ~ 85,000 nonsynonymous SNPs and all 17 endophenotypes. Nonsynonymous SNPs are located in coding regions of the genome (the exome); they are therefore exonic variants, and they also tend to be rare. The different alleles of nonsynonymous SNPs change the amino acid sequence of a protein, the effects of which can range from benign to lethal. Even in less extreme cases, however, their impact on phenotypic development is hypothesized to be greater, on average, than the impact of SNPs, which do not directly affect protein structure. This stands in sharp contrast to the (common) SNPs assessed in GWAS, which are selected for characteristics such as their ability to tag other SNPs and not necessarily for any functional relevance. We also conducted gene-based burden tests, in which the effects of individual variants within a gene are combined into a single score, similar to the VEGAS approach. Although the variants examined differ between the first five papers and the sixth, we nevertheless were able to specifically examine rare variants in the 204 NeuroSNP candidate genes from the common-variant analyses (described above).
The methods used in this paper are necessarily different from those used in the five papers on common variants, and they are described in detail in the paper itself. As in the papers on common variants, all putative endophenotypes were adjusted for the relevant covariates: gender, age cohort, chronological age, and any dummy variables representing task-specific factors that might affect observed levels. Unlike the approach adopted in the other papers, they were not adjusted for population stratification by means of the 10 PCs produced by EIGENSTRAT. Instead, a linear mixed model EMMAX (Kang et al., 2010), implemented in the program EPACTS (Kang, 2014), was used to estimate an empirical kinship matrix, analogous to the genetic relatedness matrix in GCTA. The empirical kinship matrix allowed us to adjust for familial resemblance and population stratification simultaneously.
Associations Between (Nearly) All SNPs and Endophenotypes: Whole Genome Sequencing
The final empirical article in this special issue (Vrieze, Malone, Vaidyanathan et al., 2014) represents our most comprehensive attempt to discover rare, or common, variant associations with the 17 endophenotypes. We use whole genome sequencing to search the entire genome for SNPs, whether common or rare. We found 27 million autosomal SNPs, which includes the vast majority of all SNPs genotyped on the 660W-Quad genome-wide array and the exome array described previously. Each SNP is then tested for association with each endophenotype. We also conduct gene-based burden tests, just as in the exome chip article. We describe the sequencing methodology in the sequencing article itself and do not repeat it here. Instead, we provide a brief overview of how the sequencing article complements the other articles in this special issue, and some of the challenges associated with sequence analysis.
The other six articles used fixed arrays to genotype individuals; such arrays only genotype variants that have already been discovered in other individuals. Whole genome sequencing can identify novel rare variants, never seen before in any individual. Indeed, some of the variants we describe in the sequencing article are exclusive to the MCTFR participants, and have never been reported previously, in any study. Such comprehensive genotypic information allows for comprehensive genetic association tests. It is well known, for example, that increased genotyping density increases power to discover associations, even for common variants (Y. Li, Willer, Ding, Scheet, & Abecasis, 2010). Sequencing also allows accurate genotyping of rarer variants completely missed on any commercially available array.
This wealth of genetic variation carries with it a variety of challenges. First, genotype accuracy from sequencing is strongly related to the depth of sequencing; the deeper the sequencing, the more accurate the genotypes. Very shallow sequencing of 1× or 2× (i.e., the base sequence in the genome is “read” 1 or 2 times) is sufficient to accurately capture the vast majority of common variants in the genome (Y. Li, Sidore, Kang, Boehnke, & Abecasis, 2011). However, the human genome contains some 3 billion base pairs, the “reading” of which can be expected to produce occasional genotyping error that cannot easily be differentiated from a rare variant that shows up in the occasional subject. Deep sequencing, such as 30×, provides high power to detect variants so rare that only a single copy of the minor allele is observed in the sample (i.e., appears in just one person, a “singleton”). This accuracy is achieved because reading the base pair sequence 30× makes it possible to separate sequencing errors (which might produce different values for a single base across the 30 reads) from reproducible signal (producing the same value for the base all 30 times). However, higher depth sequencing is more costly. At low depths, many participants can be sequenced for some fixed cost but high depth is more expensive per person, such that only a few individuals can be sequenced for the same fixed cost. That is, low depth sacrifices genotype precision for sample size, and high depth sacrifices sample size for precision. Our sequencing study attempted to balance these competing outcomes, simultaneously obtaining good power to accurately genotype rarer variants in a relatively large sample size. In the end, we settled on a depth of 10×, which, according to our results, provided about 75% power to discover singletons.
Second, knowledge about genomic function outside of the exome is less well developed than our knowledge of that within the exome. Noncoding function can also be highly tissue specific, and the availability of such information in relevant brain tissue is only now being released through ROADMAP (Bernstein et al., 2010; Chadwick, 2012), ENCODE (The ENCODE Project Consortium, 2012), and GTEx (GTEx Consortium, 2013). Therefore, in the present work, we refrain from using functional annotation outside of coding regions, and conduct burden tests within the exome only. However, most disease- and trait-associated SNPs are not found in coding regions (Maurano et al., 2012). Noncoding regions, which comprise over 98% of the genome, are clearly important in genome function (The ENCODE Project Consortium, 2012), and we are keen to use noncoding functional information, as it becomes available, in the future for the sequences we have generated.
Third, whole genome sequencing remains relatively expensive (over $1,000 per individual in our work), so the available sample size is only a portion of that available in the MCTFR. This limits statistical power in a sample that is likely already underpowered to detect small genetic effects. To increase our sample, while maintaining the genotype density afforded us by sequencing, we used the sequences to impute into the full available MCTFR sample. We observed an increase in imputation accuracy over that obtained through imputation with 1000 Genomes (cf. Pistis et al., 2014), the current standard in imputation. The imputation procedure allowed us to retest all 27 million sequenced variants in all 4,905 individuals with psychophysiological endophenotypes, and the increase in statistical power associated with that increased sample size.
Linear Mixed Models in Rare Variant Association Studies
In both the exome chip (Vrieze, Malone, Pankratz et al., 2014) and sequencing articles (Vrieze, Malone, Vaidyanathan et al., 2014) we used a linear mixed model called EMMAX (Kang et al., 2010) to account for population stratification and familial clustering in genetic association tests. Such models have become standard practice for these purposes (Yang, Zaitlen, Goddard, Visscher, & Price, 2014). The linear mixed model is similar to GCTA, in that one uses a kinship matrix representing all pairwise familial relationships estimated on the available genetic data. This matrix is entered as a random effect of a linear mixed model to account for variance in the phenotype due to familial and population structure. Linear mixed models are not without their pitfalls, and thus used incorrectly can lead to spurious results (Yang et al., 2014). One concern noted elsewhere that does apply, however, is in the use of linear mixed models in analysis of rare variants. When the empirical kinship matrix is computed on genome-wide common variants, it may not reflect small pockets of population stratification that are due to evolutionarily recent rare variants. In this case, there may be residual population stratification due to rare variants that are confounded with nongenetic influences (e.g., environment, cultural practices). In this case, the residual population structure can exert a spurious influence on test statistics, and this influence is not corrected for by the common variant empirical kinship matrix (Mathieson & McVean, 2013). Although rare variant stratification is theoretically possible and certainly worth scrutiny, the authors present no real-world examples of the kind of stratification they propose could be problematic. Indeed, we observe no inflation in our rare variant tests here that would lead us to believe that rare variant stratification is playing more than a negligible role in our results; neither have we observed spurious results in prior research that used linear mixed models in rare variant association tests (Vrieze et al., 2013).
Summary and Conclusion
Our approach involves applying the same set of analytic procedures to each of 17 candidate endophenotypes derived from five different psychophysiological protocols assessing constructs of broad interest in psychophysiological research. It includes elements that are both agnostic (genome-wide analyses) and hypothesis driven (plausibly relevant candidate SNPs and genes) regarding the expected results. Consistent with current convention designed to lessen the likelihood of the types of false-positive outcomes that are generally believed to be common in molecular genetic research, we adopted conservative p-value thresholds in our analyses. In reporting results, we interpreted as significant findings that exceeded these thresholds while also noting our strongest nonsignificant findings, with the hope that both are likely to be of value to investigators also interested in the genetic basis of the measures we examined. Also included are biometric analyses of the same phenotypes using the participants in the molecular genetic studies. Although our investigations are not unique in this regard, this is not a common feature of genome-wide studies of complex traits. Besides providing evidence regarding the strength of genetic influence on our measures, because MZ twins are in effect parallel forms of the same person, the MZ twin correlations provide an index of measurement reliability, also an important aspect of an endophenotype.
Although the two decades it took to acquire our study subjects produced samples that are large by standards commonly employed in psychophysiology, they are small when compared against what molecular geneticists believe are well suited to identify genetic variants associated with complex traits. However, there is no way to know what sample size is necessary to achieve success with these candidate endophenotypes in the absence of the type of evaluative investigation we have undertaken. In a review of the first 5 years of GWAS discovery for complex traits, Visscher et al. (2012) graphically showed (see their Figure 2A) that the number of discovered genetic variants was strongly correlated with sample size, such that samples of 20,000 or larger have been required to obtain hits for easy-to-measure-accurately but genetically distal traits like body mass index and height. More difficult to measure but biologically relevant traits with presumed more proximal genetic influences, like the Q–T interval in the electrocardiogram (and HDL cholesterol and bone mineral density), produced verifiable hits with sample sizes in the 2,000–5,000 range, similar to those available in the MCTFR. It is because of such findings that we launched these special issue studies with some optimism regarding the likelihood of identifying causal variants for the endophenotypes we investigated.
Supplementary Material
Acknowledgments
The research was supported by NIH grants: DA 05147, DA 13240, DA 024417, DA 036216, AA09367, DA 034606, HG 007022, and HL 117626.
Appendix 1
Endophenotype Brief Descriptions
Antisaccade Eye Tracking Error (Vaidyanathan, Malone, Donnelly et al., 2014)
Participants viewed a spot of light in the center of a computer screen that appeared to move to one side or the other of the screen (the centered dot disappeared and another appeared to the side). The participant’s task was to override the impulse to direct gaze toward the new target location and to look instead in the opposite direction, fixating on the approximate mirror image location of the target. Performance was quantified as the proportion of trials on which the participant generated a saccade in pursuit of the target instead of generating an antisaccade away from it.
EEG Measures (Malone, Burwell et al., 2014)
Electroencephalographic (EEG) activity was recorded from three electrode locations while subjects relaxed with eyes closed for 5 min. Five measures were derived from EEG at the vertex electrode Cz: Total EEG Power = total power between 0.5 and 30 Hz, Alpha EEG Power = power in the alpha band (8 to 13 Hz), Beta EEG Power = power in the beta band (13.5 to 30 Hz), Theta EEG Power = power between 4 and 7.5 Hz, and Delta EEG Power = power between 0.5 and 4 Hz. In addition, we examined two measures obtained by averaging across two bipolar occipital-parietal electrode derivations (O1–P7 and O2–P8): Alpha EEG Power 0102, defined as for Cz; and Alpha EEG Frequency 0102, defined as the dominant peak frequency in the alpha band.
P3 Event-Related Potential Amplitude Measures (Malone, Vaidyanathan et al., 2014)
To determine P3 Amplitude, subjects completed the rotated heads visual oddball task (Begleiter, Porjesz, Bihari, & Kissin, 1984). Interspersed among frequently displayed stimuli consisting of ovals were infrequently presented superior views of a stylized head displaying the nose and one ear. Subjects pressed a left button if they saw a left ear and a right button if they saw a right ear. Half of these P3-eliciting oddball targets were rotated by 180 degrees and presented with the nose facing down. An additional amplitude measure was also calculated, the P3 Genetic Factor Score, which was generated from a twin family-based factor analysis of P3 amplitude using responses recorded from three parietal electrodes. This measure captures the degree to which the covariance among P3 amplitude measures reflects the influence of shared genetic effects. Because environmental influences are not included in the genetic factor score, it should provide a stronger genetic signal than P3 amplitude, possibly conferring an advantage when searching for associated genetic variants.
Electrodermal Activity Measures (Vaidyanathan, Isen et al., 2014)
Electrodermal activity was recorded from the fingertips as part of a habituation task during which loud tones were intermittently delivered while participants viewed scenes from a closed-captioned movie. Immediately following the end of the movie presentation, participants rested with eyes closed for 5 min. Skin Conductance Level provided a measure of the participant’s tonic resting level monitored at the end of the session, when participants can be expected to be relaxed after having viewed the movie. Skin Conductance Response Frequency provided a count of the number of tones to which participants responded. Skin Conductance Response Amplitude captured mean response magnitude for trials on which participants produced an observable response. The Electrodermal Activity Factor Score provided a global measure of electrodermal activity using a factor score derived from a common factor model fit to the three skin conductance measures.
Acoustic Startle Response and Affective Startle Modulation (Vaidyanathan, Malone, Miller, McGue, & Iacono, 2014)
Three measures were derived from an affective startle modulation paradigm (Vrana, Spence, & Lang, 1988) in which participants viewed a series of well-standardized images, while their startle eye blink reactions to noise probes were recorded. Overall Startle indexed the magnitude (in μV) of the integrated electromyographic (EMG) response from the orbicularis oculi muscle averaged over all trials, regardless of image valence. Aversive Difference Startle was defined by the z score difference in mean EMG startle magnitude between aversive and neutral images and represents a measure of the degree to which startle eye blink is potentiated by aversive stimuli. Pleasant Difference Startle was defined as the z score difference in startle magnitude between pleasant and neutral images and represents a measure of the degree to which startle eye blink is attenuated by pleasant stimuli. The startle blink reflex is intensified by aversive motivational states and diminished by appetitive states.
Appendix 2 Glossary
- 1000 Genomes
Newer than the HapMap Project, the 1000 Genomes Project describes the genomes of 1,092 individuals from 14 countries and provides a validated map of 38 million single nucleotide polymorphisms (SNPs) and almost 1.5 million insertions and deletions of genetic material. It provides a reference panel for imputing SNPs that are not on a genotyping array but are in LD with SNPs on the array.
- Allelic stratification (population stratification)
This occurs if allele frequencies that vary between ethnic groups are confounded with ethnic differences in the phenotype, which can create a spurious association.
- Biometric model
Applied to family members, this statistical procedure provides estimates of the amount of variance in a phenotypic trait that is accounted for by shared genetic influences (indicating the heritability of the trait), shared environmental experience that makes family members similar to one another, and unique environmental factors that make family members different from each other.
- Common variant
A strict definition does not exist, but “common variant” often refers to SNPs whose minor (less frequently occurring) allele is present in 5% or more of the study population.
- Exons
These are the sequences of DNA found in genes that directly encode the amino acids that make up proteins. All the exons combined are referred to as the “exome” and represent perhaps 2% of the total genome sequence.
- Exome chip
Genotyping array used to identify rare nonsynonymous variants in the exome (protein coding portion of DNA).
- HapMap
A catalog of common variants derived from DNA samples from populations of African, Asian, and European ancestry. Samples were collected from individuals of Northern and Western European ancestry in the United States by the Centre d’Etude du Polymorphisme Humain (CEPH). This is the reference population for Caucasian subjects in GWAS, abbreviated CEU, and is used by the VEGAS analytic program to evaluate the strength of association of a gene with a phenotype.
- GCTA: Genome-wide complex trait analysis
A quantitative method used to estimate the degree to which SNPs in unrelated people account for their degree of phenotypic similarity. GCTA assumes genetic variance in the phenotype reflects the combined additive effect of all alleles weighted equally. When carried out on related individuals, estimates are not strictly due to measured genetic variants. Rather, they are driven by all factors that influence phenotypic similarity, including shared environment, nonadditive genetic effects, and rare variants not tagged by the genotyping array. A recent development permits modeling, and thereby accounting for, the shared environmental effects that operate within families.
- Genomic control
In GWAS, we expect the vast majority of genetic variants to have no discernible association with the phenotype of interest. Genomic control tests whether the median p value is greater or smaller than expected by chance (i.e., different from the null). If it deviates too far from expectation, there may be unknown population stratification, familial relatedness, or other problems in the sample.
- GWAS: Genome-wide association study
A molecular genetic method in which a genotyping array for hundreds of thousands of SNPs is used to examine the degree to which each is associated with a phenotype. In the current studies, we tested the degree to which each of 527,829 SNPs was associated with each psychophysiological endophenotype.
- Linkage disequilibrium
Linkage equilibrium occurs when the genotype present at one locus is independent of that at another locus. With linkage disequilibrium, there is nonrandom association between two or more alleles/SNPs, suggesting that they are inherited together and possibly functioning as a unit.
- MAC: Minor allele count
The number of times the minor allele is present in a sample or population. MAC is typically examined in studies of rare variants that occur in only a small number of people in a study sample.
- MAF: Minor allele frequency
The proportion of times the minor allele, or less frequently observed allele, is present in a sample or population. It is the MAC divided by twice the number of individuals (i.e., the MAC times the number of chromosomes in the sample).
- Manhattan plot
A plot of observed p values from a GWAS sorted by chromosome, providing a detailed picture of associations between SNPs and a phenotype. To better visualize small p values, they are scaled as −log10(p). Genome-wide significance of 5 × 10−8 is equal to 7.30 on this scale.
- Nonsynonymous
A type of genetic variant that resides within an exon and can alter the amino acid sequence of a protein, making it nonfunctional.
- Q-Q plot
Q-Q plots represent a tool for evaluating graphically the fit of observed data to a particular distribution. In GWAS, they plot observed p values against expected p values under the null distribution. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed values should conform closely to expected values, except for significant associations.
- RFGLS: Rapid feasible generalized least squares
RFGLS is a statistical package developed at the University of Minnesota to account for the correlated nature of family data in a way that is computationally efficient when running GWAS analyses.
- Rare variant
While no strict definition exists, these are often SNPs whose minor allele frequencies (MAFs) are present in fewer than 5% of those in a study sample.
- SNP: Single nucleotide polymorphism
A sequence variation in a single DNA base pair, the configuration of which varies across people.
- Tag SNP
Common SNPs may not be independent of one another due to linkage disequilibrium. For this reason, one only needs to genotype a select subset of the total number of common SNPs. If these “tag” SNPs are selected well, then a survey of only several hundred thousand SNPs provides a cost-effective way to obtain most of the information about common SNPs in the genome.
- VEGAS: Versatile gene-based association study
A quantitative method in which all the SNPs in every autosomal gene (i.e., a gene that is not on the sex chromosomes) and its surrounding region are tested in aggregate for their strength of association with a phenotype. Linkage disequilibrium among the SNPs is accounted for through simulations in generating a p value.
- Whole genome sequencing (WGS)
A genotyping method that identifies the exact sequence of bases in an entire individual genome, thus making possible the identification of rare variants.
Footnotes
Additional supporting information may be found in the online version of this article:
Appendix S1: Genome-wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokhin AP. Genetic psychophysiology: Advances, problems, and future directions. International Journal of Psychophysiology. 2014;93:173–197. doi: 10.1016/j.ijpsycho.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Bihari B, Kissin B. Event-related brain potentials in boys at risk for alcoholism. Science. 1984;225:1493–1496. doi: 10.1126/science.6474187. [DOI] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Baker TB, Biddle AK, Evans JP, Harrington H, Caspi A. Polygenic risk and the developmental progression to heavy, persistent smoking and nicotine dependence: Evidence from a 4-decade longitudinal study. JAMA Psychiatry. 2013;70:534–542. doi: 10.1001/jamapsychiatry.2013.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nature Biotechnology. 2010;28:1045–1048. doi: 10.1038/Nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Goate AM. Variants in nicotinic receptors and risk for nicotine dependence. American Journal of Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Fox J. OpenMx: An open source extended structural equation modeling framework. Psychometrika. 2011;76:306–317. doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL, Greenwood TA, Swerdlow NR, Light GA, Schork NJ Investigators of the Consortium on the Genetics of Schizophrenia. Advances in endophenotyping schizophrenia. World Psychiatry. 2008;7:11–18. doi: 10.1002/j.2051-5545.2008.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. American Journal of Human Genetics. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Computational Biology. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/Epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Bloch PJ, Ambrose-Lanci LM, Ferraro TN, Berrettini WH, Kampman KM, Lohoff FW. Further evidence for association of polymorphisms in the CNR1 gene with cocaine addiction: confirmation in an independent sample and meta-analysis. Addiction Biology. 2013;18:702–708. doi: 10.1111/j.1369-1600.2011.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Sellers TA, Schildkraut JM, Fredericksen ZS, Vierkant RA, Kelemen LE, Goode EL. Performance of amplified DNA in an Illumina GoldenGate BeadArray assay. Cancer Epidemiology, Biomarkers and Prevention. 2008;17:1781–1789. doi: 10.1158/1055-9965.EPI-07-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves LJ, Last KA, Young PA, Martin NG. Model-fitting approaches to the analysis of human behaviour. Heredity. 1978;41:249–320. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- Flint J, Munafò MR. The endophenotype concept in psychiatric genetics. Psychological Medicine. 2007;37:163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceschini N, Ardissino D, Merlini PA. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature Genetics. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, Hardiman G. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. American Journal of Psychiatry. 2011;168:930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nature Genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: Findings and implications. Biological Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, Murabito J. A genome-wide association study of depressive symptoms. Biological Psychiatry. 2013;73:667–678. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdcraft LC, Iacono WG. Cross-generational effects on gender differences in psychoactive drug abuse and dependence. Drug and Alcohol Dependence. 2004;74:147–158. doi: 10.1016/j.drugalcdep.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genetics. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG. Psychophysiologic markers of psychopathology: A review. Canadian Psychology. 1985;26:96–112. [Google Scholar]
- Iacono WG. Identifying psychophysiological risk for psychopathology: Examples from substance abuse and schizophrenia research. Psychophysiology. 1998;35:621–637. [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Taylor J, Elkins IJ, McGue M. Behavioral disinhibition and the development of substance use disorders: Findings from the Minnesota Twin Family Study. Development and Psychopathology. 1999;11:869–900. doi: 10.1017/s0954579499002369. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Lykken DT, McGue M. Psychophysiological prediction of substance abuse. In: Gordon HW, Glanz MD, editors. Individual differences in the biobehavioral etiology of drug abuse. Washington, DC: National Institute on Drug Abuse; 1996. pp. 129–160. [Google Scholar]
- Iacono WG, Malone SM. Developmental endophenotypes: Indexing genetic risk for substance abuse with the P300 brain event-related potential. Child Development Perspectives. 2011;5:239–247. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, McGue M. Minnesota Twin Family Study. Twin Research and Human Genetics. 2002;5:482–487. doi: 10.1375/136905202320906327. [DOI] [PubMed] [Google Scholar]
- Iacono WG, McGue M, Krueger RF. Minnesota Center for Twin and Family Research. Twin Research and Human Genetics. 2006;9:978–984. doi: 10.1375/183242706779462642. [DOI] [PubMed] [Google Scholar]
- Illumina. GenomeStudio Data Analysis Software [Computer software] San Diego, CA: Illumina Inc; 2008–2013. [Google Scholar]
- Insel TR, Cuthbert BN, Garvey M, Heinssen R, Pine DS, Quinn K, Wang P. Research Domain Criteria (RDoC): Toward a new classification framework for research on mental disorders. American Journal of Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas KG, Markon KE. A meta-analytic evaluation of the endophenotype hypothesis: Effects of measurement paradigm in the psychiatric genetics of impulsivity. Journal of Abnormal Psychology. doi: 10.1037/a0037094. in press. [DOI] [PubMed] [Google Scholar]
- Kang HM. Efficient and parallelizable association container toolbox (EPACTS) 2014 Retrieved from http://genome.sph.umich.edu/wiki/EPACTS.
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–354. doi: 10.1038/Ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Goate A. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Human Genetics. 2013;132:1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes MA, Malone SM, Elkins IJ, Legrand LN, McGue M, Iacono WG. The enrichment study of the Minnesota Twin Family Study: Increasing the yield of twin families at high risk for externalizing psychopathology. Twin Research and Human Genetics. 2009;12:489–501. doi: 10.1375/twin.12.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Zhou WZ, Zhang PW, Johnson C, Wei L, Uhl GR. Meta-analysis and genome-wide interpretation of genetic susceptibility to drug addiction. BMC Genomics. 2011;12:508, 1471–2164-12-508. doi: 10.1186/1471-2164-12-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Basu S, Miller MB, Iacono WG, McGue M. A rapid generalized least squares model for a genome-wide quantitative trait association analysis in families. Human Heredity. 2011;71:67–82. doi: 10.1159/000324839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: Implications for design of complex trait association studies. Genome Research. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiology. 2010;34:816–834. doi: 10.1002/Gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RJA, Rubin DB. Statistical analysis with missing data. 2. Hoboken, NJ: John Wiley & Sons, Inc; 2002. [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Marchini J. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nature Genetics. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behavior Genetics. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- Malone SM, Burwell SJ, Vaidyanathan U, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of resting EEG activity: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12344. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Luciana M, Wilson S, Sparks JC, Hunt RH, Thomas KM, Iacono WG. Adolescent drinking and motivated decision-making: A cotwin-control investigation with monozygotic twins. Behavior Genetics. 2014;44:407–418. doi: 10.1007/s10519-014-9651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Vaidyanathan U, Basu S, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of P3 event-related brain potential amplitude: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12345. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, McVean G. FaST-LMM-Select for addressing confounding from spatial structure and rare variants Reply. Nature Genetics. 2013;45:471–471. doi: 10.1038/Ng.2619. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Keyes M, Sharma A, Elkins I, Legrand L, Johnson W, Iacono WG. The environments of adopted and non-adopted youth: Evidence on range restriction from the Sibling Interaction and Behavior Study (SIBS) Behavior Genetics. 2007;37:449–462. doi: 10.1007/s10519-007-9142-7. [DOI] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, Iacono WG. A genome-wide association study of behavioral disinhibition. Behavior Genetics. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, McGue M. The Minnesota Center for Twin and Family Research genome-wide association study. Twin Research and Human Genetics. 2012;15:767–774. doi: 10.1017/thg.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP, Psychiatric GCAS. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical modeling. 6. Richmond, VA: Department of Psychiatry, Virginia Commonwealth University; 2003. [Google Scholar]
- Paaby AB, Rockman MV. Cryptic genetic variation: Evolution’s hidden substrate. Nature Reviews Genetics. 2014;15:247–258. doi: 10.1038/nrg3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Luo X, Liu X, Wu LY, Zhang Q, Wang L, Wang KS. Genome-wide association studies of maximum number of drinks. Journal of Psychiatric Research. 2013;47:1717–1724. doi: 10.1016/j.jpsychires.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistis G, Porcu E, Vrieze SI, Sidore C, Steri M, Danjou F, Sanna S. Toward optimally cost-effective designs for genotype imputation in sequencing based genome-wide association studies. 2014 Manuscript submitted for publication. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R: Foundation for Statistical Computing; Vienna, Austria: 2010. Retrieved from http://www.R-project.org. [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Sullivan PF. A mega-analysis of genome-wide association studies for major depressive disorder. Molecular Psychiatry. 2012;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Kendler K, Craddock NJ, Lee PH, Neale BM, Nurnberger JI, Sklar P. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. American Journal of Human Genetics. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nature Reviews Genetics. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Esko T, Sutin AR, de Moor MH, Meirelles O, Zhu G, Uda M. Meta-analysis of genome-wide association studies identifies common variants in CTNNA2 associated with excitement-seeking. Translational Psychiatry. 2011;1:e49. doi: 10.1038/tp.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature Genetics. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Isen JD, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of electrodermal activity: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12346. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Donnelly JM, Hammer MA, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of antisaccade eye tracking error rate: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12347. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of acoustic startle eye blink and affectively modulated startle response: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12348. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Patrick CJ, Cuthbert BN. Linking dimensional models of internalizing psychopathology to neurobiological systems: Affect-modulated startle as an indicator of fear and distress disorders and affiliated traits. Psychological Bulletin. 2009;135:909–942. doi: 10.1037/a0017222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. American Journal of Human Genetics. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana SR, Spence EL, Lang PJ. The startle probe response: A new measure of emotion? Journal of Abnormal Psychology. 1988;97:487–491. doi: 10.1037//0021-843x.97.4.487. [DOI] [PubMed] [Google Scholar]
- Vrieze SI, Feng S, Miller MB, Hicks BM, Pankratz N, Abecasis GR, McGue M. Non-synonymous exonic variants in addiction and behavioral disinhibition. Biological Psychiatry. 2013;75:783–789. doi: 10.1016/j.biopsych.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Pankratz N, Vaidyanathan U, Miller MB, Kang HM, Iacono WG. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12349. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Vaidyanathan U, Kwong A, Kang HM, Zhan X, Iacono WG. In search of rare variants: Preliminary results from whole genome sequencing of 1325 individuals with psychophysiological endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12350. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. Journal of Psychiatric Research. 2011;45:1419–1425. doi: 10.1016/j.jpsychires.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Wood AC, Neale MC. Twin studies and their implications for molecular genetic studies: Endophenotypes integrate quantitative and molecular genetics in ADHD research. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:874–883. doi: 10.1016/j.jaac.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide association studies and genomic prediction. Vol. 1019. New York, NY: Humana Press; 2013. Genome-wide complex trait analysis (GCTA): Methods, data analyses, and interpretations; pp. 215–236. [DOI] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nature Genetics. 2014;46:100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Lander ES. Searching for missing heritability: Designing rare variant association studies. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.