Abstract
We mapped ~85,000 rare nonsynonymous exonic single nucleotide polymorphisms (SNPs) to 17 psychophysiological endophenotypes in 4,905 individuals, including antisaccade eye movements, resting EEG, P300 amplitude, electrodermal activity, affect-modulated startle eye blink. Nonsynonymous SNPs are predicted to directly change or disrupt proteins encoded by genes and are expected to have significant biological consequences. Most such variants are rare, and new technologies can efficiently assay them on a large scale. We assayed 247,870 mostly rare SNPs on an Illumina exome array. Approximately 85,000 of the SNPs were polymorphic, rare (MAF < .05), and nonsynonymous. Single variant association tests identified a SNP in the PARD3 gene associated with theta resting EEG power. The sequence kernel association test, a gene-based test, identified a gene PNPLA7 associated with pleasant difference startle, the difference in startle magnitude between pleasant and neutral images. No other single nonsynonymous variant, or gene-based group of variants, was strongly associated with any endophenotype.
Descriptors: Endophenotype, Psychophysiology, Exome, Rare variant, Nonsynonymous, GWAS, P300, Startle, Antisaccade, Electrodermal, EEG
Endophenotypes have been widely proposed as useful targets for genetic association studies (Gottesman & Gould, 2003). Typically, endophenotypes are conceptualized as measures of psychological and biological processes relevant to a clinical phenotype of interest. In the causal chain from genes to disorder, endophenotypes are thought to occupy an intermediate location more directly influenced by genetic variants, which in principle makes them attractive for identifying genes that influence the clinical phenotype. In the current manuscript, we focus on identifying rare genetic variants in DNA that might be associated with such endophenotypes. We describe genetic association results for 17 psychophysiological endophenotypes, including antisaccade eye movements, measures of the resting electroencephalogram (EEG), amplitude of the P300 wave of the event-related potential, measures of electrodermal activity, and affect-modulated startle eye blink. To varying degrees, many of these measures have long been considered putative endophenotypes for disinhibitory psychopathology, substance misuse and dependence, schizophrenia, mood disorders, and other disorders. The results presented here complement results of the genome-wide association studies (GWAS) of these endophenotypes, which are described in the accompanying common variant articles in this special issue (Malone, Burwell et al., 2014; Malone, Vaidyanathan et al., 2014; Vaidyanathan, Isen et al., 2014; Vaidyanathan, Malone, Donnelly et al., 2014; Vaidyanathan, Malone, Miller, McGue, & Iacono, 2014). Detailed background information about the psychophysiological measures and their heritability is available in these other articles, which we summarize briefly here.
In these accompanying five common variant articles in this special issue, genetic association tests between these endophenotypes and common single nucleotide polymorphisms (SNPs that occur greater than 5% of the time) yielded few genome-wide significant findings. Antisaccade error was significantly associated with one SNP, located on Chromosome 2. Likewise, an aversive-neutral startle modulation difference score produced several subthreshold associations for SNPs in a small region of Chromosome 3, which are in linkage disequilibrium (LD) with each other, and therefore correlated. Gene-based analyses, in which effects for all SNPs within a gene are aggregated into a single gene-based score and tested for association with an endophenotype, yielded evidence of significant association with several endophenotypes. EEG low-frequency (delta) power was associated with a GABAA receptor gene and two others not plausibly related to brain activity. Antisaccade performance was associated with two genes on Chromosome 2—B3GNT7 and NCL—while the aversive-neutral modulation difference score was associated with the PARP14 gene on Chromosome 3.
At the same time, SNP heritability analyses of the additive aggregate effect of all available common SNPs on the Illumina 660W-Quad genotyping array, or SNPs in LD with them, account for an appreciable amount of variance in each endophenotype. That is, there is clearly a genetic signal on the array even if these five individual GWA studies did not uncover it. In addition, the SNP-based heritability does not recapture all of the heritability estimated through the twin design, suggesting that common variants do not explain all of the heritability of these endophenotypes. The idea that common SNPs only capture part of the genetic variance in a population is by now commonplace in psychiatric genetics, and has led many to consider the potential role of rare variants.
No study has yet systematically evaluated the role of rare variants in any of the endophenotypes considered in this special issue. Functional variants, such as nonsynonymous SNPs, are more likely to disrupt gene function, more likely to be rare (Tennessen et al., 2012), and are hypothesized to have greater expected impact on phenotypic development than other variants. Nonsynonymous variants are exonic, lying in the coding regions of genes, and are predicted to disrupt the gene’s coding sequence, resulting in malformed and dysfunctional protein products. A nonsynonymous variant in a critical place, such as a variant that changes an amino acid to a stop codon sequence, can cause a gene to produce a malformed protein, or no protein at all, and result in significant consequences for the organism as a whole. What is more, rare variants arose relatively recently in human evolution and are largely independent of the common variants that are most often assayed in GWAS. Therefore, the potentially causal rare variants investigated in the present article would have been missed by previous genome-wide studies, including those described in the accompanying five common variant articles of this special issue.
The increasing use of exome sequencing has made the discovery of rare exonic variants possible and even efficient. Exome chips, available from Affymetrix or Illumina, were created to genotype rare variants discovered in exome sequences from 16 studies of 12,000 total individuals. Using this sequence information, the chip was designed to assay ~250,000 nonsynonymous variants across the exome (http://genome.sph.umich.edu/wiki/Exome_Chip_Design). In the present study, we used the Illumina HumanExome BeadChip to assay rare nonsynonymous variants across the genome.
An inherent difficulty in the study of rare variants is that, by definition, very few people carry them. While only one carrier of a mutation is necessary to discover the existence of that variant, it is impossible to draw strong statistical conclusions in a single individual about the association of that variant with any phenotype. To identify many individuals who carry some rare variant requires very large sample sizes or specialized designs (e.g., extreme phenotypes, carrier pedigrees). For example, if a variant is present in only 1 in 10,000 individuals, one must genotype DNA from 100,000 individuals just to find 10 people who carry that variant. In response to this problem, the past few years have seen many new methods developed specifically for the analysis of rare variants (Asimit & Zeggini, 2010; Lee, Abecasis, Boehnke, & Lin, 2014). Perhaps the most intuitive approach is to collapse (e.g., sum) across all rare variants within a gene (called a “burden” test) and regress the phenotype on that sumscore. In the present study, we apply two complementary approaches to collapsing rare variants. First, we summed the total number of minor alleles within a given gene and tested for an association between an endophenotype and the gene sumscore (i.e., the variable threshold collapsing and multivariate count burden test [Li & Leal, 2008; Price et al., 2010]). This test suffers from low power when variants within a gene have opposite directions of effect. To overcome this potential issue, we also used the sequence kernel association test, or SKAT (Wu et al., 2011), which uses a different statistical approach to allow for variants within a gene to have different directions of effect.
In summary, we systematically evaluated the role of nonsynonymous exonic variants in ~4,000 individuals for 17 endophenotypes. We conducted association tests for individual SNPs, as well as gene-based burden tests. The present study is the largest single study of its kind and the first to comprehensively evaluate the role of rare nonsynonymous variants with psychophysiological endophenotypes.
Methods
Participants
All participants were from the Minnesota Center for Twin and Family Research (MCTFR; Iacono, McGue, & Krueger, 2006). The full MCTFR study was genotyped, but the association analyses described here were conducted only on those individuals who participated in a psychophysiology laboratory assessment: the two age cohorts of the Minnesota Twin Family Study (MTFS; Iacono & McGue, 2002) and the enrichment sample (SIBS; Keyes et al., 2009). See the accompanying methodological overview (and Figures 1 and 4 of that article) for a detailed description of the sample and study design (Iacono, Malone, Vaidyanathan, & Vrieze, 2014).
Endophenotypes
Details about each endophenotype are available in this special issue’s methodological overview (Iacono et al., 2014) and in each of the five common variant research articles. We evaluated 17 endophenotypes in all:
- P300 amplitude reduction (Malone, Vaidyanathan et al., 2014):
-
1.P3 event-related potential (P3; N = 4,166)
-
2.P3 genetic factor (gP3; N = 3,088)
-
1.
- Antisaccade tracking error rate (Vaidyanathan, Malone, Donnelly et al., 2014)
-
3.Antisaccade tracking error rate (SAC; N = 4,469)
-
3.
- Electrodermal activity (Vaidyanathan, Isen et al., 2014)
-
4.Skin conductance level (SCL; N = 3,791)
-
5.Skin conductance response frequency (fSCR; N = 4,299)
-
6.Skin conductance response amplitude (aSCR; N = 4,102)
-
7.Electrodermal activity factor (EDA; N = 4,424), a general factor derived from SCL, fSCR, and aSCR
-
4.
- Affective startle modulation (Vaidyanathan, Malone, Miller et al., 2014)
-
8.Overall startle (STRTL; N = 3,323)
-
9.Aversive difference startle (aSTRTL; N = 3,321)
-
10.Pleasant difference startle (pSTRTL; N = 3,322)
-
8.
- Resting state EEG (Malone, Burwell et al., 2014)
-
11.Total EEG power (totPower; N = 3,948)
-
12.Alpha EEG power (αPower; N = 3,948)
-
13.Beta EEG power (βPower; N = 3,948)
-
14.Theta EEG power (θPower; N = 3,948)
-
15.Delta EEG power (δPower; N = 3,948)
-
16.Alpha EEG power O1O2 (αPowerO1O2; N = 3,966)
-
17.Alpha EEG frequency O1O2 (αFreqO1O2; N = 3,966)
-
11.
Covariate Correction and Inverse Normalization
All endophenotypes were corrected for sex, chronological age, generational status (parent vs. child) and, when relevant, task-specific laboratory covariates. To account for population stratification and familial relatedness, we opted to use an empirical kinship linear mixed model approach (described below) instead of principal components. In addition, rare variant analyses can be highly sensitive to outliers, and non-normality of the phenotypic distribution which, given the small effects of genetic variants, is also very close to the residual distribution. Therefore, all endophenotypes were inverse normalized after covariate correction and prior to the association analysis. This approach reduces false positives by simultaneously eliminating non-normality and pulling in phenotypic outliers.
Genotypes
A total of 4,905 individuals in the MCTFR participated in laboratory psychophysiological testing. To improve genotype quality for the subset of individuals who participated in the genotype lab, we conducted genotype quality control using genotypes on the full sample. From the available MCTFR sample, 7,350 individuals (note this number includes only one member of each monozygotic [MZ] twin pair) were genotyped on both the Illumina HumanExome-12v1_A array and the Illumina 660W-Quad. A detailed description of quality control for the 660W-Quad is available in a prior publication (Miller et al., 2012) and in the methodological overview for this special section.
The Illumina HumanExome BeadChip includes 247,870 mostly rare single nucleotide variants (85.0% of variants have a minor allele frequency less than 1%). All 7,350 samples genotyped on this chip were clustered into the three allele groups possible for each SNP (AA, AB, and BB) using GenomeStudio (Illumina, San Diego, CA) and then imported in the GENVISIS genetic visualization software (Pankratz, 2014). Multiple quality control (QC) steps used the theta transformation, which is simply a transformation of the genotype intensity coordinates to make comparison of the genotype intensity clusters more straightforward. Markers showing evidence of poor quality (n = 15,741; see online supporting information Table S1), such as lower call rates, poor separation between genotype clusters (mean separation less than 0.30 in the theta dimension, representing allele frequency), evidence of cohybridization (mean theta deviates more than 0.20 from the expected values of 0, 0.5, and 1.0 for AA homozygotes, heterozygotes, and BB homozygotes, respectively), large theta standard deviations (> 0.025 for a homozygous cluster or > 0.07 for the heterozygous cluster), excess heterozygosity (HetExcess > 0.1 or HetExcess < −0.9), rare (0 ≤ minor allele frequency [MAF] < .0001), or monomorphic with an incomplete call rate (< 1.0) were flagged, manually reviewed, and either reclustered (n = 6,865) or discarded (n = 1,844). The remainder of manually reviewed markers were not problematic and were left undisturbed. The 246,026 variants that remained after discarding those that could not be reclustered were carried forward for standard QC testing in a sample of 3,259 unrelated white, non-Hispanic participants. Unrelated individuals were chosen because some QC metrics show bias in the presence of families (e.g., Hardy-Weinberg). A total of 422 additional markers were dropped in our standard QC screen: 236 markers still had a call rate less than 99%, 114 had a Hardy-Weinberg test p value less than the Bonferroni cutoff of 2e-7, 22 autosomal markers were associated with participant sex (p < 2e-7), and 148 variants had two or more of any of the following errors: incorrect call in a Center for the Study of Human Polymorphisms (CEPH) sample, discordant calls in a pair of duplicated samples or a monozygotic twin pair, Mendelian inconsistency, heterozygosity for an X-chromosome marker in a male sample, or nonmissing Y-chromosome genotype for a female sample. See supporting information Table S2 for a breakdown of why markers were flagged, reviewed, reclustered, and dropped. This left 245,484 exome chip markers available for analyses.
After sample QC screening, we were left with usable genotype data for 7,244 subjects, including only one member of each MZ twin pair. Samples were dropped for a no-call rate of .5% or higher (145 samples); 16 samples were dropped because of sample mix-ups; 4 samples were dropped because of consent issues; and for all duplicate samples, only the one with the highest call rate was retained. After creating records for 1,120 MZ co-twins by copying genotype data of their MZ twin, assuming MZ twins share 100% of the genotypes on the exome array, the sample size usable for GWAS increased to 8,364 subjects, including all individuals, regardless of ethnicity.
Markers from the exome chip and genome-wide chip were combined into an integrated panel of 674,930 polymorphic SNPs. We used this integrated panel primarily to generate the empirical kinship matrix for association testing, and did not consider common SNPs in these analyses, as the companion articles from this special issue present results for common SNP associations with the same endophenotypes. There were 41 individuals who passed quality control on the genome-wide chip but not the exome chip, bringing the total sample genotyped on at least one of the two arrays from 8,364 to 8,405. There were 99,195,810 overlapping genotypes between the arrays in this integrated sample (11,802 overlapping SNPs × 8,405 individuals; we included the MZ co-twins in these numbers). Of these, 98,414,738 genotypes were assayed and passed quality control on both chips (99.2%). Of these, 98,405,631 (99.9908%) were concordant. The 9,107 discordant genotypes were set to missing and not used in the association analyses.
Nonsynonymous SNPs were identified and annotated with EPACTS (Kang, 2014) using GENCODE v11 transcripts (Harrow et al., 2012). To annotate a SNP, one determines whether an allele of the SNP would result in an amino acid change or splicing disruption in one or more known transcripts.
Out of the final sample of 8,405 (for 41 of whom, rare exonic variants were available only from the Illumina 660W-Quad array), 4,905 had data on at least one psychophysiological index that is part of this study. These 4,905 subjects were utilized in the analyses described below.
Association Analysis
To correct for within-family correlation and population stratification, we used the EMMAX (Kang et al., 2010) approach as implemented in EPACTS (Kang, 2014). EMMAX uses a linear mixed model and an empirical kinship matrix to correct simultaneously for familial resemblance and population stratification (Listgarten et al., 2012). Empirical kinships are calculated from the genotype information, and represent the degree of blood relation, or consanguinity, between individuals in the study. For single variant tests we restricted analysis, and our correction for multiple testing, to variants with minor allele counts greater than 4 and minor allele frequency less than 5%. The former restriction is useful because our study does not have sufficient power to detect effects with lower minor allele frequencies, and the asymptotics of our statistical tests break down when minor allele counts are extremely low. The latter restriction is appropriate because we already comprehensively test common SNPs in the accompanying common variant articles in this special issue.
In addition to single variant tests using a score test, we also conducted two gene-based burden tests of nonsynonymous variants with MAF < .05. The first gene-based burden test was the variable threshold collapsing and multivariate count method, or VTCMC (Li & Leal, 2008; Price et al., 2010). VTCMC sums minor alleles for rare variants within a gene and tests for association between that sum and the endophenotype. The threshold for considering which variants are rare, and therefore included in the sum, is evaluated from the data. The threshold that minimizes the resulting p value is used. To avoid capitalizing on chance in selecting the optimal threshold and conducting statistical tests in the same data, we used permutation to arrive at unbiased p values. One limitation of the VTCMC approach is that it has greatest power when all variants within a gene have the same direction of effect (e.g., the rare allele always increases the endophenotype value). If rare alleles within a gene have opposite directions of effect, then VTCMC and related approaches lose power to detect effects.
Other statistical tests have been developed to allow for different directions of effect within a gene, and we use an implementation of SKAT (Wu et al., 2011) to complement the VTCMC approach. SKAT is essentially a variance component test, as it tests whether the distribution of SNP effects within a gene are overdispersed (larger than expected by chance), regardless of whether they are positive or negative.
We evaluated 204 candidate genes in more detail. These were selected from the NeuroSNP database (https://zork5.wustl.edu/nida/neurosnp.html) for their potential relevance to the endophenotypes examined here or their associated clinical phenotypes. Genes within this candidate set are known either to play a role in the major neurotransmitter or neuromodulatory systems (acetylcholine, dopamine, noradrenaline, serotonin, glutamate, GABA), to be involved in metabolizing alcohol and nicotine, or to be part of the endogenous opioid and cannabinoid systems. See the methods article in this special issue for more information about these candidate genes (Iacono et al., 2014).
For any significant single variant results, we further annotated SNPs using combined annotation dependent depletion (CADD; Kircher et al., 2014). CADD annotates variants with a variety of algorithms, including the variant effect predictor (VEP; McLaren et al., 2010), PolyPhen2 (Adzhubei et al., 2010), and sorting intolerant from tolerant (SIFT; Kumar, Henikoff, & Ng, 2009). A useful aspect of CADD is that it attempts to incorporate these and other annotation information to develop a single, integrated score for every possible single nucleotide mutation in the human genome. The algorithm used in CADD is a support vector machine (SVM), a supervised learning approach that uses known positive and negative examples of pathogenic variants to score novel variants as deleterious. CADD produces a C-score, which is a phred scaled score (phred = −10 × log10(percentile rank/100)) of the SVM prediction. For example, a score of 20 is in the 10th percentile of predicted deleteriousness, 30 in 1st percentile, and 40 in the 0.1st percentile.
Results
See Tables 1 and 2 and Figure 2 in Iacono et al. (2014) for descriptive statistics of, and correlations among, the 17 endophenotypes. Significant clustering was observed within skin conductance measures, startle, the two measures of P3, and EEG power. The full correlation matrix for the residualized endophenotypes is provided in Table S3 and displayed graphically in Figure 2 of the methods article of this special issue (Iacono et al., 2014).
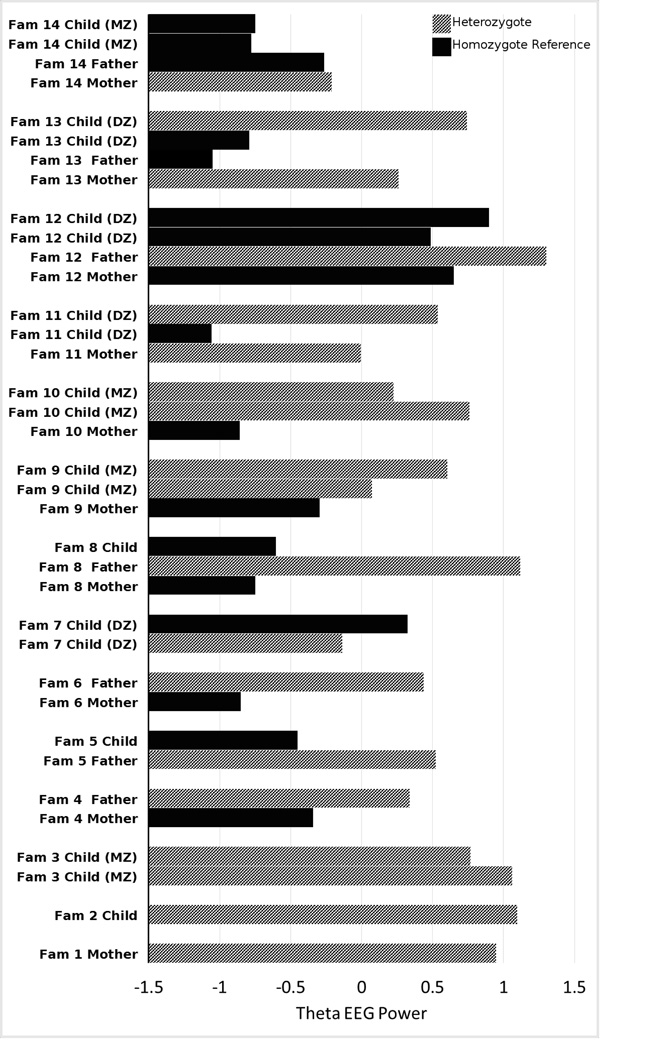
First, we tested for endophenotype associations with rare individual SNPs. Sample sizes for each endophenotype are listed in Table 1 of Iacono et al. (2014). Genomic control ranged from .99 to 1.04 considering only nonsynonymous variants with MAF < .05, suggesting minimal influence of statistical artifacts on our rare variant tests. The number of nonsynonymous variants with minor allele count (MAC) ≥ 5 and MAF < 5% ranged from 38,818 (P3 genetic factor) to 46,774 (antisaccade), resulting in per-endophenotype Bonferroni corrections from 1.29 × 10−6 to 1.07 × 10−6, respectively. At these levels of significance, one SNP was associated with significantly higher theta EEG power (β = 1.2, R2 = .6%, p = 9.5 × 10−7). The SNP is rs139550735, which is nonsynonymous with MAC of 19 (MAF = .0024), located at Chromosome 10, position 34,671,513 of the GRCh37 reference genome, with reference allele T and alternate allele C, and lies in the Par-3 family cell polarity regulator gene (PARD3). PolyPhen2 annotated the variant as “benign.” SIFT annotated it as “tolerated.” The phred-scaled CADD score was 15.21, indicating that this variant is ranked in the 3rd percentile for pathogenicity. Since the variant is rare, only a few individuals carry it, and we display theta EEG power for each family containing an individual carrying this SNP in Figure 1\f1\, which simply shows the pattern of theta EEG power in those family members who carry the rare variant and those who do not. The rs139550735 variant has been described previously in both the 1000 Genomes project and the Exome Sequencing Project. The variant had MAF = .002 in the Exome Sequencing Project, with the minor allele observed 17 times in 4,283 individuals of European ancestry in that project. These frequency estimates are highly consistent with ours. Q-Q plots and Manhattan plots for each endophenotype are available in the supporting information.
Figure 1.
Theta EEG power within each of the 14 families carrying SNP rs139550735. Bars are grouped within family. Carriers of the rare allele (heterozygotes) are in gray, individuals who do not carry the rare allele (homozygote for the reference allele) are in black. All families show a consistent direction of effect except for Family 7, where the sibling with the rare allele has lower theta power than the sibling without the rare allele.
Next, we examined VTCMC burden test association between the endophenotypes and all genes with sufficient nonsynonymous variation (i.e., gene contains nonsynonymous SNPs with MAF < .05 and has a burden allele count of 3 or more). Sample sizes for the burden tests were the same as those for single variant tests (see Methods), and the number of genes with sufficient nonsynonymous variation ranged from 14,330 (P3 genetic factor) to 14,859 (antisaccade). This resulted in per-endophenotype Bonferroni corrections of 3.4 × 10−6 and 3.5 × 10−6, depending on the number of genes tested for each phenotype. The top VTCMC-associated genes for each endophenotype are displayed in Table 1\t1\. At these levels of significance, no gene was significantly associated with any endophenotype. The PARD3 gene was the second-most significant gene for theta power EEG (p = 7.1 × 10−5, nonsignificant), but this was driven entirely by the single significant SNP rs139550735 reported in the previous paragraph.
Table 1.
Top 2 Genes for Each Endophenotype with Most Significant VTCMC Associations
| Endophenotype | Gene | Chr | No. SNPs with MAF < .05 |
Burden AC for SNPs with MAF < .05 |
Optimal burden MAC |
R2 (%) |
Direction | Bonferroni correction |
p value |
|---|---|---|---|---|---|---|---|---|---|
| P3 | TRIP10 | 19 | 9 | 20 | 4 | .41 | − | 3.4e-6 | 1.2e-4 |
| MADD | 11 | 13 | 174 | 3 | .41 | − | 3.4e-6 | 2.5e-4 | |
| gP3 | KARS | 16 | 8 | 54 | 21 | .56 | + | 3.5e-6 | 1.3e-4 |
| MADD | 11 | 13 | 125 | 3 | .6 | − | 3.5e-6 | 1.7e-4 | |
| SAC | AFTPH | 2 | 15 | 202 | 6 | .44 | + | 3.4e-6 | 8.0e-5 |
| ANXA3 | 4 | 8 | 138 | 104 | .39 | − | 3.4e-6 | 2.3e-4 | |
| SCL | DEFB116 | 20 | 2 | 166 | 163 | .44 | − | 3.4e-6 | 7.3e-5 |
| C6orf221 | 6 | 1 | 12 | 12 | .39 | + | 3.4e-6 | 1.8e-4 | |
| fSCR | PASK | 2 | 23 | 471 | 25 | .56 | − | 3.4e-6 | 1.2e-5 |
| CCT6A | 7 | 3 | 163 | 149 | .47 | − | 3.4e-6 | 2.6e-5 | |
| aSCR | KANK4 | 1 | 14 | 929 | 3 | .53 | − | 3.4e-6 | 2.2e-5 |
| SPAST | 2 | 4 | 80 | 2 | .48 | − | 3.4e-6 | 3.1e-5 | |
| EDA | ZNF691 | 1 | 2 | 10 | 6 | .40 | − | 3.4e-6 | 5.5e-5 |
| KANK4 | 1 | 14 | 998 | 3 | .44 | − | 3.4e-6 | 6.4e-5 | |
| STRTL | FBX022 | 15 | 2 | 21 | 2 | .48 | − | 3.5e-6 | 1.2e-4 |
| RAET1L | 6 | 2 | 14 | 2 | .48 | − | 3.5e-6 | 1.2e-4 | |
| aSTRTL | ANKRD13C | 1 | 2 | 35 | 7 | .50 | − | 3.5e-6 | 8.1e-5 |
| KCNA3 | 1 | 2 | 28 | 27 | .48 | − | 3.5e-6 | 1.6e-4 | |
| pSTRTL | SIL1 | 5 | 5 | 69 | 3 | .69 | + | 3.5e-6 | 7.0e-6 |
| BAI2 | 1 | 7 | 141 | 22 | .67 | + | 3.5e-6 | 1.6e-5 | |
| αPower | C17orf57 | 17 | 10 | 779 | 6 | .59 | − | 3.4e-6 | 1.0e-5 |
| OR5H2 | 3 | 7 | 125 | 52 | .57 | + | 3.4e-6 | 1.2e-5 | |
| βPower | GTPBP3 | 19 | 4 | 228 | 1 | .48 | − | 3.4e-6 | 3.4e-5 |
| C9orf9 | 9 | 2 | 5 | 4 | .47 | − | 3.4e-6 | 4.1e-5 | |
| θPower | MICAL2 | 11 | 23 | 189 | 27 | .59 | + | 3.4e-6 | 1.2e-5 |
| PARD3 | 10 | 10 | 98 | 19 | .49 | + | 3.4e-6 | 7.1e-5 | |
| δPower | FASN | 17 | 25 | 684 | 10 | .54 | + | 3.4e-6 | 5.4e-5 |
| EDC4 | 16 | 6 | 91 | 18 | .42 | − | 3.4e-6 | 1.8e-4 | |
| totPower | FASN | 17 | 25 | 684 | 10 | .49 | + | 3.4e-6 | 1.2e-4 |
| OCEL1 | 19 | 2 | 3 | 2 | .41 | − | 3.4e-6 | 1.5e-4 | |
| αPowerO1O2 | TFPI2 | 7 | 2 | 16 | 2 | .44 | − | 3.4e-6 | 1.3e-4 |
| GTSF1 | 12 | 1 | 13 | 13 | .34 | − | 3.4e-6 | 2.3e-4 | |
| αFreqO1O2 | RNF175 | 4 | 7 | 203 | 49 | .54 | − | 3.4e-6 | 2.1e-5 |
| SMAD1 | 4 | 2 | 5 | 3 | .44 | + | 3.4e-6 | 4.3e-5 |
Note. All endophenotype abbreviations are defined in the text. Genes with a burden allele count < 3 were not considered in arriving at these Bonferroni corrections. VTCMC = variable threshold collapsing and multivariate count burden test (Price et al., 2010); MAF = minor allele frequency; MAC = minor allele count; R2 = coefficient of determination, in this case variance accounted for in residualized, inverse-normalized endophenotype.
We conducted a similar analysis using the complementary SKAT gene-based test. We again evaluated genes containing SNPs with MAF < .05 and burden allele count ≥ 3. The SKAT test identified a single gene (see Table 2\t2\) as significantly associated with pleasant difference startle (pSTRTL; p value = 1.6 × 10−6). The gene, patatin-like phospholipase domain containing 7 (PNPLA7), has been implicated in adipocyte differentiation and lipid metabolism (Wilson, Gardner, Lambie, Commans, & Crowther, 2006). To better understand the individual variants that comprise the significant SKAT results, we display single variant results for all nonsynonymous variants within PNPLA7 in Table 3\t3\. It appears that the SKAT result is driven by two low-frequency variants with opposite directions of effect within the gene. Additional CADD annotation suggests that these particular variants are not highly deleterious.
Table 2.
Top Two Genes for Each Endophenotype with Most Significant SKAT Associations
| Endophenotype | Gene | Chr | No. SNPs with MAF < .05 |
Bonferroni correction |
p value |
|---|---|---|---|---|---|
| P3 | BIN3 | 8 | 2 | 3.4e-6 | 4.9e-5 |
| TRIP10 | 19 | 9 | 3.4e-6 | 6.9e-5 | |
| gP3 | KARS | 16 | 8 | 3.5e-6 | 7.7e-5 |
| ZNF560 | 19 | 10 | 3.5e-6 | 9.1e-5 | |
| SAC | ANXA3 | 4 | 8 | 3.4e-6 | 1.2e-5 |
| GALM | 2 | 4 | 3.4e-6 | 4.3e-5 | |
| SCL | DEFB116 | 20 | 2 | 3.4e-6 | 1.0e-5 |
| C6orf221 | 6 | 1 | 3.4e-6 | 1.1e-4 | |
| fSCR | CCT6A | 7 | 3 | 3.5e-6 | 9.2e-6 |
| FAM180A | 7 | 2 | 3.5e-6 | 3.3e-5 | |
| aSCR | RNF126 | 19 | 4 | 3.5e-6 | 1.0e-4 |
| MEPE | 4 | 6 | 3.5e-6 | 1.1e-4 | |
| EDA | ZNF691 | 1 | 2 | 3.4e-6 | 3.6e-5 |
| DEFB116 | 20 | 2 | 3.4e-6 | 4.1e-5 | |
| STRTL | GRID2IP | 7 | 4 | 3.4e-6 | 3.0e-5 |
| PAPSS1 | 4 | 4 | 3.4e-6 | 6.0e-5 | |
| aSTRTL | KCNA3 | 1 | 2 | 3.4e-6 | 6.7e-5 |
| C1orf43 | 1 | 2 | 3.4e-6 | 1.6e-4 | |
| pSTRTL | PNPLA7 | 9 | 22 | 3.5e-6 | 1.6e-6 |
| PHF1 | 6 | 2 | 3.5e-6 | 5.7e-5 | |
| αPower | OR5H2 | 3 | 7 | 3.4e-6 | 1.0e-5 |
| HSPB3 | 5 | 3 | 3.4e-6 | 9.3e-5 | |
| βPower | C9orf9 | 9 | 2 | 3.4e-6 | 1.9e-5 |
| SPINT1 | 15 | 11 | 3.4e-6 | 3.7e-5 | |
| θPower | CD5L | 1 | 5 | 3.4e-6 | 8.0e-5 |
| OCEL1 | 19 | 2 | 3.4e-6 | 9.6e-5 | |
| δPower | ACOT13 | 6 | 2 | 3.4e-6 | 3.1e-4 |
| KRIT1 | 7 | 3 | 3.4e-6 | 3.5e-4 | |
| totPower | OCEL1 | 19 | 2 | 3.4e-6 | 9.0e-5 |
| HSPB3 | 5 | 3 | 3.4e-6 | 2.2e-4 | |
| αPowerO1O2 | CALCOCO2 | 12 | 6 | 3.4e-6 | 8.8e-5 |
| ATF7IP2 | 16 | 6 | 3.4e-6 | 9.0e-5 | |
| αFreqO1O2 | SMAD1 | 4 | 2 | 3.4e-6 | 3.8e-5 |
| C15orf2 | 15 | 13 | 3.4e-6 | 7.3e-5 |
Note. All abbreviations are defined in Table 1. Genes with a burden allele count < 3 were not considered in arriving at this Bonferroni correction. SKAT = sequence kernel association test (Wu et al., 2011).
Table 3.
Low Frequency Nonsynonymous Variants Used in SKAT Test for Gene PNPLA7 and Pleasant Modulated Startle
| Position | REF | ALT | CADD C-score |
REF/HET/ALT | MAF | Beta (SE) | p value |
|---|---|---|---|---|---|---|---|
| 140357186 | C | T | 18.41 | 3287/6/0 | .0009 | −0.061 (.41) | .88 |
| 140357972 | A | G | 0.01 | 0/23/3270 | .0035 | 0.857 (.21) | 5.7e-5 |
| 140358896 | A | T | 2.87 | 3280/13/0 | .0020 | −0.123 (.28) | .66 |
| 140372570 | G | A | 15.64 | 3290/3/0 | .0005 | −0.483 (.59) | .41 |
| 140372588 | T | C | 0.09 | 3274/15/0 | .0023 | −0.048 (.26) | .85 |
| 140374931 | C | T | 2.96 | 3288/5/0 | .0008 | −0.124 (.45) | .78 |
| 140391737 | A | G | 14.51 | 3287/4/0 | .0006 | 0.134 (.51) | .79 |
| 140391739 | G | A | 16.22 | 3292/1/0 | .0002 | −0.160 (1.0) | .87 |
| 140392656 | T | C | 18.80 | 3292/1/0 | .0002 | 0.526 (1.0) | .60 |
| 140400464 | C | T | 10.60 | 3212/80/1 | .0125 | −0.478 (.11) | 1.9e-5 |
| 140400473 | G | C | 9.15 | 3284/9/0 | .0014 | −0.234 (.34) | .49 |
| 140409842 | G | A | 15.20 | 3286/7/0 | .0011 | −0.419 (.39) | .28 |
| 140409877 | C | A | 12.90 | 3287/6/0 | .0009 | 0.284 (.42) | .50 |
| 140409891 | G | C | 11.58 | 3287/6/0 | .0009 | 0.284 (.42) | .50 |
| 140414411 | G | A | 13.75 | 3292/1/0 | .0002 | 1.776 (1.0) | .08 |
| 140414461 | C | A | 13.01 | 3291/2/0 | .0003 | 0.447 (.72) | .53 |
| 140414474 | C | T | 12.30 | 3288/5/0 | .0008 | 0.188 (.46) | .68 |
| 140417222 | C | T | 13.65 | 3291/2/0 | .0003 | 0.212 (.71) | .77 |
| 140437936 | C | T | 13.76 | 3292/1/0 | .0002 | −0.792 (1.0) | .43 |
| 140441299 | G | A | 12.18 | 3288/5/0 | .0008 | 1.014 (.45) | .03 |
| 140441302 | C | A | 14.32 | 3289/4/0 | .0006 | 1.326 (.51) | .01 |
Note. All variants were nonsynonymous missense with no stop gain/loss or start gain/loss. The CADD C-score is phred scaled (phred = −10log10(percentile rank/100)) such that a score of 20 is in the 10th percentile of predicted deleteriousness, 30 in 1st percentile, and so on. It appears that the overall gene signal is dominated by two low-frequency variants with opposite directions of effect. Both variants are not predicted to be highly deleterious, with C-scores of < 1 and 11. PolyPhen annotation for both variants is “benign”; similarly, SIFT annotated both variants as “tolerated.” SKAT = sequence kernel association test (Wu et al., 2011); REF = reference allele on reference genome GRCh37; ALT = nonreference allele observed in the present study; REF/HET/ALT = number of individuals who are homozygous for the REFerence allele, HETerozygous, and homozygous for the ALTernate allele.
Finally, we evaluated in more detail the 204 genes in our candidate gene set. Depending on the endophenotype, 157–159 genes possessed sufficient nonsynonymous variation to be considered (more than one variant, burden allele count of 3 or more). No gene-based test was significant even when relaxing the Bonferroni significance threshold to account for 159 genes (p < .00032). The most significant gene identified by the VTCMC method was ALDH3B2 associated with alpha EEG power at O1O2 with p = .0007. The most significant gene for SKAT was total EEG power with SLC6A17 at p = .0008.
Discussion
The psychophysiological endophenotypes reported here include basic measures of brain activity (resting EEG) as well as those tapping fundamental reflexes like the startle eye blink and habituation of the electrodermal orienting response. Each is of particular interest as a possible endophenotype for a variety of psychopathology constructs. In the end, we tested 17 psychophysiological measures for association with ~85,000 exome-wide nonsynomyous SNPs from the Illumina HumanExome array. One SNP was significantly associated with theta EEG power. One gene, PNPLA7, was significantly associated with pleasant difference startle, or the difference in startle magnitude between pleasant and neutral images. None of our proposed candidate genes were significantly associated with any endophenotype, by either of the gene-based tests employed here.
The single SNP associated with theta EEG power is nonsynonymous in the PARD3 gene. This gene is involved in cell polarity regulation, expressed in the brain, and thus appears biologically plausible. A recent study reported an association between PARD3 and schizophrenia (Kim, Lee, Park, Kim, & Chung, 2012), which is sometimes associated with increased theta power (Begic et al., 2011; Clementz, Sponheim, Iacono, & Beiser, 1994; Harris, Melkonian, Williams, & Gordon, 2006; Sponheim, Clementz, Iacono, & Beiser, 1994, 2000; Sponheim, Iacono, Thuras, Nugent, & Beiser, 2003). Increased theta power is also a robust correlate of attention deficit hyperactivity disorder (ADHD; Barry, Clarke, & Johnstone, 2003), and it has been observed in adults with various forms of disinhibitory psychopathology, such as alcoholism, marijuana use, and antisocial behavior as well (Ehlers, Phillips, Gizer, Gilder, & Wilhelmsen, 2010; Rangaswamy et al., 2003; Struve et al., 1999). The companion sequencing article (Vrieze et al., 2014) found a p value of .013 for this same variant in 1,045 whole-genome-sequenced individuals (MAF = .002; MAC = 5), and a p value of 1.2 × 10−6 after imputation into the full sample of 3,948 individuals with values for this measure. The participants in the exome chip and sequencing studies are the same, so this is by no means an independent replication. It simply shows that a distinct genotyping approach reproduced the finding. PARD3 has also not been identified in previous studies of psychophysiological endophenotypes, and thus we urge replication of this finding in other data, as well as caution in interpreting the implications.
The SKAT gene-based test identified PNPLA7 as significantly associated with pleasant difference startle, which represents the degree to which startle eye blink is attenuated by pleasant stimuli. Judging by the single variant tests reported in Table 3, the SKAT association appears to be driven by two nonsynonymous variants within PNPLA7 that have opposite directions of effect. PNPLA7 has been implicated in adipocyte differentiation and lipid metabolism (Wilson et al., 2006), as well as for neurological health, particularly neuropathy (Richardson, Hein, Wijeyesakere, Fink, & Makhaeva, 2013), but has no obvious biological connection to startle psychophysiology. Using the same criteria for a nonsynonymous variant, the companion sequencing article (Vrieze et al., 2014) discovered 26 SNPs within PNPLA7 in 840 individuals and found a SKAT p value of .05. After imputation into the full sample of 3,322, the same article discovered 28 SNPs that met criteria and found a SKAT p value of 9.5 × 10−6. This is not an independent replication, but does suggest mild stability in the PNPLA7 effect in this sample across two genotyping approaches. We welcome independent replication of this finding.
In the companion GWAS paper on startle (Vaidyanathan, Malone, Miller et al., 2014), we obtained a significant association for PARP14 with modulated startle, but for aversive difference startle. The biometric and GCTA analyses included in that paper found neither of the two modulated startle measures were heritable. While this alone does not preclude the possibility of a rare variant association, it does not increase confidence in any genetic association finding for either modulated startle phenotype. In addition, although aversive difference startle has been associated with psychopathology, to our knowledge, pleasant difference startle examined alone has not. For laboratory protocols using slides drawn from the International Affective Picture System (Lang, Bradley, & Cuthbert, 1999) set we used here, the two measures are examined together, and typically variation in the pattern of modulated startle becomes the interpretative focus. Here, however, for each startle component examined in isolation, we are finding different and unrelated associations for genes with uncertain function. Taken together, these observations, in the absence of replication, further reduce confidence in the genetic findings.
Clearly, no single nonsynonymous variant, or gene-based group of variants, was strongly and indisputably associated with any psychophysiological measure. We had expected rare variants to affect these endophenotypes for two reasons. First, all but two of the endophenotypes are heritable based on the twin models reported in the companion common variant articles of this special issue. Second, also reported in the companion common variant articles, estimates of the aggregate effect of common variants (by the GCTA method) was typically less than the twin-based heritability, implicating the role of other classes of variants on these endophenotypes, such as rare variants.
Limitations and Future Directions
The nonsynonymous variants tested here are but a small fraction of all variants in the exome, which itself comprises at most 2% of the entire genome, leaving more than 99% of rare variants in these individuals untested by our analyses. This is because the exome chip is a fixed array, in that it genotypes only known nonsynonymous exonic variants based on 12,000 exome sequences. Many nonsynonymous variants that exist in our participants are missed on the exome array, although the obvious alternative of exome sequencing is far more expensive than the exome chip, and there is a balance to be struck between sample size and genotype density. While not comprehensive, tests of nonsynonymous variants, even from a fixed array, were a sensible starting point given their clearly important functionality in the genome. In a companion paper to this special issue (Vrieze et al., 2014), we report association results using whole genome sequencing, which identifies tens of millions additional rare variants that are tested for association with psychophysiological endophenotypes in a subset of 1,325 MCTFR participants. Further advances in genotype annotation continue, such that truly deleterious variants may be more precisely identified and included in gene-based tests in the future.
Future investigations will benefit from (a) increased sample size, (b) broader interrogation of rare variants, and (c) additional annotation. To increase sample size, we encourage the community of psychophysiology investigators to pool data and resources (Iacono & McGue, 2002). Indeed, such consortia have already begun and published intriguing findings in relation to MRI phenotypes (Stein et al., 2012; Thompson et al., 2014). Additional rare variation will be discovered as genome sequencing becomes more affordable, and functional annotation will continue to improve with advances in epigenomics. Our use of CADD was one attempt to leverage additional annotation to interpret our findings, but one might imagine weighting variants within a gene according to the C-score or other weights based on other annotation. In the end, the present experiment identifies a handful more potential association signals, which also require replication. Ultimately, genome sequencing will require many additional studies with large sample sizes to detect novel loci associated with the endophenotypes described here. These studies will benefit from alternative research designs and improved genotype annotation to discover and interpret associated variants and genes.
Supplementary Material
Acknowledgments
This research was supported by NIH grants DA024417, DA05147, AA09367, DA13240, DA036216, DA034606, HG007022, and HL117626. Dr. Vrieze is now at the Department of Psychology and Neuroscience, and the Institute for Behavioral Genetics, at the University of Colorado Boulder.
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article:
Figures S1–S17: QQ and Manhattan plots of association results from all 17 endophenotypes.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Sunyaev SR. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annual Review of Genetics. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- Barry RJ, Clarke AR, Johnstone SJ. A review of electrophysiology in attention-deficit/hyperactivity disorder: I. Qualitative and quantitative electroencephalography. Clinical Neurophysiology. 2003;114:171–183. doi: 10.1016/s1388-2457(02)00362-0. [DOI] [PubMed] [Google Scholar]
- Begic D, Popovic-Knapic V, Grubisin J, Kosanovic-Rajacic B, Filipcic I, Telarovic I, Jakovljevic M. Quantitative electroencephalography in schizophrenia and depression. Psychiatria Danubina. 2011;23:355–362. [PubMed] [Google Scholar]
- Clementz BA, Sponheim SR, Iacono WG, Beiser M. Resting EEG in first-episode schizophrenia patients, bipolar psychosis patients, and their first-degree relatives. Psychophysiology. 1994;31:486–494. doi: 10.1111/j.1469-8986.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Phillips E, Gizer IR, Gilder DA, Wilhelmsen KC. EEG spectral phenotypes: Heritability and association with marijuana and alcohol dependence in an American Indian community study. Drug and Alcohol Dependence. 2010;106:101–110. doi: 10.1016/j.drugalcdep.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Harris A, Melkonian D, Williams L, Gordon E. Dynamic spectral analysis findings in first episode and chronic schizophrenia. International Journal of Neuroscience. 2006;116:223–246. doi: 10.1080/00207450500402977. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Hubbard TJ. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Genome-wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview. Psychophysiology. 2014 doi: 10.1111/psyp.12343. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, McGue M. Minnesota Twin Family Study. Twin Research and Human Genetics. 2002;5:482–487. doi: 10.1375/136905202320906327. [DOI] [PubMed] [Google Scholar]
- Iacono WG, McGue M, Krueger RF. Minnesota Center for Twin and Family Research. Twin Research and Human Genetics. 2006;9:978–984. doi: 10.1375/183242706779462642. [DOI] [PubMed] [Google Scholar]
- Kang HM. Efficient and Parallelizable Association Container Toolbox (EPACTS) 2014 Retrieved from http://genome.sph.umich.edu/wiki/EPACTS. [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes MA, Malone SM, Elkins IJ, Legrand LN, McGue M, Iacono WG. The enrichment study of the Minnesota Twin Family Study: Increasing the yield of twin families at high risk for externalizing psychopathology. Twin Research and Human Genetics. 2009;12:489–501. doi: 10.1375/twin.12.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lee JY, Park HJ, Kim JW, Chung J-H. Association study between polymorphisms of the PARD3 gene and schizophrenia. Experimental and Therapeutic Medicine. 2012;3:881–885. doi: 10.3892/etm.2012.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lang PJ, Bradley MM, Cuthbert BN. International affective picture system (IAPS): Technical manual and affective ratings. Gainesville, FL: The Center for Research in Psychophysiology, University of Florida; 1999. [Google Scholar]
- Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: Study designs and statistical tests. American Journal of Human Genetics. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BS, Leal SM. Methods for detecting associations with rare variants for common diseases: Application to analysis of sequence data. American Journal of Human Genetics. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listgarten J, Lippert C, Kadie CM, Davidson RI, Eskin E, Heckerman D. Improved linear mixed models for genome-wide association studies. Nature Methods. 2012;9:525–526. doi: 10.1038/nmeth.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Burwell SJ, Vaidyanathan U, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of resting EEG activity: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12344. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Vaidyanathan U, Basu S, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of P3 event-related brain potential amplitude: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12345. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen YA, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, McGue M. The Minnesota Center for Twin and Family Research genome-wide association study. Twin Research and Human Genetics. 2012;15:767–774. doi: 10.1017/thg.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz N. Genetic visualization with GENVISIS. 2014 Manuscript in preparation. [Google Scholar]
- Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. American Journal of Human Genetics. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Choi K, Jones KA, Wang K, Begleiter H. Theta power in the EEG of alcoholics. Alcoholism, Clinical and Experimental Research. 2003;27:607–615. doi: 10.1097/01.ALC.0000060523.95470.8F. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Hein ND, Wijeyesakere SJ, Fink JK, Makhaeva GF. Neuropathy target esterase (NTE): Overview and future. Chemico-Biological Interactions. 2013;203:238–244. doi: 10.1016/j.cbi.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Sponheim SR, Clementz BA, Iacono WG, Beiser M. Resting EEG in first-episode and chronic schizophrenia. Psychophysiology. 1994;31:37–43. doi: 10.1111/j.1469-8986.1994.tb01023.x. [DOI] [PubMed] [Google Scholar]
- Sponheim SR, Clementz BA, Iacono WG, Beiser M. Clinical and biological concomitants of resting state EEG power abnormalities in schizophrenia. Biological Psychiatry. 2000;48:1088–1097. doi: 10.1016/s0006-3223(00)00907-0. [DOI] [PubMed] [Google Scholar]
- Sponheim SR, Iacono WG, Thuras PD, Nugent SM, Beiser M. Sensitivity and specificity of select biological indices in characterizing psychotic patients and their relatives. Schizophrenia Research. 2003;63:27–38. doi: 10.1016/s0920-9964(02)00385-7. [DOI] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Thompson PM. Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve FA, Straumanis JJ, Patrick G, Leavitt J, Manno JE, Manno BR. Topographic quantitative EEG sequelae of chronic marihuana use: A replication using medically and psychiatrically screened normal subjects. Drug and Alcohol Dependence. 1999;56:167–179. doi: 10.1016/s0376-8716(99)00029-0. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Drevets W. The ENIGMA Consortium: Large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior. 2014:1–30. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Isen JD, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of electrodermal activity: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12346. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Donnelly JM, Hammer MA, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of antisaccade eye tracking error rate: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12347. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of acoustic startle eye blink and affectively modulated startle response: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12348. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Vaidyanathan U, Kwong A, Kang HM, Zhan X, Iacono WG. In search of rare variants: Preliminary results from whole genome sequencing of 1,325 individuals with psychophysiological endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12350. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ. Characterization of the human patatin-like phospholipase family. Journal of Lipid Research. 2006;47:1940–1949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai TX, Li Y, Boehnke M, Lin XH. Rare-variant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.