Abstract
Biofuels and biomaterials, produced from lignocellulosic feedstock, require facile access to cellulose and hemicellulose to be competitive with petroleum processing and sugar-based fermentation. Physical-chemical barriers resulting from lignin complicates the hydrolysis biomass into fermentable sugars. Thus, the amount of lignin within a substrate is critical in determining biomass processing. The application of 13C cross-polarization, magic-angle spinning, and solid-state nuclear magnetic resonance for the direct quantification of lignin content in biomass is examined. Using a standard curve constructed from pristine lignin and cellulose, the lignin content of a biomass sample is accurately determined through direct measurement without chemical or enzymatic pre-treatment.
Keywords: lignin, biomass, solid-state NMR, cellulose
1. Introduction
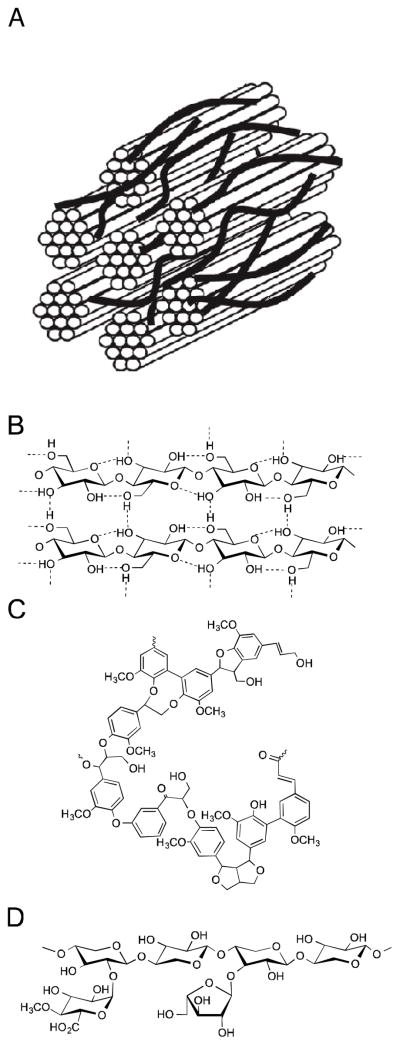
Biofuels and biomaterials produced from lignocellulosic feedstock require facile access to cellulose and hemicellulose to ensure competitive environmental and economic advantages to traditional petroleum processing and sugar fermentation pathways [1,2]. Hydrolysis of cellulose and hemicellulose into fermentable sugars is impeded by physical and chemical barriers created through the association of lignin phenylpropanoid polymers and other cell wall components of lignocellulosic biomass (Figure 1) [3]. Both covalent and non-covalent lignin-carbohydrate interactions inhibit the digestion of otherwise digestible sugars, such as cellulose and hemicellulose, by physically blocking access to these polysaccharides to cellulases and β-glucosidases. Lignin also absorbs these enzymes, resulting in another significant barrier to their action [4].
Figure 1.
Structure of lignocellulose (A) Schematic of lignocellulosic plant cell wall microstructure where the cylinders correspond to cellulose fiber bundles that are coated with hemicellulose and the dark lines to lignin (B) Structure of hydrogen-bonded cellulose polysaccharide chains comprising cellulose plant fibers (C) Major structures present in lignin (D) Xylan corresponding to a major hemicellulose structures.
Sufficient and mild delignification greatly enhances the utilization of associated saccharides, i.e., cellulose, hemicellulose, and other cell wall components [1]. Modern processing techniques [2] of lignocellulosic biomass include, physical and chemical processing methods, such as wet oxidation [5], ammonia fiber explosion [6], ozonolysis [7], and organosolvation [8], and bioprocessing where lignin is degraded using a highly efficient use of extracellular enzymes [3].
Despite lignin’s intractability in saccharification, some organisms have adapted to effectively degrade lignin in order to access the large cache of nutrition locked in the secondary cell wall of woody plants [9]. Fungi are the main organisms identified with this characteristic [3], the most proficient subset belonging to basidiomycetes (Figure 2 and Supporting Information Figure S1). Members of this group of fungi are collectively known as “white rot” because in nature, areas of wood infiltrated by their mycelia take on a whitish appearance due to depletion of brown lignin. In fact, white rot fungi are the most efficient lignin digesters observed in nature [10]. White rot fungi have evolved to secrete numerous peroxidases, such as manganese peroxidases, lignin peroxidases, and laccases that participate in ligninolysis [3]. Conversely, a class of fungi known as “brown rot” does not target lignin for degradation [11]. These fungi rely on degradation of cellulose, which leaves the appearance of brownish residue on the wood they colonize.
Figure 2.

Occurrence of brown rot and white rot in nature (A) Brown rot fungus observed in nature degrading lignocellulosic material (B) White rot fungus observed in nature degrading lignocellulosic material
Quantitative analysis of the components comprising biomass, particularly lignin, is difficult because of the limited solubility of lignocellulosics and these commonly require extensive processing prior to their analysis. 13C cross-polarization, magic-angle spinning, solid-state nuclear magnetic resonance (CP MAS ssNMR) spectroscopy may provide the ideal approach for the direct analysis of lignocellulosic feedstock [12,13,14]. CP MAS ssNMR spectroscopy was first applied to qualitatively examine cellulose, the major component of lignocellulosic feedstock [15]. It has subsequently been successfully used to investigate other biomass components, including hemicellulose and lignin [16,17]. 13C CP MAS ssNMR spectroscopy is now considered an essential tool in paper, biomaterial and biofuel processing [17]. While high-resolution 13C solution-state NMR spectroscopy can also afford a quantitative way to measure lignin content in lignocellulose biomass, this approach requires multiple steps for lignin isolation, removal of interfering proteins and carbohydrates, and lignin solubilization [18]. One way to purify lignin for analysis relies on enzymes, such as cellulases and hemicellulases, to hydrolyze cellulose and hemicellulose. Unfortunately, lignin is covalently attached to cellulosics and enzyme adsorption onto lignins, poses additional challenges for lignin isolation [19.20,21].
In the current study, 13C CP MAS ssNMR is examined to directly quantify the lignin content in lignocellulose biomass without any chemical or enzymatic pre-treatment, providing a content of lignin closer to its actual value in a biomass sample. This approach takes advantage of unique chemical structures within lignin to differentiate it from the other components of lignocellulosic biomass. Quantitative analysis of lignin in lignocellulosic biomass using 13C CP MAS ssNMR spectroscopy also required the preparation of a pristine lignin standard, to serve as a calibrant.
2. Methods
2.1 Organisms and their culturing
Fungal organisms, Fomitopsis pinicola (brown rot) and Ganoderma lucidum (white rot) were from Ecovative’s strain library but are also available from the CBS-KNAW Fungal Biodiversity Centre culture collection (http://www.cbs.knaw.nl/index.php/order). Malt extract broth (Difco, Becton, Dickinson and Co.), containing malt extract 6.0 g/L, maltose 1.8 g/L, dextrose 6.0 g/L and yeast extract 1.2 g/L, was prepared and sterilized at 15 psi for 15 min. Inoculated broth was prepared from each species separately. To inoculate, broth was transferred to a sterile blender flask and colonized millet inoculum was added at a rate of 10% (m:m). The inoculated grain was then blended thoroughly in the malt extract broth for 60 s. Approximately 25 mL of the inoculated broth was transferred to each of six sterile petri dishes to grow sheets of tissue on broth. Cultures were incubated at room temperature as fungal sheets developed on the surface. Sheets of pure tissue were harvested after 23 d of incubation and washed in deionized water sufficient to remove malt extract broth.
2.2 Preparation of biomass samples
Kenaf Biomass was acquired from Kenactiv Innovation, Inc. Kenaf pith core was collected and ground with a 0.32 cm mesh screen, then sorted over 0.21 cm mesh screen (see Supporting Information Table S1 for nutritional analysis of kenaf).
2.3. Fermentation of biomass samples
Six samples of kenaf pith (100 g each) were prepared in autoclavable spawn bags. Two bags of each of the six compositions were prepared by adding differing amounts (A-0%, 1%; B-10%, 0%; and C-10%, 1% each) of clear flour and calcium chloride, respectively (0% and 1%). These additives were used to supplement and encourage growth, particularly in the early stage as the mycelia from the inoculum begin to colonize surrounding kenaf particles. Next, water (400 mL) was added to each bag and the bags were sterilized for 60 min at 0.1 MPa pressure. Bags of substrate were inoculated at a rate of 10 % (w/w) and sealed. Inoculum consisted of yellow millet pre-colonized with cells from the relevant species. Original cultures used were acquired directly from the field in the northeast region of the United States. Samples from sets A and B (both for G. lucidum and the F. pinicola) were collected after 17 d of incubation and samples from set C were harvested after 16 d of incubation at 21 °C.
2.4 Lignin Extraction
Kenaf (50 g) was extracted with 2 L of 2% aqueous ammonium hydroxide for 2 d [22]. The extract was centrifuged (5,000 × g) for 10 min at 4°C to remove the solid kenaf residue. The supernatant was rotary evaporated with heating to remove excess aqueous ammonium hydroxide to precipitate crude lignin, which was recovered by centrifugation (5,000 × g) for 10 min at 4°C. The crude lignin was dissolved in DMSO and centrifuged (5,000 × g) 10 min at 4°C. The supernatant, which contained lignin, was collected, dried using rotary evaporator. Most of the residual DMSO could be removed by repeatedly (8-times) adding 5 mL distilled water and drying using rotary evaporation.
2.5 Solution state NMR
The solution-state NMR experiments were all performed on Bruker Advance II 800 MHz spectrometer equipped with a cryogenically cooled probe (TCI) with z-axis gradients (Bruker BioSpin, Billerica, MA). NMR data were processed and analyzed with Topsin 2.1.6 software (Bruker). The lignin was dissolved in DMSO-d6 and transferred into 5-mm NMR microtubes for one-dimensional 1H-NMR, one-dimensional 13C-NMR and two-dimensional 13C-1H heteronuclear single quantum coherence spectroscopy (HSQC) [20]. The conditions for one-dimensional 1H-NMR spectra were as follows: 32 scans, acquisition time of 0.99 s, and relaxation delay of 8.00 s. The conditions for one-dimensional 13C-NMR spectra were as follows: 4096 scans, acquisition time of 0.23 s and relaxation delay of 8 s. The conditions for two-dimensional HSQC spectra were as follows: 32 scans, acquisition time of 0.33 s and relaxation delay of 0.90 s. All experiments were carried out at 333K.
2.6 13C Cross-polarization, magic-angle spinning, solid-state NMR
The 13C cross polarization magic spinning solid-sate NMR experiment was set up according to the R.E. Taylor’s method [23]. All 13C CP MAS ssNMR experiments were performed on the 600 MHz 89 mm wide-bore Bruker Advance III spectrometer equipped with a 4 mm HXY solid-state MAS probe set up with two of the channels were configured for 1H and 13C. A reduced volume rotor (50 μL) was utilized and the active volume mapped to ensure that the active coil volume entirely contained the sample volume. Based on these findings, prior to the addition of pre-dried samples, polytetrafluoroethylene (~10 mg) was added into the bottom of the rotor in addition to a Teflon plug being positioned above the sample. The rotor was weighed before and after the addition of samples. The conditions for 13C CP MAS ssNMR were as follows: 4096 scans, spinning rate of 12.5K Hz, acquisition time of 0.02 s, and temperature of 278 k. A quantitative calibration curve was made based on peak integrals (157-142 ppm) for a series of high purity lignin samples of varying mass.
3. Results and discussion
3.1 Approach for extractive removal of lignin from biomass using room temperature ionic liquid
The widespread utilization of lignocellulosic biomass as a raw material for the production of biofuels, chemicals and materials and its inherent compositional variability makes the rapid assessment of its composition of critical importance, particularly as we move towards renewable resources [17]. A major impediment in assessing the composition and in utilizing lignocellulosic biomass is the intractable properties of lignin, which is extremely difficult to extract [18,19,20]. Until now most laboratories assess lignin content of biomass in multistep processes involving lignin extraction and closure of mass balance [24]. More recently, FTIR spectroscopy [25] has been used to estimate lignin content as well as solution-state 13C NMR spectroscopy but these methods require finding solvents to dissolve biomass samples [26]. Until now, the application of 13C CP MAS ssNMR spectroscopy has only been used for the qualitative assessment of lignin in biomass samples presumably because of the lack of available pristine, unmodified, lignin standard. The most widely available lignin is lignosulfonate, a side product of the pulp and papermaking industry, has distinctive NMR spectral characteristics making it unsuitable as a standard. Thus, we first undertook to explore methods to directly extract pristine lignin from lignocellulosic biomass using room temperature ionic liquids (RTILs) [27]. Unfortunately, 1-ethyl-3-methylimidazolium acetate RTIL [27] afforded lignin containing large amounts of cellulosics based on 13C CP MAS ssNMR (Supporting Information Figure S2).
3.2 Extraction of lignin from biomass using ammonium hydroxide
We next examined aqueous ammonium hydroxide for the extraction and recovery of pristine lignin from kenaf [28]. Pristine lignin (350 mg) was extracted from 50 g of the lignocellulosic biomass, Kenaf. The structure of the pristine lignin was characterized using solution 1D 1H-NMR and 1D 13C-NMR (Supporting Information Figures S3 and S4, respectively). The 2D HSQC spectrum of kenaf lignin allowed the assignment of the prominent signals in the 13C-NMR spectrum (Supporting Information Figure S5 and Table S2). The pristine lignin core structure matched well with that of other recent reports [18]. The recovery of lignin was low (~22 %) since no cellulase or hemicellulase was added to hydrolyze the polysaccharide and to release covalently bound lignins. However, the enzyme-free extraction reduced the level of residual proteins and provided pristine lignin of very high purity. The structure of the extracted pristine lignin was confirmed by solution NMR spectroscopy. The 1D 1H-NMR, 13C-NMR and 2D HSQC spectra (see Supporting Information) showed no protein and no polysaccharide impurities in this preparation of pristine lignin.
3.3 13C cross-polarization, magic angle spinning NMR for analysis of biomass components
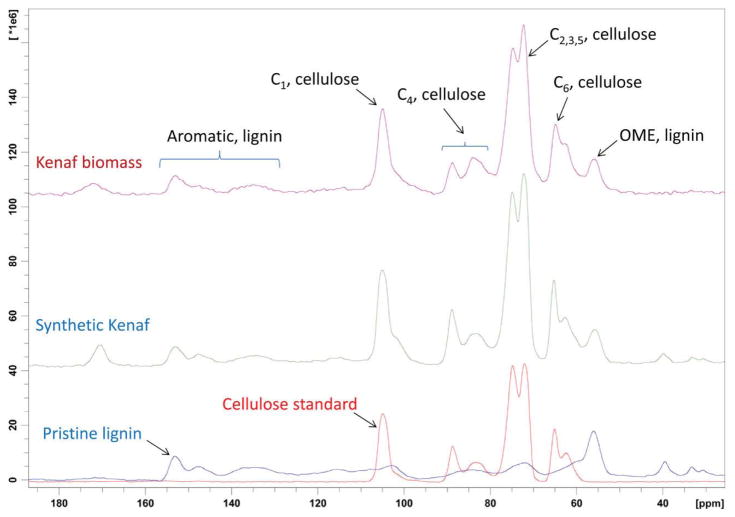
Kenaf is a typical lignocellulosic biomass, which is primarily composed of cellulose and lignin. The 13C CP/MAS spectrum of kenaf biomass could be simulated by combining the spectra of cellulose and pristine lignin (Figure 3).
Figure 3.
13C CP/MAS spectra of Kenaf biomass, synthetic kenaf, cellulose standard and pristine lignin
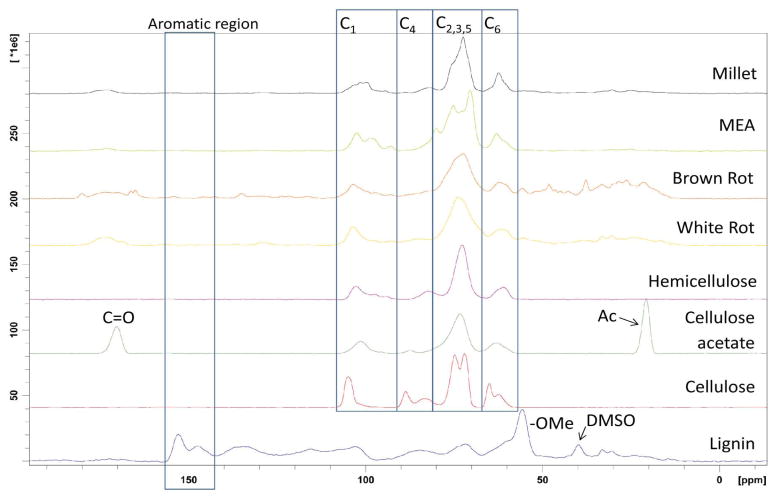
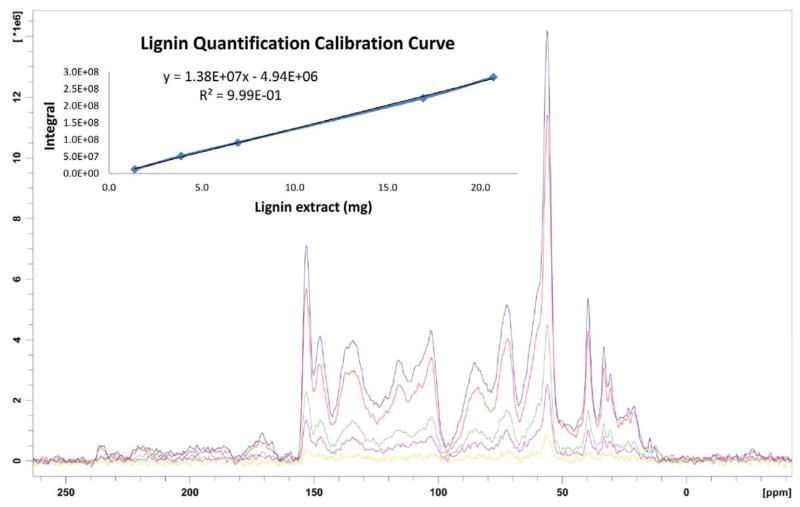
Next, standard samples of major components of lignocellulosic biomass as well as additives used in culturing fungus on biomass were examined using 13C CP MAS ssNMR (Figure 4). All biomass and culture samples, with the exception of pristine lignin extract, displayed signals corresponding to cellulosic polysaccharide. Pure cellulosics showed no signals in the aromatic region (140–160 ppm) of their spectra. The presence of aromatic signals, at 142–157 ppm, in lignin make these signals a unique identifier for lignin and the integrals of these peaks could be used for quantification. A small amount of residual DMSO could be observed in the pristine lignin sample from the signal appearing at 39 ppm. The percentage of this residual DMSO was calculated to be 7.6 wt. % by comparing the DMSO integral in this sample to the integral obtained from a spectrum of kenaf containing 14.5% DMSO (see Supporting Information). Thus, the purity of lignin was 92.4%. Using the purity determined for pristine lignin and the integrals from its spectrum (Figure 5 and Table 1), a standard curve was constructed. The excellent linearity of this standard curve allowed the accurate quantification of the lignin content in untreated biomass sample kenaf to be 32%.
Figure 4.
13C CP MAS ssNMR spectra for (top to bottom) millet, MEA brown rot fungi, white rot fungi, hemicellulose, cellulose acetate, cellulose, and lignin,
Figure 5.
13C CP/MAS ssNMR spectra of kenaf lignins of different mass and quantification calibration curve (inset)
Table 1.
Peak integrals of kenaf lignin of different mass on 157-142 ppm.
| Lignin extract (mg) | Lignina (mg) | Integral |
|---|---|---|
| 1.5 | 1.3 | 1.92 × 107 ± 2.96 × 106 |
| 4.2 | 3.6 | 5.20 × 107 ± 8.20 × 106 |
| 7.5 | 6.4 | 9.42 × 107 ± 6.50 × 106 |
| 18.3 | 15.6 | 2.27 × 108 ± 5.97 × 106 |
| 22.4 | 9.0 | 2.88 × 108 ± 4.97 × 106 |
DMSO present in lignin extract has been removed from these values.
3.4 Analysis of cellulosic biomass by 13C CP MAS ssNMR
Using the pristine lignin extracted from kenaf, we measured the lignin content in kenaf, corn stover and cotton stalks. These lignocellulosic biomasses are sources for papermaking, biofuel and bioethanol industries. Kenaf showed the highest lignin content based on 13C CP/MAS ssNMR and along the other biomass samples gave lignin values consistent with those reported in the literature (Supporting Information Table S3).
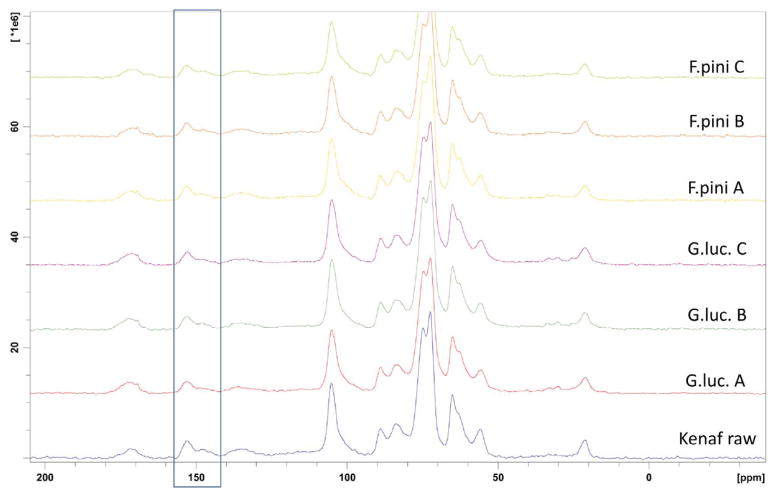
The 13C CP MAS ssNMR of kenaf on which fungi were cultured revealed the lignin content was quite different for G. lucidum (white rot) and F. pinicola (brown rot) (Figure 6, Table 2). The white rot G. lucidum fungi digested over 16% kenaf lignin reducing it to 26%, while as expected; the brown rot F. pinicola fungi did not alter the kenaf lignin.
Figure 6.
13C CP/MAS ssNMR spectra of culture samples (from top to bottom F. pinicola C, F. pinicola B, F. pinicola A, G. lucidum C, G. lucidum B, G. lucidum A, and raw kenaf.
Table 2.
Calculated lignin content of culture samples based on 13C CP/MAS ssNMR.
| Culture Samples | Lignin Content (%) |
|---|---|
| G. lucidum A | 25.8 |
| G. lucidum B | 27.4 |
| G. lucidum C | 26.5 |
| Average | 26.5 ± 0.8 |
| F. pinicola A | 32.1 |
| F. pinicola B | 32.2 |
| F. pinicola C | 31.1 |
| Average | 31.7 ± 0.6 |
Quantitative 13C-NMR has been applied to lignin qualitative and quantitative studies since the 1980s [29]. However, application of high-resolution 13C spectra has been limited due to low natural abundance of the 13C isotope, difficulties in lignin extraction, and high lignin concentrations required to achieve good signal-to-noise (S/N) ratios. The development of inverse detection in NMR (i.e., 2-D heteronuclear single quantum coherence (HSQC)) has significantly increased spectral resolution and sensitivity [26]. Pretreatment combined with cellulolytic enzyme lignin extraction [30] offers relatively higher yield with respect to the mill wood lignin extraction [31]. Unfortunately, lignin covalently attached to cellulosic moieties, and enzyme adsorption onto lignin, poses additional challenges for lignin isolation [32]. Recently, a new solution-state 2D-HSQC NMR spectroscopic method was reported to measure the acetylated plant cell wall using a whole cell wall dissolving system. Using this approach an analytical signal for lignin content with the necessity of lignin isolation. As a result, homogeneity of dissolved biomass in a solution system is required to visualize all the signals, and deduce the 1JC-H dependence of these signals. In practice, the reliability of the HSQC-based method for quantification is compromised by a number of issues, including a wide range in effective T2 relaxation times due to sample polydispersity, coupling constant deviations, homonuclear couplings, and sample dependent resonance offsets [33]. While 2D-HSQC NMR represents an innovative method ideal for investigating structural changes in lignin [34], it still has limitations.
To circumvent exhaustive extraction methodology, and diminishing results over time to determine insoluble lignin in solution, this study develops a solid-state method for quantitative and direct measurement of lignin in biomass. Quantitative solid-state NMR was first reported in middle 1990s [35], but its application was limited because of low sensitivity and resolution. With the development of cross polarization and MAS technology along with the greater availability of ultra-high field NMR magnets, the resolution and sensitivity of 13C CP/MAS ssNMR has been greatly enhanced. However, until now, 13C CP/MAS ssNMR has only been used for qualitative analysis for the insoluble components of biomass, such as cellulose [17] and lignin. An early report on structure changes in lignin and plant polysaccharides after fungal degradation using solid-state NMR first suggested the use of this method on lignocellulosic biomass [36]. The chemical structure and heterogeneity of two lignin structures from Loblolly Pine were subsequently investigated using 13C solid-state NMR [16]. This early research demonstrated that solid-state NMR was useful for lignin structural characterization. The use of a proper lignin standard in the current study clearly demonstrates that 13C CP/MAS ssNMR spectroscopy can provide fast, accurate and quantitative determination of the lignin content of lignocellulose biomass.
4. Conclusion
13C CP/MAS ssNMR spectroscopy provides a rapid and reliable method to assess the lignin content of biomass. There are several identified concerns that need to be monitored in carrying out this analysis with high accuracy. First, due to lignin structural variation among species of biomass [26], a standard lignin for quantification curve calibration should be prepared using the same biomass material to obtain a uniform standard. Second, changes in the water content of samples can result in changes in the integrals because of the 1H-13C cross polarization step employed. All samples should be dried or lyophilized and stored under the identical conditions for all measurements. Third, the standard curve should be constructed or at least a point from a past calibration repeated and subsequently used to normalize integrals from the new set of experiments to match past values every time the assay is performed. This evaluation is required in order to confirm spectrometer setup consistency along with the efficiency of the cross polarization transfer. With these parameters well understood and carefully controlled, quantitative 13C CP/MAS ssNMR method should be of great utility in paper making, biomaterials testing, tobacco analysis, the biofuel and bioethanol industry, as well as plant biology, plant metabolism and plant design, forest science and other biomass related research or industry.
Supplementary Material
HIGHLIGHTS.
Solid-state 13C-NMR for biomass analysis
Extractive preparation of pristine lignin
A direct measurement for lignin quantification
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the on line version at http: ......
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nature Biotechnology. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Shukla A, Tiwari S, Srivastava MA. Review on delignification of lignocellulosic biomass for enhancement of ethanol production potential. Renewable Sustainability Energy Review. 2014;32:713–728. [Google Scholar]
- 3.Sanchez C. Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnology Advancement. 2009;27:185–194. doi: 10.1016/j.biotechadv.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Martínez ÁT, Speranza M, Ruiz-Dueñas FJ, Ferreira P, Camarero S, Guillén F, Martínez MJ, Gutiérrez A, del Río JC. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. International Microbiology. 2005;8:195–204. [PubMed] [Google Scholar]
- 5.Palonen H, Thomsen AB, Tenkanen M, Schmidt AS, Viikari L. Evaluation of wet oxidation pretreatment for enzymatic hydrolysis of softwood. Applied Biochemistry and Biotechnology. 2004;117:1–17. doi: 10.1385/abab:117:1:01. [DOI] [PubMed] [Google Scholar]
- 6.Laureano-Pérez L, Teymouri F, Alizadeh H, Dale BE. Understanding factors that limit enzymatic hydrolysis of biomass. Applied Biochemistry and Biotechnology. 2005;121:1081–1099. doi: 10.1385/abab:124:1-3:1081. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresource Technology. 2002;83:1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 8.Papatheofanous MG, Billa E, Koullas DP, Monties B, Koukios EG. Two stage acid-catalyzed fractionation of lignocellulosic biomass in aqueous 16 ethanol systems at low temperatures. Bioresource Technology. 1995;54:305–310. [Google Scholar]
- 9.Pandey KK, Pitman AJ. FTIR studies of the changes in wood chemistry following decay by brown-rot and white-rot fungi. Industrial Biodeterioration Biodegradable. 2003;52:151–160. [Google Scholar]
- 10.Levin L, Papinutti L, Forchiassin F. Evaluation of Argentinean white rot fungi for their ability to produce lignin-modifying enzymes and decolorize industrial dyes. Bioresource Technology. 2004;94:169–176. doi: 10.1016/j.biortech.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Filley TR, Cody GD, Goodell B, Jellison J, Noser C, Ostrofsky A. Lignin demethylation and polysaccharide decomposition in spruce sapwood degraded by brown rot fungi. Organic Geochemistry. 2002;33:111–124. [Google Scholar]
- 12.Duer MJ. Introduction to solid-state NMR spectroscopy. Oxford: Blackwell; 2004. [Google Scholar]
- 13.Schmidt-Rohr K, Spiess HW. Multidimensional solid-state NMR and polymers. Academic Press; London: 1994. [Google Scholar]
- 14.Foston M. Advances in solid-state NMR of cellulose. Current Opinion in Biotechnology. 2014;27:176–184. doi: 10.1016/j.copbio.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 15.VanderHart DL, Atalla RH. Native cellulose: a composite of two distinct crystalline forms. Science. 1984;233:283–285. doi: 10.1126/science.223.4633.283. [DOI] [PubMed] [Google Scholar]
- 16.Holtman KM, Chen N, Chappell MA, Kadla JF, Xu L, Mao J. Chemical structure and heterogeneity differences of two lignins from Loblolly Pine as investigated by advanced solid-state NMR spectroscopy. Journal of Agricultural Food Chemistry. 2010;58:9882–9892. doi: 10.1021/jf101258x. [DOI] [PubMed] [Google Scholar]
- 17.Samuel R, Pu Y, Foston M, Ragauskas AJ. Solid-state NMR characterization of switchgrass cellulose after dilute acid pretreatment. Biofuels. 2010;1:85–90. [Google Scholar]
- 18.Wen JL, Sun S-L, Xue BL, Sun RC. Recent advances in characterization of lignin polymer by solution-state nuclear magnetic resonance (NMR) methodology. Materials. 2013;6:359–391. doi: 10.3390/ma6010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Zhang S, Miao S, Su Z, Wang P. Temperature sensitivity of cellulose adsorption on lignin and its impact on enzymatic hydrolysis of lignocellulosic biomass. Journal of Biotechnology. 2013;166:135–143. doi: 10.1016/j.jbiotec.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 20.Samuel R, Foston M, Jaing N, Cao S, Allison L, Studer M, Wyman C, Ragauskas AJ. HSQC (heteronuclear single quantum coherence) 13C-1H correlation spectra of whole biomass in perdeuterated pyridinium chloride-DMSO system: An effective tool for evaluating pretreatment. Fuel. 2011;90:2836–2842. [Google Scholar]
- 21.Berlin A, Balakshin M, Gilkes N, Kadla J, Maximenko V, Kubo S, Saddler J. Inhibition of cellulase, xylanase and β-glucosidase activities by softwood lignin preparations. Journal of Biotechnology. 2006;125:198–209. doi: 10.1016/j.jbiotec.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 22.Suhas P, Carrott M, Carrott R. Lignin - from natural adsorbent to activated carbon: A review. Bioresource Technology. 2007;98:2301–2312. doi: 10.1016/j.biortech.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Taylor RE. Setting up 13C CP/MAS experiments. Concepts in Magnetic Resonance Part A. 2004;22A:37–49. [Google Scholar]
- 24.Burkhardt S, Kumar L, Chandra R, Saddler J. How effective are traditional methods of compositional analysis in providing an accurate material balance for a range of softwood derived residues? Biotechnology for Biofuels. 2013;6:90. doi: 10.1186/1754-6834-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou G, Taylor G, Polle A. FTIR-ATR-based prediction and modeling of lignin and energy contents reveals independent intra-specific variation of these traits in bioenergy poplars. Plant Methods. 2011;7:9. doi: 10.1186/1746-4811-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansfield SD, Kim H, Lu F, Ralph J. Whole plant cell wall characterization using solution-state 2D NMR. Nature Protocol. 2012;7:1579–1589. doi: 10.1038/nprot.2012.064. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Mora-Pale M, Miao J, Doherty TV, Linhardt RJ, Dordick JS. Facile pretreatment of lignocellulosic biomass at high loadings in room temperature ionic liquids. Biotechnology and Bioengineering. 2011;108:2865–2875. doi: 10.1002/bit.23266. [DOI] [PubMed] [Google Scholar]
- 28.Kim TH, Lee YY. Pretreatment of corn stover by soaking in aqueous ammonia at moderate temperatures. Applied Biochemistry and Biotechnology. 2007:136–140. 81–92. doi: 10.1007/s12010-007-9041-7. [DOI] [PubMed] [Google Scholar]
- 29.Gellerstedt G, Robert D. Quantitative 13C NMR analysis of kraft lignins. Acta Chemica Scancinavica B. 1987;41:541–546. [Google Scholar]
- 30.Holtman KM, Chang HM, Kadla JF. Solution-state nuclear magnetic resonance study of the similarities between milled wood lignin and cellulolytic enzyme lignin. Journal of Agricultural Food Chemistry. 2004;52:720–726. doi: 10.1021/jf035084k. [DOI] [PubMed] [Google Scholar]
- 31.Björkman A. Isolation of lignin from finely divided wood with neutral solvents. Nature. 1954;174:1057–1058. [Google Scholar]
- 32.Faix O, Argyropoulos DS, Robert D, Neyrinck V. Determination of hydroxyl groups in lignins evaluation of 1H-, 13C-, and 31P-NMR FT-IR and wet chemical methods. Holzforschung. 1994;48:387–394. [Google Scholar]
- 33.Heikkinen S, Toikka MM, Karhunen PT, Kilpelainen LA. Quantitative 2D HSQC (Q-HSQC) via suppression of J-dependence of polarization transfer in NMR spectroscopy: Application to wood lignin. Journal of American Chemical Society. 2003;125:4362–4367. doi: 10.1021/ja029035k. [DOI] [PubMed] [Google Scholar]
- 34.Yelle DJ, Wei DS, Ralph J, Hammel KM. Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environmental Microbiology. 2011;13:1091–1100. doi: 10.1111/j.1462-2920.2010.02417.x. [DOI] [PubMed] [Google Scholar]
- 35.Yahnke MS, Rush BM, Reimer JA, Cairns EJ. Quantitative solid-state NMR spectra of CO adsorbed from aqueous solution onto a commercial electrode. Journal of American Chemistry Society. 1996;118:12250–12251. [Google Scholar]
- 36.Martínez ÁT, Gonzalez AE, Valmaseda M, Dale BE, Lambregts MJ, Haw JF. Solid-state NMR studies of lignin and plant polysaccharide degradation by fungi. Holzforschung. 1991;45:49–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.